Background: The influence of collagen glycosylation on integrin binding has not been studied previously.
Results: Glycosylation affected α3β1 integrin binding more strongly than α2β1 integrin binding.
Conclusion: Glycosylation modulated integrin/collagen interactions.
Significance: If changes in collagen glycosylation occur in malignancy, then metastasis may be altered by these changes.
Keywords: Collagen, Glycosylation, Hydroxylysine (Hyl), Integrins, Melanoma, Triple Helix
Abstract
Although type IV collagen is heavily glycosylated, the influence of this post-translational modification on integrin binding has not been investigated. In the present study, galactosylated and nongalactosylated triple-helical peptides have been constructed containing the α1(IV)382–393 and α1(IV)531–543 sequences, which are binding sites for the α2β1 and α3β1 integrins, respectively. All peptides had triple-helical stabilities of 37 °C or greater. The galactosylation of Hyl393 in α1(IV)382–393 and Hyl540 and Hyl543 in α1(IV)531–543 had a dose-dependent influence on melanoma cell adhesion that was much more pronounced in the case of α3β1 integrin binding. Molecular modeling indicated that galactosylation occurred on the periphery of α2β1 integrin interaction with α1(IV)382–393 but right in the middle of α3β1 integrin interaction with α1(IV)531–543. The possibility of extracellular deglycosylation of type IV collagen was investigated, but no β-galactosidase-like activity capable of collagen modification was found. Thus, glycosylation of collagen can modulate integrin binding, and levels of glycosylation could be altered by reduction in expression of glycosylation enzymes but most likely not by extracellular deglycosylation activity.
Introduction
Despite the continuous advances made, patients suffering from advanced stage melanoma still face a rather bleak prognosis. Melanoma remains unpredictable in its biological behavior, with a high risk of recurrence and a 50% chance to develop metastases in lymph nodes after recurrence (1). To support advances in treatment and detection, attention has turned to locating and identifying key elements that could be utilized either as possible therapeutic targets or as potential markers for the different stages of the disease. Metastasis occurs via a series of linked steps (2, 3). Tumor cells need to extravasate through the endothelium of the lymph node or blood vessel and become anchored in the local extracellular matrix (ECM)3 to initialize secondary growth formation. The principal proteins of the ECM are laminins and collagens, the latter of which serves as a lattice network to create the cellular microenvironment. Type IV collagen is the most important structural component of the basement membrane (BM), which is a specialized form of the ECM.
Metastasis requires a subtype of tumor cells capable of enduring release from the primary tumor site and traveling through the lymphatic or vasculatory system while evading killer cells and/or platelet aggregation. This process requires an altered phenotype, which allows cells to quickly adhere to and release from the BM to promote a faster migration. These phenotypic changes are most easily defined by changed expression profiles of transmembrane receptors, such as integrins, responsible for the rolling motion cells display during migration (4). Integrins are the foremost contributors in mediating cell-cell and cell-ECM adhesions. Interactions between integrins and ECM proteins, such as collagen, are crucial for adherence, migration, and invasion of tumor cells (5).
Integrins are heterodimers of noncovalently associated α and β subunits. In vertebrates, there are 18 α and 8 β subunits that can assemble into 24 different receptors with unique binding properties and tissue distributions (6, 7). Based on the structural characteristics of their α and β subunits, integrins are classified as either an I-domain or a non-I-domain, which signals a fundamentally different association mechanism between the two groups of receptor types and their respective ligands (6, 8–12). I-domain-containing integrins preferentially bind to ligands via their I-domain, which is located on the α subunit, providing a more approachable binding site and a more relaxed spatial arrangement, whereas non-I-domain integrins carry out binding partly by another portion of the α subunit and partly by the β subunit, which sterically places the ligand in a more confined space and makes the binding site less approachable and possibly less favorable (10, 12). The I-domain contains a conserved MIDAS that binds divalent metal cations. Ligand binding alters the coordination of the metal ion and shifts the I-domain from a closed, resting state to an open, active conformation, which results in increased ligand affinity and promotes subsequent integrin activation (13). Four I-domain α subunits (α1, α2, α10, and α11) associate with β1 and form a distinct collagen-binding subfamily. The structural basis of the interaction of these integrins with their ligand is a Glu residue within a collagenous Gly-Phe-Hyp-Gly-Glu-Arg motif, providing the cation coordination (9).
The β subunit plays an important role in ligand binding when α subunits lack the I-domain. Integrin β subunits contain an ion binding site homologous to MIDAS with a sequence motif of Asp-Xaa-Ser-Xaa-Ser. Mutation of any of these ion-coordinating residues within the β1, β2, β3, or β5 subunits ablated ligand binding to the respective integrins (14–16). In αβ integrin heterodimers, ligands bind to a crevice in the head domain between the αβ subunit interface. In many cases the ligand interacts with the metal ion-occupied MIDAS located within the β subunit and the propeller domain of the α subunit (17). The α3β1 integrin is a non-I-domain integrin that binds collagen and laminin (18–20). More specifically, the α3β1 integrin binds type IV collagen (21) and contributes to melanoma cell migration on this ligand (22, 23). Thus, collagen receptors include both α I-domain and non-I-domain integrins, implying differing binding mechanisms at play.
Tumor cell adhesion to the triple-helical domain of BM (type IV) collagen occurs at several different regions (24). Type IV collagen cyanogen bromide fragment 3 (CB3(IV)), which includes α1(IV) residues 388–551 and α2(IV) residues 407–570, was reported to contain the individual binding sites for the α1β1 and α2β1 integrins (25, 26). The α1β1 integrin simultaneously binds Asp441 from two α1(IV) chains and Arg458 from the α2(IV) chain (27–31). The Gly-Phe-Hyp-Gly-Glu-Arg recognition motif for the α2β1 integrin is located within the α1(IV)382–391 sequence (32). A THP model of α1(IV)382–393 binds to melanoma cells (33).
The α1(IV)531–543 sequence promotes adhesion and spreading of melanoma and other cell types (34). The α3β1 integrin was identified as the receptor that binds to type IV collagen via this sequence (35). Interestingly, peptides promoted adhesion of melanoma cells in single-stranded and triple-helical conformation (34), thus providing the first evidence for existence of triple-helix-independent integrin binding sites within the collagenous domain.
An essential characteristic of native type IV collagen is the high level of Lys hydroxylation and subsequent Hyl glycosylation present in each α chain. These post-translational modifications are carried out on almost all Lys residues present in type IV collagen, compared with the relatively low level (∼10%) of modification that is present on types I and II collagen (36, 37). Prior research conducted in our laboratory indicated an altered affinity of a cell surface proteoglycan, CD44, toward binding sites in type IV collagen based on Hyl glycosylation (38). However, prior studies have not considered how Hyl glycosylation impacts integrin recognition of collagen. To specifically examine the possible modulation of integrin function by glycosylation, THPs with Lys substituted by glycosylated Hyl for Lys543 and Lys540 from the human α1(IV)531–543 gene sequence (α3β1 integrin-specific) and Lys393 from the human α1(IV)382–393 gene sequence (α2β1 integrin-specific) were synthesized. These ligands were utilized to compare the promotion of melanoma cell adhesion, to observe the effects of ligand glycosylation. Cellular integrin concentrations were quantified utilizing immunocytochemistry. Alternative receptors were examined for recognition of glycosylated collagen. We also tested the possibility of melanoma cell modulation of collagen glycosylation by examining extracellular β-galactosidase-like activity.
MATERIALS AND METHODS
All chemicals were molecular biology or peptide synthesis grade and purchased from ThermoFisher Scientific (Waltham, MA) or Sigma-Aldrich.
Synthesis of Fmoc-d,l-Hyl[(5-O-β-Gal(Ac4))(Nϵ-Cbz)]-OPfp Building Block
The synthesis of Fmoc-d,l-Hyl[(5-O-β-Gal(Ac4))(Nϵ-Cbz)]-OPfp was performed in six steps, starting from the racemate of d,l-5-Hyl (Sigma-Aldrich). The synthetic approach has been described previously (39), and analytical data (1H NMR, 13C NMR, and mass spectra) of all intermediates and the desired product were in accordance with published ones (40). For example, MALDI-TOF MS of Fmoc-d,l-Hyl[(5-O-β-Gal(Ac4))(Nϵ-Cbz)]-OPfp yielded m/z = 1037.7535 (calculated for C49H47F5N2NaO16+, m/z = 1037.2738). RP-HPLC retention time = 19.745 min using a Vydac C18 column (5 μm, 300 Å, 150 × 4.6 mm), analytical gradient of 2–98% B in 20 min (where A was 0.1% TFA in H2O and B was 0.1% TFA in acetonitrile), with a flow rate of 1 ml/min and detection at λ = 220 and 280 nm.
Synthesis of (Glyco)peptides
The (glyco)peptide sequences were based on type IV collagen motifs possessing integrin recognition sites (see Table 1). (Glyco)peptides were synthesized by Fmoc solid phase chemistry using TentaGel S Ram resin (Advanced ChemTech, Louisville, KY) with a substitution level of 0.26 mmol/g. Peptide synthesis was carried out on the Liberty (CEM, Matthews, NC) automated microwave-assisted peptide synthesizer equipped with a Discover microwave module. Fmoc amino acids were coupled using 5 eq of each amino acid, 4.9 eq O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate, and 8 eq N-methylmorpholine (microwave power of 25 W at 50 °C, 300 s). Fmoc-d,l-Hyl[(5-O-β-Gal(Ac4))(Nϵ-Cbz)]-OPfp was incorporated manually using 3 eq of amino acid and 6 eq N,N-diisopropylethylamine with a reaction time of 17 h. The N termini of the peptides and glycopeptides were modified by coupling with n-dodecanoic acid.
TABLE 1.
Sequences, analytical data, and thermal stabilities of THPs used in this study
| Peptide designation | Sequence | Integrin binding | [M+H]+ observed (calculated) | RP-HPLC RTa | Tm |
|---|---|---|---|---|---|
| min | °C | ||||
| α1(IV)382–393 | C10- (Gly-Pro-Hyp)4-Gly-Ala-Hyp-Gly-Phe-Hyp-Gly-Glu-Arg-Gly-Glu-Lys- (Gly-Pro-Hyp)4-Tyr-NH2 | α2β1 | 3687.4666 (3687.7796) | 13.31 | 43 |
| α1(IV)382–393(Gal) | C10-(Gly-Pro-Hyp)4-Gly-Ala-Hyp-Gly-Phe-Hyp-Gly-Glu-Arg-Gly-Glu-Hyl(Gal)-(Gly-Pro-Hyp)4-Tyr-NH2 | α2β1 | 3865.1714 (3865.8273) | 11.32 | 37 |
| α1(IV)531–543 | C10- (Gly-Pro-Hyp)5-Gly-Glu-Phe-Tyr-Phe-Asp-Leu-Arg-Leu-Lys-Gly-Asp-Lys- (Gly-Pro-Hyp)5-NH2 | α3β1 | 4414.0239 (4414.1941) | 12.38 | 45 |
| α1(IV)531–543(Gal)(Gal) | C10-(Gly-Pro-Hyp)5-Gly-Glu-Phe-Tyr-Phe-Asp-Leu-Arg-Leu-Hyl(Gal)-Gly-Asp-Hyl(Gal)-(Gly-Pro-Hyp)5-NH2 | α3β1 | 4770.0703 (4770.2896) | 11.74 | 37 |
a Using a gradient of 2–70% B in 20 min under conditions given under “Materials and Methods.”
In the case of glycopeptide α1(IV)382–393(Gal), part of the peptidyl-resin was N-terminally modified with biotin containing a 20-atom PEG spacer (instead of n-dodecanoic acid). N-Biotinyl-NH-(PEG)2-COOH (EMD Millipore, San Diego, CA) was coupled manually using 3 eq molar excess along with 3 eq of O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate and 6 eq of N-methylmorpholine in DMF for 90 min. Biotinylated α1(IV)382–393(Gal) THP was utilized for β-galactosidase studies because of its favorable RP-HPLC elution profile.
Removal of side chain protecting groups and peptide-resin cleavage were carried out as reported previously (41) for 3 h in an atmosphere of ambient gas (Ar) using 7 ml of cleavage mixture (5% H2O, 5% thioanisole, 2.5% phenol, and 2.5% 1,2-ethanedithiol in TFA). Cleaved (glyco)peptides were precipitated in cold methyl tert-butyl ether, centrifuged, and lyophilized. Crude glycopeptides were subjected to Ac protecting group removal from sugar moieties using 0.1 m NaOH solution for 15 min. After this time the glycopeptide solution was neutralized with HCl and lyophilized.
Crude (glyco)peptides were purified using RP-HPLC on an Agilent 1260 Infinity series preparative HPLC equipped with a Vydac C18 column (15–20 μm, 300 Å, 250 × 22 mm). The elution gradient was 5–50% B in 60 min (where A was 0.1% TFA in H2O, and B was 0.1% TFA in acetonitrile), with a flow rate of 10 ml/min and detection at λ = 220 and 280 nm. The HPLC fractions were combined, frozen, and lyophilized.
A portion of the biotinylated glycopeptide α1(IV)382–393(Gal) (2.5 mg) was subjected to selective N-acetylation of the Hyl residue. Briefly, the glycopeptide was dissolved in 1 ml of 50 mm ammonium carbonate solution, and 100 μl of acetic anhydride was added. The reaction progress was monitored by RP-HPLC and MALDI-TOF MS, and upon completion the reaction mixture was frozen and lyophilized.
(Glyco)peptide purity was evaluated on an Agilent 1260 Infinity analytical HPLC using a Vydac C18 column (5 μm, 300 Å, 150 × 4.6 mm), analytical gradient 2–98% B in 20 min, with a flow rate of 1 ml/min and detection at λ = 220 and 280 nm. MALDI-TOF MS analysis was performed using an Applied Biosystems Voyager DE-PRO Biospectrometry work station (Carlsbad, CA) with a α-cyano-4-hydroxy-cinnamic acid/2,5-dihydroxybenzoic acid matrix.
CD Spectroscopy
Peptides were dissolved in 0.5% acetic acid and equilibrated at 4 °C (>24 h) to facilitate triple-helix formation. Peptide concentrations were determined using a Thermo Scientific NanoDrop 1000 (Waltham, MA) via absorbance at λ = 280 nm, ϵTyr = 1490 m−1 cm−1. Triple-helical structure was evaluated by near UV CD spectroscopy using a Jasco J-810 spectropolarimeter (Easton, MD) with a path length of 1 mm. Thermal transition curves were obtained by recording the molar ellipticity ([θ]) at λ = 225 nm with an increase in temperature of 20 °C/h in a range of 5–80 °C. Temperature was controlled by a JASCO PTC-348WI temperature control unit. The THP melting temperature (Tm) was defined as the inflection point in the transition region (first derivative). The spectra were normalized by designating the highest [θ]225 nm as 100% folded and the lowest [θ]225 nm as 0% folded.
Cell Culture
The M14#5 human metastatic melanoma cell line was generously provided by Dr. Barbara Mueller (Torrey Pines Institute for Molecular Studies, La Jolla, CA). The WM-115 (primary melanoma), WM-266-4 (metastatic melanoma), and SK-MEL-2 (metastatic melanoma) cell lines were obtained from American Type Culture Collection (Manassas, VA). For cell adhesion assays, cells were grown in EMEM with l-Gln (American Type Culture Collection) supplemented with 10% fetal bovine sera (HyClone), 50 units/ml penicillin, and 0.05 mg/ml streptomycin using 175-cm2 flasks. At ∼80% confluency cells were subcultured (1:3 ratio for SK-MEL-2, 1:6 for the other cell lines). For cell detachment 0.25% trypsin-EDTA solution was used (Invitrogen). Flasks were kept in a humidified incubator containing 5% CO2, and cells were passaged only eight times to avoid genetic drifts and other variations.
Immunocytochemistry
Biotinylated mouse antihuman integrin α3 subunit (CD49c, clone IA3, catalog number BAM1345) and biotinylated mouse anti-human integrin α2 subunit (CD49b, clone HAS3, catalog number BAM1233) mAbs were purchased from R&D Systems (Minneapolis, MN). Biotin-SP-conjugated ChromPure mouse IgG, whole molecule (product code 015-060-003), and mouse serum and the Cy3-conjugated (indocarbocyanine) streptavidin (product code 016-160-084) were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). Immunocytochemistry experiments were carried out according to the direct detection method employing working concentrations based on the manufacturer's recommendation. The specificities of the integrin antibodies were previously established (42, 43). Briefly, cells were plated in a 96-well Costar plate (Corning No. 3632; Fisher Scientific) at 20,000–40,000 cells/100 μl growth medium and allowed to become 70–80% confluent overnight. Growth medium was removed, and cells were rinsed with 1× PBS. Cells were fixed by incubation with 4% paraformaldehyde/PBS for 20 min at room temperature. The plate was blocked against nonspecific binding with 1% mouse serum, 1% BSA/PBS for 1 h at room temperature. Biotinylated anti-integrin α3 or α2 mAbs were diluted in 1% mouse serum, 1% BSA/PBS to 25 μg/ml, added to the cells, and incubated for 1 h at room temperature. As a negative control, biotin-SP-conjugated mouse IgG was utilized, diluted to 25 μg/ml in 1% mouse serum, 1% BSA/PBS. Following incubation with the antibodies and/or mouse IgG, cells were rinsed three times with 1 × PBS and subsequently incubated for 1 h at room temperature in the dark with Cy3-conjugated streptavidin diluted to 2 μg/ml in 1% mouse serum, 1% BSA/PBS. Background fluorescence was established by incubating the cells with the Cy3-streptavidin solution for 1 h at room temperature in the dark. The plate was washed with 1× PBS, and bound Cy3 was detected using the red filter on an Olympus IX70 inverted fluorescence microscope camera. Semi-quantitative image analyses were carried out using the Quantity One® v.4.2.2 software (Bio-Rad) on quadruplets of 16 cell/image areas. Cells were counted in selected areas using photos taken at bright light at a visible range wavelength, and then, after switching on the red filter, the selected areas were photographed to determine fluorescence. Exposure times were 250, 400, or 666 ms. Because of either cellular accumulation or entrapped dye, some areas indicated artificially high levels of fluorescence. Those “bright spots” were not utilized for quantification purposes.
Adhesion Assays
The melanoma cell adhesion assay was performed as described previously (44). Peptide ligands (see Table 1) were dissolved in 40% ethanol in PBS and further diluted to desired concentrations in PBS. Pro-BindTM 96-well plates (BD Biosciences, San Jose, CA) were coated with 100 μl of desired peptide and incubated at 4 °C overnight. Nonspecific binding sites were blocked with 2 mg/ml BSA in PBS for 2 h at 37 °C (200 μl/well). Cells were split 1–2 days before the experiment and were washed with PBS (without Ca2+ and Mg2+) and then released with Accutase (Invitrogen). Cells were then washed and resuspended to 75,000 cells/well in adhesion medium (20 mm HEPES, 2 mg/ml albumin in RPMI 1640 medium). Cell suspension was added to the plate (100 μl/well), and plates were incubated for 60 min at 37 °C. All nonattached cells were removed by washing three times with warm adhesion medium. Adherent cells were counted using CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI) and quantitated with a Synergy H4 Hybrid multimode microplate reader (BioTek, Winooski, VT).
AlphaScreen Assay
The AlphaScreen assay was performed accordingly to recently published methodology (45). His-tagged galectin-3 (1.25 μl) and biotin-ASF (1.25 μl) were added to wells containing varying concentrations of α1(IV)382–393 and α1(IV)382–393(Gal) THPs (2.5 μl, 0–1 mm final concentration) in optimized assay buffer (25 mm HEPES, 100 mm NaCl, 0.05% Tween 20, pH 7.4). Because of the low solubility of the peptides at higher concentrations, final solutions contained 1% DMSO. The nonbiotinylated ASF was used as a control. The final concentration of His-tagged galectin-3 was 100 nm and biotin-ASF 5 nm. The reaction mixture was incubated for 1 h at room temperature, and then 5 μl of nickel-chelate Acceptor and 5 μl streptavidin-conjugate Donor beads were simultaneously added to a final concentration of 25 μg/ml. Incubation proceeded for 1 h in the dark at room temperature, and the assay plate was subsequently read at 22 °C in the AlphaScreen mode on a Synergy H4 Hybrid multimode microplate reader. AlphaScreen signal counts (cps) versus log [ligand] (M) were expressed as the mean of five replicate measurements. The IC50 values were obtained by nonlinear regression analysis using the Graph Pad Prism 5.04 software.
Molecular Modeling
To generate a model of α1(IV)382–393(Gal) THP interacting with the α2β1 integrin, a homology modeling approach was utilized. Briefly, starting structure 1DZI was used as a template (9). Collagen-like peptide residues were mutated manually in PyMOL (46) and UCSF Chimera software (47). Residues were mutated using Dunbrack backbone-dependent rotamer library (48). Charges were added using AMBER ff12SB force field, and for unknown residues (Gal) were calculated using AM1-BCC model (49). Mutated residues were subjected to minimization using the antechamber program (50) included in Chimera.
The α3β1 integrin model was built using the α5β1 integrin x-ray crystallographic structure (51). The α5 subunit was replaced with α3 by homology modeling using the Modeler program (52) and subsequent minimization steps of α3/β1 interface residues using the antechamber module of the Chimera package. Next, docking of α1(IV)531–543 single-stranded peptide was performed using Autodock Vina (53). Because the geometry of the α1(IV)531–543 peptide backbone is unknown, we have selected three different combinations of φ/ψ torsion angles within the polyproline type II family, namely φ/ψ of −60°/150°, −70°/160°, and −75°/175°. Three separate docking runs were performed and compared. In each docking the peptide backbone was kept rigid, and side chains contained rotatable bonds. The docking site was chosen arbitrarily and contained the top of the α3β1 interface along with the MIDAS site in the β1 subunit.
β-Galactosidase Activity Assessment
Isolated β-Galactosidase with Synthetic Substrate
Escherichia coli β-galactosidase (EC 3.2.1.23, grade VIII) was purchased from Sigma-Aldrich. Enzymatic assays were performed in 100 mm phosphate buffer, pH 7.2, supplemented with 10 mm MgCl2 and 5 mm 2-mercaptoethanol (added freshly before the assay). The enzyme activity was determined using the fluorogenic substrate MUG (Sigma-Aldrich) at λexcitation = 365 nm and λemission = 445 nm. β-Galactosidase activity was measured using the Synergy H4 Hybrid Multi-Mode Microplate Reader over a period of 1 h, with occasional shaking to assure even substrate distribution.
Isolated β-Galactosidase with Peptide Substrates
α1(IV)382–393(Gal) THP was selected as a model putative substrate. To determine the influence of the Hyl ϵ-NH2 group on β-galactosidase activity, an acetylated version of α1(IV)382–393(Gal) THP, α1(IV)382–393(Gal)-Ac THP, was prepared (see earlier description).
α1(IV)382–393(Gal) and α1(IV)382–393(Gal)-Ac THPs (46 μg each) were incubated at 37 °C with 100 U of β-galactosidase in 100 mm phosphate buffer, pH 7.2, supplemented with 10 mm MgCl2 and 5 mm 2-mercaptoethanol. After 4 and 24 h, aliquots were taken and analyzed using RP-HPLC/MALDI-TOF MS.
Melanoma Cells with Synthetic Substrate
Extracellular β-galactosidase activity was assessed using a whole cell assay with primary and metastatic melanoma cells. The WM-115 and WM-266-4 cell lines were plated in 24-well plate format at 50,000 cells/well (Corning CellBIND, Corning) and cultured overnight (24 h) using four different media types: EMEM, EMEM supplemented with HI-FBS, OptiMEM, and OptiMEM with HI-FBS. The HI-FBS concentration was 5% (v/v), and the total volume of the media was 500 μl. After 24 or 48 h, MUG was added to a final concentration of 30 μm, and β-galactosidase activity was measured using the Synergy H4 Hybrid multimode microplate reader over a period of 1 h, with occasional shaking to assure even substrate distribution.
Melanoma Cells with Peptide Substrates
The WM-115 and WM-266-4 cell lines were plated in 24-well format at 50,000 cells/well and cultured overnight (24 h) using four different media types: EMEM, EMEM supplemented with HI-FBS, OptiMEM, and OptiMEM with HI-FBS. The HI-FBS concentration was 5% (v/v), and the total volume of the media was 500 μl. Cells were grown overnight, and THPs were added to a final concentration of 35 μm. The incubation was carried out for 24 and 48 h. Aliquots of media were taken, filtered through a 0.22-μm HPLC filter, and subjected to RP-HPLC analysis. All fractions were then collected and analyzed using MALDI-TOF MS.
Released Melanoma Cells with Synthetic and Peptide Substrates
CUG synthetic substrate (Invitrogen) was diluted to a 60 μm in 1 × PBS. To test cell suspensions, subconfluent cells were rinsed with 1 × PBS and released with 5 mm EDTA/PBS. Cells were washed and rediluted to 2 × 10−5–1 × 10−6 cells/ml in PBS containing 10 mm MgCl2 and 5 mm 2-mercaptoethanol. 100 μl of cells were combined with 50 μl of CUG, and enzymatic activity was measured for 30–90 min, with recurring agitation to maintain the cells in suspension, at λexcitation = 400 nm and λemission = 450 nm on a SpectraMAX GeminiEM 96-well plate spectrafluorometer and quantified by the SoftMax Pro 4.3LS software. As a positive control, 50 μl of β-galactosidase and 50 μl of CUG were added to 100 μl of PBS containing the activators and tested simultaneously.
RESULTS
Synthesis of Galactosylated Hyl Building Block
To prepare a galactosylated Hyl building block, we utilized a synthetic approach developed by Kihlberg and co-workers (39), in which 9-BBN simultaneously protects the carboxyl and amino functionalities of amino acids (54). 9-BBN was used for regioselective protection of the α-amino and α-carboxyl groups of d,l-5-Hyl. The resulting 9-BBN complex was then employed in transformations such as ϵ-amino group protection and O-glycosylation. Further manipulations led to preparation of the Fmoc-d,l-Hyl[(5-O-β-Gal(Ac4))(Nϵ-Cbz)]-OPfp building block, suitable for direct use in peptide synthesis under standard Fmoc chemistry conditions. The Cbz group was chosen instead of tert-butyloxycarbonyl for ϵ-amino group protection of Hyl because it is more acid stable during O-glycosylation conditions. Fmoc-d,l-Hyl[(5-O-β-Gal(Ac4))(Nϵ-Cbz)]-OPfp was obtained in six steps, with a yield comparable with previously published ones (39).
(Glyco)peptide Synthesis and Characterization
d,l-Hyl[(5-O-β-Gal(Ac4))(Nϵ-Cbz)] was incorporated into THPs possessing sequences from the α1(IV) collagen chain recognized by α2β1 and α3β1 integrins. We prepared two sets of peptides containing either the Hyl(O-Gal) residue or its Lys counterpart (Table 1). The Hyl(O-Gal) residue was incorporated manually, whereas other amino acids were incorporated using an automated synthesizer under microwave conditions. The N termini of all (glyco)peptides were modified with n-dodecanoic acid to ensure triple-helical character of the (glyco)peptides and to facilitate their attachment to plastic surfaces during the adhesion assay (55, 56).
All peptides were characterized by RP-HPLC and MALDI-TOF MS (Table 1), with appropriate purity and mass values observed. The triple-helical character of the peptides was analyzed by CD spectroscopy, in the range of λ = 250–180 nm (Fig. 1). All peptides had characteristic triple-helical spectra, with a positive peak at λ = 222 nm and a negative peak at λ = 205 nm. Thermal transition curves were obtained by recording molar ellipticity ([θ]) at λ = 225 nm as a function of increasing temperature (Fig. 2). The melting point (Tm) was defined as the inflection point in the transition region (Table 1). All peptides exhibited good triple-helix stability, with Tm values ranging from 37 to 45 °C. Galactosylation of Hyl had a destabilizing effect on the triple-helix, because the glycopeptides had Tm values 6–8 °C lower than the corresponding nonglycosylated peptides. This could be caused by the presence of racemic d,l-5-Hyl used in the present study.
FIGURE 1.

CD spectra of THPs. THPs were dissolved in 0.1% acetic acid to a final concentration of 50 μm. Scans were taken over the range of λ = 180–250 nm.
FIGURE 2.

Thermal transition curves of THPs obtained by recording the molar ellipticity ([θ]) at λ = 225 nm with a temperature change of 20 °C/h over the range of 5–80 °C and normalized to fraction folded. The THP melting temperature (Tm) was defined as the inflection point in the transition region (first derivative).
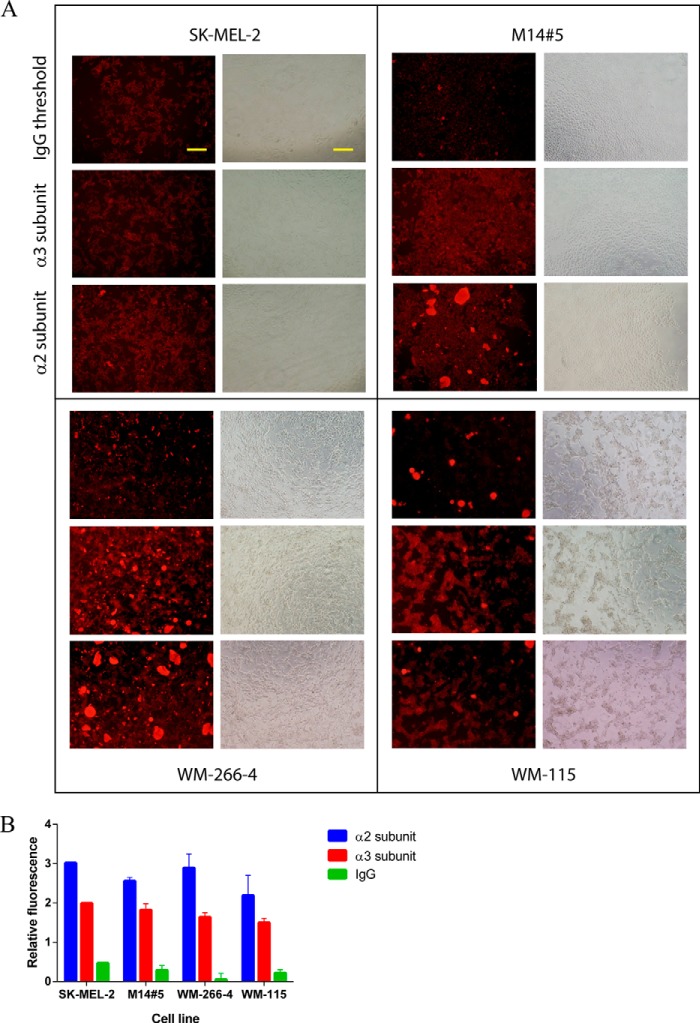
Immunocytochemistry
The cell surface concentrations of the α2 and α3 subunits of the α2β1 and α3β1 integrins were evaluated for all melanoma cell lines by immunocytochemistry. Image analysis for each cell line for both receptor subunits (Fig. 3A) provided semiquantitative numerical information on cellular integrin concentrations. Numerical values reflected relative fluorescence intensities of the same cell number, normalized by area measured, and by subtraction of the autofluorescence of the cells (Fig. 3B). All four cell lines showed abundant levels of both integrin subunits, with somewhat higher levels for the α2 subunit compared with the α3 subunit (Fig. 3B). The primary cell line (WM-115) had lower levels of the α2 and α3 subunits compared with the metastatic cell lines. Overall, integrin levels were sufficient to investigate melanoma-ligand interactions.
FIGURE 3.
The cell surface concentrations of the α2 and α3 subunits of the α2β1 and α3β1 integrins evaluated for melanoma cell lines by immunocytochemistry. A, image analysis for each cell line for both receptor subunits and IgG background. Bars indicate 100 μm. B, semiquantitative numerical values for cellular integrin concentrations, obtained by relative fluorescence intensities of the same number of cells, normalized by area measured and by subtraction of the autofluorescence of the cells. Conditions are given under “Materials and Methods.”
Melanoma Cell Adhesion
The influence of glycosylation on melanoma cell adhesion was examined over a THP concentration range of 0–50 μm (Fig. 4). All melanoma cell lines exhibited similar binding curves to each of the nonglycosylated peptides, α1(IV)382–393 THP and α1(IV)531–543 THP (Fig. 4, top left and bottom left). Adhesion to α1(IV)382–393 THP was observed at the lowest peptide concentration tested (0.1 μm), whereas adhesion to α1(IV)531–543 THP initiated at 1.0 μm and reached a maximum at 10 μm (Fig. 4, top left and bottom left). The α1(IV)531–543 THP dose dependence mirrors that reported previously (34). Although adhesion to α1(IV)382–393 THP has been reported previously (32, 33), the present results are the first dose dependences.
FIGURE 4.
Adhesion of melanoma cells as a function of α1(IV)382–393 THP (top left), α1(IV)382–393(Gal) THP (top right), α1(IV)531–543 THP (bottom left), and α1(IV)531–543(Gal)(Gal) THP (bottom right) concentration. Cells were allowed to adhere for 1 h at 37 °C. All assays were repeated in triplicate. Conditions are given under “Materials and Methods.”
In the case of α1(IV)382–393 THP, glycosylation of Hyl393 had modest influence on cell adhesion (Fig. 4, top right). Nonglycosylated α1(IV)382–393 THP promoted cell adhesion over the whole range of concentrations studied (0.1–50 μm). The glycosylated α1(IV)382–393(Gal) THP showed reduced adhesion levels in the lower THP concentration range (5 μm and below) with the most pronounced difference for the M14#5 metastatic cell line (Fig. 4, top right).
All cell lines were very sensitive toward the glycosylation of the α1(IV)531–543 sequence (Fig. 4, bottom right). Cells were adherent to α1(IV)531–543 THP in a range of 1–50 μm (Fig. 4, bottom left). The glycosylation of Hyl540 and Hyl543 dramatically decreased adhesion (Fig. 4, bottom right). Cells were adherent to α1(IV)531–543(Gal)(Gal) THP only at the highest concentration tested (50 μm).
Galectin-3 Interaction with THPs
Galectin-3 is known to mediate cell binding to galactosylated ligands (57). Thus, we examined the possibility that glycosylation may result in a switch of receptor binding, in that galectin-3 may mediate binding to glycosylated THPs. Binding was tested using a galectin-3 AlphaScreen assay (45) with α1(IV)382–393 and α1(IV)382–393(Gal) THPs along with ASF as a control (Fig. 5). Only nonspecific binding of the THPs with galectin-3 was observed (IC50 in the millimolar range) with no difference between glycosylated and nonglycosylated ligands. Thus, galectin-3 does not appear to be involved in binding to these ligands.
FIGURE 5.
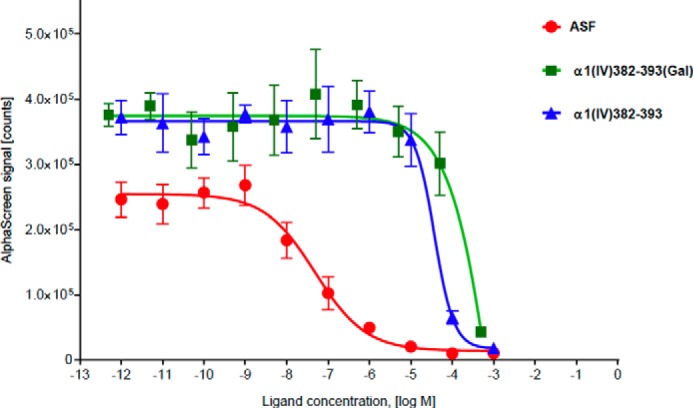
Results of AlphaScreen assay for determination of the binding of α1(IV)382–393 and α1(IV)382–393(Gal) THPs to galectin-3. ASF was used as positive control.
Molecular Modeling of integrin·THP Complexes
To further investigate the influence of glycosylation of α1(IV)382–393 and α1(IV)531–543 THPs on binding to their respective integrins, molecular modeling was performed. Models for both integrin·THP complexes were prepared (Figs. 6 and 7).
FIGURE 6.

Molecular modeling of α1(IV)382–393(Gal) THP interaction with the α2 integrin subunit I-domain.
FIGURE 7.
Molecular modeling studies of α3β1 integrin with α1(IV)531–543 peptide. A, a small deviation of φ/ψ angles (10°) caused by flexibility of this region results in strand separation. B, docking studies of α1(IV)531–543 peptide using three different sets of φ/ψ angles. Docked peptide (backbone in gray, Lys in blue, and Asp/Glu in yellow) binds across the α/β integrin interface (magenta/cyan, respectively). C, refined docking simulations where only Gly541 and/or Gly544 were allowed to be flexible.
For the α2β1 integrin, a model was generated using the x-ray crystallographic structure of the α2 I-domain in complex with a THP (PDB: 1DZI) (9). The Hyl393 glycosylation site is located four residues away from the Glu389 responsible for binding to the MIDAS of the I-domain. It appears that the Hyl393 site is at the outer interface of the integrin interaction site, and thus mono-glycosylation does not impact binding significantly (Fig. 6). When the disaccharide-containing residue (Glc-Gal)Hyl is considered, glycosylation of Hyl393 will interfere with binding to the α2 integrin because the binding site is masked by the sugar moiety (data not shown). Thus, the effect of glycosylation on α2β1 integrin binding to type IV collagen may very well depend on whether monosaccharide or disaccharide glycosylation has occurred.
The recognition site for the α3β1 integrin is much different from α2β1 because there is no homology between the peptide ligand sequences. Also, the α1(IV)531–543 sequence does not have the classic -Gly-Xaa-Yaa- repeat required for stabilization of the triple-helix, but rather has a noncollagen-like insertion/break region of n = 7 (58). The break region within α1(IV)531–543 is anticipated to have some strand separation, based on prior studies of break regions within THPs (59, 60). Conversely, the α1(IV)531–543 break region does not possess either Pro or Hyp and nor do the flanking regions, and thus the break is not anticipated to significantly affect the triple-helical structure of the remainder of the THP (61, 62). In addition, the presence of hydrophobic residues within the break region further aids in the stability of the THP (62).
For molecular modeling studies, it was assumed that the α1(IV)531–543 region possesses a polyproline type II-like structure (Fig. 7A). Single-stranded α1(IV)531–543 was used for molecular docking purposes. Three different sets of polyproline type II-like torsion angles, φ/ψ = −60°/150°, −70°/160°, and −75°/175°, were considered, and the peptide backbone was kept rigid upon docking (Fig. 7B). Previous studies revealed the crucial role of both Asp residues of the α1(IV)531–543 sequence for binding to the α3β1 integrin (34). Taking this into account, another set of docking was performed where the Gly541 and/or Gly544 residues were allowed flexibility around the Cα atom. In each docking result the lowest energy ligands were obtained by having the Asp542 residue in close proximity of the MIDAS (Fig. 7C). The docking studies revealed that glycosylation of Hyl540 and Hyl543 occurred right in the middle of key electrostatic/metal binding interactions and thus would dramatically impact the binding of the α3β1 integrin to the α1(IV)531–543 THP.
β-Galactosidase Activity Evaluation
Knowing that glycosylation could negatively impact integrin binding to type IV collagen, we next examined whether melanoma cells could modulate O-glycosylation of the microenvironment. Initially, E. coli β-galactosidase was tested with the α1(IV)382–393(Gal) THP. Different enzyme concentrations (1–100 units) were compared using incubation at 37 °C for up to 72 h. At certain time points aliquots were taken and subjected to RP-HPLC/MALDI-TOF MS analysis. No hydrolysis of Gal by E. coli β-galactosidase was observed (data not shown). Activity of the β-galactosidase was confirmed with MUG fluorogenic substrate (data not shown).
The influence of the ϵ-amino group of Hyl on β-galactosidase activity was then tested. Prior studies indicated that β-galactosidase was effective in cleaving the Gal moiety from (Gal)Hyl only if the ϵ-amino group was acetylated (63–65). No cleavage of the sugar moiety was observed when α1(IV)382–393(Gal) THP, in either nonacetylated or acetylated (α1(IV)382–393(Gal)-Ac THP) form, was treated with β-galactosidase (data not shown). Although this result is in contrast to the results obtained by Spiro (63–65), the prior study tested the enzyme activity on isolated (Gal)Hyl moiety only.
Whole cell assays were next performed. Two cell lines were selected: primary (WM-115) and metastatic (WM-266–4) melanoma obtained from the same patient. Different cell culture media (EMEM and OptiMEM with or without HI-FBS) were also tested. In the first experiment, cells were grown for 24 or 48 h, and then MUG was added and activity monitored for 1 h (Fig. 8). There was no activity present in EMEM and OptiMEM media (Fig. 8, A and C). In contrast, media containing HI-FBS possessed some β-galactosidase activity (Fig. 8, B and D). In the case of OptiMEM + HI-FBS, the activity was associated with the presence of serum, because control (without cells) also exhibited this activity (Fig. 8D). However, both WM-115 and WM-266-4 exhibited some activity toward MUG above background in EMEM + HI-FBS (Fig. 8B). The results from 48 h were identical (data not shown).
FIGURE 8.
Evaluation of extracellular β-galactosidase activity using the fluorogenic substrate MUG with WM-115 and WM-266–4 melanoma cells. After initial 24 h growth, cells were incubated with MUG, and activity was monitored using λexcitation = 365 nm and λemission = 445 nm. Cell culture media tested were EMEM (A), EMEM + HI-FBS (B), OptiMEM (C), and OptiMEM + HI-FBS (D). RFU, relative fluorescent units.
Next, WM-115 and WM-266-4 were incubated with α1(IV)382–393 THP, α1(IV)382–393(Gal) THP, α1(IV)531–543 THP, and α1(IV)531–543(Gal,Gal) THP. After 24 or 48 h of incubation, an aliquot of culture media was taken and subjected to HPLC/MALDI-TOF MS analysis. The nonglycosylated peptides served as internal controls for determination of the hydrolysis pattern. The RP-HPLC profiles of peptides incubated with media served as a control. Interestingly, RP-HPLC analysis showed that all peptides were stable in media containing HI-FBS (data not shown).
All peptides were clearly identified during RP-HPLC/MALDI-TOF MS analysis of aliquots (Fig. 9). Under the employed conditions, neither of the glycosylated THPs was deglycosylated, as confirmed by MS analyses (data not shown). It was also possible to perform MALDI-TOF MS analyses of crude aliquots (without HPLC separation), and these results confirmed that glycopeptides were unmodified (Fig. 10). The results with cells in suspension were identical, in that (a) β-galactosidase activity could be observed with CUG and inhibited by phenylethyl β-d-thiogalactopyranoside (a selective β-galactosidase inhibitor) and (b) no degalactosylation of the THPs was found (data not shown).
FIGURE 9.
Representative example of RP-HPLC profile of crude media filtrate after incubation of WM-115 cells with THPs for 24 h in EMEM + HI-FBS medium. α1(IV)382–393(Gal) THP (A), biotinylated α1(IV)382–393(Gal) THP (B), α1(IV)531–543 THP (C), and α1(IV)531–543(Gal)(Gal) THP (D) were identified by retention time (indicated with arrow) following MALDI-TOF MS analysis. Similar profiles were obtained for all other tested media.
FIGURE 10.

A, MALDI-TOF MS profile of α1(IV)531–543(Gal)(Gal) THP collected from the indicated peak in Fig. 9D. An arrow indicates the m/z corresponding to the glycopeptide, [M+H]+ = 4772.38 (theoretical 4770.29). B, MALDI-TOF MS profile of crude WM-115 cell filtrate after 24 h of incubation with α1(IV)382–393(Gal) THP in EMEM + HI-FBS medium. An arrow indicates the m/z corresponding to the biotinylated glycopeptide, [M+H]+ = 4255.30 (theoretical 4254.97).
DISCUSSION
Tumor cells interact with type IV collagen at the site of extravasation through distinct cellular receptors, including the α1β1, α2β1, and α3β1 integrins. Integrins contribute to the ability of melanoma cells to migrate, invade, and metastasize to secondary sites (6, 66). Because they play a pivotal role in both inside-out and outside-in signaling (67), integrins affect most aspects of cell behavior, including shape, motility, differentiation, proliferation, and survival. Thus, it is not surprising that these receptors are also known to be differentially expressed in tumors relative to normal cells, depending on tumor type and stage of progression (4, 6, 66, 68, 69).
The types and concentration of cellular receptors has long been a focus for finding indicators of disease progression. Testing of 10 different human melanoma cell lines found that the α2, α3, and β1 integrin subunits were expressed on all of them (70). It is interesting to note that the α3 subunit showed the highest expression profile, with subtle differences in regards to the invasive profile of a given cell line. The same variation was observed for the α2 and β1subunits, showing higher expression levels in more invasive tumor types, although the overall concentrations were somewhat lower than that of the α3 subunit. In light of these prior results, the role of the associative relationships between type IV collagen and α2β1 and α3β1 integrins with regards to melanoma progression was examined here.
Prior research conducted in our laboratory indicated an altered affinity of a cell surface proteoglycan, CD44, toward binding sites in type IV collagen based on glycosylation (38). This result led to us to consider whether hydroxylation/glycosylation of Lys residues modulates ligand binding by other receptors, such as integrins. Previous studies have not considered how Hyl glycosylation impacts on integrin recognition of collagen. To specifically examine the possible modulation of integrin function by glycosylation, THPs with Lys substituted by glycosylated Hyl for Lys393 from the human α1(IV)382–393 gene sequence (α2β1 integrin-specific), and Lys543 and Lys540 from the human α1(IV)531–543 gene sequence (α3β1 integrin-specific) were synthesized. These ligands were utilized to compare the promotion of cell adhesion.
Collagen glycosylation was found to modulate integrin binding. The integrins were affected differently, with only modest inhibition of α2β1 binding (with the primary melanoma cell line being least affected) and significant inhibition for α3β1 interaction.
Molecular modeling studies of the α2β1 integrin in complex with the glycosylated ligand α1(IV)382–393(Gal) THP indicated that (Gal)Hyl393 was at the outer interface of the integrin interaction site, and thus galactosylation only slightly diminished binding (Fig. 6). However, for the cell lines tested herein, the effects of ligand glycosylation on binding varied, with the smallest effect on the primary melanoma cell line (WM-115) and the largest effect on the highly metastatic M14#5 cell line (Fig. 4, top right). Because the relative amount of α2β1 integrin on each cell surface was similar (Fig. 3B), variations in activity could be due to interactions of the glycosylated ligand with different activation states of the α2β1 integrin or with different cell surface complexes that incorporate the α2β1 integrin.
The simulations of interactions of the α3β1 integrin with α1(IV)531–543 THP indicated that the relatively flexible 531–543 region is capable of binding across the α3β1 interface with Asp542 binding to the MIDAS motif within the β1 subunit (Fig. 7). Because the MIDAS motif is located in the αβ interface groove on β subunit, complexation of the Mg2+ cation by the Asp542 side chain is a primary driving force for binding the peptide to the receptor (Fig. 7C). This model is with agreement with previously published data identifying Asp542 as a critical residue for α3β1 integrin binding to α1(IV)531–543 (34) and consistent with several integrin x-ray crystallographic structures including αIIbβ3 (71), α5β1 (51), and αvβ3 (17). Glycosylation within the α1(IV)531–543 sequence results in significant inhibition of integrin binding, mostly likely because of the proximity of the galactosylated residues (Hyl540 and Hyl543) to the key electrostatic/metal binding interactions via Asp542. Although inhibition caused by glycosylation is an uncommon phenomenon, the presence of sialic acid on sialoglycoprotein P2B reduced the binding of tumor cells to type IV collagen (72, 73).
The reduced binding of integrins caused by ligand glycosylation presents a possible “cryptic sites” mechanism by which tumor cells may invade the BM (38). In the native, glycosylated state, regions within type IV collagen may have minimal interaction with receptors such as the α3β1 integrin and CD44. After tumor cells bind to type IV collagen (presumably via the α2β1 integrin), cell surface or secreted glycosidases could liberate the collagen-bound carbohydrates. This process would expose cryptic sites for interaction with the α3β1 integrin, CD44/CSPG, and/or other cell surface receptors.
Extracellular removal of carbohydrates could also occur under other circumstances. Numerous bacterial pathogens bind to collagen (74), with binding occurring at several sites within the triple helix (75). The collagen binding protein from Staphylococcus aureus has been identified as CNA, and its mode of binding has been determined (76). Upon binding to collagen, bacteria could secrete β-galactosidases that facilitate deglycosylation. Reduced glycosylation could impact integrin interactions, as well as other collagen-binding proteins. The endocytic collagen receptor urokinase plasminogen activator receptor-associated protein mediates glycosylated collagen turnover (77). DDR1 binds to type IV collagen (78), and this binding may be mediated by ligand glycosylation (79).
Ultimately, glycosylation could be modulated extracellularly in similar fashion to intracellular protein dynamic glycosylation/phosphorylation (80, 81). The post-translational modification of Hyl is catalyzed by two groups of collagen glycosyltransferases, Hyl galactosyltransferase (EC 2.4.1.50) and (Gal)Hyl glucosyltransferase (EC 2.4.1.66), resulting in the formation of (Gal)Hyl and (Glc-Gal)Hyl, respectively (82). The Hyl galactosyltransferase activity has been ascribed to the multifunctional enzyme LH3 (83, 84) and/or GLT25D1 and GLT25D2 (85). LH3 appears responsible for the (Gal)Hyl glucosyltransferase activity (84, 86–89). LH3 can function extracellularly, glycosylating native, triple-helical collagens (90, 91). In fact, extracellular glycosyltransferase activity of LH3 is vital for the cell growth and viability (92). A cell surface galactosyltransferase functions as a type IV collagen adhesion molecule and galactosylates type IV collagen (93). Platelets can supply sugar donor substrates for extracellular glycosylation (94). Collagen Hyl residues can also be phosphorylated (95), and phosphorylation of collagen can occur extracellularly (96).
For dynamic modification of collagen to occur, carbohydrates would need to be removed from type IV collagen extracellularly. An age-dependent increase in β-galactosidase activity (at pH 6) has been reported (97), and a cell surface inactive β-galactosidase functions as an elastin and laminin receptor (98). An α-glucosidase that removes glucose from (Glc-Gal)Hyl has been characterized (99). However, we found no evidence that deglycosylation could be performed extracellularly, because triple-helical glycopeptides were not substrates for purified β-galactosidase or melanoma cells. Thus, although a deglycosylation/cryptic site mechanism provides interesting speculation, it should also be noted that glycosylation is not 100% efficient; collagen O-glycosylation sites are found as mixtures of Lys, Hyl, (Gal)Hyl, and (Glc-Gal)Hyl (36, 89, 100, 101). Thus, receptor interaction may just occur with the subpopulation of type IV collagen that does not contain carbohydrate.
Alternatively, tumor cell binding may be mediated by differential glycosylation that is tissue-specific. For example, LH3 is found extracellularly in kidney, spleen, and muscle (91). Thus, α2β1 and α3β1 integrin binding may be regulated by different levels of type IV collagen glycosylation as determined by LH3 activity. It is also possible that Hyl glycosylation enzymes are decreased in cancer, in similar fashion to β3-N-acetylglucosaminyltransferase-1 (102), and this in turn could enhance receptor association with type IV collagen.
The present study has focused on cellular interactions with glycosylated collagen models. When one considers the BM in vivo, variations in collagen glycosylation are generally unknown. If the levels of glycosylation are indeed modulated, tumor interactions with and response to the BM may be altered or simply compensated for by adherence to other BM ligands (i.e. laminin). Future studies may consider the complexities of cell-BM interactions and the role of glycosylation within that microenvironment.
Acknowledgment
We thank Dr. Janelle Lauer for assistance with the immunocytochemistry.
This work was supported, in whole or in part, by National Institutes of Health Grant CA77402 (to G. B. F.).
- ECM
- extracellular matrix
- Ac
- acetyl
- ASF
- asialofetuin
- BBN
- 9-borabicyclo[3.3.1]nonane
- BM
- basement membrane
- Cbz
- benzyloxycarbonyl
- CUG
- 3-carboxyumbeliferyl-β-d-galactopyranoside
- EMEM
- Eagle's minimum essential medium
- Fmoc
- 9-fluorenylmethoxycarbonyl
- HI-FBS
- heat-inactivated FBS
- Hyl
- 5-hydroxy-l-lysine
- Hyp
- 4-hydroxy-l-proline
- LH3
- lysyl hydroxylase 3
- MIDAS
- metal-ion-dependent adhesive site
- MUG
- 4-methylumbelliferyl β-d-galactopyranoside
- Pfp
- pentafluorophenyl
- THP
- triple-helical peptide
- RP
- reversed phase.
REFERENCES
- 1. Hoekstra H. J. (2008) The European Approach to In-transit Melanoma Lesions. Int. J. Hyperthermia 24, 227–237 [DOI] [PubMed] [Google Scholar]
- 2. Fidler I. J. (1990) Critical factors in the biology of human cancer metastasis: twenty-eighth G.H.A. Clowes memorial award lecture. Cancer Res. 50, 6130–6138 [PubMed] [Google Scholar]
- 3. Cairns R. A., Khokha R., Hill R. P. (2003) Molecular mechanisms of tumor invasion and metastasis: an integrated view. Curr. Mol. Med. 3, 659–671 [DOI] [PubMed] [Google Scholar]
- 4. Hood J. D., Cheresh D. A. (2002) Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2, 91–100 [DOI] [PubMed] [Google Scholar]
- 5. Schwartz M. A. (2001) Integrin signaling revisited. Trends Cell Biol. 11, 466–470 [DOI] [PubMed] [Google Scholar]
- 6. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 7. Barczyk M., Carracedo S., Gullberg D. (2010) Integrins. Cell Tissue Res. 339, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hynes R. O. (1987) Integrins: a family of cell surface receptors. Cell 48, 549–554 [DOI] [PubMed] [Google Scholar]
- 9. Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Structural basis of collagen recognition by integrin α2β1. Cell 101, 47–56 [DOI] [PubMed] [Google Scholar]
- 10. Gullberg D. E., Lundgren-Akerlund E. (2002) Collagen-binding I domain integrins: what do they do? Prog. Histochem. Cytochem. 37, 3–54 [DOI] [PubMed] [Google Scholar]
- 11. Shimaoka M., Takagi J., Springer T. A. (2002) Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 31, 485–516 [DOI] [PubMed] [Google Scholar]
- 12. Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 13. Liddington R. C., Ginsberg M. H. (2002) Integrin activation takes shape. J. Cell Biol. 158, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Puzon-McLaughlin W., Takada Y. (1996) Critical residues for ligand binding in an I domain-like structure of the integrin β1 subunit. J. Biol. Chem. 271, 20438–20443 [DOI] [PubMed] [Google Scholar]
- 15. Lin E. C., Ratnikov B. I., Tsai P. M., Carron C. P., Myers D. M., Barbas C. F., 3rd, Smith J. W. (1997) Identification of a region in the integrin β3 subunit that confers ligand binding specificity. J. Biol. Chem. 272, 23912–23920 [DOI] [PubMed] [Google Scholar]
- 16. Lin E. C., Ratnikov B. I., Tsai P. M., Gonzalez E. R., McDonald S., Pelletier A. J., Smith J. W. (1997) Evidence that the integrin β3 and β5 subunits contain a metal ion-dependent adhesion site-like motif but lack an I domain. J. Biol. Chem. 272, 14236–14243 [DOI] [PubMed] [Google Scholar]
- 17. Xiong J. P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D. L., Joachimiak A., Goodman S. L., Arnaout M. A. (2001) Crystal structure of the extracellular segment of integrin αVβ3. Science 294, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruoslahti E. (1991) Integrins as receptors for extracellular matrix. In Cell Biology of the Extracellular Matrix (May E. D., ed) pp. 343–363, Plenum Press, New York [Google Scholar]
- 19. Hynes R. O. (1992) Integrins: versitality, modulation, and signaling in cell adhesion. Cell 69, 11–25 [DOI] [PubMed] [Google Scholar]
- 20. Kühn K., Eble J. (1994) The structural bases of integrin-ligand interactions. Trends Cell Biol. 4, 256–261 [DOI] [PubMed] [Google Scholar]
- 21. Takada Y., Wayner E. A., Carter W. G., Hemler M. E. (1988) Extracellular matrix receptors, ECMRII and ECMRI, for collagen and fibronectin correspond to VLA-2 and VLA-3 in the VLA family of heterodimers. J. Cell. Biochem. 37, 385–393 [DOI] [PubMed] [Google Scholar]
- 22. Yoshinaga I. G., Vink J., Dekker S. K., Mihm M. C., Jr., Byers H. R. (1993) Role of α3β1 and α2β1 integrins in melanoma cell migration. Melanoma Res. 3, 435–441 [DOI] [PubMed] [Google Scholar]
- 23. Melchiori A., Mortarini R., Carlone S., Marchisio P. C., Anichini A., Noonan D. M., Albini A. (1995) The α3β1 integrin is involved in melanoma cell migration and invasion. Exp. Cell Res. 219, 233–242 [DOI] [PubMed] [Google Scholar]
- 24. Parkin J. D., San Antonio J. D., Pedchenko V., Hudson B., Jensen S. T., Savige J. (2011) Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum. Mutat. 32, 127–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vandenberg P., Kern A., Ries A., Luckenbill-Edds L., Mann K., Kühn K. (1991) Characterization of a type IV collagen major cell binding site with affinity to the α1β1 and the α2β1 integrins. J. Cell Biol. 113, 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kern A., Eble J., Golbik R., Kühn K. (1993) Interaction of type IV collagen with the isolated integrins α1β1 and α2β1. Eur. J. Biochem. 215, 151–159 [DOI] [PubMed] [Google Scholar]
- 27. Eble J. A., Golbik R., Mann K., Kühn K. (1993) The α1β1 integrin recognition site of the basement membrane collagen molecule α1(IV)2α2(IV). EMBO J. 12, 4795–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Golbik R., Eble J. A., Ries A., Kühn K. (2000) The spatial orientation of the essential amino acid residues arginine and aspartate within the α1β1 integrin recognition site of collagen IV has been resolved using fluorescence resonance energy transfer. J. Mol. Biol. 297, 501–509 [DOI] [PubMed] [Google Scholar]
- 29. Saccà B., Sinner E.-K., Kaiser J., Lübken C., Eble J. A., Moroder L. (2002) Binding and docking of synthetic heterotrimeric collagen type IV peptides with α1β1 integrin. ChemBioChem 3, 904–907 [DOI] [PubMed] [Google Scholar]
- 30. Renner C., Saccà B., Moroder L. (2004) Synthetic heterotrimeric collagen peptides as mimics of cell adhesion sites of the basement membrane. Biopolymers 76, 34–47 [DOI] [PubMed] [Google Scholar]
- 31. Barth L., Sinner E. K., Cadamuro S. A., Renner C., Oesterhelt D., Moroder L. (2009) Homotrimeric collagen petpides as model systems for cell adhesion studies. In Peptides for Youth: The Proceedings of the 20th American Peptide Symposium (Escher E., Lubell W. D., Del Valle S., eds) pp. 295–296, Springer Science, New York: [DOI] [PubMed] [Google Scholar]
- 32. Knight C. G., Morton L. F., Peachey A. R., Tuckwell D. S., Farndale R. W., Barnes M. J. (2000) The collagen-binding A-domains of integrin α1β1 and α2β1 recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J. Biol. Chem. 275, 35–40 [DOI] [PubMed] [Google Scholar]
- 33. Baronas-Lowell D., Lauer-Fields J. L., Borgia J. A., Sferrazza G. F., Al-Ghoul M., Minond D., Fields G. B. (2004) Differential modulation of human melanoma cell metalloproteinase expression by α2β1 integrin and CD44 triple-helical ligands derived from type IV collagen. J. Biol. Chem. 279, 43503–43513 [DOI] [PubMed] [Google Scholar]
- 34. Miles A. J., Skubitz A. P., Furcht L. T., Fields G. B. (1994) Promotion of cell adhesion by single-stranded and triple-helical peptide models of basement membrane collagen α1(IV)531–543: evidence for conformationally dependent and corformationally independent type IV collagen cell adhesion sites. J. Biol. Chem. 269, 30939–30945 [PubMed] [Google Scholar]
- 35. Miles A. J., Knutson J. R., Skubitz A. P., Furcht L. T., McCarthy J. B., Fields G. B. (1995) A peptide model of basement membrane collagen α1(IV)531–543 binds the α3β1 integrin. J. Biol. Chem. 270, 29047–29050 [DOI] [PubMed] [Google Scholar]
- 36. Babel W., Glanville R. W. (1984) Structure of human-basement-membrane (type IV) collagen: complete amino-acid sequence fo a 914-residue-long pepsin fragment from the α1(IV) chain. Eur. J. Biochem. 143, 545–556 [DOI] [PubMed] [Google Scholar]
- 37. Seyer J. M., Hasty K. A., Kang A. H. (1989) Covalent structure of collagen: amino acid sequence of an arthritogenic cyanogen bromide peptide from type II collagen of bovine cartilage. Eur. J. Biochem. 181, 159–173 [DOI] [PubMed] [Google Scholar]
- 38. Lauer-Fields J. L., Malkar N. B., Richet G., Drauz K., Fields G. B. (2003) Melanoma cell CD44 interaction with the α1(IV)1263–1277 region from basement membrane collagen is modulated by ligand glycoslyation. J. Biol. Chem. 278, 14321–14330 [DOI] [PubMed] [Google Scholar]
- 39. Syed B. M., Gustafsson T., Kihlberg J. (2004) 9-BBN as a convenient protecting group in functionalisation of hydroxylysine. Tetrahedron 60, 5571–5575 [Google Scholar]
- 40. Holm B., Broddefalk J., Flodell S., Wellner E., Kihlberg J. (2000) An improved synthesis of a galactosylated hydroxylysine building block and its use in solid-phase glycopeptide synthesis. Tetrahedron 56, 1579–1586 [Google Scholar]
- 41. Andersson I. E., Andersson C. D., Batsalova T., Dzhambazov B., Holmdahl R., Kihlberg J., Linusson A. (2011) Design of glycopeptides used to investigate class II MHC binding and T-cell responses associated with autoimmune arthritis. PLoS One 6, e17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tenchini M. L., Adams J. C., Gilberty C., Steel J., Hudson D. L., Malcovati M., Watt F. M. (1993) Evidence against a major role for integrins in calcium-dependent intercellular adhesion of epidermal keratinocytes. Cell Adhes. Commun. 1, 55–66 [DOI] [PubMed] [Google Scholar]
- 43. Anbazhagan R., Bartkova J., Stamp G., Pignatelli M., Gusterson B. (1995) Expression of integrin subunits in the human infant breast correlates with morphogenesis and differentiation. J. Pathol. 176, 227–232 [DOI] [PubMed] [Google Scholar]
- 44. Lauer J. L., Gendron C. M., Fields G. B. (1998) Effect of ligand conformation on melanoma cell α3β1 integrin-mediated signal transduction events: implications for a collagen structural modulation mechanism of tumor cell invasion. Biochemistry 37, 5279–5287 [DOI] [PubMed] [Google Scholar]
- 45. Yegorova S., Chavaroche A. E., Rodriguez M. C., Minond D., Cudic M. (2013) Development of an AlphaScreen assay for discovery of inhibitors of low-affinity glycan-lectin interactions. Anal. Biochem. 439, 123–131 [DOI] [PubMed] [Google Scholar]
- 46. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 47. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Dunbrack R. L., Jr. (2002) Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 12, 431–440 [DOI] [PubMed] [Google Scholar]
- 49. Jakalian A., Jack D. B., Bayly C. I. (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. parameterization and validation. J. Comput. Chem. 23, 1623–1641 [DOI] [PubMed] [Google Scholar]
- 50. Wang J., Wang W., Kollman P. A., Case D. A. (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 [DOI] [PubMed] [Google Scholar]
- 51. Nagae M., Re S., Mihara E., Nogi T., Sugita Y., Takagi J. (2012) Crystal structure of α5β1 integrin ectodomain: atomic details of the fibronectin receptor. J. Cell Biol. 197, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fiser A., Sali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 53. Trott O., Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dent W. H., 3rd, Erickson W. R., Fields S. C., Parker M. H., Tromiczak E. G. (2002) 9-BBN: an amino acid protecting group for functionalization of amino acid side chains in organic solvents. Org. Lett. 4, 1249–1251 [DOI] [PubMed] [Google Scholar]
- 55. Fields G. B., Lauer J. L., Dori Y., Forns P., Yu Y.-C., Tirrell M. (1998) Proteinlike molecular architecture: biomaterial applications for inducing cellular receptor binding and signal transduction. Biopolymers 47, 143–151 [DOI] [PubMed] [Google Scholar]
- 56. Yu Y.-C., Tirrell M., Fields G. B. (1998) Minimal lipidation stabilizes protein-like molecular architecture. J. Am. Chem. Soc. 120, 9979–9987 [Google Scholar]
- 57. Yongye A. B., Calle L., Ardá A., Jiménez-Barbero J., André S., Gabius H. J., Martínez-Mayorga K., Cudic M. (2012) Molecular recognition of the Thomsen-Friedenreich antigen-threonine conjugate by adhesion/growth regulatory galectin-3: nuclear magnetic resonance studies and molecular dynamics simulations. Biochemistry 51, 7278–7289 [DOI] [PubMed] [Google Scholar]
- 58. Long C. G., Thomas M., Brodsky B. (1995) Atypical Gly-X-Y sequences surround interruptions in the repeating tripeptide pattern of basement membrane collagen. Biopolymers 35, 621–628 [DOI] [PubMed] [Google Scholar]
- 59. Li Y., Brodsky B., Baum J. (2007) NMR shows hydrophobic interactions replace glycine packing in the triple helix at a natural break in the (Gly-X-Y)n repeat. J. Biol. Chem. 282, 22699–22706 [DOI] [PubMed] [Google Scholar]
- 60. Hwang E. S., Thiagarajan G., Parmar A. S., Brodsky B. (2010) Interruptions in the collagen repeating tripeptide pattern can promote supramolecular association. Protein Sci. 19, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mohs A., Popiel M., Li Y., Baum J., Brodsky B. (2006) Conformational features of a natural break in the type IV collagen Gly-X-Y repeat. J. Biol. Chem. 281, 17197–17202 [DOI] [PubMed] [Google Scholar]
- 62. Thiagarajan G., Li Y., Mohs A., Strafaci C., Popiel M., Baum J., Brodsky B. (2008) Common interruptions in the repeating tripeptide sequence of non-fibrillar collagens: sequence analysis and structural studies on triple-helix peptide models. J. Mol. Biol. 376, 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spiro R. G. (1967) The structure of the disaccharide unit of the renal glomerular basement membrane. J. Biol. Chem. 242, 4813–4823 [PubMed] [Google Scholar]
- 64. Spiro R. G. (1969) Characterization and quantitative determination of the hydroxylysine-linked carbohydrate units of several collagens. J. Biol. Chem. 244, 602–612 [PubMed] [Google Scholar]
- 65. Spiro M. J., Spiro R. G. (1971) Studies on the biosynthesis of the hydroxylysine-linked disaccharide unit of basement membranes and collagens II: kidney galactosyltransferase. J. Biol. Chem. 246, 4910–4918 [PubMed] [Google Scholar]
- 66. Haass N. K., Smalley K. S., Li L., Herlyn M. (2005) Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 18, 150–159 [DOI] [PubMed] [Google Scholar]
- 67. Giancotti F. G., Ruoslahti E. (1999) Integrin signaling. Science 285, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 68. Humphries M. J., Mould A. P. (2001) Structure. An anthropomorphic integrin. Science 294, 316–317 [DOI] [PubMed] [Google Scholar]
- 69. van der Flier A., Sonnenberg A. (2001) Function and interactions of integrins. Cell Tissue Res. 305, 285–298 [DOI] [PubMed] [Google Scholar]
- 70. Marshall J. F., Nesbitt S. A., Helfrich M. H., Horton M. A., Polakova K., Hart I. R. (1991) Integrin expression in human melanoma cell lines: heterogeneity of vitronectin receptor composition and function. Int. J. Cancer 49, 924–931 [DOI] [PubMed] [Google Scholar]
- 71. Springer T. A., Zhu J., Xiao T. (2008) Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J. Cell Biol. 182, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dennis J., Waller C., Timpl R., Schirrmacher V. (1982) Surface sialic acid reduces attachment of metastatic tumour cells to collagen type IV and fibronectin. Nature 300, 274–276 [DOI] [PubMed] [Google Scholar]
- 73. Laferté S., Dennis J. W. (1988) Glycosylation-dependent collagen-binding activities of two membrane glycoproteins in MDAY-D2 tumor cells. Cancer Res. 48, 4743–4748 [PubMed] [Google Scholar]
- 74. Allen B. L., Katz B., Höök M. (2002) Streptococcus anginosus adheres to vascular endothelium basement membrane and purified extracellular matrix proteins. Microb. Pathog. 32, 191–204 [DOI] [PubMed] [Google Scholar]
- 75. Patti J. M., Boles J. O., Höök M. (1993) Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry 32, 11428–11435 [DOI] [PubMed] [Google Scholar]
- 76. Zong Y., Xu Y., Liang X., Keene D. R., Höök A., Gurusiddappa S., Höök M., Narayana S. V. (2005) A “collagen hug” model for Staphylococcus aureus CNA binding to collagen. EMBO J. 24, 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Madsen D. H., Ingvarsen S., Jürgensen H. J., Melander M. C., Kjøller L., Moyer A., Honoré C., Madsen C. A., Garred P., Burgdorf S., Bugge T. H., Behrendt N., Engelholm L. H. (2011) The non-phagocytic route of collagen uptake: A distinct degradation pathway. J. Biol. Chem. 286, 26996–27010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Valiathan R. R., Marco M., Leitinger B., Kleer C. G., Fridman R. (2012) Discoidin domain receptor tyrosine kinases: new players in cancer progression. Cancer Metastasis Rev. 31, 295–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vogel W., Gish G. D., Alves F., Pawson T. (1997) The discoidin domain receptor tyrosine kinases are activated by collagen. Mol. Cell 1, 13–23 [DOI] [PubMed] [Google Scholar]
- 80. Zeidan Q., Hart G. W. (2010) The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 123, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kivirikko K. I., Myllylä R. (1982) Posttranslational enzymes in the biosynthesis of collagen: intracellular enzymes. Methods Enzymol. 82, 245–304 [DOI] [PubMed] [Google Scholar]
- 83. Wang C., Luosujärvi H., Heikkinen J., Risteli M., Uitto L., Myllylä R. (2002) The third activity for lysyl hydroxylase 3: galactosylation of hydroxylysyl residues in collagens in vivo. Matrix Biol. 21, 559–566 [DOI] [PubMed] [Google Scholar]
- 84. Rautavuoma K., Takaluoma K., Passoja K., Pirskanen A., Kvist A.-P., Kivirikko K. I., Myllyharju J. (2002) Characterization of three fragments that constitute the monomers of the human lysyl hydroxylase isoenzymes 1–3. J. Biol. Chem. 277, 23084–23091 [DOI] [PubMed] [Google Scholar]
- 85. Schegg B., Hülsmeier A. J., Rutschmann C., Maag C., Hennet T. (2009) Core glycosylation of collagen is initiated by two β(1-O)galactosyltransferases. Mol. Cell. Biol. 29, 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Heikkinen J., Risteli M., Wang C., Latvala J., Rossi M., Valtavaara M., Myllylä R. (2000) Lysyl hydroxylase 3 is a multifunctional protein possessing collagen glucosyltransferase activity. J. Biol. Chem. 275, 36158–36163 [DOI] [PubMed] [Google Scholar]
- 87. Ruotsalainen H., Sipilä L., Vapola M., Sormunen R., Salo A. M., Uitto L., Mercer D. K., Robins S. P., Risteli M., Aszodi A., Fässler R., Myllylä R. (2006) Glycosylation catalyzed by lysyl hydroxylase 3 is essential for basement membranes. J. Cell Sci. 119, 625–635 [DOI] [PubMed] [Google Scholar]
- 88. Sricholpech M., Perdivara I., Nagaoka H., Yokoyama M., Tomer K. B., Yamauchi M. (2011) Lysyl hydroxylase 3 glucosylates galactosylhydroxylysine residues in type I collagen in osteoblast culture. J. Biol. Chem. 286, 8846–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sricholpech M., Perdivara I., Yokoyama M., Nagaoka H., Terajima M., Tomer K. B., Yamauchi M. (2012) Lysyl hydroxylase 3-mediated glucosylation in type I collagen: molecular loci and biological significance. J. Biol. Chem. 287, 22998–23009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Salo A. M., Wang C., Sipilä L., Sormunen R., Vapola M., Kervinen P., Ruotsalainen H., Heikkinen J., Myllylä R. (2006) Lysyl hydroxylase 3 (LH3) modifies proteins in the extracellular space, a novel mechanism for matrix remodeling. J. Cell. Physiol. 207, 644–653 [DOI] [PubMed] [Google Scholar]
- 91. Myllylä R., Wang C., Heikkinen J., Juffer A., Lampela O., Risteli M., Ruotsalainen H., Salo A., Sipilä L. (2007) Expanding the lysyl hydroxylase toolbox: new insights into the localization and activities of lysyl hydroxylase 3 (LH3). J. Cell. Physiol. 212, 323–329 [DOI] [PubMed] [Google Scholar]
- 92. Wang C., Kovanen V., Raudasoja P., Eskelinen S., Pospiech H., Myllylä R. (2009) The glycosyltransferase activities of lysyl hydroxylase 3 (LH3) in the extracellular space are important for cell growth and viability. J. Cell. Mol. Med. 13, 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Babiarz B., Cullen E. (1992) 3T3 cell surface galactosyltransferase is a calcium-dependent adhesion molecule for collagen type IV. Exp. Cell Res. 203, 276–279 [DOI] [PubMed] [Google Scholar]
- 94. Lee M. M., Nasirikenari M., Manhardt C. T., Ashline D. J., Hanneman A. J., Reinhold V. N., Lau J. T. (2014) Platelets support extracellular sialylation by supplying the sugar donor substrate. J. Biol. Chem. 289, 8742–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Urushizaki Y., Seifter S. (1985) Phosphorylation of hydroxylysine residues in collagen synthesized by cultured aortic smooth muscle cells. Proc. Natl. Acad. Sci. U.S.A. 82, 3091–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zimina E. P., Fritsch A., Schermer B., Bakulina A. Y., Bashkurov M., Benzing T., Bruckner-Tuderman L. (2007) Extracellular phosphorylation of collagen XVII by ecto-casein kinase 2 inhibits ectodomain shedding. J. Biol. Chem. 282, 22737–22746 [DOI] [PubMed] [Google Scholar]
- 97. Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Privitera S., Prody C. A., Callahan J. W., Hinek A. (1998) The 67-kDa enzymatically inactive alternatively spliced variant of β-galactosidase is identical to the elastin/laminin-binding protein. J. Biol. Chem. 273, 6319–6326 [DOI] [PubMed] [Google Scholar]
- 99. Sternberg M., Spiro R. G. (1979) Studies on the catabolism of hydroxylysine-linked disaccharide units of basement membranes and collagens. J. Biol. Chem. 254, 10329–10336 [PubMed] [Google Scholar]
- 100. Nayak B. R., Spiro R. G. (1991) Localization and structure of the asparagine-linked oligosaccharides of type IV collagen from glomerular basement membrane and lens capsule. J. Biol. Chem. 266, 13978–13987 [PubMed] [Google Scholar]
- 101. Taga Y., Kusubata M., Ogawa-Goto K., Hattori S. (2012) Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry. Mol. Cell. Proteomics 10.1074/mcp.M111.010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bao X., Kobayashi M., Hatakeyama S., Angata K., Gullberg D., Nakayama J., Fukuda M. N., Fukuda M. (2009) Tumor suppressor function of laminin-binding α-dystroglycan requires a distinct β3-N-acetylglucosaminyltransferase. Proc. Natl. Acad. Sci. U.S.A. 106, 12109–12114 [DOI] [PMC free article] [PubMed] [Google Scholar]