Background: Death receptor 5 (DR5) triggers cell death upon binding to its ligand TRAIL.
Results: ATF3 promotes DR5 induction and apoptotic cell death upon zerumbone or celecoxib treatment in human p53-deficient colorectal cancer cells.
Conclusion: ATF3 is an essential transcription factor for p53-independent DR5 induction through the ROS-ER stress pathway.
Significance: ATF3 may be a useful biomarker for TRAIL-based anticancer therapy.
Keywords: Apoptosis, Cancer Therapy, Colon Cancer, ER Stress, Gene Regulation, Reactive Oxygen Species (ROS), ATF3, DR5, p53-independent
Abstract
Death receptor 5 (DR5) is a death domain-containing transmembrane receptor that triggers cell death upon binding to its ligand, TNF-related apoptosis-inducing ligand (TRAIL), and a combination of TRAIL and agents that increase the expression of DR5 is expected to be a novel anticancer therapy. In this report, we demonstrate that the stress response gene ATF3 is required for endoplasmic reticulum stress-mediated DR5 induction upon zerumbone (ZER) and celecoxib (CCB) in human p53-deficient colorectal cancer cells. Both agents activated PERK-eIF2α kinases and induced the expression of activating transcription factor 4 (ATF4)-CCAAT enhancer-binding protein (C/EBP) homologous protein, which were remarkably suppressed by reactive oxygen species scavengers. In the absence of ATF3, the induction of DR5 mRNA and protein was abrogated significantly, and this was associated with reduced cell death by cotreatment of TRAIL with ZER or CCB. By contrast, exogenous expression of ATF3 caused a more rapid and elevated expression of DR5, resulting in enhanced sensitivity to apoptotic cell death by TRAIL/ZER or TRAIL/CCB. A reporter assay demonstrated that at least two ATF/cAMP response element motifs as well as C/EBP homologous protein motif at the proximal region of the human DR5 gene promoter were required for ZER-induced DR5 gene transcription. Taken together, our results provide novel insights into the role of ATF3 as an essential transcription factor for p53-independent DR5 induction upon both ZER and CCB treatment, and this may be a useful biomarker for TRAIL-based anticancer therapy.
Introduction
Programmed cell death by apoptosis serves as a natural barrier to cancer development, and its deregulation contributes frequently and critically to tumorigenesis (1). Therefore, the resistance of cancer cells to apoptosis provides a logical target for potential therapeutic intervention. In colorectal cancers, increased cell death is well correlated with tumor regression after radio- or chemotherapy (2, 3). Therefore, understanding the molecular pathways that are implicated in cancer cell death is essential for developing novel therapeutic approaches.
Death receptors (DRs)3 4 and 5 are members of the TNF receptor superfamily (4–7) and are expressed in a broad range of cancer cells. The binding of the death ligand TNF-related apoptosis-inducing ligand (TRAIL/APO-2L) to death receptors causes the trimer formation of DRs, followed by recruitment of the adaptor protein FAS-associated protein with death domain (FADD) and the formation of death-inducing signal complex (DISC). Then, the initiator caspase 8 or 10 is recruited and triggers the apoptotic signal while having low toxicity toward normal cells (8). However, resistance against TRAIL-mediated cancer cell death occurs at various points in the apoptotic pathway (9). To overcome this resistance, agents that up-regulate the expression of TRAIL receptors or down-regulate antiapoptotic proteins may have the potential to sensitize cells to TRAIL. Indeed, the induction of p53-dependent DR5 or DR4 expression correlates with an increased responsiveness of cancer cells to TRAIL upon DNA damage of cancer cells (6, 10–12).
The endoplasmic reticulum (ER) is a specialized organelle required for crucial roles such as protein folding and Ca2+ storage. A variety of agents that induce the accumulation of misfolded or unfolded proteins or Ca2+ release cause ER stress (13, 14), and it has also become apparent that protein folding and reactive oxygen species (ROS) in the ER are closely linked (15, 16). To restore ER function, cells activate an adaptation program called the unfolded protein response (UPR). UPR signaling is mediated through at least three major pathways that are initiated by the stress sensors IRE1, PERK, or ATF6, coordinating a temporal shutdown of protein translation and a complex gene transcriptional program to safeguard cell survival. However, in cases of excessive stress, the UPR executes apoptosis to eliminate faulty cells (13, 14). In cancer cells, the UPR is activated broadly and plays a role in cancer cell death, dormancy, and aggressive growth as well as sensitivity to chemotherapeutic agents (17). Indeed, agents inducing ER stress could be exploited as potential antitumor drugs that overcome TRAIL resistance in part through the p53-independent activation of DR5 gene transcription, and transcription factors such as CHOP (18–20) and SP1 (21) have been shown to play a role in the activation of the DR5 gene. However, it still remains elusive which factor(s) or mechanism(s) affect(s) the efficacy of the up-regulation of DR5 by ER stress.
The stress response gene ATF3 is one of the ATF/CREB family transcription factors and facilitates apoptotic cell death upon stress response (22). Remarkably, ATF3 has been shown to be a direct target of p53 (23–25). We have reported previously that, upon DNA damage of human colon cancer cells, ATF3 sensitizes cells to TRAIL-mediated apoptosis by activating the DR5 gene promoter through cooperation with p53 (12). Moreover, ATF3 is also integral to the PERK/eIF2α signaling branch of the UPR (26). Indeed, it has been reported that many of the signals that cause the ER/UPR pathway also induce ATF3 (20, 27, 28).
Zerumbone (ZER), a bioactive sesquiterpene purified from the Zingiber zerumbet Smith, has an antiproliferative activity against several cancer cells, including colorectal cancer (29–31). The cytotoxicity of ZER is reported to be mediated through the induced expression of DR4/5 (30). However, the underlying mechanism of the transcriptional activation of the DR gene is not fully understood. Celecoxib (CCB), a selective inhibitor of cyclooxygenase 2 (COX-2), has been approved as a nonsteroidal anti-inflammatory drug. However, CCB also exhibits additional biological activities and targets. For instance, it up-regulates the expression of DR5 and sensitizes tumor cells to TRAIL-induced apoptosis, and its COX-2 inhibition is dispensable for antitumor effects (32–35).
Here we show that ZER and CCB activated the ER/UPR pathway by ROS production and up-regulated ATF3 and CHOP to induce the expression of DR5. ATF3 enhanced the sensitization of cancer cells to TRAIL-mediated apoptosis, providing insights into the role of ATF3 in the stress response of p53-deficient human colon cancer cells. ATF3 may represent a novel biomarker or therapeutic target for TRAIL-based therapeutic approaches.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
The expression vector encoding human ATF3 (pCI-ATF3) and the retrovirus vector for human ATF3 have been detailed elsewhere (12). The expression vector for CHOP was constructed by subcloning human CHOP cDNA into the pCIneo vector. ZER was obtained as described previously (36, 37), and CCB was from Sigma-Aldrich (St. Louis, MO). Recombinant APO2/TRAIL was purchased from PeproTech (Rocky Hill NJ). The antibodies used were as follows. Biotinylated anti-DR5 antibody DJR2–2 was provided by Dr. Yagita (Juntendo University). Anti-ATF3 (C-19), anti-CHOP (B-3 and R-20), anti-PERK (c-16), anti-phosphorylated PERK (Thr-981), anti-eIF2α (FL-315), anti-DR4 (H-130), and anti-DR5 (N-19) were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-DR5 (catalog no. 2019) was from Prosci (San Diego, CA). Anti-ATF3 (catalog no. HPA001562) and anti-β-actin (catalog no. AC-74) were from Sigma-Aldrich. Anti-cleaved caspase 3 (Asp-175) and anti-phosphorylated eIF2α (catalog no. 9721) were from Cell Signaling Technology (Danvers, MA). Anti-PARP (catalog no. C2-10) was from Trevigen (Gaithersburg, MD), and anti-KDEL (catalog nos. GRP78 and SPA-827) were from Stressgen (Victoria, Canada). N-acetylcysteine, l-glutathione (GSH), 2′,7′-dichlorodihydrofluorescein diacetate, and DAPI were purchased from Nacalai Tesque (Kyoto, Japan), Sigma-Aldrich, Invitrogen, and Dojindo (Kumamoto, Japan), respectively. Other chemicals were of reagent grade.
Cell Culture and Media
HCT116-p53null cells were a gift from Dr. Vogelstein and cultured in McCoy medium supplemented with 10% FBS and 1% penicillin/streptomycin. SW480 cells were purchased from the ATCC and cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. MEFs were isolated by trypsinization of embryos dissected on day 13.5 of gestation and cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% l-glutamate.
Generation of Atf3 Null MEFs in a p53-deficient Background
Atf3 knockout mice were generated as described previously (12). These mice were crossed with p53 gene knockout mice (38), and Atf3 null/p53 null double knockout (DKO) mice were obtained. Wild-type or mutant Atf3 gene loci were determined by genomic PCR using primer sets of 5′-TGCAAAAGGAAACTGACCAAG-3′ (#4 forward) and 5′-CTGGCCTGGATGTTGAAGCAT-3′ (#4 reverse) or 5′-GGTGTGTTTACCTTCTTCATT-3′ (#3 forward) and 5′-TCTTGATCTTCCTGTTTCAGT-3′ (#3 reverse), respectively. For the p53 gene, primer sets of 5′-ACACACCTGTAGCTCCAGCAC-3′ (#int4 forward) and 5′-AGCGTCTCACGACCTCCGTC-3′ (#ex5 reverse) or 5′-GTGTTCCGGCTGTCAGCGCA-3′ (#neo forward) and 5′-AGCGTCTCACGACCTCCGTC-3′ (#ex5 reverse) were used for wild-type or mutant loci, respectively. All animal work was approved and conducted according the guidelines of Committees of Animal Experiments (License Number 0140155A) and Recombinant DNA Experiments (License Number 2010-205C) of Tokyo Medical and Dental University.
Whole Cell Extracts and Western Blot Analysis
Whole cell extracts were prepared and subjected to Western blotting as described previously (12). Protein bands were developed by using Luminol reagent (catalog no. sc-2048, Santa Cruz Biotechnology) or ECL (GE Healthcare), and images were captured by Las 500 (GE Healthcare).
Cell Surface Expression of DR5 by FACS
Cells (1.0 × 106) treated as indicated were resuspended in ice-cold PBS containing 1% BSA and 0.1% NaN3 and then incubated on ice for 30 min with saturating concentrations of 10 μg/ml biotinylated anti-DR5 antibody (DJR2-2). After further incubation with 10 μg/ml streptavidin-phycoerythrin (PE) (Cappel, Aurora, OH) or control IgG-PE (Nacalai Tesque) on ice for 30 min, cells were analyzed by flow cytometry (BD Biosciences).
Measurement of Intracellular ROS
Cells treated as indicated were harvested and incubated with 10 μm of 2′,7′-dichlorodihydrofluorescein diacetate for 30 min at 37 °C, and fluorescence signals were measured by flow cytometry.
Trypan Blue Exclusion Assay and DAPI Staining
Trypan blue exclusion assays were performed as described previously (39). For morphological analysis, cells were fixed in methanol, incubated with 2.5 μg/ml of DAPI, and then analyzed at 420 nm using a fluorescent microscope (Olympus, Tokyo, Japan).
Quantitative Reverse Transcription (qRT) PCR
Total RNA was isolated using an RNeasy mini kit (Qiagen) and assayed for mRNAs of various genes by quantitative RT-PCR using a kit from Qiagen. The following primer pairs were used: human ATF3, 5′-GGAGTGCCTGCAGAAAGAGT-3′ (forward) and 5′-CCATTCTGAGCCCGGACAAT-3′ (reverse); human DR5, 5-CAGGTGTCAACATGTTGTCC-3′ (forward) and 5′-ATCGAAGCACTGTCTCAGAG-3′ (reverse); human DNA-damage-inducible transcript 3 (DDIT3, CHOP), 5′-AGCTGGAACCTGAGGAGAGA-3′ (forward) and 5′-GGGTCAAGAGTGGTGAAGAT-3′ (reverse); human ATF4, 5′-TGAAGGAGTTCGACTTGGATGCC-3′ (forward) and 5′-CAGAAGGTCATCTGGCATGGTTTC-3′ (reverse); human GAPDH, 5′-GAGTCAACGGATTTGGTCGT-3′ (forward) and 5′-TTGATTTTGGAGGGATCTCG-3′ (reverse); mouse ATF3, 5′-TTACCGTCAACAACAGACCC-3′ (forward) and 5′-TCAGCTCAGCATTCACACTC-3′ (reverse); mouse DR5, 5′-GAGGCAATGGTTGCTCTGTA-3′ (forward) and 5′-CTATGTCCGAACAATACTCG-3′ (reverse); and mouse GAPDH, 5′-CGTCCCGTAGACAAAATGGT-3′ (forward) and 5′-TTGATGTTAGTGGGGTCTCG-3′ (reverse). GAPDH was used as an internal control. Data represent means ± S.E. of three independent experiments.
Knockdown of ATF3 and ATF4
The retrovirus vectors siATF3-363, siATF3-493, and siATF3-500 were constructed by subcloning an oligonucleotides 363 (5′-TGGAAAGTGTGTGAATGCTGAACT-3′), 491 (5′-AAGATGAGAGAAACCTCTTTA-3′), and 500 (5′-GAAACCTCTTTATCCAACAGATA-3′) into pMX-puroII-U6, and corresponding retroviruses were prepared in Plat E cells (40). Human colon cancer cells were first transfected with pcDNA-mCAT encoding the ecotropic retrovirus receptor, followed by infection with a mixture of retroviruses siATF3-363, siATF3-493, and siATF3-500 or siGFP as a control. ATF3 cells that were knocked down were selected by culturing in 10–40 μg/ml puromycin for several days. For silencing ATF4, the siRNA oligo for ATF4 (catalog no. sc-35112) was purchased from Santa Cruz Biotechnology. Transfection of siRNA was performed using X-tremeGENE siRNA transfection reagent (Roche) according to the instructions of the manufacturer. Briefly, 10 μl of X-tremeGENE siRNA transfection reagent was mixed with 90 μl of Opti-MEM (Invitrogen) and then incubated with a mixture of 15 μl of 10 μm siRNA and 85 μl of Opti-MEM for 20 min at room temperature. The complexes were then applied to the cells cultured on a 6-well plate. After incubation for 24 h, the medium was removed, and cells were treated with ZER or CCB.
Luciferase Assay
Various 5′ deletions of human DR5 gene reporter plasmids and pDR5Luc-552 plasmids containing CHOP, Elk-1, and NF-κB site mutants were gifts from Dr. T Sakai (Kyoto Prefectural University, Japan) and Dr. H. G. Wang (Pennsylvania State University), respectively. A set of pDR5Luc-448 reporter plasmids containing mutations of ATF3–2 at −442 to −415, −3 at −275 to −268, and −4 at −183 to −175 relative to the transcriptional start site of the human DR5 gene were prepared by an overlap extension PCR protocol and contained sequences of 5′-AGGAGACA-3′ (ATF3-2), 5′-TGTCTAGA-3′ (ATF3-3), and 5′-TGTCTCCT-3′ (ATF3-4) compared with wild-type sequences of 5′-AGGCGTCA-3′ (ATF3-2), 5′-TGACGAGA-3′ (ATF3-3), and 5′-TGACGCCT-3′ (ATF3-4), respectively. Each reporter plasmid of the human DR5 gene was transfected and assayed as described previously (12), and the luciferase activity was measured using a Dual-Luciferase reporter assay system from Promega. Data shown are mean ± S.E. of three independent experiments.
ChIP Assay
ChIP assays were performed as described previously (12). Briefly, cells were cross-linked by treatment with 1% formaldehyde for 20 min. Equal aliquots of isolated chromatin were pulled down by anti-ATF3, anti-CHOP antibodies, or control IgG. The chromatins that were pulled down by each antibody were reverse-cross-linked, and DNAs were purified. Then, PCR was performed using the following primer pairs: ATF3-BS3, 5′-AAGGTTAGTTCCGGTCCTTC-3′ (forward) and 5′-TTCCACCACAGGTTGGTGAC-3′ (reverse); ATF3-BS4, 5′-GCAGTTGCACATTGGATCTG-3′ (forward) and 5′-TATGTGTCCAGGCTGACTTG-3′ (reverse); CHOP-BS, 5′-ACCCAGAAACAAACCACAGC-3′ (forward) and 5′-ACTGCAAATTCCACCACAGG-3′ (reverse); and GAPDH, 5′-CTTGACTCCCTAGTGTCCTTC-3′ (forward) and 5′-AAGGTCTTGAGGCCT-3′(reverse). The resulting PCR products were analyzed by 2% agarose gel electrophoresis.
RESULTS
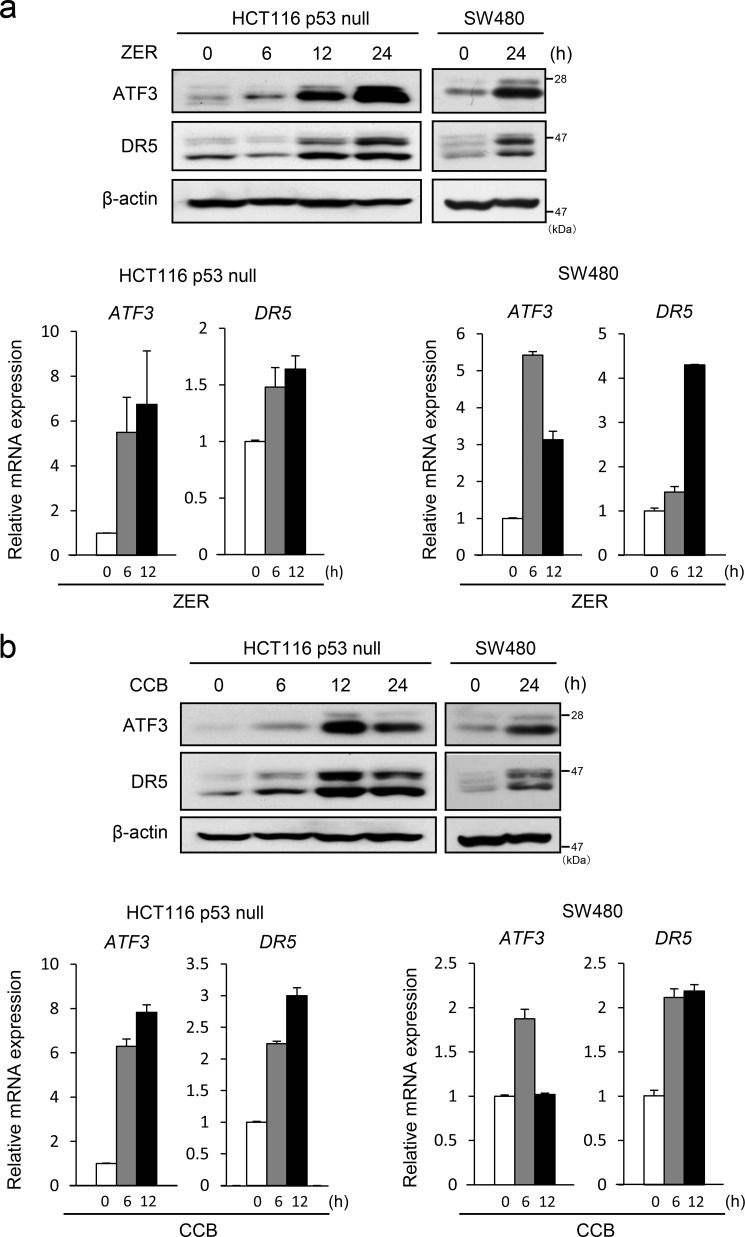
ZER and CCB Induce the Expression of ATF3 and DR5 in HCT116-p53 Null and SW480 Cells
We have shown previously that ATF3 promotes efficient DR5 induction upon DNA damage in a synergistic action with p53 (12). To further study the role of ATF3 in DR5 expression in p53-deficient cells, we treated HCT116 and SW480 cells that harbor a null or mutated p53 gene, respectively, with ZER. As shown in Fig. 1a, the expression of ATF3 and DR5 proteins was up-regulated in these cells, and this was associated with increased expression of mRNA for each gene. We further stimulated cells with CCB, known as a DR5 inducer. CCB also strongly induced the expression of ATF3 and DR5 proteins and mRNAs (Fig. 1b). It should be noted here that the bands for DR5 on Western blot analysis appear to be more than one, probably because of splice variants and/or O-glycosylation of the DR5 protein (41). These results indicate that both ZER and CCB are capable of inducing ATF3 and DR5 expression in p53-deficient human colon cancer cells and that this is, at least in part, caused by transcriptional activation of each gene.
FIGURE 1.
ZER and CCB induce the expression of DR5 and ATF3 in p53-deficient human colorectal cancer cells. a, HCT116-p53null cells or SW480 cells were treated with 20 μm ZER for the indicated times, and their whole cell extracts were assayed for DR5 and ATF3 proteins by Western blotting (top panel). β-actin was used as a loading control. The expression of ATF3 and DR5 mRNAs was also analyzed by qRT-PCR and normalized to the level of GAPDH (bottom panel). b, HCT116-p53null cells or SW480 cells were treated with 50 μm CCB, and the expression of DR5 and ATF3 was analyzed for both mRNA and protein as in a.
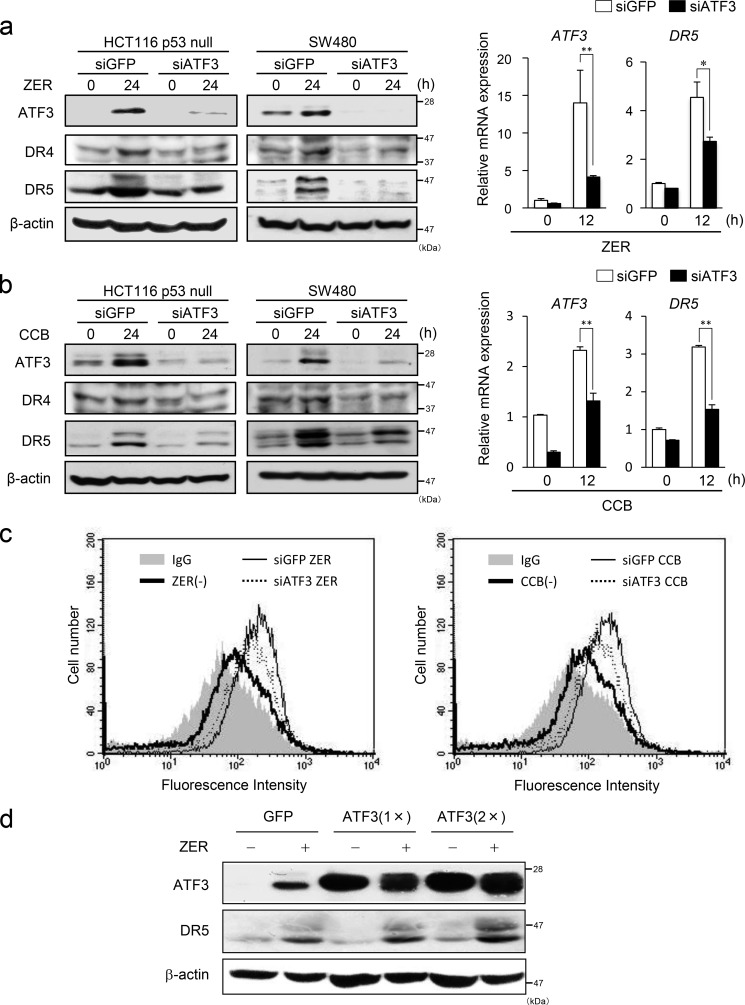
ATF3 Is Required for Efficient DR5 Induction by ZER and CCB in p53-deficient Cells
Next, we addressed whether ATF3 has a role in the induction of DR5 in p53-deficient cells. To this end, we knocked down the ATF3 gene in these cells and treated them with ZER or CCB. As shown in Fig. 2, a and b, gene silencing of ATF3 significantly suppressed the induction of DR5 protein and mRNA by ZER and CCB compared with control cells. Because ZER has been reported to induce DR4 (30), we also examined the expression of DR4 protein. DR4 was induced by both ZER and CCB, and its induction was partly suppressed in ATF3 knockdown cells (Fig. 2, a and b, left panels). We next examined the DR5 expression on the cell surface of these cells. As in Fig. 2c, ATF3 knockdown significantly reduced the level of induction of DR5 protein on the surfaces of these cells. We further examined whether the overexpression of ATF3 might affect DR5 induction. The induction of DR5 by ZER was enhanced significantly in cells overexpressing a different amount of ATF3 (Fig. 2d). Taken together, this indicates that ATF3 plays a role as an activator for the efficient induction of DR5 by ZER and CCB in p53-deficient cells. Furthermore, it also suggests that ATF3 may take part in the induction of DR4. However, the impact of its induction by CCB or ZER was smaller than that of DR5. Therefore, we focused on DR5 from this point forward.
FIGURE 2.
ATF3 is required for DR5 induction by ZER or CCB in p53-deficient cells. a, ATF3 was knocked down in HCT116-p53null cells or SW480 cells, and both cells were treated with 20 μm ZER for the indicated times. DR4, DR5, and ATF3 proteins were analyzed by Western blotting. β-actin was used as a loading control (left panel). The expression of ATF3 and DR5 mRNAs in HCT116-p53null cells was also analyzed by qRT-PCR and normalized to the level of GAPDH (right panel). b, ATF3 knockdown cells were treated with 50 μm CCB, and the expression of DR4, DR5 and ATF3 was assayed as in a. c, cell surface expression of DR5 was measured by flow cytometry. ATF3 knockdown HCT116-p53null cells or siGFP HCT116-p53null cells were treated with ZER (left panel) or CCB (right panel) for 24 h as under “Experimental Procedures.” Shaded no line, vehicle with control IgG; thick line, vehicle with anti-DR5 antibody; thin line, ZER or CCB of siGFP cells with anti-DR5 antibody; dashed line, ZER or CCB of siATF3 cells with anti-DR5 antibody. d, HCT116-p53null cells stably expressing FLAG-ATF3 or GFP were treated with 20 μm ZER for 24 h, and their whole cell extracts were analyzed for DR5 and ATF3 proteins by Western blotting using β-actin as a loading control. *, p < 0.05; **, p < 0.01.
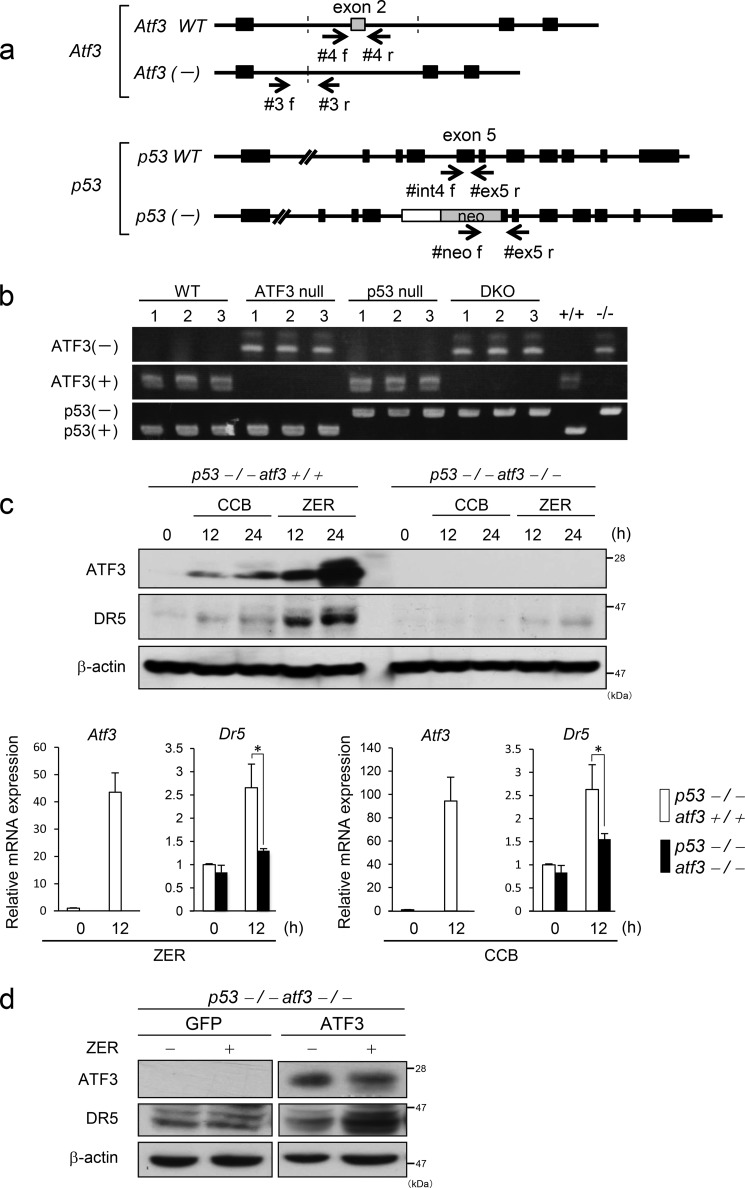
Loss of Atf3 Strongly Inhibits the Induction of DR5 by ZER and CBB in MEFs
To further clarify the role of ATF3 for the p53-independent induction of DR5 by ZER and CCB, we generated Atf3/p53 DKO mice by crossing Atf3 null mice with p53 null mice, as shown in Fig. 3, a and b. The resultant DKO MEFs were treated with ZER or CCB, and the expression of DR5 was examined by Western blotting and qRT-PCR. As shown in Fig. 3c, the induction of DR5 by ZER or CCB was suppressed significantly in DKO MEFs compared with Atf3+/+p53−/− MEFs at both mRNA and protein levels. Next, ATF3 was reintroduced into DKO MEFs, and these cells were tested to see whether the DR5 induction by ZER was rescued. Western blotting showed that the DR5 induction was recovered significantly (Fig. 3d). In the case of mice, a number of DR5 bands on the Western blot appear to be more closely positioned than human DR5. These data support the hypothesis that ATF3 plays a role in efficient DR5 expression by ZER and CCB in p53-deficient MEFs.
FIGURE 3.
Generation of Atf3/p53 knockout mice and the loss of Atf3 impairs DR5 induction by ZER or CBB in p53 null MEFs. a, schematic of the generation of Atf3 and p53 knockout mice. The positions of the primers for the genomic PCR are indicated. b, genomic DNA from newborn mice was extracted and subjected to PCR using the primer sets listed under “Experimental Procedures.” PCR products were analyzed with 2% agarose gel electrophoresis. C, Atf3+/+p53−/− or DKO MEFs were treated with 50 μm CCB or 20 μm ZER for the indicated times, and the expression of DR5 and ATF3 was analyzed by Western blotting (top panel) and qRT-PCR (bottom panel), respectively. d, ATF3 was stably expressed by reintroduction into DKO MEFs, and these cells were treated with ZER. The expression of ATF3 and DR5 proteins was measured by Western blotting using β-actin as a loading control. *, p < 0.05.
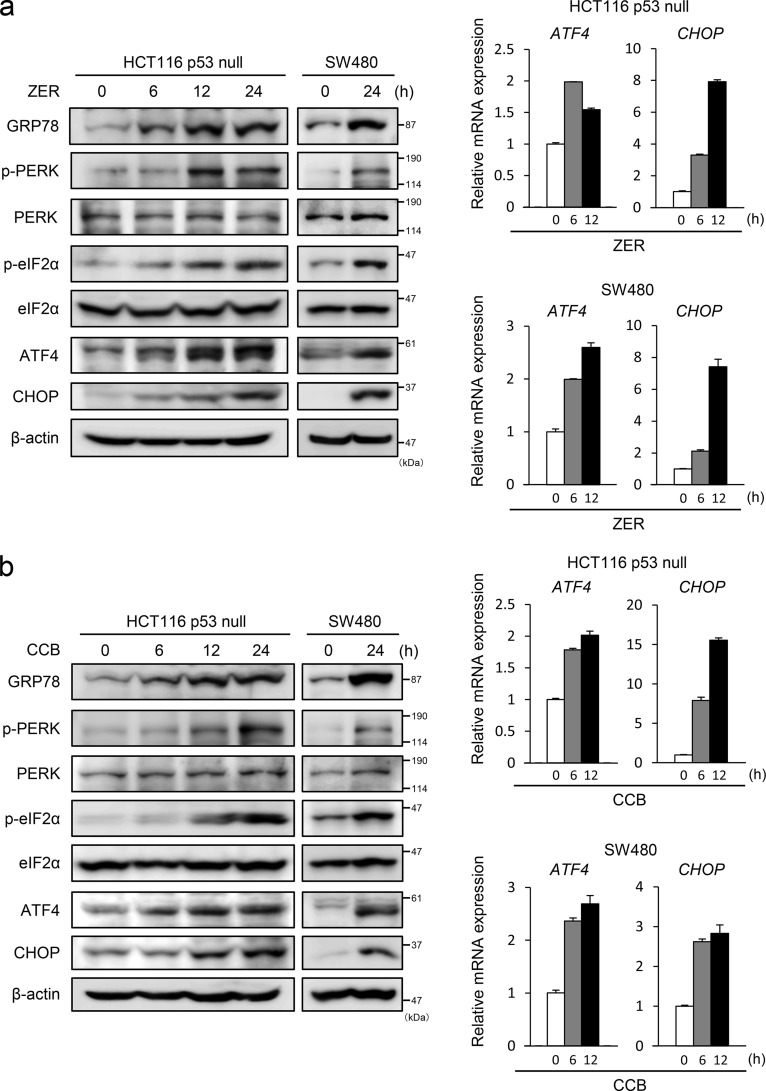
ZER and CCB Cause the Activation of e ER/UPR Pathway and Promote the Expression of ATF4 and CHOP through the Generation of ROS
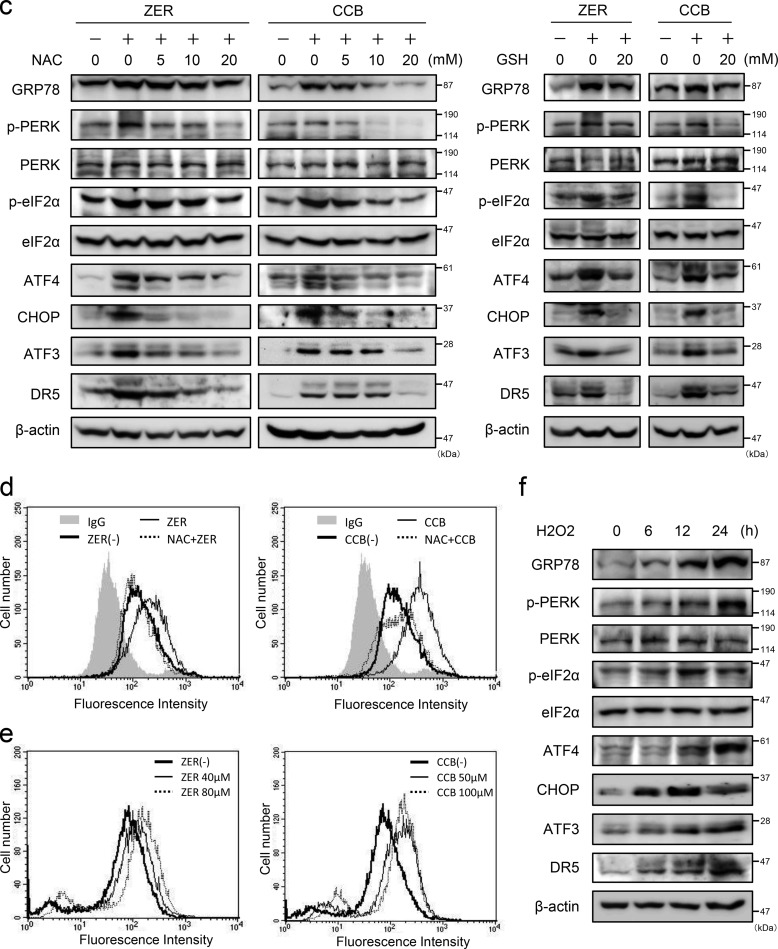
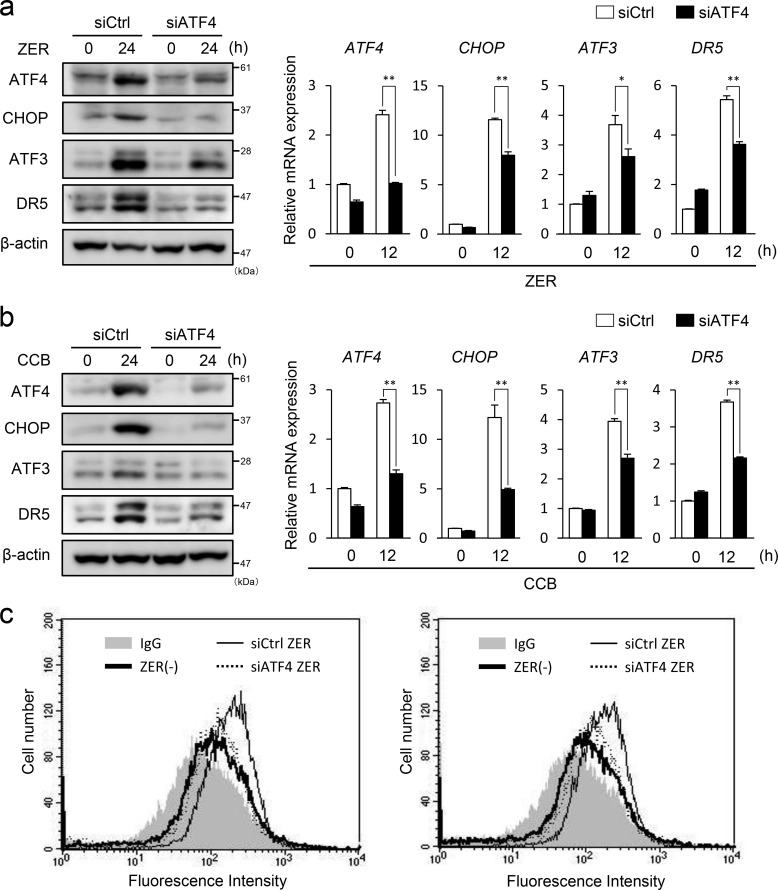
Recently, it has been reported that some agents can induce DR5-dependent apoptosis through ER stress (18–20). Therefore, we examined whether ZER and CCB cause ER stress and induce the expression of ER stress markers. As shown in Fig. 4, a and b, the level of GRP78 and phosphorylation of PERK and eIF2α were elevated by ZER or CCB treatment, indicating that the ER/UPR pathway was activated by these agents. ATF4 and CHOP proteins and their mRNAs were also up-regulated in these cells. These data strongly suggest that ZER and CCB cause ER stress and activate the PERK-eIF2α arm of the UPR to induce the expression of ATF4, CHOP, ATF3, and DR5 (Fig. 1). We next addressed whether ROS play a role in the ER stress induced by ZER or CCB because these agents have been reported to generate ROS (30, 31). As shown in Fig. 4c, left panel, the induction of GRP78 and activation of the PERK-eIF2α pathway were suppressed, and the expression of ATF4, ATF3, CHOP, and DR5 was inhibited by pretreatment of N-acetylcysteine (NAC) in a dose-dependent manner. We further examined the effect of another ROS scavenger, GSH, on this pathway and found that pretreatment of GSH partly suppressed the induction of the ER stress markers ATF3 and DR5 (Fig. 4c, right panel). Importantly, the induction of cell surface expression of DR5 by ZER or CCB was abrogated remarkably in NAC-treated cells (Fig. 4d). To ascertain the involvement of ROS, we measured intracellular ROS levels by FACS analysis and found that ZER and CCB actually induced intracellular ROS (Fig. 4e). Additionally, we treated the cells with H2O2 to examine whether direct application of ROS by H2O2 can stimulate the same pathway. As shown in Fig. 4f, H2O2 also induced the ER stress markers ATF3 and DR5. These results strongly suggested that ZER and CCB provoke ER stress, at least in part, via generation of ROS. To further clarify the pathway of DR5 induction, we examined the role of ATF4. As shown in Fig. 5, in ATF4 knockdown cells, the induction of CHOP, ATF3, and DR5 by ZER or CCB was reduced significantly at both the protein and mRNA levels (Figs. 5, a and b). The induced expression of DR5 on the cell surface was also reduced in these cells (Fig. 5c). By contrast, ATF3 gene silencing did not show a significant effect on the induction of ATF4 or CHOP by ZER or CCB, whereas it suppressed DR5 induction (data not shown and Fig. 2). These results clearly support that ER stress is caused by ZER and CCB, followed by the activation of the ATF4/ATF3/CHOP pathway and DR5 gene transcription.
FIGURE 4.
The UPR is activated by ZER and CCB, and ROS scavengers inhibit it activation and cell surface expression of DR5. HCT116-p53null cells or SW480 cells were treated with 20 μm ZER (a) or 50 μm CCB (b) for the indicated times, and the expression of GRP78, PERK, eIF2α, ATF4, and CHOP proteins was examined by Western blotting. β-actin was used as a loading control (left panel). ATF4 and CHOP mRNAs were also measured by qRT-PCR and normalized to GAPDH (right panel). c, HCT116-p53null cells were incubated with the indicated concentrations of NAC (left panel) or GSH (right panel) for 1 h, followed by ZER or CCB treatment. Cell extracts were assayed as in a. d, cell surface expression of DR5 of NAC-treated HCT116-p53null cells was measured following treatment with 20 μm ZER (left panel) or 50 μm CCB (right panel) for 24 h as under “Experimental Procedures.” Shaded no line, vehicle with control IgG; thick line, vehicle with anti-DR5 antibody; thin line, ZER or CCB of control cells with anti-DR5 antibody; dashed line, ZER or CCB of NAC-treated cells with anti-DR5 antibody. e, after treatment with the indicated concentrations of ZER (left panel) or CCB (right panel) for 24 h, HCT116-p53null cells were labeled with 2′,7′-dichlorodihydrofluorescein diacetate, and intracellular ROS levels were measured by flow cytometry. f, HCT116-p53null cells were incubated with 0.2 mm H2O2 for 30 min, and then cells were collected at the indicated time points and assayed as in a.
FIGURE 5.
ATF4 knockdown suppresses ZER- or CCB-induced expression of CHOP, ATF3, and DR5. ATF4 was knocked down in HCT116-p53null cells, and these cells were treated with 40 mm ZER (a) or 50 mm CCB (b) for 24 h. Expression of ATF4, CHOP, ATF3, and DR5 was measured by Western blotting (left panel) and qRT-PCR (right panel), respectively. siCtrl, control siRNA. c, cell surface expression of DR5 of control or siATF4 cells was measured after ZER (left panel) or CCB (right panel) treatment as under “Experimental Procedures.” Shaded no line, vehicle with control IgG; thick line, vehicle with anti-DR5 antibody; thin line, ZER or CCB of control cells with anti-DR5 antibody; dashed line, ZER or CCB of siATF4 cells with anti-DR5 antibody. *, p < 0.05; **, p < 0.01.
ATF3 and CHOP Bind and Activate the DR5 Gene Promoter in Response to ZER
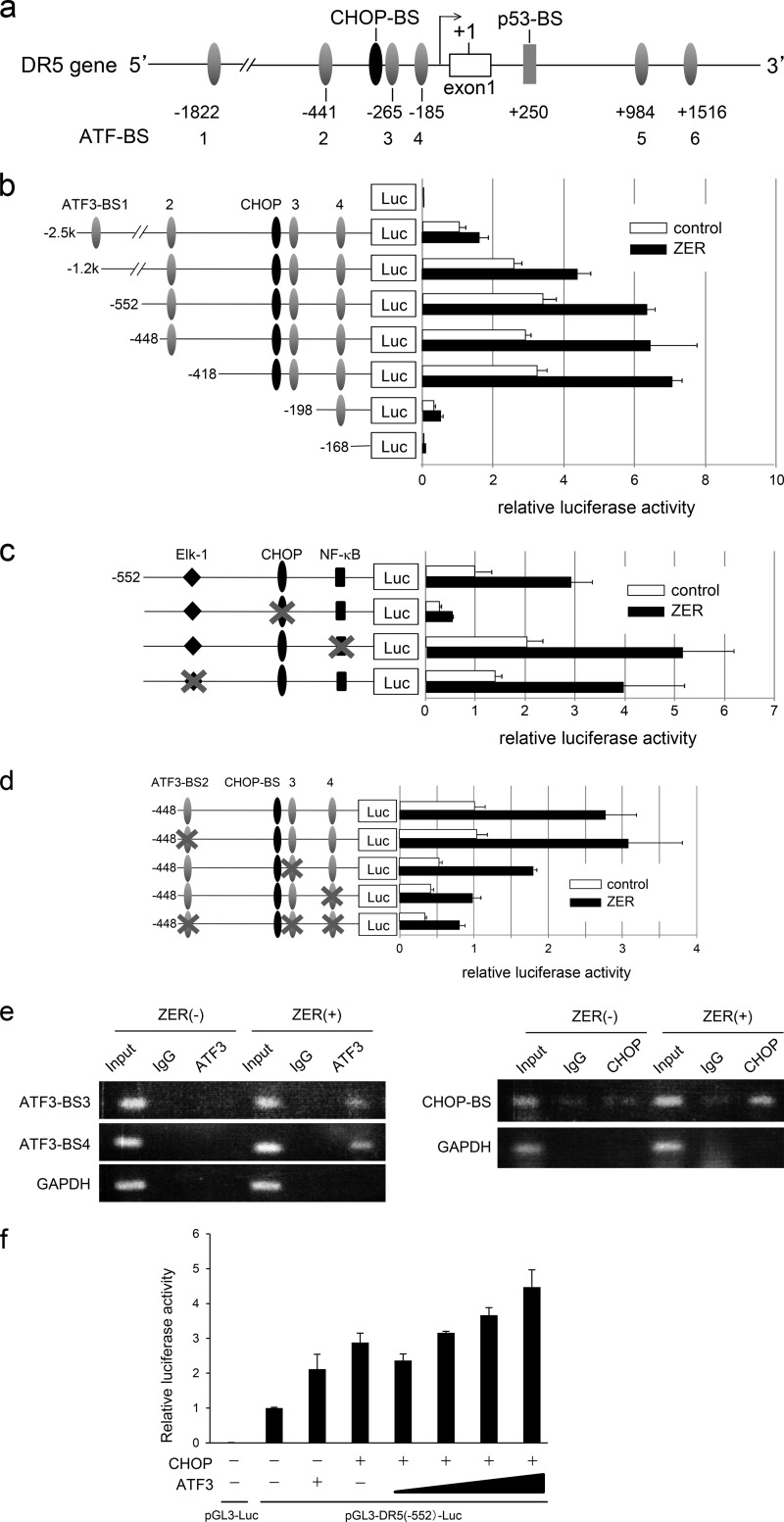
The data above strongly support the hypothesis that ATF3 plays a role in p53-independent activation of DR5 gene transcription upon ZER or CCB treatment. We have shown previously that ATF3 was recruited onto the DR5 promoter in response to DNA damage and induced DR5 gene luciferase activity in collaboration with p53 (12). Thus, the role of ATF3 or other factor(s) in DR5 gene activation was explored. As shown in Fig. 6a, the human DR5 gene promoter contains at least six ATF/CRE binding sites (ATF-BS1 to ATF-BS6) (12), each CHOP, Elk-1, and NF-κB site in the proximal region and a p53 binding site in the first intron. We first analyzed the human DR5 gene luciferase activity using reporter plasmids that lack the p53 site to exclude its effect. Reporter constructs containing various 5′ deletions down to −418 were clearly activated by ZER. However, further deletion to −198 remarkably reduced the activation, and −168 showed only background activity (Fig. 6b), indicating that p53-independent ZER responsive element(s) is/are localized between −418 and −168. This region contains ATF-BS3 and ATF-BS4 and putative sites for Elk-1, CHOP, and NF-κB. To test the role of these sites, we measured the activity of reporters with a mutation of each binding site. As shown in Fig. 6c, the CHOP mutation significantly reduced the activation, whereas neither the Elk1 nor the NF-κB mutants showed significant effects. We further examined the effect of mutations of ATF-BS sites. Among the mutants tested, ATF-BS4 showed a remarkable suppression and BS3 exhibited a modest but significant reduction, whereas ATF-BS2 had no effect (Fig. 6d). These data strongly support the hypothesis that both the CHOP and ATF3 sites play roles in activating the DR5 gene promoter in response to ZER. Thus, we addressed whether ATF3 and CHOP are recruited onto the corresponding motifs of the DR5 gene promoter in response to ZER treatment. As in Fig. 6e, ATF3 protein was indeed recruited onto ATF-BS3 and ATF-BS4. Under these conditions, CHOP protein also bound to the CHOP element region. Further, the effect of overexpression of ATF3 and CHOP on reporter activity was examined to address whether ATF3 and CHOP cooperatively activate the DR gene promoter. As shown in Fig. 6f, either ATF3 or CHOP alone produced a modest activation of reporter activity. In the presence of both proteins, however, reporter activity was activated further. Collectively, this clearly indicates that ATF3 and CHOP play a role in activating DR5 gene expression in response to ZER treatment.
FIGURE 6.
ATF3 and CHOP are required for ZER-induced activation of the DR5 gene promoter. a, schematic of the human DR5 gene promoter region. Six ATF/CREB binding sites (ATF-BS1 through ATF-BS6) and binding sites for CHOP (CHOP-BS) and p53 (p53-BS) are depicted. b, HCT116-p53null cells were treated with ZER for 12 h, and a ChIP assay was carried out using anti-ATF3 or anti-CHOP antibodies as under “Experimental Procedures.” Luc, luciferase. c, each of the 5′ deletion mutants of the human DR5 gene reporter plasmid was transfected into HCT116-p53null cells. 24 h post-transfection, cells were treated with 20 μm ZER for 12 h, and luciferase activity was measured. Relative luciferase activity represents the fold induction compared with that of non-stimulated cells transfected with pDR5-Luc(-2.5k). d, cells were transfected with pDR5-Luc(-552) reporters harboring mutations at binding sites for CHOP, NF-κB, or Elk1 and treated as in c. Relative luciferase activity represents the fold induction compared with that of untreated pDR5-Luc(-552). e, cells were transfected with pDR5-Luc(-448) containing mutations at ATF-BS2, ATF-BS3, or ATF-BS4 and treated with ZER as in c. Relative luciferase activity represents the fold induction compared with that of untreated pDR5-Luc(-448). f, cells were cotransfected with pDR5-Luc(-552) and each of the expression plasmids for ATF3 or CHOP. Relative luciferase activity represents the fold induction compared with that of cells transfected with empty vector. Data are mean ± S.E. of three independent experiments.
ATF3 Sensitizes p53-deficient Colorectal Cancer Cells to Apoptosis by Combined Treatment with ZER/TRAIL
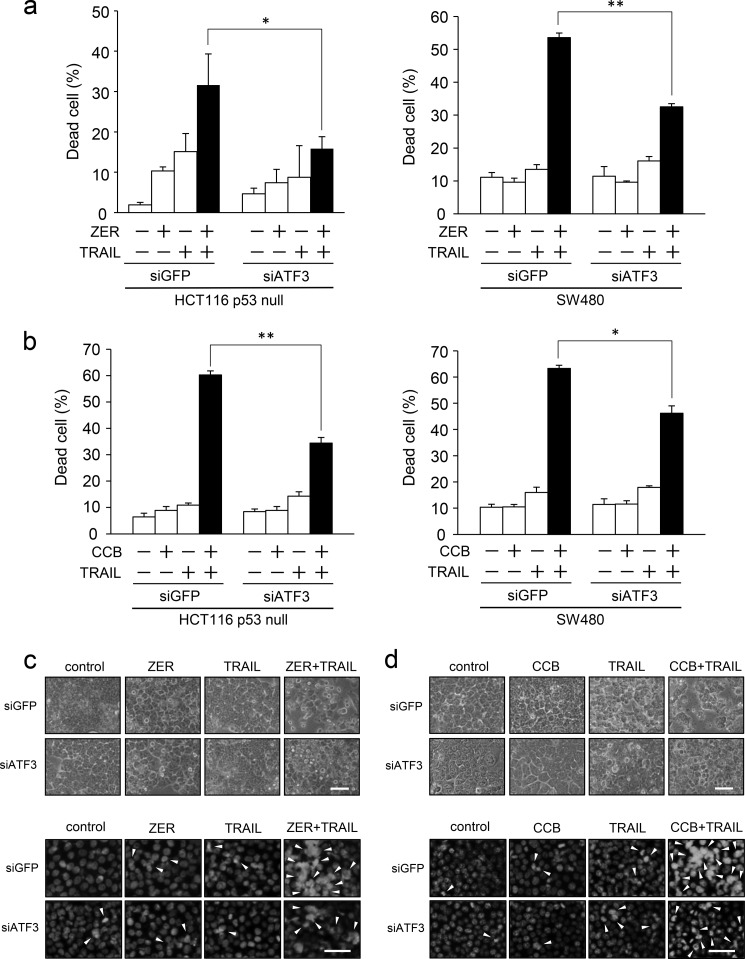
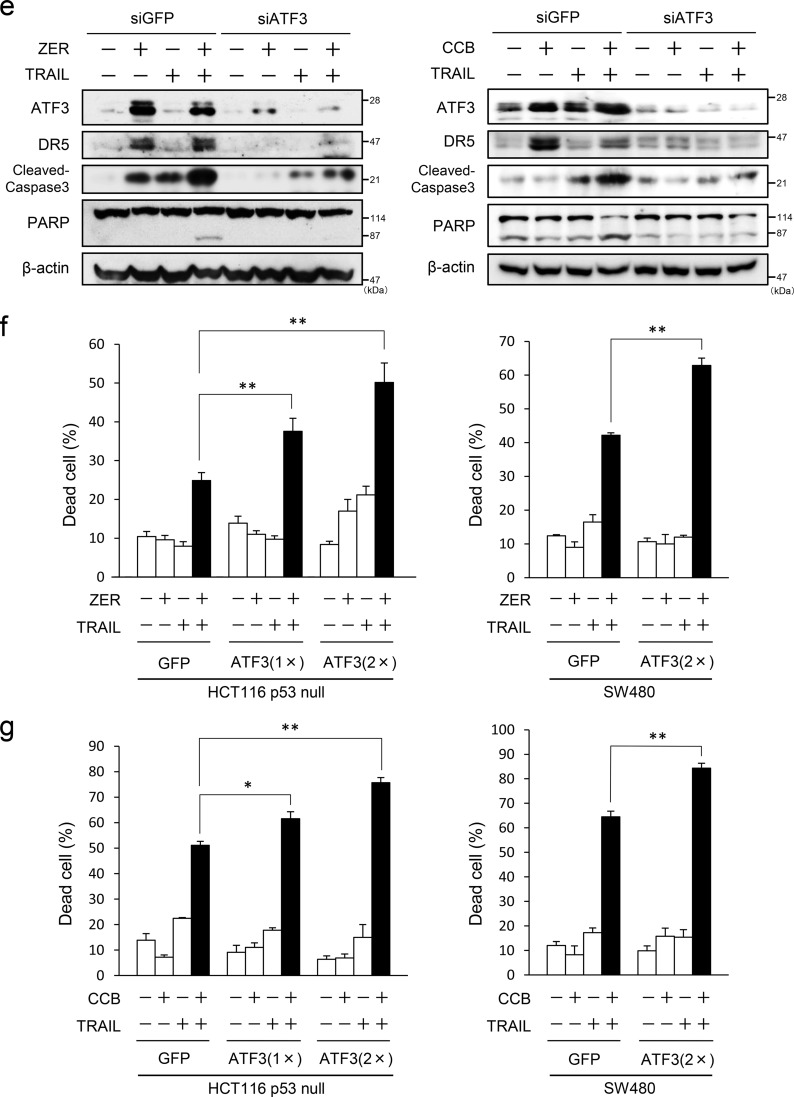
By the data above it was demonstrated that ATF3 regulates the cell surface expression of DR5 upon ZER or CCB treatment in p53-deficient human colorectal cells. Therefore, we examined the effect of ATF3 on cancer cell death by the combination of TRAIL and ZER or CCB. To this end, ATF3 was knocked down in HCT116-p53null and SW480 cells, and these cells were stimulated with ZER or CCB in combination with TRAIL. As shown in Fig. 7a, ZER or TRAIL alone showed a modest increase in cell death. Cotreatment of control siGFP cells with ZER and TRAIL markedly increased cell death. However, the increase in cell death was inhibited significantly in ATF3 knockdown cells. The results of cotreatment with CCB showed the same tendency (Fig. 7b). A morphological study showed that the combination of TRAIL and ZER (Fig. 7c) or CCB (Fig. 7d) increased the number of control cells with the property of apoptosis but that the appearance of these cells was reduced significantly in ATF3 knockdown cells. Biochemical analysis also showed that ATF3 knockdown inhibited not only the induction of DR5 but the cleavage of caspase 3 and poly (ADP-ribose) polymerase (PARP), markers of apoptotic cell death compared with siGFP cells (Fig. 7e). Next, the effect of ATF3 overexpression on the induction of cell death was also examined. Cell death by cotreatment of ZER/TRAIL (Fig. 7f) or CCB/TRAIL (Fig. 7g) was enhanced significantly by exogenous ATF3 expression compared with the GFP control. Taken together, this indicates that ATF3 induction by ZER or CCB is required for increased sensitivity of apoptosis to the combined treatment with TRAIL in p53-deficient colon cancer cells.
FIGURE 7.
ATF3 promotes the ZER or CCB/TRAIL-induced apoptosis of p53-deficient colorectal cancer cells. a, ATF3 was knocked down in HCT116-p53null cells (left panel) or SW480 cells (right panel), and cells were treated with 30 μm ZER and/or 2.5 ng/ml TRAIL for 24 h. Cell death was measured using a trypan blue exclusion assay. The proportions of dead cells from three independent experiments are shown. b, ATF3 knockdown cells were treated with 75 μm CCB and/or 2.5 ng/ml TRAIL for 24 h. Cell death was measured as in a. HCT116-p53null cells were treated with 20 μm ZER (c) or 50 μm CCB (d) and/or 2.5 ng/ml TRAIL for 24 h. The cells were examined by phase-contrast microscopy (top panel) or fluorescence microscope after staining with DAPI (bottom panel). Arrowheads indicate cells with condensation or fragmentation of nuclei. Scale bars = 50 μm. e, whole cell extracts were prepared from cells treated as in a and analyzed by Western blotting for cleaved caspase 3, cleaved PARP, ATF3, and DR5 proteins. β-actin was used as a loading control. f, HCT116-p53null cells (left panel) or SW480 cells (right panel) stably expressing FLAG-ATF3 or GFP were treated with 30 μm ZER and/or 2.5 ng/ml TRAIL for 24 h, followed by a trypan blue exclusion assay as in a. g, ATF3 overexpressed cells as in f were treated with 75 μm CCB and/or 2.5 ng/ml TRAIL for 24 h, followed by a trypan blue exclusion assay as in a. Data are mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01.
DISCUSSION
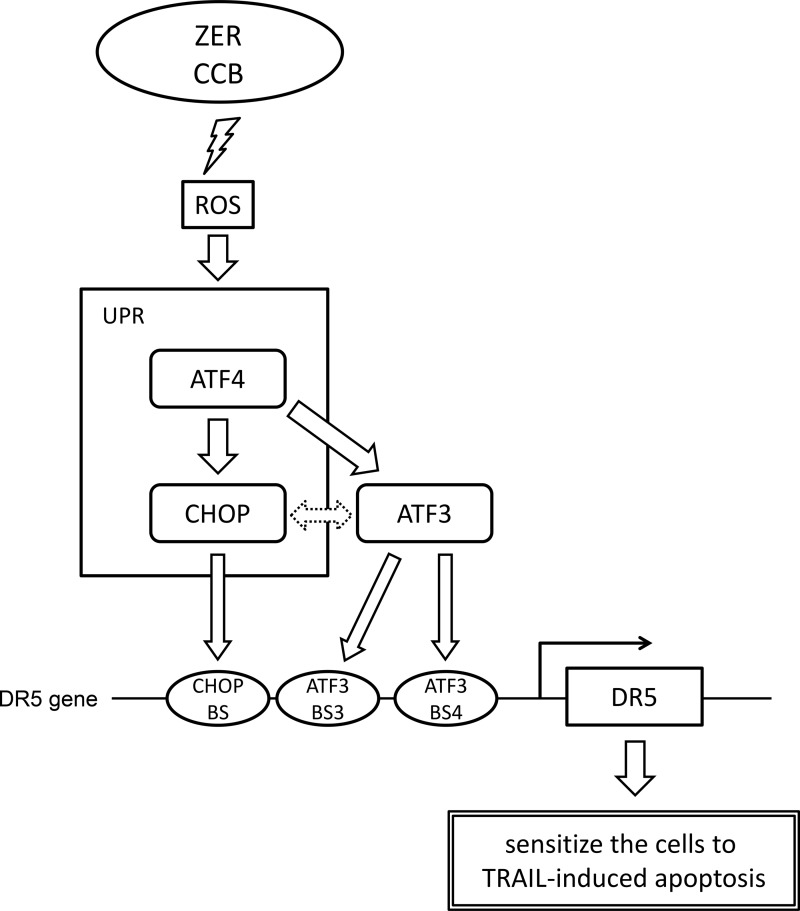
We have shown previously that stress-induced transcription factor ATF3, a direct target of p53, trans-activates DR5 expression upon DNA damage through cooperative binding on the chromatin and promotes the TRAIL-mediated cell death of human colon cancer cells (12). In this study, ZER and CCB, which are known to induce the expression of DR5 (30, 32–35), cause ER stress, activate the PERK/eIF2α branch of the UPR, and lead to the induction of the ATF4/CHOP/ATF3/DR5 transcriptional cascade through the generation of ROS in p53-deficient human colon cancer cells. The expression of DR5 protein on the cell surface is induced, and the combined treatment with TRAIL sensitizes cells to apoptosis, as shown in Fig. 8. This model highlights the role of the transcription factors ATF3 and CHOP to trans-activate the DR5 gene transcription in p53-deficient cells, whereas ATF3 cooperates with p53 upon DNA damage in cells with wild-type p53 (12).
FIGURE 8.
Proposed mechanism of ATF3 in the p53-independent induction of DR5 gene expression by ZER or CCB. In cells with p53 mutation, ZER or CCB provokes ER stress via generation of ROS and activates the PERK-eIF2α branch of the UPR, followed by the cascaded transcriptional program of ATF4/CHOP/ATF3. ATF3 and CHOP cooperatively transactivate DR5 gene transcription through their recruitment onto ATF3-BS3/4 and CHOP-BS of the promoter. The dashed line with the arrow between ATF3 and CHOP represents no mutual negative regulation, as mentioned under “Discussion.” The enhanced expression of DR5 on the cell surface sensitizes colon cancer cells to TRAIL-induced apoptosis. BS, binding site.
ATF3 is a downstream target gene of p53 but also an integral component of the PERK/eIF2α kinase and IRE1/TRAF2 pathways of the UPR (26–28, 42), and it was highly induced in ZER- or CCB-treated, p53-deficient human colon cancer cells along with CHOP (Figs. 1 and 4). It should be noted here that there is a mutual negative regulation between ATF3 and CHOP. That is, ATF3 and CHOP form an inactive heterodimer to repress ATF3 function (43), and ATF3 represses the expression of CHOP through its binding to the CHOP promoter (44). Moreover, a direct interaction between ATF3 and CHOP to form a heterocomplex has also been reported by a combinatorial analysis of transcription factors (45). In this study, however, we neither detected the up-regulation of CHOP mRNA and protein by ZER or CCB in ATF3 knockdown cells nor observed the measurable ATF3-CHOP complex by immunoprecipitation in ZER-treated cells (data not shown). By contrast, both ATF3 and CHOP were recruited concomitantly onto the DR5 gene promoter in a ChIP assay (Fig. 6b), and the presence of both factors further activated the reporter activity of the DR5 gene (Fig. 6e). Therefore, it is likely that ATF3 and CHOP do not function as mutual repressors but cooperatively up-regulate DR5 transcription, at least in cells treated with ZER or CCB. Furthermore, many other factors may also play roles in ZER- or CCB-induced DR5 gene induction because it has been reported that the DR5 gene promoter is activated by SP1 (21), NF-κB (46, 47), or Elk1 (34).
ZER has both anti-inflammatory and anticancer activities (31). The inhibition of NF-κB has been proposed as a major mechanism for these effects (48), whereas it has been reported that NF-κB activates the DR5 gene in immune or stress response (46, 47). In this study, however, the induction of the DR5 reporter gene by ZER was not affected by mutation of the NF-κB site (Fig. 6d) but was enhanced by cooperative activation by ATF3 and CHOP (Fig. 6e). Therefore, it seems unlikely that NF-κB plays a role, at least in ZER-induced DR5 up-regulation. By contrast, the inhibition of NF-κB activity by ZER (48) might be implicated in overcoming TRAIL resistance. Indeed, NF-κB can induce the transcription of antiapoptotic genes such as c-FLIP, cIAP1/2, XIAP, and Bcl-KL, and its activation may cause TRAIL resistance (9, 49). In this regard, ATF3 can interact with NF-κB, especially with the p65 subunit, and suppresses the transcription of NF-κB target genes such as cIAP2 and XIAP (50–52). Therefore, ATF3 not only up-regulates the TRAIL receptor but also inhibits the NF-κB activity to overcome TRAIL resistance.
This study also showed that ZER-induced DR5 expression was abolished by ROS scavengers, consistent with a previous report (30, 53, 54). More recently, it has been reported that ZER induces cellular proteostress through its binding to cellular proteins to activate the ubiquitin-proteasome system or heat shock response (36, 37). This may provide another mechanism by which ZER causes ER stress and activates the UPR through the generation of ROS, as reported in this study.
CCB is reported to have several COX-2-independent anticancer activities (55). Among those, CCB inhibits the ER Ca2+ pump to aggravate ER stress and the UPR via PERK-mediated phosphorylation of eIF2α along with greatly increased expression of GRP78 and CHOP (56, 57). Consistent with this, CCB activated the PERK-eIF2α arm of the UPR in human colon cancer cells, and this response resulted in the up-regulation of the ATF4-ATF3-CHOP-DR5 pathway (Figs. 1 and 4, a and b). These effects were abolished by ROS scavengers, suggesting the crucial role of ROS in CCB and ZER effects (Fig. 4, c and d). Both ZER and CCB have been reported to enhance ROS generation (30, 31). CCB increases the mitochondrially derived rather than NADPH-dependent ROS independently of COX2 activity (58). Although the molecular detail of the induction of ROS by ZER is still obscure, ZER has been found to bind directly with Keap1, which regulates Nrf2, a transcription factor related to the antioxidant response (59). Recent work with non-coxib analogs such as dimethyl-celecoxib (57) and OSU-03012 (60) has revealed their stronger cytotoxic efficacy compared with its parental compound, partly because of ER stress/UPR by these compounds. Indeed, OSU-03012 has been reported to induce ROS-mediated cytotoxicity (60), which suggests a role of ER stress/UPR in the action of OSU-03012 because ER stress and oxidative stress constitute a vicious cycle in the UPR (15, 16). It would be interesting to elucidate whether non-coxib agents cause ER stress/UPR to up-regulate the expression of DR5 and sensitize cancer cell death.
It has been suggested that cancer cells become TRAIL-resistant through the deterioration of DR5 cell surface transport even though the total amount of DR5 is comparable (61). In addition, large amounts of DR5 were localized in the nucleus in HeLa and HepG2 cells (62). We did confirm that CCB or ZER induces the increased expression of DR5 on the cell surface. However, the stability of DR5 mRNA or protein has not been investigated. To maximize the effect of TRAIL therapy, efficient cell surface transport and prolonged half-life of DR5 seem to be essential. We must await further studies to clarify the details of the regulation of DR5 localization and half-life induced by CCB or ZER.
In summary, this study showed that ZER and CCB, which have anticancer activity, cause ER stress and activate the UPR in p53-deficient human colon cancer cells. More importantly, this study specifically details a molecular mechanism of stress response gene ATF3, which up-regulates DR5 independently of p53 and therefore induces ultimate TRAIL sensitization of cancer cells. Because the ER stress response or its aggravation by agents can be utilized to overcome TRAIL resistance in advanced cancer, ATF3 might be also a target or biomarker for novel therapies for cancers with p53 mutation.
Acknowledgments
We thank Drs. T. Kitamura, H. G. Wang, and T. Sakai for plasmids.
This study was supported by JSPS KAKENHI Grants 24590373 and 25134708 (to S. K.) and 24118002 (to J. K.).
- DR
- death receptor
- TRAIL
- TNF-related apoptosis-inducing ligand
- ER
- endoplasmic reticulum
- ROS
- reactive oxygen species
- UPR
- unfolded protein response
- PERK
- protein kinase-like endoplasmic reticulum kinase
- ATF
- activating transcription factor
- CHOP
- C/EBP homologous protein
- CREB
- cAMP response element-binding protein
- ZER
- zerumbone
- CCB
- celecoxib
- MEF
- mouse embryonic fibroblast
- DKO
- double knockout
- qRT-PCR
- quantitative reverse transcription PCR
- NAC
- N-acetylcysteine.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2011) The hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 2. Farczádi E., Szántó J., Kaszás I., Benyó I., Bodnár Z., Szlobodnyik J., Szende B. (1999) Changes in apoptotic and mitotic activity in rectal carcinoma after short-term cytostatic therapy as possible predictive factors. Neoplasma 46, 219–223 [PubMed] [Google Scholar]
- 3. Scott N., Hale A., Deakin M., Hand P., Adab F. A., Hall C., Williams G. T., Elder J. B. (1998) A histopathological assessment of the response of rectal adenocarcinoma to combination chemo-radiotherapy: relationship to apoptotic activity, p53 and bcl-2 expression. Eur. J. Surg. Oncol. 24, 169–173 [DOI] [PubMed] [Google Scholar]
- 4. Pan G., Ni J., Wei Y. F., Yu G., Gentz R., Dixit V. M. (1997) An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277, 815–818 [DOI] [PubMed] [Google Scholar]
- 5. Walczak H., Degli-Esposti M. A., Johnson R. S., Smolak P. J., Waugh J. Y., Boiani N., Timour M. S., Gerhart M. J., Schooley K. A., Smith C. A., Goodwin R. G., Rauch C. T. (1997) TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 16, 5386–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu G. S., Burns T. F., McDonald E. R., 3rd, Jiang W., Meng R., Krantz I. D., Kao G., Gan D. D., Zhou J. Y., Muschel R., Hamilton S. R., Spinner N. B., Markowitz S., Wu G., el-Deiry W. S. (1997) KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 17, 141–143 [DOI] [PubMed] [Google Scholar]
- 7. Chaudhary P. M., Eby M., Jasmin A., Bookwalter A., Murray J., Hood L. (1997) Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-κB pathway. Immunity 7, 821–830 [DOI] [PubMed] [Google Scholar]
- 8. Gonzalvez F., Ashkenazi A. (2010) New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene 29, 4752–4765 [DOI] [PubMed] [Google Scholar]
- 9. Dimberg L. Y., Anderson C. K., Camidge R., Behbakht K., Thorburn A., Ford H. L. (2013) On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 32, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sheikh M. S., Burns T. F., Huang Y., Wu G. S., Amundson S., Brooks K. S., Fornace A. J., Jr., el-Deiry W. S. (1998) p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor α. Cancer Res. 58, 1593–1598 [PubMed] [Google Scholar]
- 11. Xiang H., Fox J. A., Totpal K., Aikawa M., Dupree K., Sinicropi D., Lowe J., Escandón E. (2002) Enhanced tumor killing by Apo2L/TRAIL and CPT-11 co-treatment is associated with p21 cleavage and differential regulation of Apo2L/TRAIL ligand and its receptors. Oncogene 21, 3611–3619 [DOI] [PubMed] [Google Scholar]
- 12. Taketani K., Kawauchi J., Tanaka-Okamoto M., Ishizaki H., Tanaka Y., Sakai T., Miyoshi J., Maehara Y., Kitajima S. (2012) Key role of ATF3 in p53-dependent DR5 induction upon DNA damage of human colon cancer cells. Oncogene 31, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 13. Malhotra J. D., Kaufman R. J. (2007) The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 15. Malhotra J. D., Kaufman R. J. (2007) Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid. Redox Signal 9, 2277–2293 [DOI] [PubMed] [Google Scholar]
- 16. Higa A., Chevet E. (2012) Redox signaling loops in the unfolded protein response. Cell. Signal. 24, 1548–1555 [DOI] [PubMed] [Google Scholar]
- 17. Ma Y., Hendershot L. M. (2004) The role of the unfolded protein response in tumour development: friend or foe? Nat. Rev. Cancer 4, 966–977 [DOI] [PubMed] [Google Scholar]
- 18. Yamaguchi H., Wang H. G. (2004) CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 279, 45495–45502 [DOI] [PubMed] [Google Scholar]
- 19. Moon D. O., Park S. Y., Choi Y. H., Ahn J. S., Kim G. Y. (2011) Guggulsterone sensitizes hepatoma cells to TRAIL-induced apoptosis through the induction of CHOP-dependent DR5: involvement of ROS-dependent ER-stress. Biochem. Pharmacol. 82, 1641–1650 [DOI] [PubMed] [Google Scholar]
- 20. Xu L., Su L., Liu X. (2012) PKCδ regulates death receptor 5 expression induced by PS-341 through ATF4-ATF3/CHOP axis in human lung cancer cells. Mol. Cancer Ther. 11, 2174–2182 [DOI] [PubMed] [Google Scholar]
- 21. Kim Y. H., Park J. W., Lee J. Y., Kwon T. K. (2004) Sodium butyrate sensitizes TRAIL-mediated apoptosis by induction of transcription from the DR5 gene promoter through Sp1 sites in colon cancer cells. Carcinogenesis 25, 1813–1820 [DOI] [PubMed] [Google Scholar]
- 22. Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., Sivaprasad U. (1999) ATF3 and stress responses. Gene Expr. 7, 321–335 [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang C., Gao C., Kawauchi J., Hashimoto Y., Tsuchida N., Kitajima S. (2002) Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem. Biophys. Res. Commun. 297, 1302–1310 [DOI] [PubMed] [Google Scholar]
- 24. Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., Ruan Y. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 25. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 26. Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. (2004) Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai Y., Zhang C., Nawa T., Aso T., Tanaka M., Oshiro S., Ichijo H., Kitajima S. (2000) Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH(2)-terminal kinase and promoter response element. Blood 96, 2140–2148 [PubMed] [Google Scholar]
- 28. Gjymishka A., Su N., Kilberg M. S. (2009) Transcriptional induction of the human asparagine synthetase gene during the unfolded protein response does not require the ATF6 and IRE1/XBP1 arms of the pathway. Biochem. J. 417, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim M., Miyamoto S., Yasui Y., Oyama T., Murakami A., Tanaka T. (2009) Zerumbone, a tropical ginger sesquiterpene, inhibits colon and lung carcinogenesis in mice. Int. J. Cancer 124, 264–271 [DOI] [PubMed] [Google Scholar]
- 30. Yodkeeree S., Sung B., Limtrakul P., Aggarwal B. B. (2009) Zerumbone enhances TRAIL-induced apoptosis through the induction of death receptors in human colon cancer cells: Evidence for an essential role of reactive oxygen species. Cancer Res. 69, 6581–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prasannan R., Kalesh K. A., Shanmugam M. K., Nachiyappan A., Ramachandran L., Nguyen A. H., Kumar A. P., Lakshmanan M., Ahn K. S., Sethi G. (2012) Key cell signaling pathways modulated by zerumbone: role in the prevention and treatment of cancer. Biochem. Pharmacol. 84, 1268–1276 [DOI] [PubMed] [Google Scholar]
- 32. Liu X., Yue P., Zhou Z., Khuri F. R., Sun S. Y. (2004) Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J. Natl. Cancer Inst. 96, 1769–1780 [DOI] [PubMed] [Google Scholar]
- 33. He Q., Luo X., Jin W., Huang Y., Reddy M. V., Reddy E. P., Sheikh M. S. (2008) Celecoxib and a novel COX-2 inhibitor ON09310 up-regulate death receptor 5 expression via GADD153/CHOP. Oncogene 27, 2656–2660 [DOI] [PubMed] [Google Scholar]
- 34. Oh Y. T., Liu X., Yue P., Kang S., Chen J., Taunton J., Khuri F. R., Sun S. Y. (2010) ERK/ribosomal S6 kinase (RSK) signaling positively regulates death receptor 5 expression through co-activation of CHOP and Elk1. J. Biol. Chem. 285, 41310–41319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grösch S., Tegeder I., Niederberger E., Bräutigam L., Geisslinger G. (2001) COX-2 independent induction of cell cycle arrest and apoptosis in colon cancer cells by the selective COX-2 inhibitor celecoxib. FASEB J. 15, 2742–2744 [DOI] [PubMed] [Google Scholar]
- 36. Ohnishi K., Ohkura S., Nakahata E., Ishisaka A., Kawai Y., Terao J., Mori T., Ishii T., Nakayama T., Kioka N., Matsumoto S., Ikeda Y., Akiyama M., Irie K., Murakami A. (2013) Non-specific protein modifications by a phytochemical induce heat shock response for self-defense. PloS ONE 8, e58641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohnishi K., Nakahata E., Irie K., Murakami A. (2013) Zerumbone, an electrophilic sesquiterpene, induces cellular proteo-stress leading to activation of ubiquitin-proteasome system and autophagy. Biochem. Biophys. Res. Commun. 430, 616–622 [DOI] [PubMed] [Google Scholar]
- 38. Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr., Butel J. S., Bradley A. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 [DOI] [PubMed] [Google Scholar]
- 39. Kawauchi J., Zhang C., Nobori K., Hashimoto Y., Adachi M. T., Noda A., Sunamori M., Kitajima S. (2002) Transcriptional repressor activating transcription factor 3 protects human umbilical vein endothelial cells from tumor necrosis factor-α-induced apoptosis through down-regulation of p53 transcription. J. Biol. Chem. 277, 39025–39034 [DOI] [PubMed] [Google Scholar]
- 40. Morita S., Kojima T., Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 41. Wagner K. W., Punnoose E. A., Januario T., Lawrence D. A., Pitti R. M., Lancaster K., Lee D., von Goetz M., Yee S. F., Totpal K., Huw L., Katta V., Cavet G., Hymowitz S. G., Amler L., Ashkenazi A. (2007) Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 13, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 42. Zhang C., Kawauchi J., Adachi M. T., Hashimoto Y., Oshiro S., Aso T., Kitajima S. (2001) Activation of JNK and transcriptional repressor ATF3/LRF1 through the IRE1/TRAF2 pathway is implicated in human vascular endothelial cell death by homocysteine. Biochem. Biophys. Res. Commun. 289, 718–724 [DOI] [PubMed] [Google Scholar]
- 43. Chen B. P., Wolfgang C. D., Hai T. (1996) Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell Biol. 16, 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolfgang C. D., Chen B. P., Martindale J. L., Holbrook N. J., Hai T. (1997) Gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell Biol. 17, 6700–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ravasi T., Suzuki H., Cannistraci C. V., Katayama S., Bajic V. B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N., Carninci P., Daub C. O., Forrest A. R., Gough J., Grimmond S., Han J. H., Hashimoto T., Hide W., Hofmann O., Kamburov A., Kaur M., Kawaji H., Kubosaki A., Lassmann T., van Nimwegen E., MacPherson C. R., Ogawa C., Radovanovic A., Schwartz A., Teasdale R. D., Tegnér J., Lenhard B., Teichmann S. A., Arakawa T., Ninomiya N., Murakami K., Tagami M., Fukuda S., Imamura K., Kai C., Ishihara R., Kitazume Y., Kawai J., Hume D. A., Ideker T., Hayashizaki Y. (2010) An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140, 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ravi R., Bedi G. C., Engstrom L. W., Zeng Q., Mookerjee B., Gélinas C., Fuchs E. J., Bedi A. (2001) Regulation of death receptor expression and TRAIL/Apo2L-induced apoptosis by NF-κB. Nat. Cell Biol. 3, 409–416 [DOI] [PubMed] [Google Scholar]
- 47. Shetty S., Graham B. A., Brown J. G., Hu X., Vegh-Yarema N., Harding G., Paul J. T., Gibson S. B. (2005) Transcription factor NF-κB differentially regulates death receptor 5 expression involving histone deacetylase 1. Mol. Cell Biol. 25, 5404–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takada Y., Murakami A., Aggarwal B. B. (2005) Zerumbone abolishes NF-κB and IκBα kinase activation leading to suppression of antiapoptotic and metastatic gene expression, up-regulation of apoptosis, and downregulation of invasion. Oncogene 24, 6957–6969 [DOI] [PubMed] [Google Scholar]
- 49. Gyrd-Hansen M., Meier P. (2010) IAPs: from caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 50. Kaszubska W., Hooft van Huijsduijnen R., Ghersa P., DeRaemy-Schenk A. M., Chen B. P., Hai T., DeLamarter J. F., Whelan J. (1993) Cyclic AMP-independent ATF family members interact with NF-κB and function in the activation of the E-selectin promoter in response to cytokines. Mol. Cell Biol. 13, 7180–7190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hua B., Tamamori-Adachi M., Luo Y., Tamura K., Morioka M., Fukuda M., Tanaka Y., Kitajima S. (2006) A splice variant of stress response gene ATF3 counteracts NF-κB-dependent anti-apoptosis through inhibiting recruitment of CREB-binding protein/p300 coactivator. J. Biol. Chem. 281, 1620–1629 [DOI] [PubMed] [Google Scholar]
- 52. Gilchrist M., Thorsson V., Li B., Rust A. G., Korb M., Roach J. C., Kennedy K., Hai T., Bolouri H., Aderem A. (2006) Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178 [DOI] [PubMed] [Google Scholar]
- 53. Shin J. W., Ohnishi K., Murakami A., Lee J. S., Kundu J. K., Na H. K., Ohigashi H., Surh Y. J. (2011) Zerumbone induces heme oxygenase-1 expression in mouse skin and cultured murine epidermal cells through activation of Nrf2. Cancer Prev. Res. (Phila.) 4, 860–870 [DOI] [PubMed] [Google Scholar]
- 54. Sobhan P. K., Seervi M., Deb L., Varghese S., Soman A., Joseph J., Mathew K. A., Raghu G., Thomas G., E. S., S. M., R. S. K. (2013) Calpain and reactive oxygen species targets Bax for mitochondrial permeabilisation and caspase activation in zerumbone induced apoptosis. PloS ONE 8, e59350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grösch S., Maier T. J., Schiffmann S., Geisslinger G. (2006) Cyclooxygenase-2 (COX-2)-independent anticarcinogenic effects of selective COX-2 inhibitors. J. Natl. Cancer Inst. 98, 736–747 [DOI] [PubMed] [Google Scholar]
- 56. Tsutsumi S., Namba T., Tanaka K. I., Arai Y., Ishihara T., Aburaya M., Mima S., Hoshino T., Mizushima T. (2006) Celecoxib upregulates endoplasmic reticulum chaperones that inhibit celecoxib-induced apoptosis in human gastric cells. Oncogene 25, 1018–1029 [DOI] [PubMed] [Google Scholar]
- 57. Kardosh A., Golden E. B., Pyrko P., Uddin J., Hofman F. M., Chen T. C., Louie S. G., Petasis N. A., Schönthal A. H. (2008) Aggravated endoplasmic reticulum stress as a basis for enhanced glioblastoma cell killing by bortezomib in combination with celecoxib or its non-coxib analogue, 2,5-dimethyl-celecoxib. Cancer Res. 68, 843–851 [DOI] [PubMed] [Google Scholar]
- 58. Hamdulay S. S., Wang B., Birdsey G. M., Ali F., Dumont O., Evans P. C., Haskard D. O., Wheeler-Jones C. P., Mason J. C. (2010) Celecoxib activates PI-3K/Akt and mitochondrial redox signaling to enhance heme oxygenase-1-mediated anti-inflammatory activity in vascular endothelium. Free Radic. Biol. Med. 48, 1013–1023 [DOI] [PubMed] [Google Scholar]
- 59. Ohnishi K., Irie K., Murakami A. (2009) In vitro covalent binding proteins of zerumbone, a chemopreventive food factor. Biosci. Biotechnol. Biochem. 73, 1905–1907 [DOI] [PubMed] [Google Scholar]
- 60. Gao M., Yeh P. Y., Lu Y. S., Hsu C. H., Chen K. F., Lee W. C., Feng W. C., Chen C. S., Kuo M. L., Cheng A. L. (2008) OSU-03012, a novel celecoxib derivative, induces reactive oxygen species-related autophagy in hepatocellular carcinoma. Cancer Res. 68, 9348–9357 [DOI] [PubMed] [Google Scholar]
- 61. Jin Z., McDonald E. R., 3rd, Dicker D. T., El-Deiry W. S. (2004) Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J. Biol. Chem. 279, 35829–35839 [DOI] [PubMed] [Google Scholar]
- 62. Kojima Y., Nakayama M., Nishina T., Nakano H., Koyanagi M., Takeda K., Okumura K., Yagita H. (2011) Importin β1 protein-mediated nuclear localization of death receptor 5 (DR5) limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced cell death of human tumor cells. J. Biol. Chem. 286, 43383–43393 [DOI] [PMC free article] [PubMed] [Google Scholar]