Background: The p38 MAPK pathway plays a pivotal role in controlling cell responses to stress.
Results: FBXO31 fine-tunes p38 MAPK signaling and protects cancer cells from genotoxic stress by promoting degradation of the p38 activator MKK6.
Conclusion: FBXO31 is a novel negative regulator of MKK6-p38 signaling.
Significance: This report sheds new light on the protective mechanism of FBXO31 in stress-induced apoptosis.
Keywords: Apoptosis, Cancer, p38 MAPK, Protein Degradation, Stress Response, Ubiquitination, F-box Protein, FBXO31, MKK6
Abstract
The p38 MAPK signal transduction pathway plays an important role in inflammatory and stress responses. MAPKK6 (MKK6), a dual specificity protein kinase, is a p38 activator. Activation of the MKK6-p38 pathway is kept in check by multiple layers of regulations, including autoinhibition, dimerization, scaffold proteins, and Lys-63-linked polyubiquitination. However, the mechanisms underlying deactivation of MKK6-p38, which is crucial for maintaining the magnitude and duration of signal transduction, are not well understood. Lys-48-linked ubiquitination, which marks substrates for proteasomal degradation, is an important negative posttranslational regulatory machinery for signal pathway transduction. Here we report that the accumulation of F-box only protein 31 (FBXO31), a component of Skp1·Cul1·F-box protein E3 ligase, negatively regulated p38 activation in cancer cells upon genotoxic stresses. Our results show that FBXO31 binds to MKK6 and mediates its Lys-48-linked polyubiquitination and degradation, thereby functioning as a negative regulator of MKK6-p38 signaling and protecting cells from stress-induced cell apoptosis. Taken together, our findings uncover a new mechanism of deactivation of MKK6-p38 and substantiate a novel regulatory role of FBXO31 in stress response.
Introduction
Ubiquitination is an important posttranslational modification that plays crucial roles in cell cycle regulation, DNA repair, apoptosis, innate immune response, and neuron degeneration (1–5). In canonical ubiquitination, Lys-48-linked polyubiquitin chains label substrates for proteasome-dependent degradation. Recognition and ubiquitination of substrates are achieved by sequential action of three enzymes, namely E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase). The Skp1·Cul1·F-box protein complex belongs to the cullin RING E3 ligase superfamily. Within this complex, the F-box domain of F-box protein interacts with S phase kinase-associated protein 1 (Skp1) which, in turn, binds with cullin 1 (Cul1). F-box proteins confer substrate specificity by recognizing a discrete number of phosphorylated substances through protein-protein interaction domains located carboxyl-terminal to the F-box domain (6). F-box only protein 31 (FBXO31) belongs to an F-box protein family with no recognizable substrate binding domains. FBXO31 is a new player in the DNA damage response (7), and the only substrate reported to date is cyclin D1 (8). Given the diversity of substrates of F-box proteins, it is expected that FBXO31 may have other undiscovered substrates and regulatory functions when responding to genotoxic stresses.
p38 MAPK is the mammalian ortholog of yeast Hog1p MAPK and is activated by growth factors, proinflammatory cytokines, and environmental stresses, therefore playing a pivotal role in inflammatory responses and stress responses (9, 10). Activation of p38 MAPK is by dual phosphorylation on the threonine and tyrosine residues of the “TGY” motif by MAPKK3 (MKK3), MAPKK6 (MKK6), and MAPKK4 (MKK4). Among the p38 activators, MKK6 can activate all p38 family members, including p38α, p38β, p38γ, and p38δ, in response to a variety of external stimuli (11). Under physiological conditions, activation of the MKK6-p38 signaling pathway is transient and well balanced by deactivation (12). Long term activation of p38 can induce cell apoptosis (13–15). However, compared with MKK6-p38 activation, little is known about how deactivation of MKK6-p38 is regulated. Phosphorylation and ubiquitination are two important posttranslational modifications that regulate protein kinase activity and signal pathway transduction. Dephosphorylation by phosphatases has been found to play an important role in deactivating the MKK6-p38 signaling pathway (16), but involvement of ubiquitination in deactivation of the MKK-p38 cascade is barely known.
Here we identify FBXO31 as a negative regulator of the p38 signal transduction pathway. By interacting with phosphorylated MKK6 and triggering Lys-48-linked ubiquitination and degradation of MKK6, FBXO31 exerts antiapoptotic effects on cancer cells responding to stress stimuli.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
Myc-FBXO31 plasmids were generated by PCR amplification from a breast cDNA library and cloned into the pBabe-retro vector at the EcoRI and SalI restriction sites. FLAG-tagged, full-length FBXO31 and FBXO31 truncation mutants were obtained by PCR amplification and cloned into the pEGFPC2 vector (Clontech, Palo Alto, CA). GST-FBXO31 plasmids were prepared using PCR and cloned into the pGEX-4T-1 vector (Amersham Biosciences). The HA-MKK3, HA-MKK6, HA-MKK6A, and FLAG-p38α plasmids were provided by Prof. Jiahuai Han (Xiamen University, China). The myc-tagged ubiquitin WT, K48R, K63R, and K48/63R plasmids were provided by Prof. X. M. Zhang (Institute of Basic Medical Sciences, National Center of Biomedical Analysis, Beijing, China). PLKO.1 scrambled shRNA (plasmid 1864), FLAG-MKK6 (plasmid 13517), and FLAG-MKK6 (S207E/T211E) (plasmid 13518) were obtained from Addgene (Cambridge, MA) (17, 18). FLAG-MKK6 (S207A) and FLAG-MKK6 (T211A) were generated using a site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mission FBXO31 shRNAs were purchased from Sigma-Aldrich (St. Louis, MO). The sequences of shRNA#1 and shRNA#2 targeting the 3′ UTR and coding sequence of FBXO31, respectively, were CCGGGCCTCAGTGCATTTGGCAAATCTCGAGATTTGCCAAATGCACTGAGGCTTTTTG and CCGGGTTCATCTACACCAGTCAGTACTCGAGTACTGACTGGTGTAGATGAACTTTTTTG.
Human TNFα and EGF were purchased from PeproTech (Rocky Hill, NJ). Doxorubicin and cycloheximide were purchased from Sigma-Aldrich. MG132 was obtained from Merck Bioscience (Darmstadt, Germany). λ Phosphatase was obtained from New England Biolabs (Ipswich, MA).
Cell Culture and Establishment of Stable Cell Lines
The esophageal squamous cell carcinoma cell lines KYSE150, KYSE510, KYSE30, and KYSE410 (19), obtained from DSMZ (Braunschweig, Germany), were cultured in RPMI 1640 medium (Sigma) containing 10% FBS (Invitrogen). The ESCC2 cell line T.Tn was cultured in DMEM/F12 (Invitrogen) supplemented with 10% FBS (20). HeLa cells, HEK293 cells, and the packaging cells 293 Phoenix and 293T were cultured in DMEM containing 10% FBS. KYSE410 and T.Tn cells were used for a stable overexpression study, whereas KYSE150 and KYSE510 cells were used to construct stable FBXO31 knockdown cell lines. The 293T cells were first transfected with either scrambled shRNA or FBXO31 shRNA-containing plasmids using Lipofectamine 2000 (Invitrogen) following the instructions of the manufacturer. The viral supernatant was then collected and used to infect KYSE150 and KYSE510 cells. The stable integrants were selected with puromycin (Sigma). The same protocol was used to generate FBXO31 stable overexpression cell lines selected with G418 (Invitrogen).
Immunoblotting, Immunoprecipitation, and Antibodies
Preparation of whole cell lysates and immunoblotting were performed as described previously (21). For the immunoprecipitation assay, cells transfected with the indicated plasmids were harvested with NETN buffer containing 1 mm PMSF, 2.5 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm activated sodium orthovanadate, and then cell lysates were incubated with the appropriate antibody (1 μg) for 4 h at 4 °C, followed by 2 h of incubation with protein-A-Sepharose beads (GE Healthcare). The beads were washed in four changes of lysis buffer and eluted in 20 μl of 2× SDS/PAGE sample buffer for immunoblotting. Anti-FBXO31 antibody (catalog no. ab86137) was purchased from Abcam (Cambridge, UK). Antibodies against cyclin D1, phospho-p38α, and total p38α were from BD Biosciences. Pan-poly(ADP-ribose) polymerase and cleaved caspase 3 antibodies were obtained from Cell Signaling Technology (Beverly, MA). Anti-myc (9E10), anti-myc (A-14), and anti-actin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HA (rabbit) and anti-FLAG (M2) antibodies were purchased from Sigma-Aldrich. Anti-HA (mouse) and anti-MKK6 were purchased from Covance (Berkeley, CA) and Gentaur (Brussels, Belgium), respectively. Anti-Lys-48Ub and anti-K63Ub antibodies were from Millipore (Billerica, MA). Experiments involving immunoblotting and immunoprecipitation were performed at least three times (i.e. three biological replicates). For immunoblotting, cell samples in each experiment were pooled from triplicate wells of 6-well culture plates for protein extraction. The most representative set of immunoblots is presented in the figures.
In Vivo Ubiquitination
HEK293 cells were transfected with the indicated plasmids. After 24 h, cells were treated with 10 μm MG132 for 6 h and then harvested with 100 μl of cell lysis buffer (1% SDS, 150 mm NaCl, 10 mm Tris-HCl (pH 8.0) with 2 mm sodium orthovanadate, 50 mm sodium fluoride, and protease inhibitors). The cell lysates were then boiled immediately for 10 min, followed by brief sonication. An aliquot of 900 μl of dilution buffer (10 mm Tris-HCl (pH 8.0), 150 mm NaCl, 2 mm EDTA, and 1% Triton) was added to the lysate, and the mixture was incubated at 4 °C for 30–60 min with rotation. Then, samples were centrifuged at 20,000 × g for 30 min at 4 °C. Supernatants were collected and incubated with HA antibody (1 μg) overnight at 4 °C with rotation. Protein-A-Sepharose beads (GE Healthcare) were added to the mixture the following day. After incubation for 2 h, the beads were washed four times with lysis buffer and eluted in 20 μl of 2× SDS/PAGE sample buffer for immunoblotting.
GST Pulldown Assay
GST and GST-FBXO31 fusion proteins were expressed and purified according to the instructions of the manufacturer (Amersham Biosciences Pharmacia). The GST pulldown assay was performed as described previously (22).
Flow Cytometry
About 1–2 × 106 single cells pooled from replicate cultures of the same experiment were harvested and washed in cold PBS twice and then fixed in 70% ethanol overnight. The next day, cells were washed once in cold PBS and then incubated in propidium iodide buffer (PBS containing 40 μg/ml propidium iodide and 100 μg/ml RNase) at 37 °C for 30 min prior to analysis by flow cytometry (BD FACSCanto II Analyzer, BD Biosciences). The percentage of sub-G1 population indicative of cell death was analyzed with FlowJo using Dean-Jett-Fox methods. The mean value was calculated from three independent experiments.
Colony Survival Assay
Cancer cells were seeded in 6-well plates at 0.5–1 × 104 cells/well in triplicates and then subjected to 50 J/m2 UV irradiation the next day, after cells were attached to the plate. After 10 days, the colonies were fixed, stained with crystal violet, and counted. The mean value was obtained from three independent experiments.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 15 min at room temperature and then permeabilized with 0.25% Triton X-100 for 5 min. The cells were incubated with 1:500 anti-FBXO31 antibody (Abcam) and anti-HA (Sigma) for 1 h, followed by three washes in PBS, and then incubated with 1:2000 secondary antibodies (Alexa Fluor 488 donkey anti-rabbit IgG or Alexa Fluor 594 donkey anti-mouse IgG, Invitrogen) for 1 h. Immunostaining of cells was visualized using confocal microscopy (LSM 700, Carl Zeiss, NY) with a ×63 objective.
Statistical Analysis
The results were analyzed using SPSS (Aspire Software International, Leesburg, VA). Means ± S.E. were calculated from at least three independent experiments and compared by analysis of variance. All statistical tests were two-sided, and p < 0.05 was deemed statistically significant.
RESULTS
Accumulation of FBXO31 Inhibits Sustained p38 Phosphorylation in Response to Genotoxic Stress
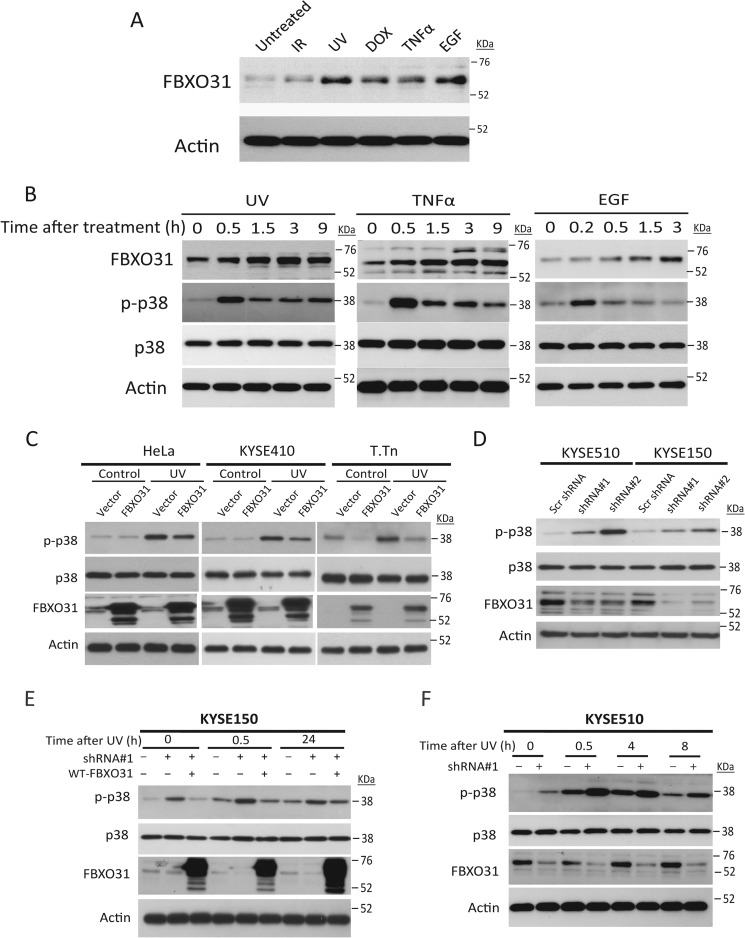
Because FBXO31 has been reported to be a DNA damage-responsive protein (8), we first investigated the FBXO31 expression level under genotoxic stresses. Induction of FBXO31 was observed upon ionizing radiation, UV irradiation, doxorubicin, TNFα, and EGF treatment (Fig. 1A). Temporal changes of FBXO31 and p38 in HeLa cells after treatment with UV irradiation, TNFα, and EGF showed a transient increase of phosphorylated p38 (p-p38), indicating p38 activation, followed by a drop in p-p38 that coincided with FBXO31 accumulation (Fig. 1B).
FIGURE 1.
Accumulation of FBXO31 inhibits sustained p38 activation in response to genotoxic stress. A, FBXO31 responded to genotoxic stress. HeLa cells were treated with ionizing radiation (IR, 5 gray), UV irradiation (50 J/m2), doxorubicin (DOX, 1 μg/ml, 6 h), TNFα (100 ng/ml, 6 h), or EGF (10 ng/ml, 3 h). FBXO31 and actin protein levels in cell lysates were determined by immunoblotting. B, HeLa cells were exposed to various stressors that included 5 gray of ionizing radiation (followed by culture for 3 h), 50 J/m2 UV irradiation (followed by culture for 3 h), doxorubicin (1 μg/ml for 6 h), TNFα (100 ng/ml for 6 h), or EGF (10 ng/ml for 3 h) and then harvested to immunoblot for FBXO31, p-p38, total p38, and actin expression. C, FBXO31 suppresses p38 activation. Serum-starved HeLa cells and the ESCC cell lines KYSE410 and T.Tn with stable expression of FBXO31 or empty vector were harvested 30 min after treatment with or without 50 J/m2 UV exposure for Western blot analysis. p38 MAPK pathway activation was compared using p-p38α antibody. D, cell lysates from serum-starved KYSE150 and KYSE510 cells with stable expression of the indicated shRNA were subjected to immunoblotting for p-p38, total p38, FBXO31, and actin expression. Scr, scrambled. E, ectopic expression of wild-type FBXO31 in the FBXO31 knockdown cell line reversed p38α phosphorylation. F, Western blot analysis showing temporal changes in p38 MAPK activation in KYSE510 cells with or without stable FBXO31 knockdown over a period of 8 h after 50 J/m2 UV irradiation.
Gain of function and loss of function studies were performed to determine whether FBXO31 has a regulatory role in p38 MAPK signaling. Overexpression of FBXO31 led to suppressed p38 phosphorylation in serum-starved cells with or without UV stress (Fig. 1C), whereas knockdown of FBXO31 induced the expression of p-p38 (Fig. 1D). Furthermore, shRNA rescue experiment showed that ectopic expression of wild-type FBXO31 in a stable knockdown cell line expressing shRNA#1, which targets the 3′ UTR of FBXO31 mRNA, could abolish p38α phosphorylation induced by FBXO31 knockdown (Fig. 1E), therefore confirming that FBXO31 can negatively regulate p38 signaling. Because both the amplitude and duration of p38 MAPK activation can affect cell fate, we compared the temporal changes in p-p38 expression between FBXO31 knockdown cells and vector control cells after UV exposure. The results indicated that, unlike control cells in which p38 phosphorylation peaked at 0.5 h, cells with stable FBXO31 knockdown showed enhanced and sustained p38 phosphorylation up to 8 h after UV exposure (Fig. 1F).
FBXO31 Interacts with MKK6
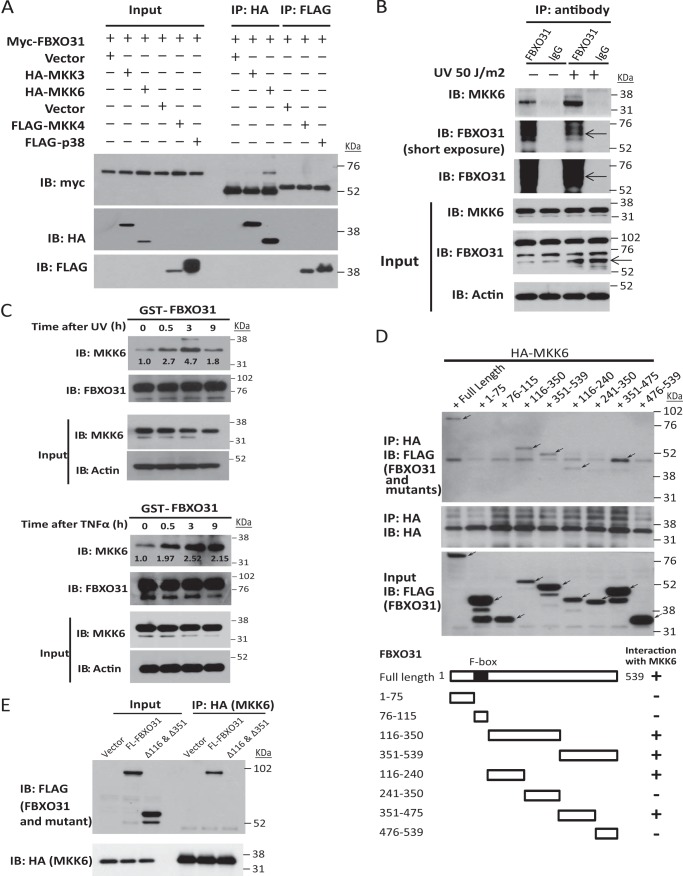
To explore the underlying mechanism by which FBXO31 regulates the p38 MAPK pathway, we investigated the interaction of FBXO31 with p38α and its upstream MAPKKs. Coimmunoprecipitation results showed that myc-FBXO31 could interact with HA-MKK6 but not FLAG-p38 or other tagged MAPKKs, namely MKK3 and MKK4 (Fig. 2A). Importantly, we verified the association of endogenously expressed FBXO31 and MKK6 using immunoprecipitation and found that it was stimulated by UV irradiation (Fig. 2B). A GST pulldown assay was performed to study the temporal changes in the interaction between MKK6 and FBXO31 upon genotoxic stress. The results showed a robust interaction between GST-FBXO31 and endogenous MKK6 that reached maximum at 3 h under UV irradiation and TNFα treatment (Fig. 2C), indicating that FBXO31 recognizes and binds with MKK6 upon stresses. Substrate recognition and binding domains for most F-box proteins are localized at the COOH terminus downstream of the F-box motif (23). Because FBXO31 belongs to the F-box only family of proteins that have no recognizable substrate binding domains, we next truncated FBXO31 to study which domains are required for its interaction with MKK6. Domain mapping results showed that amino acids 116–240 and 351–475, which located at the COOH terminus of the F-box motif of FBXO31, were essential for MKK6 binding (Fig. 2D). This was confirmed by the diminished interaction between MKK6 and FBXO31 upon deletion of amino acids 116–240 and 351–475 (Fig. 2E, Δ116 and Δ351) in FBXO31.
FIGURE 2.
FBXO31 interacts with MKK6. A, HEK293 cells were transfected with myc-FBXO31 and other indicated plasmids. Whole cell lysates were immunoprecipitated (IP) with HA antibody (for MKK3 and MKK6) or FLAG antibody (for MKK4 and p38), and then myc antibody was used to immunoblot (IB) for FBXO31 binding. B, untransfected HEK293 cells were serum-starved for 24 h and then treated with or without UV irradiation, followed by MG132 for 6 h. Cell lysates were immunoprecipitated with anti-FBXO31 antibody, which efficiently pulled down endogenous MKK6. Normal IgG was used as a negative control. FBXO31-bound proteins and whole cell lysates (input) were immunoblotted. C, HEK293 cells were serum-starved for 24 h and then treated with 50 J/m2 UV irradiation or 100 ng/ml TNFα. Cell lysates were harvested at the indicated times and incubated with GST-FBXO31-bound beads. The endogenous MKK6 binding was then analyzed by immunoblotting. The intensity of the MKK6 bands was quantified using ImageJ (National Institutes of Health, Bethesda, MD) and normalized to captured GST-FBXO31. D, HEK293 cells were transfected with the HA-MKK6 plasmid and FLAG-FBXO31 or its truncation vectors. Cell lysates were immunoprecipitated using HA antibody, and the bound proteins were analyzed by immunoblotting with anti-FLAG antibody to map the MKK6-binding regions on FBXO31. Arrows indicate the truncated FBXO31 proteins. The schematic in the bottom panel shows the truncated mutants of FBXO31. E, control empty vector, full-length FBXO31 (FL-FBXO31), or amino acids 116–240 and the 351–475 deletion mutant (Δ116 & Δ351) were cotransfected with HA-MKK6 in HEK293 cells. Whole cell lysates were immunoprecipitated with anti-HA antibody, and then FLAG antibody was used to immunoblot for MKK6 binding.
Because F-box proteins generally require a posttranslational modification of the substrate, we investigated whether recognition of MKK6 as substrate by FBXO31 is phosphorylation-dependent. λ Phosphatase abrogated the interaction between GST-FBXO31 and endogenous MKK6 in the absence of the phosphatase inhibitor sodium orthovanadate (Fig. 3A), suggesting that FBXO31 binds with phosphorylated MKK6. It could be seen that serum-starved cells without UV treatment also expressed a low level of p-MKK6 (Fig. 3A, first lane). This phenomenon helps explain the p-MKK6 expression and background FBXO31-MKK6 interaction observed in Fig. 2, A–E, which were induced by serum starvation or transfection reagents in cells without UV irradiation. Because activation of MKK6 occurs through dual phosphorylation on Ser-207 and Thr-211 (24), we investigated which phosphorylation sites are required for FBXO31 binding. We found that the phosphorylation mimic FLAG-MKK6 (S207E/T211E) showed a marked increase in FBXO31 binding, whereas mutation of S207A or T211A decreased the interaction (Fig. 3B). These results indicate that FBXO31 can recognize the activated form of MKK6.
FIGURE 3.
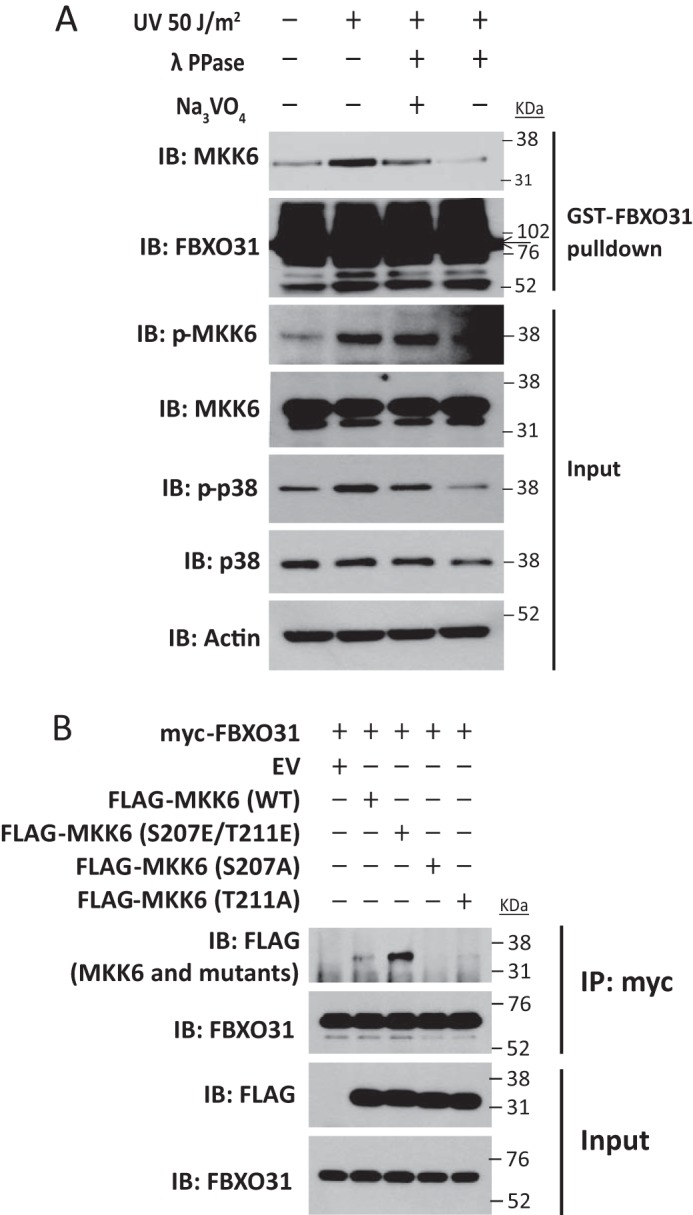
FBXO31 binds with phosphorylated MKK6. A, serum-starved HEK293 cells were treated with or without UV irradiation and then cultured for 3 h. Lysates from cells treated with UV irradiation were subjected to λ phosphatase treatment in the absence or presence of sodium orthovanadate (Na3VO4) at 30 °C for 30 min. Then, cell lysates were incubated with GST-FBXO31-bound beads at 4 °C overnight. Endogenous MKK6 binding was analyzed by immunoblotting (IB). B, HEK293 cells were transfected with myc-FBXO31 plasmids and plasmids encoding an empty vector (EV) or FLAG-tagged MKK6 derivatives (WT, S207E/T211E, S207A, and T211A). Whole cell lysates were immunoprecipitated (IP) with myc antibody, and then FBXO31 and FLAG antibodies were used for immunoblotting.
FBXO31 Induces Lys-48-linked Polyubiquitination and Degradation of MKK6
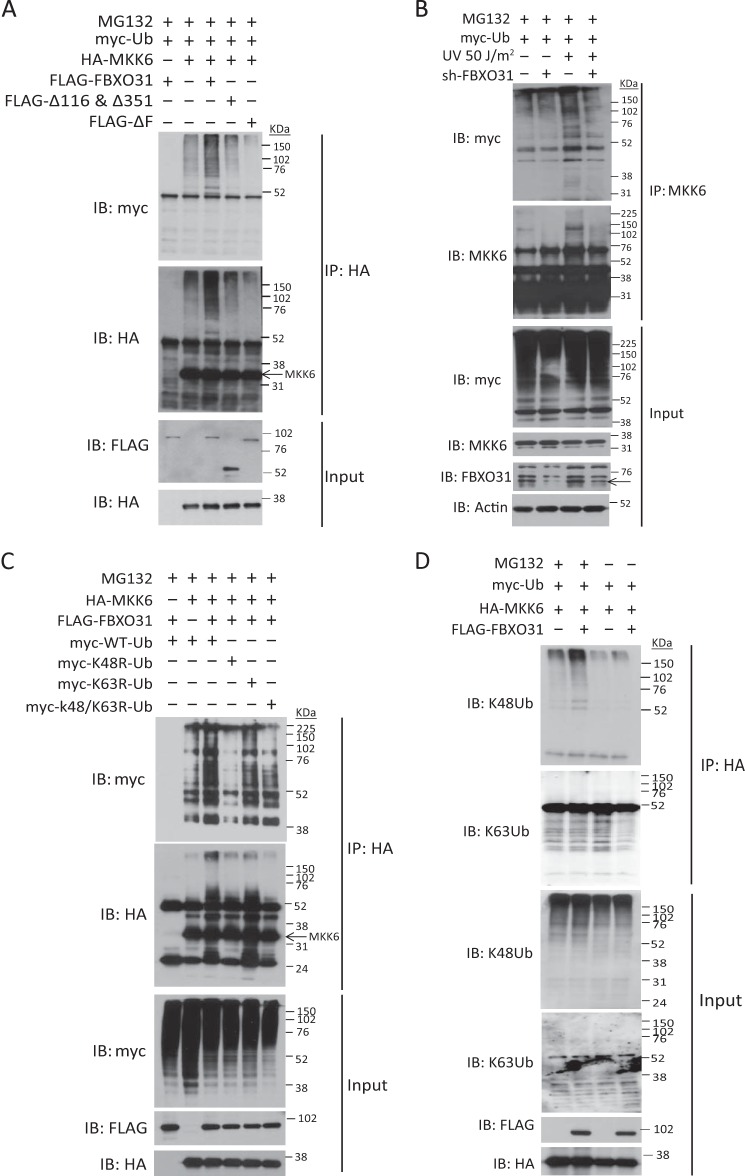
Because FBXO31 has been reported to interact with Cul1 and Skp1 at its F-box domain (25) and because our results showed that FBXO31 could bind with p-MKK6, it raised the possibility that MKK6 is a potential substrate for Skp1·Cul1·F-box proteinFBXO31 E3 ligase and that FBXO31 may mediate MKK6 ubiquitination. To explore this possibility, an FBXO31 mutant with F-box deletion (ΔF) was constructed. An in vivo ubiquitination assay using HEK293 cells cotransfected with myc-tagged ubiquitin expression vectors and different combinations of HA-MKK6, FLAG-tagged WT-FBXO31, Δ116 and Δ351-FBXO31, and ΔF-FBXO31 showed that only WT-FBXO31, but not its mutants, could induce polyubiquitination of HA-MKK6 under proteasomal inhibition with MG132 (Fig. 4A). To further confirm this effect, endogenous ubiquitination of MKK6 in the FBXO31 knockdown cells and control cells with and without UV irradiation were compared. The results showed that UV irradiation enhanced the ubiquitination on MKK6 compared with the untreated cells, suggesting posttranslational regulation of MKK6 upon stress. Knockdown of FBXO31 decreased the stress-induced MKK6 ubiquitination (Fig. 4B). To determine which type of ubiquitin linkage was covalently attached to MKK6 by FBXO31, the ubiquitin mutant plasmids K48R, K63R, and K48/63R, which block the generation of polyubiquitin chain through Lys-48, Lys-63, and both Lys-48 and Lys-63, respectively, were used. The results showed that overexpression of FBXO31 promotes Lys-48-linked, rather than Lys-63-linked, polyubiquitination of HA-MKK6 (Fig. 4C). The role of FBXO31 in mediating Lys-48-linked polyubiquitination of MKK6 was further confirmed by the increase in Lys-48 ubiquitin but not Lys-63 ubiquitin conjugates on HA-MKK6 in FBXO31-overexpressing cells (Fig. 4D). These results substantiate that FBXO31 induces Lys-48-linked polyubiquitination of MKK6.
FIGURE 4.
FBXO31 induces Lys-48-linked polyubiquitination of MKK6. A, HEK293 cells were transfected with the indicated plasmids for 24 h and then treated with 10 μm MG132 or vehicle for 6 h. Cell lysates were immunoprecipitated (IP) with anti-HA antibody. The bound proteins and whole cell lysates were analyzed by immunoblotting (IB) with anti-myc for ubiquitin (myc-Ub), anti-HA for MKK6 (HA-MKK6), and anti-FLAG for FBXO31 (FLAG-FBXO31) and its mutants. B, KYSE150 cells expressing scrambled shRNA or FBXO31 shRNA#1 were transfected with myc-Ub plasmids for 24 h and then subjected to UV irradiation, followed by treatment with 20 μm MG132 for 4 h. Cell lysates were immunoprecipitated with anti-MKK6 antibody. C and D, HEK293 cells transfected with indicated plasmids were treated as described in A. The HA-bound proteins and whole cell lysates in D were immunoblotted for Lys-48 (K48) and Lys-63 (K63) linkage-specific polyubiquitin using specific antibodies.
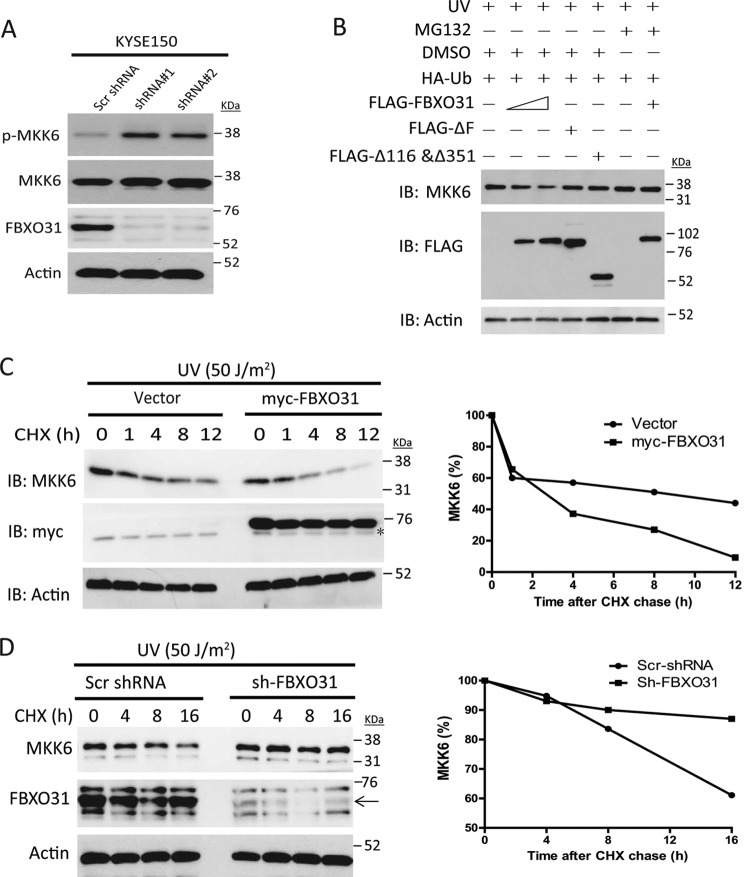
Because Lys-48-linked polyubiquitin predominantly marks substrates for proteasomal degradation (26), we proceeded to study whether FBXO31 affects MKK6 stability. Knockdown of FBXO31 in KYSE150 cells led to up-regulation of endogenous phosphorylated and MKK6 expression compared with cells expressing scrambled shRNA after 24 h of serum starvation (Fig. 5A). Transient overexpression of FLAG-FBXO31 decreased endogenous MKK6 expression in a dose-dependent manner, whereas the ΔF and Δ116 and Δ351 mutants had no effect (Fig. 5B). MG132 treatment reversed the FLAG-FBXO31-mediated reduction in MKK6 protein, suggesting that UV-induced MKK6 degradation is via the ubiquitin-proteasome system (Fig. 5B). Cycloheximide chase experiments showed that overexpression of FBXO31 decreased the turnover of endogenous MKK6 (Fig. 5C), whereas knockdown of FBXO31 delayed the UV-induced MKK6 reduction (Fig. 5D).
FIGURE 5.
FBXO31 regulates MKK6 turnover. A, serum-starved indicated stable cell lines were harvested for detection of endogenous phosphorylated and total MKK6 expression. Scr, scrambled. B, HEK293 cells were transiently transfected with 0.25 μg HA-Ub and FLAG-FBXO31 (0.5 or 1 μg) or 1 μg of mutant plasmids and then incubated for 24 h. The next day, cells were subjected to 50 J/m2 UV irradiation followed by treatment with dimethyl sulfoxide (DMSO) or MG132 for 9 h. IB, immunoblot. C, left panel, lysates from T.Tn cells expressing myc-FBXO31 or an empty vector were treated with 50 μg/ml cycloheximide (CHX) for the indicated times and subjected to immunoblotting for endogenous MKK6 expression. The asterisk indicates nonspecific bands. The intensity of the MKK6 bands was normalized to the actin loading control and quantified using ImageJ (National Institutes of Health) to study the turnover of MKK6 (right panel). D, KYSE150 cells expressing scrambled shRNA or FBXO31 shRNA were subjected to UV irradiation followed by treatment with 50 μg/ml cycloheximide for the indicated durations. Endogenous MKK6 expressions were analyzed using Western blotting (left panel), and the intensity of the MKK6 bands was quantified to study MKK6 turnover (right panel). The arrow in the left panel indicates the FBXO31 band.
FBXO31 Suppresses Stress-induced Cell Apoptosis by Modulating the MKK6-p38 Pathway
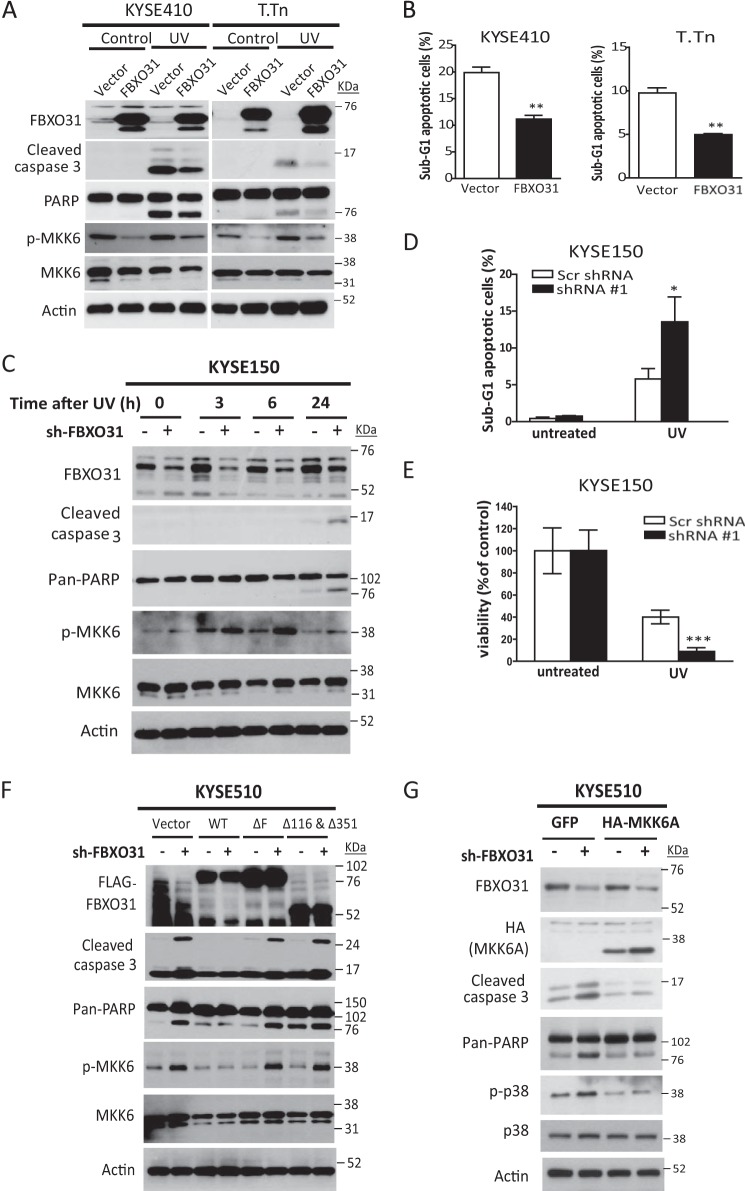
Gain of function experiments showed that overexpression of FBXO31 decreased MKK6 and p-MKK expression and protected cells from UV-induced apoptosis, as evidenced by lower expressions of cleaved caspase 3 and poly(ADP-ribose) polymerase (Fig. 6A) and the lower sub-G1 populations (Fig. 6B) compared with control cells expressing the empty vector. Because our data showed that FBXO31 accumulation occurred after p38 activation (Fig. 1B) and that increased FBXO31 could recognize activated MKK6 and facilitate MKK6 proteasomal degradation (Figs. 3 and 5), we hypothesized that FBXO31 may suppress stress-induced cell apoptosis by modulating the MKK6 pathway. Higher expressions of cleaved caspase 3 and cleaved poly(ADP-ribose) polymerase, accompanied by up-regulation of MKK6 and p-MKK6, were detected in ESCC cell lines with stable FBXO31 knockdown after UV irradiation compared with cells expressing scrambled shRNA (Fig. 6C). Flow cytometry and a colony survival assay also confirmed that knockdown of FBXO31 sensitized ESCC cells to UV stress, resulting in an increased sub-G1 population (Fig. 6D) and less colony formation (Fig. 6E). In addition, overexpression of WT-FBXO31, but not the F-box deletion mutant (Fig. 6F, ΔF) or MKK6-binding deletion mutant (Fig. 6F, Δ116 & Δ351), in stable FBXO31-knockdown cells reversed the up-regulation of MKK6 and p-MKK6 and rescued the apoptosis induced by knockdown of FBXO31, indicating that the effect of FBXO31 on apoptosis is dependent on an intact F-box and interaction with MKK6. To further confirm that regulation of cell apoptosis by FBXO31 is mediated via MKK6, a dominant negative form of MKK6 (MKK6A) (24) was overexpressed in the stable FBXO31 knockdown cell line KYSE510-shRNA#1. The data showed that apoptosis induced by knockdown of FBXO31 was abrogated by MKK6A (Fig. 6G). Collectively, these results indicate that MKK6 is a major mediator through which FBXO31 regulates cell apoptosis.
FIGURE 6.
FBXO31 suppresses stress-induced cell apoptosis by modulating the MKK6-p38 pathway. A and B, serum-starved ESCC cells with stable overexpression of FBXO31 or an empty vector were treated with or without 50 J/m2 UV irradiation and incubated for 24 h. Then, cell lysates were extracted for Western blot analysis to detect changes in apoptosis markers, and endogenous phosphorylated MKK6 and MKK6 (A) or cells were harvested for flow cytometry analysis (B). The mean percentages of sub-G1 cells from three independent experiments are shown. **, p < 0.01 compared with control cells with empty vector expression. PARP, poly(ADP-ribose) polymerase. C, ESCC cells with FBXO31 knockdown were serum-starved for 24 h and then subjected to UV irradiation. Cell lysates collected at the indicated time points were immunoblotted for analysis of apoptosis and MKK6 activation. D, KYSE150 cells with the indicated shRNA expression were treated with 50 J/m2 UV irradiation for 24 h, and then cells were analyzed by flow cytometry. The mean percentages of sub-G1 cells from three independent experiments are shown. *, p < 0.05 compared with control cells expressing scrambled (Scr) shRNA. E, colony formation of UV-irradiated KYSE150 cells expressing scrambled shRNA or FBXO31-shRNA#1 cells. The bar graphs show the number of colonies relative to untreated cells. Data were calculated from three independent experiments. Error bars represent mean ± S.E. ***, p < 0.001 compared with control cells expressing Scr shRNA after UV treatment. F and G, KYSE510 cells expressing scrambled shRNA or FBXO31 shRNA#1 were transfected with the indicated plasmids and then treated with UV irradiation. Cell lysates were harvested after 6 h for Western blot analysis.
DISCUSSION
We identified FBXO31 as a novel negative regulator that deactivates MKK6-p38 signaling when cancer cells respond to genotoxic stresses. Under normal conditions, cells exposed to environmental stress typically exhibit rapid activation and decay of MAPK activity (27). It is believed that the deactivation of the MAPK pathway is a consequence of negative feedback loops that regulate kinase activity, abundance, and cellular localization through changes in kinase phosphorylation and ubiquitination (28–31). Degradation of kinases within the MAPK cascade by the ubiquitin-proteasome pathway to attenuate MAPK signaling has been reported for certain MAPKK kinases (MEKK1 and MEKK2) (32, 33), MAPKs (ERK2 and ERK7) (34, 35), and MAPKKs such as MKK4, which is ubiquitinated by E3 ligase itch (a homologous to the E6-AP carboxyl terminus (HECT) domain-containing Nedd4-like ubiquitin protein ligase) and degraded to negatively regulate JNK activation (31). Whether degradation of MKK6 is involved in deactivation of p38 signaling has not been defined. Here our discovery that FBXO31 mediates Lys-48-linked ubiquitination and degradation of MKK6 to down-regulate p38 activation adds an important piece to the puzzle. Our data show that rapid activation of p38 precedes FBXO31 accumulation (Fig. 1B). This delicate temporal regulation may serve as a mechanism that allows the downstream effects of p38 activation, such as transcriptional regulation on stress-responsive genes, to take place in a fine-tuned manner, with accumulation of FBXO31 (which recognizes phosphorylated MKK6 and degrades MKK6) acting as a “brake” to prevent long term or overactivation of MKK6-p38 signaling. Therefore, our findings support that FBXO31 provides a mechanism other than dephosphorylation to facilitate deactivation of MKK6-p38 signaling and maintain cellular homeostasis. Phosphorylation of MKK6 on Ser-207 and Thr-211 was found to be important for FBXO31 binding (Fig. 3B). Because these two residues are dually phosphorylated by MAPKK kinases upon stresses and essential for activation of MKK6 and, in turn, p38 phosphorylation, it is likely that FBXO31 has the ability to sense the initial target kinase activity.
Our discovery that FBXO31 accumulates and mediates MKK6 degradation in cancer cells responding to genotoxic stresses shed new light on its role as a protective mechanism against stress-induced cell apoptosis. The positive correlation between FBXO31 expression and apoptosis of UV-irradiated ESCC cells demonstrated in this study is consistent with the findings of Santra et al. (8) showing that RNAi-mediated knockdown of FBXO31 can markedly sensitize melanoma cells to γ irradiation. However, they found that this effect is mediated by deregulation and proteolysis of cyclin D1, which serves as a substrate for FBXO31. Interestingly, knockdown of FBXO31 did not affect cyclin D1 expression, nor did it rescue DNA damage-induced cyclin D1 degradation in ESCC cell lines.3 Considering that F-box proteins can bind with a diversity of substrates and that each substrate may be regulated by many different E3 ligases, depending on cell type and substrate modification (such as phosphorylation) (36), we speculate that, for ESCC cells, cyclin D1 is not the major substrate of FBXO31 and that it does not mediate the antiapoptotic effect of FBXO31. On the contrary, our data in Fig. 6, F and G, strongly implicate MKK6 as a major mediator through which FBXO31 regulates cell apoptosis.
Taken together, we identified FBXO31 as a novel negative regulator that deactivates MKK6-p38 signaling when cells respond to genotoxic stresses. This provides a mechanism other than dephosphorylation to facilitate the deactivation of stress-activated signaling.
Acknowledgments
We thank Prof. R. J. Davis and Dr. D. M. Sabatini for the plasmids obtained from Addgene, Prof. K. Orth (Department of Molecular Biology, University of Texas Southwestern Medical Center) for His-MKK6 plasmids, and Dr. M. R. Green (Howard Hughes Medical Institute, University of Massachusetts Medical School) for the S278A FBXO31 plasmid. We also thank the University of Hong Kong Li Ka Shing Faculty of Medicine Faculty Core Facility for assistance with flow cytometry and imaging services.
J. Liu, unpublished observations.
- ESCC
- esophageal squamous cell carcinoma
- Ub
- ubiquitin
- p-p38
- phosphorylated p38
- p-MKK6
- phosphorylated MKK6.
REFERENCES
- 1. Yang W. L., Zhang X., Lin H. K. (2010) Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene 29, 4493–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang T. T., D'Andrea A. D. (2006) Regulation of DNA repair by ubiquitylation. Nat. Rev. Mol. Cell Biol. 7, 323–334 [DOI] [PubMed] [Google Scholar]
- 3. Hoeller D., Dikic I. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 [DOI] [PubMed] [Google Scholar]
- 4. Bhoj V. G., Chen Z. J. (2009) Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 [DOI] [PubMed] [Google Scholar]
- 5. Giasson B. I., Lee V. M. (2003) Are ubiquitination pathways central to Parkinson's disease? Cell 114, 1–8 [DOI] [PubMed] [Google Scholar]
- 6. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 7. Shiloh Y. (2009) FBXO31: a new player in the ever-expanding DNA damage response orchestra. Sci. Signal. 2, e73. [DOI] [PubMed] [Google Scholar]
- 8. Santra M. K., Wajapeyee N., Green M. R. (2009) F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 459, 722–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ashwell J. D. (2006) The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat. Rev. Immunol. 6, 532–540 [DOI] [PubMed] [Google Scholar]
- 10. Ono K., Han J. (2000) The p38 signal transduction pathway: activation and function. Cell. Signal. 12, 1–13 [DOI] [PubMed] [Google Scholar]
- 11. Cuenda A., Rousseau S. (2007) p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773, 1358–1375 [DOI] [PubMed] [Google Scholar]
- 12. Asthagiri A. R., Lauffenburger D. A. (2001) A computational study of feedback effects on signal dynamics in a mitogen-activated protein kinase (MAPK) pathway model. Biotechnol. Prog. 17, 227–239 [DOI] [PubMed] [Google Scholar]
- 13. Tobiume K., Matsuzawa A., Takahashi T., Nishitoh H., Morita K., Takeda K., Minowa O., Miyazono K., Noda T., Ichijo H. (2001) ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang W. H., Grégori G., Hullinger R. L., Andrisani O. M. (2004) Sustained activation of p38 mitogen-activated protein kinase and c-Jun N-terminal kinase pathways by hepatitis B virus X protein mediates apoptosis via induction of Fas/FasL and tumor necrosis factor (TNF) receptor 1/TNF-α expression. Mol. Cell. Biol. 24, 10352–10365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ventura J. J., Hübner A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006) Chemical genetic analysis of the time course of signal transduction by JNK. Mol. Cell 21, 701–710 [DOI] [PubMed] [Google Scholar]
- 16. Keyse S. M. (2000) Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr. Opin. Cell Biol. 12, 186–192 [DOI] [PubMed] [Google Scholar]
- 17. Raingeaud J., Whitmarsh A. J., Barrett T., Dérijard B., Davis R. J. (1996) MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16, 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 19. Shimada Y., Imamura M., Wagata T., Yamaguchi N., Tobe T. (1992) Characterization of 21 newly established esophageal cancer cell lines. Cancer 69, 277–284 [DOI] [PubMed] [Google Scholar]
- 20. Kawamata H., Furihata T., Omotehara F., Sakai T., Horiuchi H., Shinagawa Y., Imura J., Ohkura Y., Tachibana M., Kubota K., Terano A., Fujimori T. (2003) Identification of genes differentially expressed in a newly isolated human metastasizing esophageal cancer cell line, T. Tn-AT1, by cDNA microarray. Cancer Sci. 94, 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hui C. M., Cheung P. Y., Ling M. T., Tsao S. W., Wang X., Wong Y. C., Cheung A. L. (2006) Id-1 promotes proliferation of p53-deficient esophageal cancer cells. Int. J. Cancer 119, 508–514 [DOI] [PubMed] [Google Scholar]
- 22. Pan X., Zhao J., Zhang W. N., Li H. Y., Mu R., Zhou T., Zhang H. Y., Gong W. L., Yu M., Man J. H., Zhang P. J., Li A. L., Zhang X. M. (2009) Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc. Natl. Acad. Sci. U.S.A. 106, 3788–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 24. Han J., Lee J. D., Jiang Y., Li Z., Feng L., Ulevitch R. J. (1996) Characterization of the structure and function of a novel MAP kinase kinase (MKK6). J. Biol. Chem. 271, 2886–2891 [DOI] [PubMed] [Google Scholar]
- 25. Kumar R., Neilsen P. M., Crawford J., McKirdy R., Lee J., Powell J. A., Saif Z., Martin J. M., Lombaerts M., Cornelisse C. J., Cleton-Jansen A. M., Callen D. F. (2005) FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 65, 11304–11313 [DOI] [PubMed] [Google Scholar]
- 26. Pickart C. M. (2000) Ubiquitin in chains. Trends Biochem. Sci. 25, 544–548 [DOI] [PubMed] [Google Scholar]
- 27. Raman M., Chen W., Cobb M. H. (2007) Differential regulation and properties of MAPKs. Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 28. Owens D. M., Keyse S. M. (2007) Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 29. Johnson G. L., Dohlman H. G., Graves L. M. (2005) MAPK kinase kinases (MKKKs) as a target class for small-molecule inhibition to modulate signaling networks and gene expression. Curr. Opin. Chem. Biol. 9, 325–331 [DOI] [PubMed] [Google Scholar]
- 30. Morrison D. K., Davis R. J. (2003) Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19, 91–118 [DOI] [PubMed] [Google Scholar]
- 31. Ahn Y. H., Kurie J. M. (2009) MKK4/SEK1 is negatively regulated through a feedback loop involving the E3 ubiquitin ligase itch. J. Biol. Chem. 284, 29399–29404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu W. H., Lai M. Z. (2005) Deltex regulates T-cell activation by targeted degradation of active MEKK1. Mol. Cell. Biol. 25, 1367–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamashita M., Ying S. X., Zhang G. M., Li C., Cheng S. Y., Deng C. X., Zhang Y. E. (2005) Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu Z., Xu S., Joazeiro C., Cobb M. H., Hunter T. (2002) The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9, 945–956 [DOI] [PubMed] [Google Scholar]
- 35. Kuo W. L., Duke C. J., Abe M. K., Kaplan E. L., Gomes S., Rosner M. R. (2004) ERK7 expression and kinase activity is regulated by the ubiquitin-proteosome pathway. J. Biol. Chem. 279, 23073–23081 [DOI] [PubMed] [Google Scholar]
- 36. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]