Background: Tom40 has been modeled as a 19-strand β-barrel after the three-dimensional structure of porin. However, a 13-strand model for porin also exists.
Results: Several β-strands were mapped in Tom40, all are predicted by the 19-strand model.
Conclusion: Data support the 19-strand model for Tom40.
Significance: Predictions relating the Tom40 structure and function can be made with more confidence.
Keywords: Membrane Protein, Mitochondria, Neurospora, Protein Structure, Transmembrane Domain, SCAM, Tom40, β-Barrel Protein
Abstract
Most proteins found in mitochondria are translated in the cytosol and enter the organelle via the TOM complex (translocase of the outer mitochondrial membrane). Tom40 is the pore forming component of the complex. Although the three-dimensional structure of Tom40 has not been determined, the structure of porin, a related protein, has been shown to be a β-barrel containing 19 membrane spanning β-strands and an N-terminal α-helical region. The evolutionary relationship between the two proteins has allowed modeling of Tom40 into a similar structure by several laboratories. However, it has been suggested that the 19-strand porin structure does not represent the native form of the protein. If true, modeling of Tom40 based on the porin structure would also be invalid. We have used substituted cysteine accessibility mapping to identify several potential β-strands in the Tom40 protein in isolated mitochondria. These data, together with protease accessibility studies, support the 19 β-strand model for Tom40 with the C-terminal end of the protein localized to the intermembrane space.
Introduction
Mitochondria of eukaryotic cells contain a small genome that encodes a few proteins necessary for complete mitochondrial function. However, the vast majority of mitochondrial proteins are encoded in the nucleus, synthesized on cytosolic ribosomes, and imported into the organelle. Most nuclear-encoded mitochondrial proteins enter the organelle via the translocase of the outer mitochondrial membrane (TOM)2 complex (1–7). The TOM complex of both Saccharomyces cerevisiae and Neurospora crassa consists of five core proteins (Tom40, Tom22, Tom7, Tom6, and Tom5) and two receptors (Tom70 and Tom20) that are more loosely associated. The TOM complex of animals is quite similar to the fungal version, whereas the plant complex contains several different subunits (8, 9). Although some of the components of the TOM complex may vary in different organisms, the Tom40 protein is highly conserved (10–13). The protein forms the actual pore through which mitochondrial proteins traverse the outer membrane from the cytosol (14–17) and has been shown to be essential for the viability of both S. cerevisiae and N. crassa (18, 19). Tom40 is thought to exist as a β-barrel within the outer membrane and secondary structure analysis has confirmed a high level of β-sheet structure (14, 17, 20, 21). Although the overall appearance of the TOM complex has been revealed through electron microscopy studies (15, 22–24), no three-dimensional structure of Tom40 has been determined.
The most common β-barrel protein in the mitochondrial outer membrane is the voltage-dependent anion channel, also known as porin. The three-dimensional structure of human and mouse porin has been determined by three independent groups utilizing NMR and x-ray crystallography (25–27). Each of these studies resulted in a model of porin as a 19-stranded β-barrel structure with an N-terminal α-helix plugging the pore in the closed state. Comparative studies have revealed a phylogenetic relationship between Tom40 and porin (10–13, 25). The relationship between the two proteins, coupled with extensive bioinformatic analysis and other experimental approaches, has led to the development of 19 β-strand models of Tom40 based on the known structure of porin (11–13, 28). Tom40 is also thought to have an α-helical region near the N terminus, preceding the β-strands.
There is general acceptance of the 19-strand porin structure (hereafter known as the “three-dimensional model”) in the literature (13, 29, 30). However, it has been pointed out that the three-dimensional model conflicts with conclusions based on many previous biochemical and functional studies of porin (31–39). As a result, it has been suggested (36) that the three-dimensional structure was actually determined on a non-native form of the protein that arose during its purification for structural analysis. An alternate model of porin structure (hereafter referred to as the “biochemical” or BIO model), based on the functional data, suggests that the porin channel exists in a structure containing 13 β-strands with an α-helical region near the N terminus (36).
Because the current model for Tom40 structure is largely derived by modeling on the porin structure, any controversy regarding the structure of the latter will affect the reliability of the Tom40 model. In this report we describe the use of the substituted cysteine accessibility method (SCAM) to provide evidence for the location of several potential β-strands within N. crassa Tom40 (NcTom40). To determine the orientation of the protein in the membrane, we also inserted protease factor Xa sites at various positions of the protein for protease accessibility studies in both intact mitochondria and crude mitoplasts. Our data support the existence of at least six of the β-strands predicted in the 19-strand three-dimensional model. Two of these six would not be predicted if Tom40 were modeled after the 13-strand BIO model of porin. Because our SCAM study was performed on Tom40 protein present in intact mitochondria it seems likely that the protein was in its native conformation. Thus, our results favor the modeling of Tom40 as a structure containing 19 β-strands. Previous studies on NcTom40 (19, 40) that described the effects of mutations on its function, assembly, and stability have been re-examined in light of the current 19 β-strand model of the Tom40 structure.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
N. crassa was routinely grown in Vogel's medium at 30 °C as described (41). The N. crassa strains used as controls, or as parents to develop strains containing specific tom40 mutations, are listed in Table 1. Strains expressing Tom40 proteins with individual site-directed mutations were generated by transformation with plasmids carrying tom40 genes, specifically altered by site-directed mutagenesis, into one of two sheltered heterokaryons. Each of the heterokaryons is composed of one nucleus with a non-functional allele of tom40 and another nucleus with a wild type version of the gene. The first heterokaryon (RIP40het) was isolated (19) using the technique of sheltered RIP (repeat-induced point mutation). The second (Tom40 KO-5) was developed by the method of sheltered disruption in heterokaryon HP1 as described previously for mutants of Tob55 (42). The nuclei of the heterokaryons can be differentiated and selected using differing auxotrophic and drug-resistance markers. Thus, transformants of the tom40 knock-out nucleus can be directly selected as viable transformants when mutant (but still functional) alleles of tom40 are introduced. Following selection and purification, homokaryotic strains expressing a specific version of the Tom40 protein are obtained.
TABLE 1.
Strains used in this study
| Strain (isolate used) | Genotype | Origin or reference |
|---|---|---|
| 76-26 | his-3 mtrR a (mtrR imparts flourophenylalanine resistance) | R. L. Metzenberg |
| 71-18 | pan-2 bmlR a (bmlR imparts benomyl resistance) | R. L. Metzenberg |
| HP1 | Heterokaryon of 76–26 and 71–18 | Nargang Lab. |
| Tom40 KO-5 | Sheltered heterokaryon. As HP1, but with replacement of tom40 gene in 76–26 nucleus with a hygromycin resistance (hygR) cassette | Transformation of HP1 with split marker fragments for tom40 knockout. |
| HostV | cyh-2 lys-2 leu-5 mei-2 a | Fungal Genetics Stock Center number 7255 |
| MateV | am132 inl inv mei-2 A | Fungal Genetics Stock Center number 7265 |
| 40Dup1 | cyh-2 lys-2 leu-5 mei-2 a contains an ectopic 1.8-kb copy of tom40. Hygromycin resistant. | Transformation of HostV with tom40 pRIP-4 plasmid (19) |
| RIP40het (F40-6) | Sheltered heterokaryon: (cyh-2 lys-2 leu-5 mei-2 tom40RIP + am132 inl inv mei-2) both nuclei contain the ectopic tom40RIP | MateV x 40Dup1 (19) |
| C294A (170-6) | cyh-2 lys-2 leu-5 mei-2 tom40RIP contains ectopic copy of tom40 with sole Cys residue 294 changed to Ala. | Transformation of RIP40het with plasmid pC8 |
| L10C (10-2-1) | cyh-2 lys-2 leu-5 mei-2 tom40RIP contains ectopic copy of tom40 with Cys-294 changed to Ala and Leu-10 residue changed to Cys. | Transformation of RIP40het with specific SCAM plasmid construct |
| L322C (322-5-1) | cyh-2 lys-2 leu-5 mei-2 tom40RIP contains ectopic copy of tom40 with Cys-294 changed to Ala and Leu-322 residue changed to Cys. | Transformation of RIP40het with specific SCAM plasmid construct |
| S344C (344-4-1) | cyh-2 lys-2 leu-5 mei-2 tom40RIP contains ectopic copy of tom40 with Cys-294 changed to Ala and Ser-344 residue changed to Cys. | Transformation of RIP40het with specific SCAM plasmid construct |
A total of 103 strains containing changes for individual residues in Tom40 for SCAM analysis were constructed in either RIP40het or Tom40 KO-5. Selection for transformants made in RIP40het (mutations to Cys residues at positions 144 to 182 and 316 to 339) was for the Host V genotype so that they have the genetic background cyh-2 lys-2 leu-5 mei-2 a. Those created in Tom40 KO-5 (Cys mutations at residues 90 to 120 and 135 to 143) were selected for the 76-26 nucleus and these have the genetic background his-3 mtrR a. All the selected transformants also carry an ectopic copy of tom40 containing a single codon change to give Cys at the desired position. Seven strains with triple factor Xa cleavage sites inserted at specific sites of the Tom40 protein were constructed using heterokaryon Tom40 KO-5. Transformants of the 76-26 nucleus were selected so that they have the genotype his-3 mtrR a. These strains also contain an ectopic copy of tom40 containing a triple factor Xa site at one specific location as described under “Results.”
All Cys-substituted versions of Tom40 were created in plasmid pC8, which carries bleomycin resistance and a tom40 gene whose lone endogenous Cys codon had been changed to encode Ala. The triple factor Xa containing versions of tom40 were created in plasmid pB3, which is identical to pC8 except that the endogenous codon for the Cys residue of tom40 was not mutated. Single factor Xa sites (Ile-Glu-Gly-Arg encoded by ATCGAGGGGAGG) were added sequentially using three successive rounds of site-directed mutagenesis to create triple factor Xa sites at the desired location of the protein. All genes modified by site-directed mutagenesis were sequenced to confirm the desired change and ensure that no other random mutations had been introduced.
Mitochondrial Isolation
Mycelium was grown at 30 °C and harvested by filtration. For SCAM analysis the cells were ground using a mortar and pestle in the presence of sand and PBSSP isolation buffer (phosphate-buffered salts, sucrose, phenylmethylsulfonyl fluoride (PMSF): 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4, 0.25 m sucrose, 1 mm PMSF). Mitochondria were isolated by differential centrifugation as described previously (43, 44). For all other experiments, mitochondria were isolated using buffers described previously (43).
SCAM Biotin Maleimide Labeling
Maleimide labeling reagents react with the thiol group of Cys residues in an aqueous environment (45). We reasoned that β-strands in membrane-embedded β-barrel proteins would be expected to give an alternating label/no label pattern of labeling. This would reflect the orientation of R-groups in the amino acid residues of a β-strand, which would alternate between existing in the aqueous lumen of the pore and being embedded in the membrane. SCAM was performed with mitochondria that were either freshly isolated or that had been frozen at −80 °C immediately after isolation. Mitochondria (50–200 μg of protein) were suspended in 500 μl of PBSSP. 10 μl of labeling reagent (EZ-linkTM maliemide-PEG2-biotin, Thermo Fisher Scientific, Rockford IL), dissolved at a concentration of 40 mm in PBSSP, was added and the samples were rocked at room temperature for 2 h. Mitochondria were re-isolated and washed twice with PBSSP containing 1% β-mercaptoethanol to quench excess labeling reagent.
Because Tom40 is an integral membrane protein, an alkaline extraction was performed, as described previously (44), following the second wash to minimize background signal from contaminating soluble proteins. The pellets were resuspended in lysis buffer (50 mm Tris, pH 7.4, 1% sodium deoxycholate, 0.1% Triton X-100) and rocked at 4 °C for 30 min. The sample was then clarified by centrifugation at 11,000 × g for 30 min at 4 °C. The clarified supernatant was collected for Tom40 immunoprecipitation. Immunoprecipitation was performed overnight with gentle rocking at 4 °C using Protein A-agarose beads that had been cross-linked (using dimethyl pimelimidate) to IgGs from rabbit antiserum directed against N. crassa Tom40. Beads were purified, following the cross-linking step, by centrifugation at 11,000 × g for 4 min at 4 °C and three washes with lysis buffer.
Cracking buffer (0.06 m Tris-HCl, pH 6.8, 2.5% SDS, 5% β-mercaptoethanol, 5% sucrose) was added to the beads with bound Tom40 and the mixture was shaken for 30 min at room temperature followed by incubation for 20 min at 95 °C. The beads were then removed by centrifugation at 11,000 × g for 4 min at 4 °C. The Tom40 containing supernatant was electrophoresed through a 10% SDS-PAGE gel. Proteins were then transfered to a nitrocellulose membrane. The membrane was blocked with 3% bovine serum albumin (BSA) in TBS-Tween (150 mm NaCl, 12.5 mm Tris, pH 7.5, 0.5% Tween 20) and then probed via incubation for 2 h with either streptavidin-HRP (horseradish peroxidase) conjugate or streptavidin-biotinylated HRP complex in 3% BSA/TBS-Tween 20 (both obtained from Amersham Biosciences). Both of these reagents bind with the free biotin moieties carried on the SCAM labeling reagent if it reacted with a given Cys-substituted Tom40 protein. Following three washes in TBS-Tween and one in TBS, the membranes were saturated with enzyme-catalyzed light reagents (KPL, Gaithersburg, MD) and signal was detected via exposure to x-ray film (Biomax XAR, Eastman Kodak Co.).
General Procedures
Mitochondrial proteins were analyzed by either SDS-PAGE (46) or blue native gel electrophoresis (BNGE) (47, 48). Western blotting (49), import and assembly of proteins into isolated mitochondria (50), analysis of TOM complex stability (51), and transformation of N. crassa (42) were performed as described previously. Factor Xa protease was obtained from New England Biolabs (Ipswich, MA) and used in accordance with the suppliers instructions. When required, mitochondria were converted to “crude mitoplasts” as described previously (52). This procedure ruptures the outer membrane but does not remove it. This results in the release of the contents of the intermembrane space (IMS) and makes both the cytosolic and IMS sides of the outer membrane accessible to added proteases. In some cases irrelevant lanes were electronically removed from gel blots or autoradiograms.
RESULTS
To establish a known anchor point for the topology of the predicted structure of NcTom40 we wished to obtain experimental evidence for the existence of the most C-terminal predicted β-strand (strand 19 of the three-dimensional model), which has been shown to contain the β-signal for insertion of the protein into the mitochondrial outer membrane by the sorting and assembly machinery) complex (28, 53), and also verify that the C terminus of the protein exists in the IMS. Establishing these positions would allow firm predictions for the location of loops between strands as being in either the cytosol or IMS in any model of the protein.
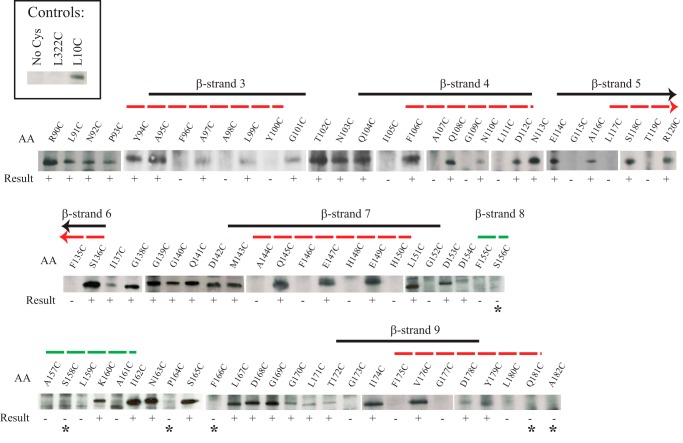
A Cys-scanning approach was used to provide evidence for the location of β-strand 19 of the NcTom40 protein. Although we are not aware of previous attempts to map membrane spanning β-strands by SCAM, we reasoned that residues of a β-strand whose R-groups reside in the aqueous environment of the pore lumen would be able to react with the maleimide labeling reagent, whereas those that are embedded in the hydrophobic bilayer would be unable to react. Given that the R-groups of residues comprising β-strands in a membrane-localized β-barrel should alternate between the pore lumen and the membrane, an alternating pattern of label/no label was expected for residues in a β-strand. This pattern should often be found to correspond to an alternating pattern of hydrophilic and hydrophobic residues. To validate the Cys-scanning approach for Tom40 we used a version of the tom40 gene with the endogenous Cys changed to Ala, to introduce new Cys codons at either amino acid positions 10 or 344, near the N and C terminus of the 349-residue protein, respectively. These residues should act as positive controls for the labeling procedure because, to our knowledge, neither of these residues has been predicted to be in a β-strand, or any other membrane spanning domain, in any model of the fungal protein published to date. Both residues were shown to be accessible to the labeling reagent, whereas the control Tom40 protein in which the only endogenous Cys residue had been changed to Ala, was not labeled (Fig. 1A).
FIGURE 1.
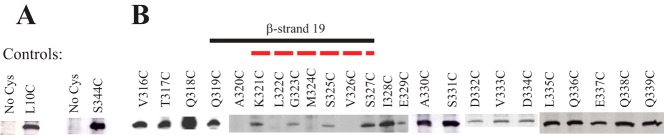
Defining the position of the C-terminal β-strand of NcTom40. A, SCAM analysis of control strains. Strains were developed that expressed Tom40 protein that lacked any Cys residues (No Cys) or contained single Cys residues near the N or C terminus of the protein (L10C and S344C) at positions predicted to be labeled by the maleimide reagent. Mitochondria were isolated from the strains and exposed to the labeling reagent, Tom40 was immunoprecipitated, and Western blot analysis using streptavidin-HRP to detect biotin maleimide-labeled Tom40 molecules was performed as described under “Experimental Procedures.” The appearance of a band at a given position indicates that the Cys residue is present in an aqueous environment and is able to react with the biotin maleimide. B, as in A, except that mitochondria for SCAM analysis were isolated from strains expressing Tom40 with single Cys substitution mutations at the indicated positions. The region that conforms to the predicted alternating labeling pattern for β-strand 19 is indicated by a solid black line. The region predicted as β-strand 19 for NcTom40 by modeling (12) is shown by a red dotted line. The results for the Cys mutant strains were not all derived from the same gel/Western blot. Controls were present on each gel/blot, but these and other irrelevant lanes have been removed to allow linear presentation of data.
Individual mutant versions of the tom40 gene encoding Tom40 proteins with Cys residues at each of positions 316 to 339 were then constructed in a copy of the gene in which the lone endogenous Cys codon had been changed to Ala. The resulting individual Cys-mutant forms were transformed into a sheltered heterokaryon as described under “Experimental Procedures” to yield homokaryotic strains expressing different Cys-mutant versions of the protein. All of the Cys substitution constructs gave rise to viable homokaryotic strains in the appropriate nuclear background. Because Tom40 is essential for viability (19), this suggests that no gross structural or functional abnormalities resulted from the Cys substitutions. Mitochondria were then isolated from each of the mutant strains and the ability of residues 316 to 339 to react with biotin maleimide was examined. The predicted alternating pattern of label/no label for a β-strand was observed for residues 319 to 327 (Fig. 1B). The results also demonstrate that residues 328 to 339, which are C-terminal to the β strand, and residues 316 to 318, which are N-terminal to the β strand, were labeled (Fig. 1B). These data provide good experimental support for the existence of the predicted β-strand 19 and for the approach of Cys scanning to detect membrane spanning β-strands. The results are also in agreement with cross-linking data suggesting the existence of β-strand 19 in S. cerevisiae (28).
The two regions immediately flanking β-strand 19 must exist on opposite sides of the mitochondrial outer membrane. To demonstrate that the C terminus occurs in the IMS, as suggested by previous studies (14, 16), we constructed tom40 variants encoding three tandem factor Xa protease sites between residues 316 and 317 (site Xa5), 317 and 318 (site Xa6), and following the C-terminal residue 349 (site Xa8). The position of these and other factor Xa sites created (described below) relative to the predicted 19 β-strands, IMS loops, and cytosolic loops of the three-dimensional model is shown in Fig. 2A. Mitochondria isolated from strains expressing these Tom40 proteins were treated with externally added factor Xa protease. A description of fragments expected to be produced upon factor Xa cleavage of Tom40 containing these sites is given in Fig. 2B. The site at the C terminus of the protein (Xa8) was found to be resistant to cleavage, whereas the two Tom40 proteins carrying sites just preceding the β-strand (Xa6 and Xa5) were accessible to the protease (Fig. 2C). Converting mitochondria that contain site Xa8 to crude mitoplasts makes the site accessible to the added protease as shown by the slight increase in electrophoretic mobility that occurs as the result of the loss of factor Xa sites at the very C terminus of the protein (Fig. 2C). This result eliminates the possibility that the site is simply in a conformation that makes it unavailable for factor Xa digestion. Taken together these data strongly suggest that the C terminus of NcTom40 is present in the IMS and protected from externally added protease, whereas the region just prior to the β-strand is exposed on the cytosolic side of the mitochondrial outer membrane. We found that factor Xa cleavage of Tom40 was almost never complete under the conditions used in these experiments.
FIGURE 2.
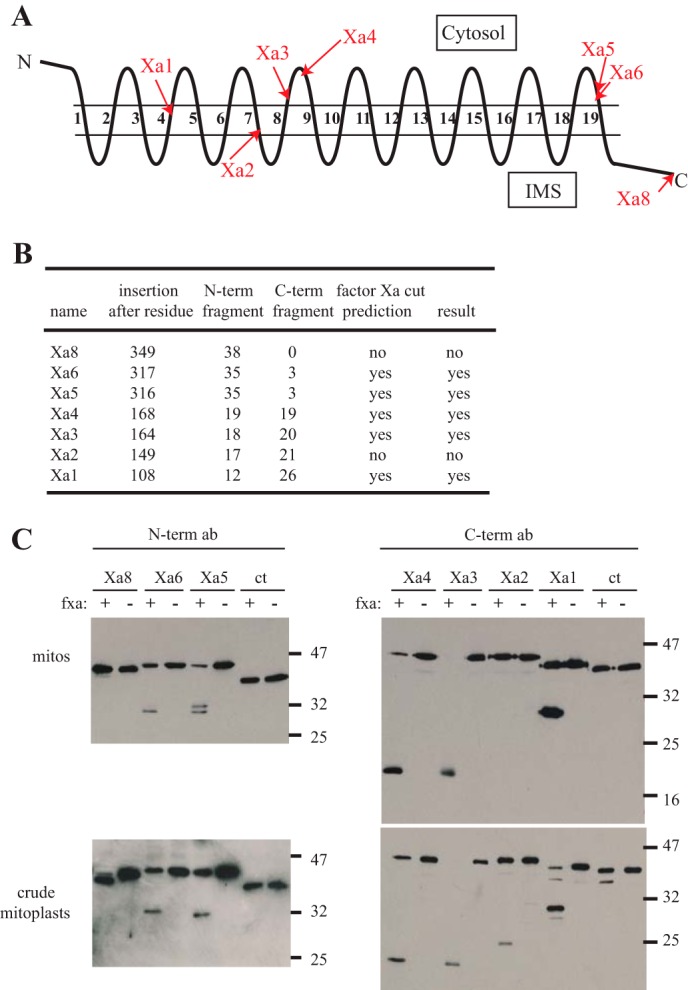
Protease accessibility of triple factor Xa sites introduced into NcTom40. A, representation of NcTom40 in the outer membrane with the relative position of predicted cytosolic loops, membrane spanning β-strands (numbered), and IMS loops shown as predicted for the 19-strand model (12). The locations of triple factor Xa sites introduced into the protein in individual strains are indicated by red arrows. B, the sites where triple factor Xa sites were inserted into the NcTom40 protein, the predicted sizes (kDa) of the N- and C-terminal (N-term and C-term) fragments that would be produced by cleavage at those sites, and the prediction of cleavage by factor Xa treatment of intact mitochondria, based on the 19 β-strand structural model, are shown. The predictions assume that the C terminus exists in the IMS and that triple factor Xa sites beginning within a β-strand have at least one or two of the Xa sites exposed in the compartment that is at the C-terminal end of the insertion site. The actual result of cleavage by factor Xa protease, as shown in panel C, is also given. C, the two top panels show Western blots of isolated, mitochondria (mitos) containing Tom40 protein with factor Xa sites inserted at the positions indicated in panel A. Mitochondria were treated with factor Xa protease (fxa) or left untreated (as indicated) and then subjected to SDS-PAGE and Western blotting. The two lower panels are identical to the upper panels, except that the mitochondria were subjected to a procedure that creates crude mitoplasts, prior to treatment with factor Xa. This allows the protease access to the IMS and exposes both sides of the outer membrane to the protease. The two left panels were immunodecorated with antibody produced to a peptide corresponding to the first 12 amino acids of NcTom40 (N-term ab). The two right panels were decorated with antibody produced to a peptide for the last 12 amino acids of NcTom40 (C-term ab). The ct lanes are mitochondria isolated from a control strain (76-26) containing no factor Xa sites. The positions of standard molecular weight markers are shown on the right side of the blots. The C-terminal antibody detects an unknown band in certain lanes, just below the full-length proteins.
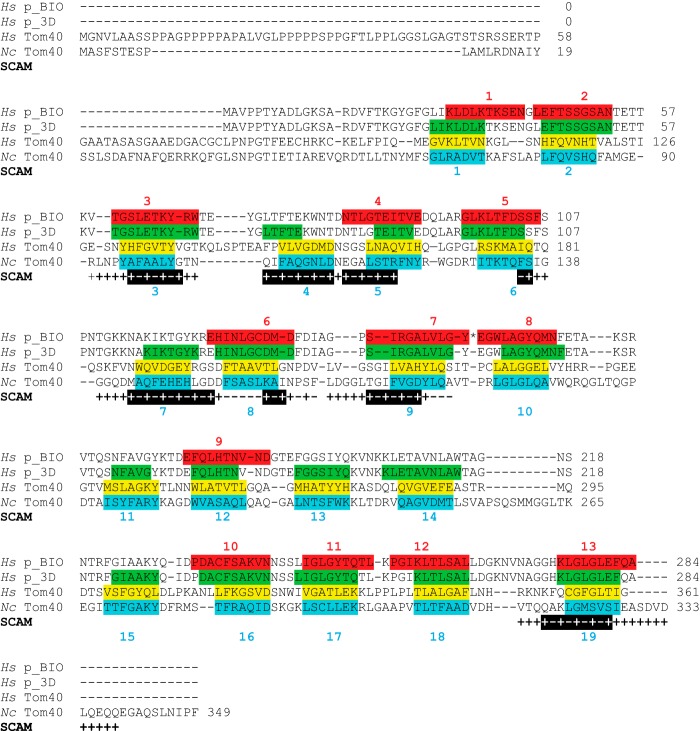
We continued our examination of the model for NcTom40 structure by Cys scanning and factor Xa site accessibility. The regions predicted to contain β-strands 3, 4, 5, 7, 8, and 9 of the three-dimensional model (12) were examined by Cys scanning. Residues near the C-terminal end of strand 6 were also examined. Good evidence for predicted strands 3, 4, 5, 7, and 9 was obtained as residues 95–101 (strand 3), 104–112 (strand 4), 114–120 (strand 5), 143–153 (strand 7), and 172–178 (strand 9) display an alternating pattern of labeled and unlabeled residues (Fig. 3). β-Strand 6 is predicted to occur from residues 130 to 136. We examined only three residues in this region, but see one unlabeled residue (135) and two labeled residues (136, 137) that may correspond to the C-terminal end of the strand. β-Strand 8 is predicted to exist from residues 155 to 161. Here we observed the alternating pattern only for residues 160–162 (Fig. 3). The remaining N-terminal residues predicted to be in the strand are unlabeled. A possible explanation for the lack of labeling in the N-terminal region of strand 8 may be that it interacts with the N-terminal α-helix of the protein. In the porin three-dimensional structure, an N-terminal α-helix was found to reside within the pore when the protein was in the closed state (25–28). One study identified a small hydrophobic patch of the pore lumen involving β-strands 9 and 10 that interact with the defined residues of the α-helix (26). Another study suggested that the α-helix was orientated adjacent to β-strands 8 to 19 with specific hydrogen bonds identified between the α-helix and residues at the N-terminal end of β-strand 8 (27). This correlates to the region of strand 8 lacking the anticipated labeling in our SCAM data (Fig. 3). Thus, if the predicted α-helix at the Tom40 N terminus behaves in a similar manner, it might shield residues in strand 8 from the labeling reagent. Interestingly, cross-linking studies have shown the S. cerevisiae Tom40 N-terminal α-helix resides within the TOM complex channel in close proximity to β-strand 9 (28). Our evidence for β-strand placement is summarized (Fig. 4) relative to the predictions for the placement of β-strands in the N. crassa and human Tom40 proteins and in comparison to the three-dimensional and BIO models for the porin structure (11–13, 36).
FIGURE 3.
SCAM analysis of different regions of NcTom40. Results and presentation are as described in the legend to Fig. 1. Controls are shown in the upper left box and include L322C as a negative control based on the results in Fig. 1B. β-Strands are as indicated as in Fig. 1. The arrowheads for β-strands 5 and 6 indicate that the strand should extend further in the indicated direction, but no SCAM data were obtained for the region. The labeling result is shown below each position. * indicates positions where labeling was expected, due to the position of a predicted strand or loop, but not observed. The dashed green line indicates the predicted position of β-strand 8 (12) that was not confirmed by experimental results (see text).
FIGURE 4.
Alignment of human porin, human Tom40, and NcTom40 with SCAM results. The amino acid sequence of porin from Homo sapiens (Hs) is shown in the top two sequences. In the first (Hs p_BIO) the position of the 13 β-strands predicted by the BIO model (36) are shown (highlighted and numbered in red). An asterisk (*) represents a sharp (zero residue) hairpin loop between β-strands 7 and 8 in the BIO model. The second sequence (Hs p_3D) shows the position (green) of the 19 β-strands from the three-dimensional structures of porin. The third sequence (Hs Tom40) shows the position (yellow) of the 19 β-stands predicted to exist in human Tom40 (11–13). The bottom sequence (Nc Tom40) represents the NcTom40 showing the predicted 19 β-strands (highlighted and numbered in teal). The position and alignment of strands in the last three sequences are as previously described (12). The SCAM results from this study are displayed below the NcTom40 sequence. Positive (+) and negative (−) labeling signals are indicated. SCAM results that suggest the position of a β-strand are shown as highlighted black bars below the sequence.
Additional factor Xa sites were inserted into the gene between residues 168 and 169 (site Xa4), residues 164 and 165 (site Xa3), residues 149 and 150 (site Xa2), and residues 108 and 109 (site Xa1) of the protein. The location of these sites and the predictions for cutting and fragment size are shown on Fig. 2, A and B, with respect to the 19-strand model. As expected, cleavage occurred in whole mitochondria at sites Xa3 and Xa4, which should be accessible from the cytosolic side of the membrane. We were uncertain if the constructs containing sites Xa1 and Xa2 would give rise to viable transformants because the sites of insertion occur within predicted β strands (strands 4 and 7, respectively) but transformants were obtained. For site Xa1, the protein was cleaved by factor Xa in whole mitochondria. We interpret this as a triple factor Xa site (12 residues in total) that begins within a β-strand and emerges in the cytosol where it is accessible to externally added protease (Fig. 2A). On the other hand, for site Xa2, the cleavage site would be predicted to emerge within the IMS and be inaccessible to added factor Xa protease. In agreement with this interpretation, the Xa2 site was not cut by the enzyme in whole mitochondria but was cut when mitochondria were converted to crude mitoplasts (Fig. 2C). As mentioned above for mapping Factor Xa sites near the C terminus, cleavage was not complete under the conditions used.
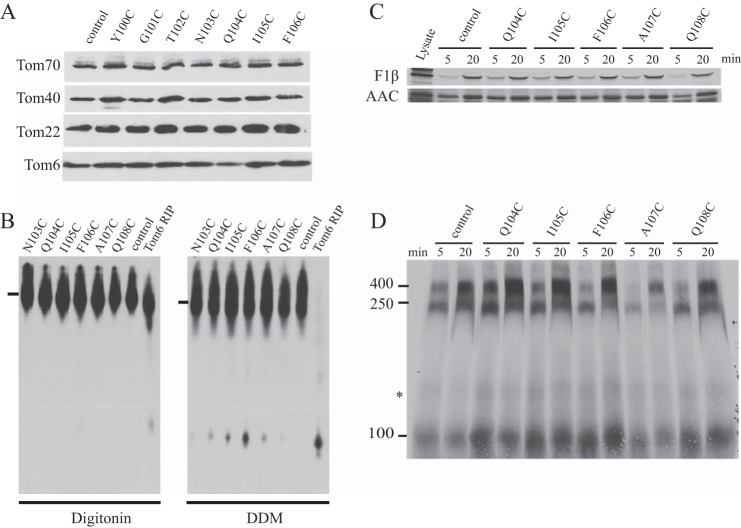
The fact that all Cys and factor Xa mutants created are viable demonstrates that the mutant Tom40s are functional, because the protein is known to be essential for viability (19). However, to evaluate possible minor alterations in protein function we examined various aspects of Tom40 function in a few selected mutants. Western blot analysis was performed on mitochondria isolated from each of the Cys-mutants from residues 94 to 106 to determine the levels of various TOM complex components (Fig. 5A). Despite slight differences in protein level between certain strains, it appears that TOM components are not severely altered in any of the mutants. TOM complex stability in Cys substitution strains 103 to 108 was analyzed via BNGE following solubilization of mitochondria in either digitonin or dodecylmaltoside. All mutant mitochondria maintain near wild type TOM complex stability (Fig. 5B).
FIGURE 5.
Analysis of TOM complex composition and function in mitochondria from selected Tom40 Cys-mutant strains. A, cells from the indicated strains (top of panel) were grown in liquid culture, and mitochondria were isolated. Mitochondrial proteins were subjected to SDS-PAGE followed by transfer to nitrocellulose, and immunodecoration with the antibodies indicated on the left. The control strain was 76-26, which contains wild type Tom40. B, isolated mitochondria from the strains indicated (top of panel) were dissolved in 1% digitonin or 1% n-dodecyl-β-d-maltoside (DDM) for analysis of complex stability using BNGE. Blots were decorated with Tom40 antibody. The controls are Tom6 RIP (a strain lacking Tom6 that shows severely reduced TOM complex stability (51)) and 76-26 (control), which contains wild type TOM complex. C, radiolabeled matrix precursor F1β and inner membrane precursor ADP/ATP carrier (AAC) were incubated (for 5 or 20 min, as indicated) with mitochondria isolated from the Tom40 Cys-substitution strains or the 76-26 control. Following import, mitochondria were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membrane, and import was analyzed by autoradiography. Lysate represents 33% of the radiolabeled lysate added to each import reaction. D, radiolabeled Tom40 precursor was incubated for 5 and 20 min with mitochondria isolated from the Tom40 Cys-substitution strains indicated (top of panel) as well as the 76-26 control strain. Mitochondria were dissolved in 1% digitonin and subjected to BNGE. The proteins were transferred to PVDF membrane and analyzed by autoradiography. The size of the mature TOM complex (400 kDa), and assembly intermediates I (250 kDa) and II (100 kDa) are indicated on the left. * indicates an undefined band.
To evaluate TOM complex function in Cys-mutants 104 to 108, we analyzed in vitro import of the radiolabeled F1β (β-subunit of the F1 portion of ATP synthase) and ADP/ATP carrier precursors into isolated mitochondria. The import of these precursors into mitochondria confirm that the Tom40 Cys-mutant TOM complexes examined maintain their function (Fig. 5C). We also examined the ability of Cys-mutant TOM complexes to assemble wild type Tom40 precursor into the TOM complex. The presence of the precursor in two Tom40 assembly intermediates and the fully assembled TOM complex, as described previously (19, 55), suggests that the Tom40 assembly pathway is functional in the mutant strains examined (Fig. 5D). Some variability among the mutant strains was observed. In particular Cys-mutant A107C has an overall reduction in Tom40 assembly compared with control and other strains examined. However, both intermediates and the final complex are observed at both time points suggesting a normal, but slightly delayed assembly process. Thus, despite quantitative differences, all Tom40 mutants are functional and give rise to viable cells.
DISCUSSION
In general, our approach of analyzing N. crassa Tom40 using SCAM has proven successful. Formally, our SCAM data do not prove the existence of a β-strand in a given region, rather they simply demonstrate that the predicted pattern of labeling occurs. Conceivably, this pattern could occur in non β-strand regions where alternate residues might be shielded from interaction with the labeling reagent by mechanisms such as protein-protein interactions. However, this seems unlikely as a random occurrence. Furthermore, our data are generally congruent with the predicted location of β-strands in Tom40 from bioinformatics and modeling studies (11–13). Because labeling was performed in whole mitochondria, it is likely that the protein was in its native conformation. The predicted alternating pattern of labeling for a β-strand in a membrane-bound β-barrel protein was seen in at least six regions (strands 3, 4, 5, 7, 9, and 19) of the three-dimensional model, with portions of two others (strands 6 and 8) also observed. All of the strands detected correlate well with strands predicted in the recent models of the Tom40 structure that were modeled after the three-dimensional structure of the related β-barrel protein, porin.
Eloquent arguments have been made for and against each of the two current models for porin structure: the 13-strand BIO model and the 19-strand three-dimensional model (29, 36). Our data do not directly relate to this discussion. However, given the compelling evidence for a phylogenetic relationship between Tom40 and porin, and the likelihood that both contain the same number of β-strands, our data indirectly support the three-dimensional model of porin, at least with respect to the regions analyzed. Specifically, residues within strands 4 and 7 of the three-dimensional model are not predicted to be membrane spanning β-strands in the BIO model of porin but these regions did display the pattern of labeling predicted for a β-strand in our study (Figs. 3 and 4). It should also be noted that if Tom40 were modeled after the BIO model of porin, the residues comprising β-strand 4 of the three-dimensional model and the location of our factor XaI site would be predicted to occur within a loop located in the IMS and would only be accessible to the protease in crude mitoplasts. As shown in Fig. 2, this site was cleaved in intact mitochondria, in agreement with the prediction using the 19-strand model. Our results using SCAM analysis of Tom40 in whole mitochondria suggest that direct insight into the debate about porin structure could be gained using a similar approach.
Although there is generally good correspondence between the location of the predicted strands for NcTom40 (12) and those suggested by the SCAM-mapping approach, the edges of the mapped strands often do not precisely correspond to the predicted strands. Conceivably, the availability for labeling of residues at the edges of a strand may be influenced by their interaction with the bilayer, phosphate head groups of the bilayer, and/or residues that begin or end non-membrane spanning loops of the protein. Slight differences in the start and end points of the Tom40 β-strands have also occurred in the different modeling studies (11–13). Stretches of positive labeling were observed for some regions flanking β-strands, as expected for cytosolic or IMS loops (for example, residues 328–339 following β-strand 19). However, certain regions and residues (for example residues 164, 166, and 180–182) appeared as unexpectedly unlabeled residues. The SCAM results suggest that if strand 9 ends at residue 178, then residues 180–182 should be accessible to labeling as part of the exposed loop between β-strands 9 and 10. However, it has been noted that residues of small loops between membrane spanning α-helices can sometimes be inaccessible to Cys-labeling reagents (56). A similar explanation may hold for the lack of labeling at residues 164 and 166. An alternative possibility is that certain residues may be shielded from labeling via interactions of Tom40 with other proteins that make up the TOM complex.
One unexpected observation was that triple factor Xa cleavage sites inserted at sites expected to disrupt strands 4 and 7 still gave rise to viable transformants. It seemed likely that disruption of the strands would result in loss of Tom40 assembly and subsequent lethality, because the protein is essential for viability. This is especially true for strand 4 where factor Xa sites were inserted near the middle of the β strand identified in our study (after residue 108). It is conceivable that the beginning of the strand was sufficient for assembly and/or that one or more residues of the first factor Xa cleavage site allowed completion of a transmembrane region. Interestingly, the sequence of the factor Xa cleavage site (Ile-Glu-Gly-Arg) occurs in an alternating hydrophobic/hydrophilic pattern following a hydrophilic residue at the site of insertion for both strands 4 and 7. Conceivably, one or more of these amino acids might be included as part of a β-strand because their physical properties are appropriate. This might be sufficient to allow assembly. Interestingly, a S. cerevisiae porin construct containing a FLAG tag in predicted β-strand 14 of the three-dimensional model was also found to assemble (37).
To gain further insight into the possibility of maintaining Tom40 assembly and function when β strands are interrupted, we re-examined previous work (19, 40) on the effect of specific mutations on the assembly and function of NcTom40. These studies were completed prior to the appearance of the 19 β-strand three-dimensional structure for porin and the modeled Tom40 structure. We were particularly interested in any mutations that could now be placed in the predicted β-strand regions of the current model. The position of relevant mutations within NcTom40, and relative to the location of the 19 β-strands, is shown in Fig. 6.
FIGURE 6.
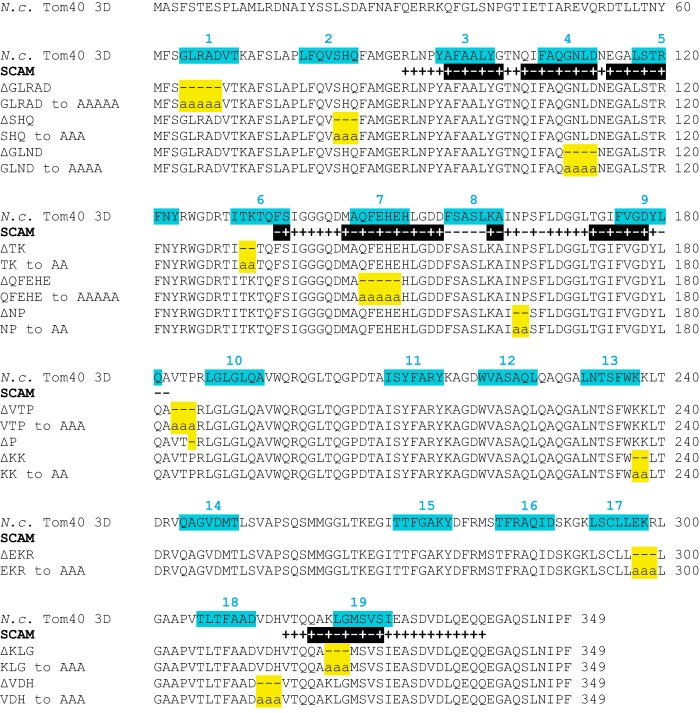
Previously studied mutants of N. crassa Tom40 aligned with the three-dimensional model and SCAM results. The top sequence is the wild type NcTom40 protein with the 19 modeled β-strands shown (highlighted and numbered in teal). All SCAM results (from Figs. 1 and 3) are indicated below the NcTom40 sequence. Black shading indicates the regions with alternating label (+) and no label (−) signals that are likely to represent β-strands. Below the SCAM results is a depiction of the Tom40 protein of each mutant with the region that is discussed in the text containing the amino acid deletions (−) or alanine substitutions (a) indicated (19, 40). The region affected in each mutant is highlighted in yellow.
The most C-terminal β-strand of Tom40 houses the β-signal (XpXGXXhXh: p, polar residue; h, hydrophobic residue) for recognition of incoming β-barrel precursors by the sorting and assembly machinery complex (53, 54). NcTom40 strand 19 contains a match (AKLGMSVSI) to the proposed signal. Examination of the effect of mutations created in this region (19) generally validates the existence of a β-strand containing the signal. Both a deletion (ΔKLG) of residues in the signal region and a change of KLG to AAA were examined. In vitro import of a ΔKLG Tom40 precursor into wild type mitochondria revealed that the mutant protein could not be inserted into the membrane. However, the KLG to AAA version displayed a near wild type assembly phenotype. This suggests that the polar and glycine residues of the conserved sequence are not mandatory for insertion and assembly of Tom40, but that the number of residues in that region of the strand is critical. Alternatively, deletion of an odd number of residues would also break the alternating hydrophobic/hydrophilic pattern of residues in a strand and this could prevent assembly. The ΔKLG construct was also unable to rescue a tom40 null nucleus, whereas KLG to AAA did rescue. Mitochondria isolated from the AAA mutant strain showed normal import phenotypes for the precursors tested. However, the TOM complex in the KLG to AAA mutant strain showed reduced stability (19) suggesting that the integrity of Tom40 in the TOM complex, and/or interactions with other members of the complex, are dependent on these residues in β-strand 19.
Another study characterized several additional mutants involving deletion or Ala substitution of conserved Tom40 sequences (40). When examined in the context of the N. crassa Tom40 19-strand model, 14 of the mutants have deletions or Ala substitutions of residues affecting predicted β-strands (Fig. 6). Three of the deletion constructs ΔSHQ (strand 2), ΔKK (strand 13), and ΔEKR (strand 17) affected the C-terminal end of the predicted β strands and failed to rescue the Tom40 null nucleus. In vitro assays revealed defects of assembly into the TOM complex for all three deletion proteins. Ala substitution mutants of the same residues did rescue the Tom40 null nucleus. The Ala substitutions for SHQ and KK showed minimal assembly defects in vitro, whereas the EKR substitution showed slow progression past the 100-kDa intermediate II stage. Examination of the TOM complex in isolated mitochondria from the Ala substitution mutants revealed moderate to severe stability defects, again suggesting the importance of those residues for either Tom40 stability or TOM complex interactions.
Surprisingly, four additional deletion constructs affecting residues in the predicted β-strands 1 (ΔGLRAD), 4 (ΔGLND), 6 (ΔTK), and 7 (ΔQFEHE) of the 19-strand model were able to rescue the tom40 null nucleus. However, the resulting rescued mutant strains displayed reduced growth phenotypes. The Ala substitution versions of these mutants demonstrated little to no growth phenotype. The TOM complex of the strains containing deletions of GLRAD, GLND, or QFEHE also showed severe stability defects, whereas complex stability in the TK deletion was only mildly affected. In vitro assembly assays for the Tom40 protein of these rescuing β-strand deletion mutants into wild type mitochondria revealed that all stalled at an intermediate assembly position with very little precursor progressing to the fully assembled TOM complex. The Ala substitution mutants for GLRAD and QFEHE also showed relatively severe assembly defects, whereas those for GLND and TK displayed only a mild assembly phenotype.
The ability of at least some deletion mutants, and all Ala substitution mutants affecting β-strands, to rescue Tom40 function in vivo suggests that Tom40 β-strand/barrel topology may be flexible to a certain extent and that some mutant proteins have the ability to assemble and function as a viable import pore, despite inefficiencies and structural instability. Particularly interesting is the fact that some mutants lacking portions of predicted β-strands can still assemble into a functional pore (strains ΔGLRAD, ΔGLND, ΔTK, and ΔQFEHE). Further analysis of both Tom40 and porin mutants may shed further light on the requirements for function and assembly, including the features necessary to allow assembly in mutants affected in β-strands.
Acknowledgment
We are grateful for the assistance of the fungal genetics stock center with strains used in this study.
This work was supported by grants from the Canadian Institutes of Health Research and Natural Sciences and Engineering Research Council (to F. E. N.).
- TOM
- translocase of the outer mitochondrial membrane
- BNGE
- blue native gel electrophoresis
- F1β
- β-subunit of the F1 portion of ATP synthase
- IMS
- intermembrane space
- RIP
- repeat induced point mutation
- SCAM
- substituted cysteine accessibility method.
REFERENCES
- 1. Neupert W., Herrmann J. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 2. Endo T., Yamano K. (2009) Multiple pathways for mitochondrial protein traffic. Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 3. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmidt O., Pfanner N., Meisinger C. (2010) Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 [DOI] [PubMed] [Google Scholar]
- 5. Becker T., Böttinger L., Pfanner N. (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci. 37, 85–91 [DOI] [PubMed] [Google Scholar]
- 6. Dudek J., Rehling P., van der Laan M. (2013) Mitochondrial protein import: common principles and physiological networks. Biochim. Biophys. Acta 1833, 274–285 [DOI] [PubMed] [Google Scholar]
- 7. Harbauer A. B., Zahedi R. P., Sickmann A., Pfanner N., Meisinger C. (2014) The protein import machinery of mitochondria-A regulatory hub in metabolism, stress, and disease. Cell Metab. 19, 357–372 [DOI] [PubMed] [Google Scholar]
- 8. Hoogenraad N. J., Ward L. A., Ryan M. T. (2002) Import and assembly of proteins into mitochondria of mammalian cells. Biochim. Biophys. Acta 1592, 97–105 [DOI] [PubMed] [Google Scholar]
- 9. Perry A. J., Rimmer K. A., Mertens H. D., Waller R. F., Mulhern T. D., Lithgow T., Gooley P. R. (2008) Structure, topology and function of the translocase of the outer membrane of mitochondria. Plant Physiol. Biochem. 46, 265–274 [DOI] [PubMed] [Google Scholar]
- 10. Pusnik M., Charrière F., Mäser P., Waller R. F., Dagley M. J., Lithgow T., Schneider A. (2009) The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol. Biol. Evol. 26, 671–680 [DOI] [PubMed] [Google Scholar]
- 11. Zeth K. (2010) Structure and evolution of mitochondrial outer membrane proteins of β-barrel topology. Biochim. Biophys. Acta 1797, 1292–1299 [DOI] [PubMed] [Google Scholar]
- 12. Gessmann D., Flinner N., Pfannstiel J., Schlösinger A., Schleiff E., Nussberger S., Mirus O. (2011) Structural elements of the mitochondrial preprotein-conducting channel Tom40 dissolved by bioinformatics and mass spectrometry. Biochim. Biophys. Acta 1807, 1647–1657 [DOI] [PubMed] [Google Scholar]
- 13. Bay D. C., Hafez M., Young M. J., Court D. A. (2012) Phylogenetic and coevolutionary analysis of the β-barrel protein family comprised of mitochondrial porin (VDAC) and Tom40. Biochim. Biophys. Acta 1818, 1502–1519 [DOI] [PubMed] [Google Scholar]
- 14. Hill K., Model K., Ryan M. T., Dietmeier K., Martin F., Wagner R., Pfanner N. (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395, 516–521 [DOI] [PubMed] [Google Scholar]
- 15. Künkele K. P., Heins S., Dembowski M., Nargang F. E., Benz R., Thieffry M., Walz J., Lill R., Nussberger S., Neupert W. (1998) The preprotein translocation channel of the outer membrane of mitochondria. Cell 93, 1009–1019 [DOI] [PubMed] [Google Scholar]
- 16. Künkele K. P., Juin P., Pompa C., Nargang F. E., Henry J. P., Neupert W., Lill R., Thieffry M. (1998) The isolated complex of the translocase of the outer membrane of mitochondria. Characterization of the cation-selective and voltage-gated preprotein-conducting pore. J. Biol. Chem. 273, 31032–31039 [DOI] [PubMed] [Google Scholar]
- 17. Ahting U., Thieffry M., Engelhardt H., Hegerl R., Neupert W., Nussberger S. (2001) Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J. Cell Biol. 153, 1151–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baker K. P., Schaniel A., Vestweber D., Schatz G. (1990) A yeast mitochondrial outer membrane protein is essential for protein import and cell viability. Nature 348, 605–609 [DOI] [PubMed] [Google Scholar]
- 19. Taylor R. D., McHale B. J., Nargang F. E. (2003) Characterization of Neurospora crassa Tom40-deficient mutants and effect of specific mutations on Tom40 assembly. J. Biol. Chem. 278, 765–775 [DOI] [PubMed] [Google Scholar]
- 20. Becker L., Bannwarth M., Meisinger C., Hill K., Model K., Krimmer T., Casadio R., Truscott K. N., Schulz G. E., Pfanner N., Wagner R. (2005) Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J. Mol. Biol. 353, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 21. Mager F., Gessmann D., Nussberger S., Zeth K. (2011) Functional refolding and characterization of two Tom40 isoforms from human mitochondria. J. Membr. Biol. 242, 11–21 [DOI] [PubMed] [Google Scholar]
- 22. Ahting U., Thun C., Hegerl R., Typke D., Nargang F. E., Neupert W., Nussberger S. (1999) The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Model K., Prinz T., Ruiz T., Radermacher M., Krimmer T., Kühlbrandt W., Pfanner N., Meisinger C. (2002) Protein translocase of the outer mitochondrial membrane: role of the import receptors in the structural organization of the TOM complex. J. Mol. Biol. 316, 657–666 [DOI] [PubMed] [Google Scholar]
- 24. Model K., Meisinger C., Kühlbrandt W. (2008) Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J. Mol. Biol. 383, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 25. Bayrhuber M., Meins T., Habeck M., Becker S., Giller K., Villinger S., Vonrhein C., Griesinger C., Zweckstetter M., Zeth K. (2008) Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. U.S.A. 105, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hiller S., Garces R. G., Malia T. J., Orekhov V. Y., Colombini M., Wagner G. (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321, 1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ujwal R., Cascio D., Colletier J. P., Faham S., Zhang J., Toro L., Ping P., Abramson J. (2008) The crystal structure of mouse VDAC1 at 2.3-Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U.S.A. 105, 17742–17747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu J., Wenz L. S., Zerbes R. M., Oeljeklaus S., Bohnert M., Stroud D. A., Wirth C., Ellenrieder L., Thornton N., Kutik S., Wiese S., Schulze-Specking A., Zufall N., Chacinska A., Guiard B., Hunte C., Warscheid B., van der Laan M., Pfanner N., Wiedemann N., Becker T. (2013) Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 154, 596–608 [DOI] [PubMed] [Google Scholar]
- 29. Hiller S., Abramson J., Mannella C., Wagner G., Zeth K. (2010) The 3D structures of VDAC represent a native conformation. Trends Biochem. Sci. 35, 514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Summers W. A., Court D. A. (2010) Origami in outer membrane mimetics: correlating the first detailed images of refolded VDAC with over 20 years of biochemical data. Biochem. Cell Biol. 88, 425–438 [DOI] [PubMed] [Google Scholar]
- 31. Blachly-Dyson E., Peng S., Colombini M., Forte M. (1990) Selectivity changes in site-directed mutants of the VDAC ion channel: structural implications. Science 247, 1233–1236 [DOI] [PubMed] [Google Scholar]
- 32. Thomas L., Kocsis E., Colombini M., Erbe E., Trus B. L., Steven A. C. (1991) Surface topography and molecular stoichiometry of the mitochondrial channel, VDAC, in crystalline arrays. J. Struct. Biol. 106, 161–171 [DOI] [PubMed] [Google Scholar]
- 33. Thomas L., Blachly-Dyson E., Colombini M., Forte M. (1993) Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc. Natl. Acad. Sci. U.S.A. 90, 5446–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song J., Colombini M. (1996) Indications of a common folding pattern for VDAC channels from all sources. J. Bioenerg. Biomembr. 28, 153–161 [DOI] [PubMed] [Google Scholar]
- 35. Song J., Midson C., Blachly-Dyson E., Forte M., Colombini M. (1998) The topology of VDAC as probed by biotin modification. J. Biol. Chem. 273, 24406–24413 [DOI] [PubMed] [Google Scholar]
- 36. Colombini M. (2009) The published 3D structure of the VDAC channel: native or not? Trends Biochem. Sci. 34, 382–389 [DOI] [PubMed] [Google Scholar]
- 37. McDonald B. M., Wydro M. M., Lightowlers R. N., Lakey J. H. (2009) Probing the orientation of yeast VDAC1 in vivo. FEBS Lett. 583, 739–742 [DOI] [PubMed] [Google Scholar]
- 38. Colombini M. (2012) Mitochondrial outer membrane channels. Chem. Rev. 112, 6373–6387 [DOI] [PubMed] [Google Scholar]
- 39. Colombini M. (2012) VDAC structure, selectivity, and dynamics. Biochim. Biophys. Acta 1818, 1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sherman E. L., Taylor R. D., Go N. E., Nargang F. E. (2006) Effect of mutations in Tom40 on stability of the translocase of the outer mitochondrial membrane (TOM) complex, assembly of Tom40, and import of mitochondrial preproteins. J. Biol. Chem. 281, 22554–22565 [DOI] [PubMed] [Google Scholar]
- 41. Davis R. H., De Serres F. J. (1970) Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17A, 79–143 [Google Scholar]
- 42. Hoppins S. C., Go N. E., Klein A., Schmitt S., Neupert W., Rapaport D., Nargang F. E. (2007) Alternative splicing gives rise to different isoforms of the Neurospora crassa Tob55 protein that vary in their ability to insert β-barrel proteins into the outer mitochondrial membrane. Genetics 177, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nargang F. E., Rapaport D. (2007) Neurospora crassa as a model organism for mitochondrial biogenesis. in Mitochondria, Practical Protocols (Leister D. L., Herrmann J., eds) pp. 107–123, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 44. Wideman J. G., Go N. E., Klein A., Redmond E., Lackey S. W., Tao T., Kalbacher H., Rapaport D., Neupert W., Nargang F. E. (2010) Roles of the Mdm10, Tom7, Mdm12, and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol. Biol. Cell 21, 1725–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bogdanov M., Zhang W., Xie J., Dowhan W. (2005) Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): application to lipid-specific membrane protein topogenesis. Methods 36, 148–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 47. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 48. Schägger H., Cramer W. A., von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 49. Good A. G., Crosby W. L. (1989) Anaerobic induction of alanine aminotransferase in barley root tissue. Plant Physiol. 90, 1305–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harkness T. A., Nargang F. E., van der Klei I., Neupert W., Lill R. (1994) A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J. Cell Biol. 124, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sherman E. L., Go N. E., Nargang F. E. (2005) Functions of the small proteins in the TOM complex of Neurospora crassa. Mol. Biol. Cell 16, 4172–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hoppins S. C., Nargang F. E. (2004) The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279, 12396–12405 [DOI] [PubMed] [Google Scholar]
- 53. Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Krüger V., Prinz C., Meisinger C., Guiard B., Wagner R., Pfanner N., Wiedemann N. (2008) Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132, 1011–1024 [DOI] [PubMed] [Google Scholar]
- 54. Imai K., Gromiha M. M., Horton P. (2008) Mitochondrial β-barrel proteins, an exclusive club? Cell 135, 1158–1159; author reply 1159–1160 [DOI] [PubMed] [Google Scholar]
- 55. Model K., Meisinger C., Prinz T., Wiedemann N., Truscott K. N., Pfanner N., Ryan M. T. (2001) Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8, 361–370 [DOI] [PubMed] [Google Scholar]
- 56. Loo T. W., Clarke D. M. (1995) Membrane topology of a cysteine-less mutant of human P-glycoprotein. J. Biol. Chem. 270, 843–848 [DOI] [PubMed] [Google Scholar]