Background: DNA polymerase η is a specialized, error-prone DNA polymerase capable of synthesis past bulky DNA adducts.
Results: Sphingosine and sphinganine stimulate the activity of Pol η.
Conclusion: Sphingosine modulates DNA lesion bypass in addition to controlling cell proliferation following DNA damage.
Significance: There are no known stimulators of DNA polymerases. Stimulation by sphingosine represents a novel mode of modulating Pol η activity.
Keywords: DNA Damage, DNA Polymerase, Drug Screening, Genomic Instability, Sphingolipid, Sphingosine, Translesion
Abstract
Translesion (TLS) DNA polymerases are specialized, error-prone enzymes that synthesize DNA across bulky, replication-stalling DNA adducts. In so doing, they facilitate the progression of DNA synthesis and promote cell proliferation. To potentiate the effect of cancer chemotherapeutic regimens, we sought to identify inhibitors of TLS DNA polymerases. We screened five libraries of ∼3000 small molecules, including one comprising ∼600 nucleoside analogs, for their effect on primer extension activity of DNA polymerase η (Pol η). We serendipitously identified sphingosine, a lipid-signaling molecule that robustly stimulates the activity of Pol η by ∼100-fold at low micromolar concentrations but inhibits it at higher concentrations. This effect is specific to the Y-family DNA polymerases, Pols η, κ, and ι. The addition of a single phosphate group on sphingosine completely abrogates this effect. Likewise, the inclusion of other sphingolipids, including ceramide and sphingomyelin to extension reactions does not elicit this response. Sphingosine increases the rate of correct and incorrect nucleotide incorporation while having no effect on polymerase processivity. Endogenous Pol η activity is modulated similarly as the recombinant enzyme. Importantly, sphingosine-treated cells exhibit increased lesion bypass activity, and sphingosine tethered to membrane lipids mimics the effects of free sphingosine. Our studies have uncovered sphingosine as a modulator of TLS DNA polymerase activity; this property of sphingosine may be associated with its known role as a signaling molecule in regulating cell proliferation in response to cellular stress.
Introduction
TLS4 DNA polymerases are an evolutionarily conserved family of specialized, error-prone DNA polymerases. They are distinguished from other DNA polymerases by the presence of capacious active site binding pockets that can accommodate and enable DNA synthesis past bulky DNA adducts (1–3). In so doing, TLS DNA polymerases help prevent the stalling and collapse of replication forks and ensuing DNA breaks/rearrangements. The largest class of such DNA polymerases is the Y-family, which includes, in human cells, Pols η, κ, and ι, and Rev1. DNA Pol η has garnered the most attention as mutations in POLH are causally linked to the variant form of Xeroderma pigmentosum (4), an inherited disease associated with sunlight sensitivity and increased incidence of skin cancers (5). Pol η has been shown in vitro to efficiently and accurately bypass the UV-induced lesion, cyclobutane pyrimidine dimer (CPD) (6). It is believed that in the absence of Pol η, error-prone translesion synthesis across CPD lesions by other specialized DNA polymerases, including Pol ι, results in increased mutagenesis and carcinogenesis. Pol η has also been shown to bypass cisplatin-induced cross-linked adducts (7) and oxidative lesions (8). In addition, by virtue of its error-prone nature, Pol η has been implicated in immunoglobulin gene somatic hypermutation (9).
Although no specific disorders are associated with the other Y-family TLS DNA polymerases, each of them has unique attributes. Pol ι, an ortholog of Pol η, is the only polymerase that is reported to use Hoogsteen base pairing to efficiently mispair dG residues across template dT (10). Pol κ specializes in the bypass of benzo[a]pyrene diol-epoxide-DNA adducts; it copies past these lesions in vitro, and cells depleted for Pol κ are sensitive to benzo[a]pyrene diol-epoxide (11, 12). Additionally, it has been reported that mutagenic bypass activity by Pol κ homologs (DinB) consists predominantly of −1 frameshifts, which arise from the extra-helical displacement of DNA lesions (13). REV1, or deoxcytidyl transferase, is restricted to inserting dC residues opposite template lesions, including abasic sites and N2-dG adducts (14). An additional non-catalytic, scaffolding role for REV1 that facilitates polymerase switch during TLS has also been proposed (15).
Due to the mutagenic nature of DNA synthesis by TLS DNA polymerases, cells employ an array of regulatory mechanisms to limit TLS activity, including post-translational modifications, relocalization upon DNA damage, and binding to accessory proteins (1, 13). Some cancer cells take advantage of error-prone TLS to bypass DNA adducts generated by chemotherapeutic drugs (16). One manner in which they do so is by up-regulating the level of TLS DNA polymerases. For example, Pol κ levels are reported to be elevated in lung cancer (17), whereas Pol ι levels are increased in breast cancer (18). In addition, cisplatin or oxaliplatin treatment has been shown to induce Pol η in non-small-cell lung cancer and gastric adenocarcinoma (19, 20). Overexpression of translesion DNA polymerases may reduce the effectiveness of chemotherapy regimens and render cancer cells resistant to chemotherapy (21). Consistent with this, Doles et al. (22) and Xie et al. (23) demonstrated that suppression of REV3, the catalytic subunit of the B-family TLS polymerase Pol ζ, sensitizes drug-resistant lung tumors to chemotherapy.
We screened several collections of small molecule compounds to identify inhibitors of TLS DNA polymerases that could potentiate the effect of cancer chemotherapeutic agents. We identified and report here on the surprising discovery of a modulator, which both stimulates and inhibits TLS DNA polymerases. We identified the compound as the biologically active sphingolipids, sphingosine, and dihydrosphingosine (sphinganine). We show that of all sphingolipids evaluated, sphingosine and sphinganine alone manifest this effect. Although low concentrations markedly stimulate the rate of DNA primer extension as well as the extent of nucleotide misincorporation and misextension by Pol η, high concentrations inhibit polymerase activity. These effects of sphingosine are specific to the Y-family TLS polymerases, Pols η, κ, and ι. Notably, sphingosine increases lesion bypass activity in cells, and sphingosine bound to model liposomes manifests similar effects as the free compound. Our studies have uncovered a novel mode of modulation of Pol η and have linked two responses of cells to stress, namely DNA lesion bypass by TLS DNA polymerases and sphingolipid-mediated signaling to control cell proliferation.
EXPERIMENTAL PROCEDURES
Reagents and Enzymes
Five small molecule libraries were analyzed; four were obtained from the National Institutes of Health (NCI-approved Oncology Drug Set, Mechanistic Set, Diversity Set II, and Natural Product Set II), and one consisting of 622 compounds enriched for nucleoside analogs, was from the Open Innovation Center for Drug Discovery, The University of Tokyo, Japan. The compounds were stored as 10 mm stock solutions in dimethyl sulfoxide at −80 °C. d-erythro-Sphinganine, d-erythro-sphingosine, d-erythro-sphingosine-1-phosphate, and natural brain ceramide and sphingomyelin were purchased from Avanti Polar Lipids Inc (Alabaster, AL). l-erythro-Sphingosine was obtained from Sigma-Aldrich. Gel-purified synthetic oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). The sequence of the primer-template (P/T1) used for measuring the effect of small molecules on DNA polymerase activity was 5′-CGCGCCGAATTCCCGCTAGCAAT-3′ and 5′-GCGCGGAAGCTTGGCTGCAGAAGATTGCAGCGGGAATTCGGCGCG-3′. The sequence of the primer-template (P/T2) used for evaluating CPD bypass activity in vitro was 5′-CACTGACTGTATGA-3′ and 5′-CTCGTCAGCATCT-TCATCATACAGTCAGTG-3′, whereas that (P/T3) used for measurements of Vmax and Km values was 5′-CACTGACTGTATGATG-3′ and 5′-CTCGTCAGCATCT-TCATCATACAGTCAGTG-3′,where T-T indicates the cyclobutane pyrimidine dimer. The T-T DNA template was generously provided by S. Iwai (Osaka University, Osaka, Japan). A template containing two consecutive T residues in place of the CPD lesion was used in control reactions.
Human DNA polymerase η (1–511 amino acids) was purified from insect cells as described (6). Full-length Pols η, κ, and ι were purchased from Enzymax (Lexington, KY). Human Pols β and δ were expressed and purified as described (24),5 whereas human Pol γ was purified from baculovirus-infected insect cells.6 Calf thymus Pol α-primase complex and human Pol ϵ were generous gifts from F. Perrino (Wake Forest Park, NC) and S. Linn (Berkeley, CA), respectively.
Sample Fractionation and Mass Spectrometry
Samples containing the modulator were separated on an Agilent 1290 Infinity System. A Thermo Hypersil Gold UPLC column coupled to a C18 guard cartridge was used for fractionation employing a gradient of water and methanol as the mobile phase. Fractions were collected at 1-min intervals for a total duration of 20 min, dried under N2, resuspended in dimethyl sulfoxide, and evaluated in polymerase activity assays. Peak fractions were subject to a second round of ultra high-pressure liquid chromatography (UPLC) fractionation; the identity of the compound in the single fraction that tested positive for stimulatory activity was determined by mass spectrometry using a Waters Synapt G1 QTOF instrument.
Primer Extension Assays
5′-32P-labeled DNA primers (P) were hybridized to a 2-fold molar excess of complementary template (T) DNA. 10 nm of the P/T DNA substrate was extended by limiting amounts (∼0.2 nm) of DNA polymerase η in the absence or presence of the indicated sphingolipid at 37 °C for 10 min, unless stated otherwise. Reactions were carried out in buffer containing 40 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 10 mm DTT, 60 mm KCl, and 2.5% glycerol. Control reactions for sphinganine or sphingosine (SO) contained 10% dimethyl sulfoxide, whereas those for ceramide, sphingomyelin, or sphingosine-1-phosphate (S1P) included 10% methanol. Aliquots of the terminated reactions were electrophoresed through 14% polyacrylamide-urea gels; extension products were visualized by PhosphorImager analyses (GE Healthcare) and quantified using ImageJ software (NIH).
Measurements of the kinetic constants for single nucleotide incorporation by Pol η were carried out as described (25). Briefly, we first systematically varied the enzyme concentration and reaction time using saturating concentrations of dNTPs. These experiments established the requisite conditions for subsequent kinetic analysis, i.e. linearity of incorporation with time during a fixed incubation period. Extension was then monitored as a function of dNTP and sphingosine concentrations.
Immunoprecipitation
∼ 2 × 107 human embryonic kidney epithelial cells (293T) were lysed by incubation in 500 μl of lysis buffer (200 mm Hepes-KOH, pH 7.4, 155 mm KCl, 1.5 mm MgCl2, 2 mm DTT, 0.5% Triton X-100, 0.2 mm PMSF, and 10 μg/ml of aprotinin, pepstatin, and leupeptin) on ice for 10 min. The suspension was centrifuged at 2000 × g for 10 min, and the clarified supernatant was used for immunoprecipitating Pol η; ∼500 μg of total protein were incubated with the Pol η-specific monoclonal antibody, 5C6 for 1 h at 4 °C (26). A 25% suspension of Protein A/G agarose beads was added to precipitate the complex, and incubation was carried out as described above. The enzyme-antibody complex coupled to beads was washed extensively with buffer containing 20 mm Tris-HCl buffer, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, and 10% glycerol, and resuspended in 15 μl of 25 mm Tris-HCl buffer, pH 8.0, 0.5 mm EDTA, 1 mm DTT, 0.05% Nonidet P-40, and 25% glycerol. Aliquots of the resuspended immune complex were diluted and assayed for CPD bypass activity in the absence or presence of 8 μm sphingosine.
CPD Bypass Activity in Cells
CPD bypass activity in cells was measured using a modification of our recently established oligonucleotide retrieval assay (27, 28). A synthetic oligonucleotide containing a site-specific CPD lesion was extended by ligating oligonucleotides, 5′-AmMC6ACGGAGGGAATCGGAGGTCGC-3′ and 5′-CCTTCCACCTCCCATTCCTGATTCAGTCACTG-ddC-3′, at the 5′ and 3′ termini, respectively. Complementary oligomers to form double-stranded ligation sites at the 5′ and 3′ termini were 5′-TGCTGACGAGGCGACCTCCG-3′ and 5′-AGGTGGAAGGCACTGACTGT-3′, respectively. After ligation, excess free oligomers were degraded by λ exonuclease. The ligation efficiency (20%) was calculated from the ratio of amplification of the ligated versus unligated template and used to normalize substrate concentrations. The template was then hybridized to a biotinylated primer, 5′-biotin-GCACGTCAGGCACGGCGTCCAGTGACTGAATCAGGAATGGGAGGTGGAAGGCACTGACTGTATGATG-3′, to form the partial duplex DNA substrate.
This construct was transfected into SV40-immortalized fibroblast cells (GM0639) using calcium phosphate; the final concentration was 0.2 nm. After incubation for 2 h, oligonucleotides were retrieved via magnetic streptavidin bead capture by virtue of the 5′-biotin tag on the primer strand. Bypass efficiency was quantified by qPCR using the Brilliant III Ultra-Fast SYBR Green qPCR Master Mix (Agilent) in a DNA Engine Opticon 2 machine (MJ Research). The PCR primer pairs for measuring unextended and extended/bypassed products were a/b (5′-GCACGTCAGGCACGGCGTC-3′ and 5′-CATCATACAGTCAGTGCCTTCCACCTCC-3′) and a/c (5′-GCACGTCAGGCACGGCGTC-3′ and 5′-GGAGGTCGCCTCGTCAGCATC-3′), respectively. % Bypass was calculated as the ratio of (amplification by primers a/c over primers a/b) × 100.
To test for a difference in CPD bypass activity in SO-treated versus control cells, the experiment effect (shift in overall means on different experimental days seen in Fig. 5) and unequal variance among the different experiments were taken into account using a log transformation of the outcome with weighted least squares regression (29), including a variable for experiment in the model. Specifically, % bypass activity values were log-transformed, the variance of the transformed observations was calculated within each experiment, and this variance was used to inversely weight the observations in the least square regression. Variables for both treatment (variable of interest) and experiment (confounder) were included in the weighted least squares regression model. This resulted in normally distributed residuals as required for an unbiased test statistic with this small sample size.
FIGURE 5.
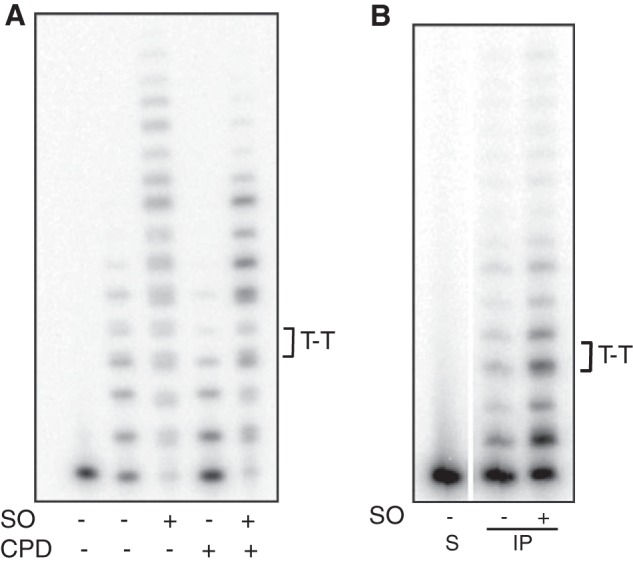
SO stimulates bypass activity of Pol η. CPD bypass activity of recombinant Pol η (A) or endogenous native enzyme immunoprecipitated from human cells in culture (B) was monitored using P/T2 DNA substrate in the absence or presence of 8 μm sphingosine as described under “Experimental Procedures.” The control template contained two tandem dT residues instead of the lesion; the sequence was identical otherwise. S, substrate (−) enzyme.
Liposome Preparation
Liposomes were generated by using 2.5 mg of the synthetic phospholipid, 1-oleoyl-2-palmitoyl-sn-glycero-3-phosphocholine (99% pure; Avanti Polar Lipids) either by itself or mixed with sphingosine or sphingomyelin at 20 mol % with respect to 1-oleoyl-2-palmitoyl-sn-glycero-3-phosphocholine. Lipids, dissolved in chloroform or methanol, were dispensed in glass tubes, and the organic solvents were first evaporated under a stream of argon and further dried under vacuum. Lipid films were hydrated by the addition of 0.25 ml of 1× Pol η reaction buffer. The lipid suspension was extruded through a 100-nm polycarbonate filter at room temperature (using the lipid extruder from Avanti Polar Lipids) to yield homogenous unilamellar vesicles. The vesicles were stored at 4 °C and used within 3 days of preparation. Fraction SO and SM bound to liposomes (f) was calculated using the formula, f = K × Cl/1 + K × Cl, where K = 1/CMC (critical micelle concentration), and Cl is the lipid concentration; we estimate that > 96% of SO and SM were bound to liposomes.
RESULTS
Identification of the Modulator
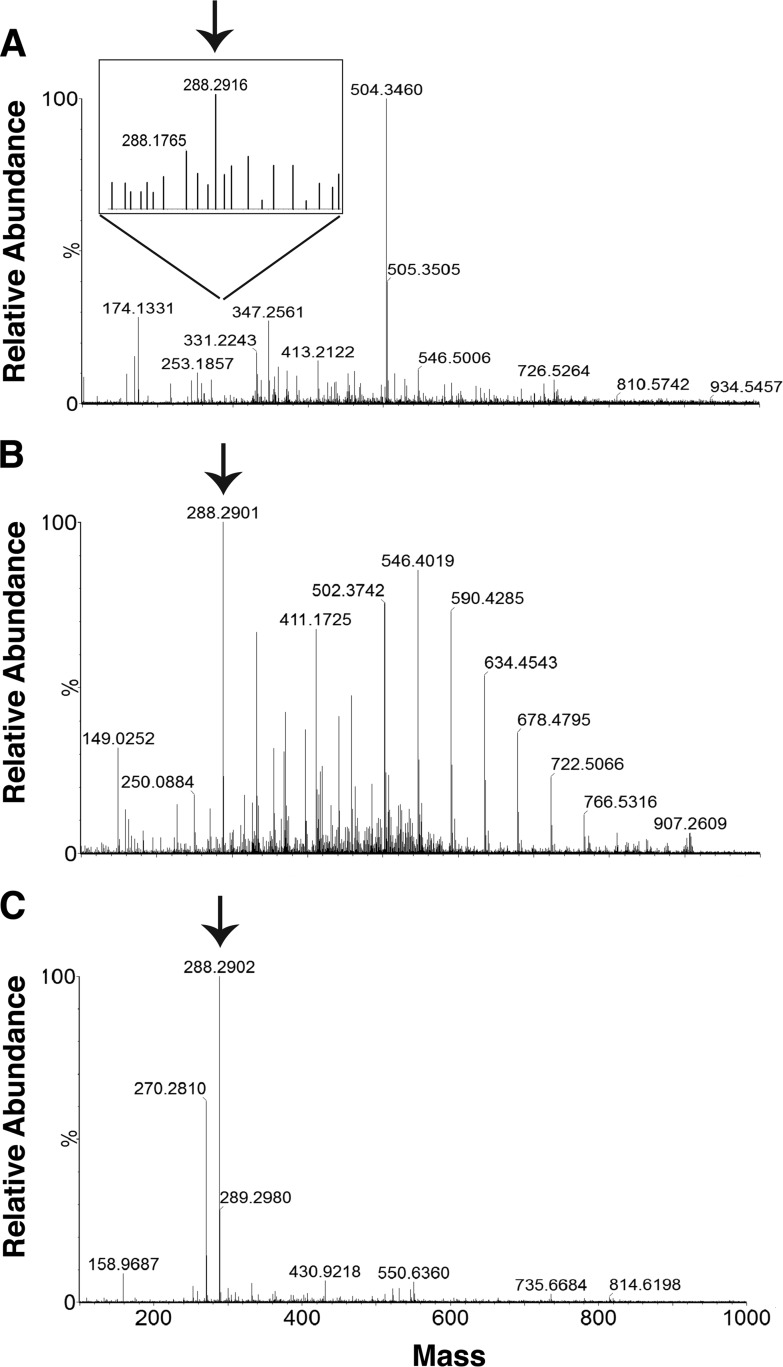
We screened multiple libraries of small molecules for inhibitors of DNA polymerases as potential anti-tumor agents. To facilitate screening, we pooled the compounds into groups of four and evaluated their effects on the polymerase activity of DNA Pol η by monitoring the extension of an end-labeled primer. We identified one pool from the library of nucleoside analogs that markedly stimulated the polymerase activity of Pol η. Upon deconvolution, the stimulatory compound was identified as 6-chloropurine-tetrahydropyran (2-(6-aminopurin-9-yl)-6-(hydroxymethyl)-2H-3,4,5,6-tetrahydro-pyran-3,4,5-triol). This compound was obtained from two independent commercial sources. Mass spectrometric and NMR analyses revealed that one of the two preparations was homogenous but lacked stimulatory activity. However, the second preparation that stimulated Pol η activity had multiple components (Fig. 1A). To separate the stimulatory factor from other components, we fractionated the second preparation by chromatography through two sequential reverse-phase UPLC columns and assayed individual fractions for their effect on Pol η activity. A single fraction containing a compound with an m/z of 288.2902 (Fig. 1B) stimulated primer extension by Pol η (data not shown). This mass did not correspond to that of the starting compound (expected mass, 238.7). A search of the Scripps-Metlin database for molecules with a mass of 288.2902 ± 5 ppm offered three possibilities, two of which were deuterium compounds; the third was the sphingolipid, C-17 sphinganine (SA; Fig. 1C).
FIGURE 1.
Mass spectrometric analyses of the stimulator. The compound (from the library of nucleoside analogs) containing stimulatory activity was enriched by fractionation through two reverse phase ultra-pressure columns. Individual fractions were assayed for their effect on the polymerase activity of DNA Pol η. Shown are the mass spectrometric profiles of the starting compound (A), the single fraction obtained after two rounds of UPLC purification that stimulated Pol η activity (B), and the C17-sphinganine standard (C). Note the presence of the peak (arrow) with mass 288.2902, corresponding to that of C17-sphinganine, which increased in abundance following UPLC (B). The scales used for the mass spectrometric profiles in A and B are 1.48 × 104 and 2.61 × 105, respectively. Peaks other than that corresponding to mass 288.2902 in B were also present in the mobile phase.
Sphinganine Uniquely Stimulates the Primer Extension Activities of DNA Pols η, κ, and ι
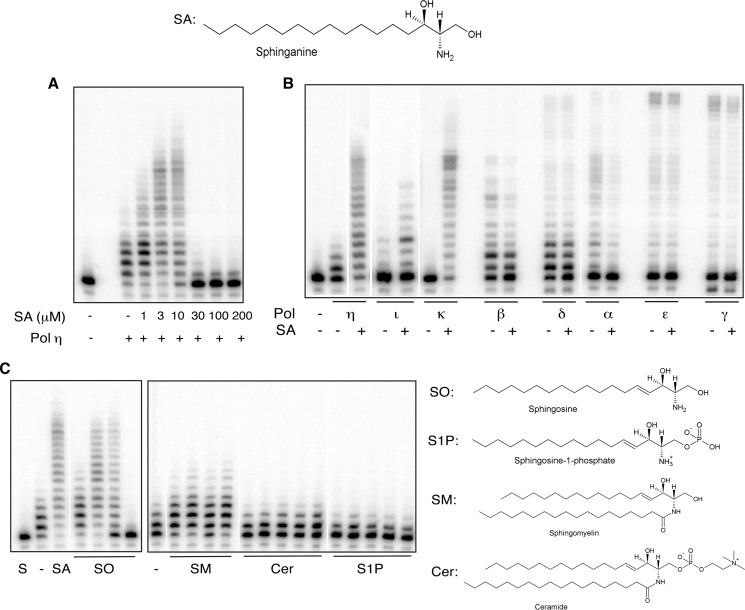
To evaluate the effect of the identified sphingolipid, homogenous preparations of C-17 sphinganine were added to primer extension reactions. Surprisingly, we observed that it stimulated Pol η activity; the increase in nucleotide incorporation was linear with respect to SA concentrations from 1 to 10 μm; primer extension was enhanced by ∼100-fold (Fig. 2A). At higher concentrations, however, the activity declined precipitously. The decrease in activity could be related to the critical micelle concentration, reported to be in the low μm range (30), which could block the accessibility of the enzyme active site to deoxynucleotide substrates and/or the DNA primer-template. Stimulation was indifferent to the order of addition of Pol η and SA and was comparable with both truncated and full-length Pol η, indicating that the C-terminal 202 amino acids, which include the proliferating cell nuclear antigen-binding motifs, are dispensable for stimulation by SA (data not shown). Furthermore, stimulation was independent of DNA sequence context, occurring with a multitude of primer-template DNA substrates, including templates containing cyclobutane pyrimidine dimers (data not shown and see Fig. 5A). Notably, the stimulatory effect was specific to Pol η and to Pols κ and ι, members of the Y-family DNA polymerases. Although the extent of stimulation of Pol κ paralleled that of Pol η, it was approximately an order of magnitude lower with Pol ι. In contrast, SA did not stimulate the primer extension activities of an array of other mammalian DNA polymerases, including nuclear and mitochondrial replicative DNA polymerases, Pols α, β, γ, δ, and ϵ (Fig. 2B).
FIGURE 2.
Sphinganine and sphingosine uniquely stimulate the extension activity of Y-family TLS DNA polymerases, Pols η, ι, and κ. Homogenous preparations of sphinganine were used at the indicated concentrations in primer extension reactions with Pol η (A) or at 10 μm concentrations with Pols η, ι, κ, α, β, γ, δ, and ϵ (B). SA (10 μm), SO (0.5, 2, 10, and 30 μm); SM (0.5, 2, 10, 50, and 100 μm); ceramide (0.5, 2, 10, 50, and 100 μm); and S1P (0.7, 2.8, 14, 70, and 140 μm) were evaluated for specificity in primer extension reactions with Pol η, (C). Control reactions for SO and SA contained 10% dimethyl sulfoxide, whereas those for SM, ceramide (Cer), and S1P included 10% methanol. Extension reactions and gel electrophoresis were carried out as described under “Experimental Procedures.” S, substrate (−) enzyme.
Pol η Activity Is Stimulated by Sphingosine, but Not by Other Sphingolipids
Sphinganine (dihydrosphingosine) is identical to SO except for a single saturated bond. Because the common form of SO has an 18-carbon chain length, we examined the effect of C-18 SO on Pol η activity in this and all subsequent experiments carried out with SO. Similarly to C-17 SA, C-18 SO also elicited the dual response by Pol η (Fig. 2C and data not shown); fold increase and peak stimulatory concentration of SO, both d- and l-stereoisomers, were equivalent to that of SA. In addition, similarly to SA, it inhibited polymerase activity at concentrations exceeding 10 μm. SA and SO are involved in the de novo synthesis of ceramide and production of S1P, respectively (31). In contrast to SO, neither ceramide nor S1P, even at 10–50-fold higher concentrations than SA and SO, exhibited stimulatory or inhibitory effects on DNA synthesis by Pol η (Fig. 2C). Furthermore, the robust stimulation observed with SO was unaffected when it was present together with S1P at a 1:1 ratio, suggesting that S1P likely does not interact functionally or interfere with interactions between SO and Pol η (data not shown). Free SO in the cell is generated from the hydrolysis of sphingomyelin (32). Consistent with the stimulatory effect of SO, we observed subtle yet reproducible stimulation of Pol η activity by sphingomyelin (Fig. 2C). A short chain (C4) derivative of SO, threoninol, had no effect on the activity of Pol η (data not shown), implicating the importance of both the carbon chain length as well as the functional head group of SO in eliciting the response by Pol η. As a control, we also included the non-ionic detergent, Triton X-100 in extension reactions. Again, we observed no stimulation or inhibition of Pol η activity (data not shown), further corroborating the specificity of SA and SO.
Sphingosine Increases the Rate of Nucleotide Incorporation by Pol η and Is Non-competitive with dNTP Binding
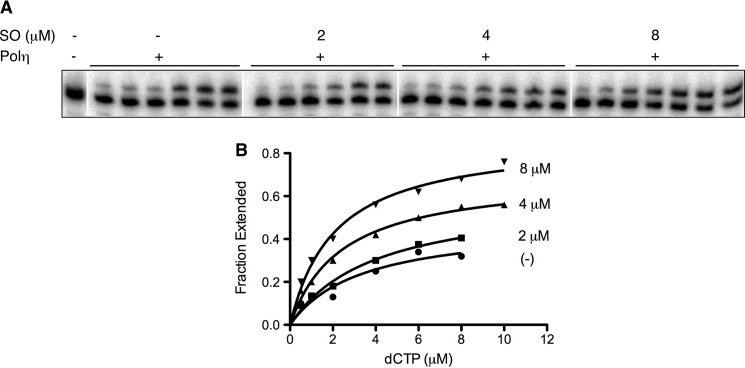
Kinetic studies were carried out to interpret the basis of stimulation. Processivity experiments, which evaluate the number of nucleotides incorporated before the polymerase disengages from the DNA, revealed that Pol η is distributive. It incorporates between 4–5 nt at each nucleotide addition step; this remained unchanged upon the addition of sphingosine, suggesting that stimulation by SO is not the result of increased processivity by Pol η (data not shown). We also measured the rate of incorporation of the initiating dNTP as a function of dNTP and sphingosine concentration. Incorporation kinetics were measured in the presence of increasing concentrations of dCTP (0.5–10 μm) without or with, 2, 4, or 8 μm SO. As presented in Fig. 3 and Table 1, there was a concentration-dependent increase in the rate of incorporation of the initiating nucleotide with a 3-fold increase in the kcat value with 8 μm SO. A similar analysis, which examined the rate of incorporation of dATP across a CPD lesion, also showed an elevated (5-fold) kcat upon the addition of SO (data not shown). However, we did not observe changes in the Km for dCTP in the presence of SO, consistent with SO interactions being non-competitive with respect to dNTP binding. SO could bind within a hydrophobic pocket of Pol η to induce conformational changes that render the enzyme more proficient. Attempts to visualize direct SO-Pol η interactions by x-ray crystallography, however, have been unsuccessful thus far.7
FIGURE 3.
Kinetic analysis of single nucleotide incorporation by Pol η. Steady state reactions to measure kinetic constants were carried out as described under “Experimental Procedures.” A representative gel (A) and the corresponding Michealis-Menten graphs (B) show the incorporation of the correct nucleotide, dCTP (concentrations ranging from 0.5–10 μm) by Pol η as a function of the indicated concentrations of SO. 9, 6, and 4.5 fmol of Pol η were used in reactions with 0, 2, and 4/8 μm SO, respectively.
TABLE 1.
Steady-state kinetic parameters for nucleotide incorporation in the absence or presence of sphingosine
Km and kcat values for the incorporation of dCTP across from template dG by Pol η were measured under steady state conditions as described under “Experimental Procedures.” Reactions were carried out at 37 °C for 5 min in the absence or presence of indicated concentrations of sphingosine. Data were fit by nonlinear regression using GraphPad Prism software (version 5). Data are the average ± S.D. of two to three independent measurements.
| SO | Km | kcat |
|---|---|---|
| μm | μm | min−1 |
| 0 | 3.6 ± 1.7 | 0.7 ± 0.3 |
| 2 | 4.1 ± 0.2 | 1.2 ± 0.2 |
| 4 | 4.3 ± 2.2 | 1.5 ± 0.4 |
| 8 | 3.2 ± 1.6 | 2.3 ± 0.6 |
Sphingosine Increases Misincorporation of dNTPs
To examine whether SO affects primer extension when one or more dNTPs is absent, reactions were carried out either in the presence of single dNTPs, or with three of the four dNTPs. In single nucleotide addition experiments, SO stimulated the incorporation of both the correct nucleotide, dCTP, and the incorrect nucleotides to comparable extents (Fig. 4A; 4- to 7-fold increase). Steady-state reactions examining nucleotide misincorporation kinetics also revealed an increase (2-fold) in the median kcat value in the presence of 10 μm SO, with no change in the median Km value (data not shown). Similarly, SO stimulated extension reactions in the presence of three dNTPs. Strong pause sites were observed immediately preceding or at the position corresponding to the missing nucleotide; however, in every case, read-through products were readily discernible in the presence of SO, accounting for an ∼90% increase compared with reactions lacking SO (Fig. 4B). In preliminary experiments, we again observed a 2-fold increase in the rate of extension of a 3′-terminal mismatched nucleotide (data not shown). By enhancing the rate of nucleotide misincorporation and misextension, SO can likely increase the rate of mutagenesis by Pol η.
FIGURE 4.
SO increases nucleotide misinsertion and misextension by Pol η. Pol η (0.6 nm) was incubated with DNA P/T1 (10 nm) in the presence of single dNTPs (either correct (dCTP) or incorrect (dATP, dGTP, dTTP) (A)) or three of four dNTPs (B), each at 100 μm concentrations. Extension reactions were carried out at 37 °C for 10 min in the absence (−) or presence (+) of 8 μm sphingosine. Note the strong pause sites at or immediately preceding the missing dNTP. S, substrate (−) enzyme; N, control with all four dNTPs. The sequence of the DNA template is shown on the left.
Sphingosine Stimulates Primer Extension Activity of Endogenous Pol η
To ascertain that native Pol η responds to SO similarly as the recombinant protein, we isolated Pol η from human cells by using a Pol η-specific monoclonal antibody and assayed primer extension activity with the immunoprecipitated enzyme. To ensure that the observed activity is due to Pol η, we assayed TLS across a CPD lesion in the absence or presence of SO. We observed efficient thymine dimer bypass activity in the immunoprecipitated sample, which was concentration-dependent (data not shown); this activity of endogenous Pol η increased upon the addition of SO (Fig. 5B), suggesting that Pol η can be regulated by SO in vivo. The magnitude of stimulation of native Pol η was not as high as that observed with the recombinant protein (Fig. 5A); this could be due to the presence of endogenous SO in the immunoprecipitate (data not shown).
Sphingosine Increases Lesion Bypass Activity in Cells
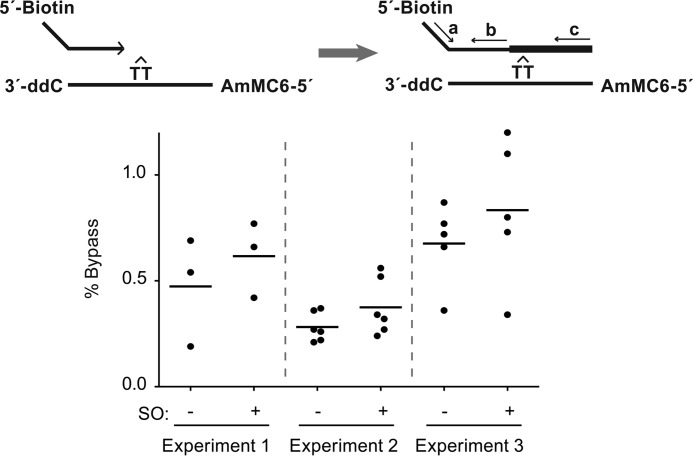
To examine whether SO increases translesion activity in cells, we carried out initial experiments with a partial duplex DNA oligonucleotide construct containing a site-specific CPD lesion. We transfected this substrate into SV40-immortalized fibroblasts (27, 28) and monitored bypass activity in the absence or presence of SO treatment by qPCR, as described under “Experimental Procedures.” The apoptotic response triggered by SO precluded the use of high concentrations. Nonetheless, cells treated with low concentrations (2–5 μm) of SO exhibited 28% higher bypass activity compared with untreated cells; these differences in SO-treated samples were consistent across three independent experiments (p = 0.046 (95% confidence interval, 2–62% higher) (Fig. 6). While low, the fold increase observed in our experiments is as follows: a) statistically significant, b) consistent across three independent experiments, and c) similar in magnitude to the increase in overall DNA synthesis observed by Zhang et al. (33) in SO-treated cells.
FIGURE 6.
Sphingosine increases CPD lesion bypass activity in cells. GM0639 cells were transfected with a partial duplex DNA oligonucleotide construct containing a site-specific cyclobutane pyrimidine dimer. Following a 2-h incubation period, the oligonucleotide was retrieved, and the extent of lesion bypass was quantified by qPCR using the primer pairs, a/b and a/c, as shown on the schematic. % Bypass observed in untreated (−) or SO-treated ((+); 2 μm (experiment 1) or 5 μm (experiments 2 and 3)) cells in three independent trials is presented. ddC, dideoxycytidine; AmMC, 5′-amino modifier C6.
Membrane-bound Sphingosine Mimics the Effects of Free Sphingosine
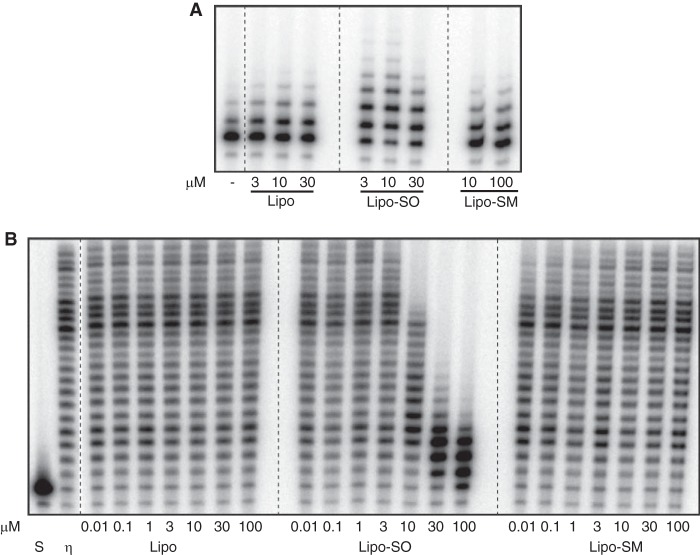
SO is a natural constituent of cells. It could be tethered to membrane lipids in vivo via its hydrophobic alkyl chain. To determine whether membrane-bound SO modulates Pol η activity similarly as unbound SO, we prepared model unilamellar lipid vesicles using the synthetic phospholipid, phosphatidylcholine. Independent preparations contained either SO or SM. The liposome preparations were diluted such that the concentrations of membrane-bound SO and SM were comparable with those of free SO and SM used in experiments thus far, and their effects were evaluated in primer extension reactions containing Pol η. Membrane-bound SO, but not PC vesicles or membrane-bound SM, specifically modulated Pol η activity (Fig. 7). Membrane-bound SO both stimulated (Fig. 7A) and inhibited (Fig. 7B) DNA extension activity, albeit to a lower extent than observed with free SO. Furthermore, the concentrations that elicited this response were equivalent to those of free SO. For example, whereas 10 μm bound-SO stimulated extension activity, 30 μm and higher concentrations elicited strong inhibition. We estimate that >96% of SO is membrane-bound under our conditions, and we have shown that concentrations corresponding to ≤4% free SO have no effect on TLS polymerase activity. Thus, both the specificity and response elicited by SO in solution are recapitulated with membrane-bound SO.
FIGURE 7.
Membrane-bound SO mimics the effect of free SO. Liposomes prepared with 1-oleoyl-2-palmitoyl-sn-glycero-3-phosphocholine, either alone or mixed with SO or SM, were added at increasing concentrations to primer extension reactions to evaluate stimulatory (A) or inhibitory (B) effects. Visualization of maximal stimulation and inhibition by liposomes with SO necessitated the use of different Pol η amounts in the extension reactions presented in A and B. Pol η (2 fmol) was incubated with 3, 10, or 30 μm liposomes (Lipo) or liposomes with SO (Lipo-SO) or with 10 or 100 μm liposomes with sphingomyelin (Lipo-SM) in A, or 9 fmol of Pol η were incubated with 0.01, 0.1, 1, 3, 10, 30, or 100 μm of each liposome preparation in B. Primer extension reactions with P/T1 were carried out as described, and products were visualized by PhosphorImager analysis of denaturing gels. S, substrate (−) enzyme.
DISCUSSION
We set out to identify small molecule inhibitors of TLS DNA polymerases. Instead, we serendipitously uncovered two related stimulators, the lipid-signaling molecules, sphingosine and dihydrosphingosine. Only about a dozen mechanistically well-characterized enzyme activators have been identified thus far, but none of them target DNA metabolic enzymes, let alone DNA polymerases (34). To our knowledge, sphingolipids represent the first example of naturally occurring lipid compounds that markedly stimulate DNA polymerase activity and, uniquely, that of the Y-family TLS DNA polymerases, Pols η, κ, and ι.
Sphingosine, the simplest of all sphingolipids, is recognized for its role in cell signaling to control cell proliferation (31, 35–37). Our studies, however, have uncovered a hitherto unknown and unreported function of sphingosine, namely, its ability to stimulate the DNA polymerase activity of Y-family TLS DNA polymerases. We show that SA and SO, but not the phosphorylated form S1P or other sphingolipids, stimulate in vitro the DNA primer extension activity of TLS DNA polymerases in a sequence-independent manner (Figs. 2 and 5A). Stimulation is specific to Pols η, κ, and ι; the polymerase activities of family A, B, and X DNA polymerases are unaffected (Fig. 2, B and C). The effect of SO is non-competitive with respect to dNTPs resulting in increased rates of dNTP incorporation (Fig. 3 and Table 1). By increasing reaction rates, SO also promotes increased misincorporation and misextension by Pol η (Fig. 4, A and B). Furthermore, we show that endogenous Pol η isolated from cells in culture is stimulated by SO similarly as the recombinant enzyme (Fig. 5, A and B). Notably, we show that 1) SO increases CPD lesion bypass activity in cells (Fig. 6) and that 2) membrane-bound SO has similar effects as free SO i.e. stimulation at low concentrations and inhibition at higher concentrations (Fig. 7, A and B).
Sphingolipids such as ceramide, SO, and S1P are bioactive signaling molecules that regulate cellular processes including growth, proliferation, senescence, and apoptosis (31, 36, 37). Of these three compounds, only SO uniquely stimulates the activity of Y-family TLS DNA polymerases. Endogenous free SO is generated from the hydrolysis of sphingomyelin, which is present in plasma and nuclear membranes, and subnuclear membrane fractions tightly associated with chromatin. It is plausible that SO is also anchored to nuclear membrane fractions that are in proximity to sites of DNA transactions involving DNA polymerases. It has been reported that UV irradiation increases ceramide levels and, presumably, sphingosine levels by the action of ceramidase (38). The resulting SO could regulate Pol η activity in a UV and SO dose-dependent manner in vivo.
Sphingosine has been reported to regulate the activity of a number of protein kinases including PKC, PKB/AKT, MAPK, and v/c-Src kinase (39), all implicated in cell proliferation; in each case, SO inhibits kinase activity to potentially limit cell proliferation. SO was also shown to inhibit the RNA primase activity of the replicative DNA polymerase, Pol α (32) and inhibit transcription of CYP17 by binding to the nuclear receptor, steroidogenic factor-1, essential for steroid hormone biosynthesis (40). Our studies on SO are unique in that rather than the reported inhibitory effects, we show stimulation of enzyme activity at intracellular concentrations of SO (32). Interestingly, the expression of Pol κ is highest in the adrenal cortex- the site of steroid biosynthesis (41), and Pol κ has also been implicated in bypassing DNA adducts generated during steroid biosynthesis (42). Thus, SO serves to both regulate steroid hormone synthesis as well as facilitate bypass of resultant steroid adducts by Pol κ.
Together with ceramide and S1P, SO is a part of the rheostat that decides whether cells grow and divide or die. Although ceramide controls cell growth inhibition and apoptosis induction, S1P contributes to cell proliferation, metastasis, and resistance to apoptosis (37). The effect of SO on the other hand differs depending on its levels; low levels are reported to promote cell proliferation, whereas high levels are proapoptotic (35). We propose that up-regulation of TLS DNA polymerase activity by SO may be necessary to prevent replication fork stalling in response to cellular stress and, therefore, to promote cellular proliferation. It is certainly consistent with our observations where low levels of both free and membrane-bound SO that stimulate TLS Pol activity promote cellular proliferation, whereas high levels, which are proapoptotic, inhibit activity.
In bacteria, blockage of fork progression induces transcription of error-prone DNA polymerases. This process, referred to as “the SOS response,” is induced to limit the potential detrimental effects of fork stalling, i.e. DNA breaks and ensuing rearrangements and deletions (43). Likewise, there are reports of transcriptional up-regulation of TLS DNA polymerases in human cells (41, 44, 45) as well as evidence for post-translational modification and cellular relocalization upon DNA damage (46). Furthermore, there are reports that TLS DNA polymerase protein levels are elevated in a number of cancers, presumably, to handle the overload of DNA adducts generated by chemotherapeutic drugs (17, 19, 20). Overexpression of translesion DNA polymerases may reduce the effectiveness of chemotherapy and render cancer cells chemoresistant (21). Modulation of polymerase activity by sphingolipids following cellular stress may be yet another mechanism by which TLS DNA polymerases is regulated. Our studies provide a novel link between two processes involved in the stress response of cells: error-prone DNA synthesis to avoid replication collapse and signaling by lipid molecules to control cellular proliferation.
Acknowledgments
We thank Ayu Rahardjo for help in screening the small molecule libraries, Drs. Alexey Merz, Margaret Lo, and Sharona Gordon for assistance with liposome preparations, and members of our laboratories for helpful comments and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants P01-CA 077852 and P01-AG 001751 (to L. A. L.).
J.-C. Shen, unpublished data.
M. W. Schmitt, unpublished data.
W. Yang, personal communication.
- TLS
- translesion
- Pol η
- DNA polymerase η
- SA
- sphinganine
- SO
- sphingosine
- S1P
- sphingosine-1-phosphate
- SM
- sphingomyelin
- CPD
- cyclobutane pyrimidine dimer
- P
- primer
- T
- template
- qPCR
- quantitative PCR
- UPLC
- ultra high-pressure liquid chromatography.
REFERENCES
- 1. Guo C., Kosarek-Stancel J. N., Tang T. S., Friedberg E. C. (2009) Y-family DNA polymerases in mammalian cells. Cell Mol. Life Sci. 66, 2363–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., Walker G. C. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lange S. S., Takata K., Wood R. D. (2011) DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 5. Cleaver J. E., Afzal V., Feeney L., McDowell M., Sadinski W., Volpe J. P., Busch D. B., Coleman D. M., Ziffer D. W., Yu Y., Nagasawa H., Little J. B. (1999) Increased ultraviolet sensitivity and chromosomal instability related to P53 function in the xeroderma pigmentosum variant. Cancer Res. 59, 1102–1108 [PubMed] [Google Scholar]
- 6. Masutani C., Kusumoto R., Iwai S., Hanaoka F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 19, 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vaisman A., Masutani C., Hanaoka F., Chaney S. G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry 39, 4575–4580 [DOI] [PubMed] [Google Scholar]
- 8. Kino K., Ito N., Sugasawa K., Sugiyama H., Hanaoka F. (2004) Translesion synthesis by human DNA polymerase η across oxidative products of guanine. Nucleic Acids Symp. Ser. 48, 171–172 [DOI] [PubMed] [Google Scholar]
- 9. Pavlov Y. I., Rogozin I. B., Galkin A. P., Aksenova A. Y., Hanaoka F., Rada C., Kunkel T. A. (2002) Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase η during copying of a mouse immunoglobulin κ light chain transgene. Proc. Natl. Acad. Sci. U.S.A. 99, 9954–9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nair D. T., Johnson R. E., Prakash S., Prakash L., Aggarwal A. K. (2004) Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature 430, 377–380 [DOI] [PubMed] [Google Scholar]
- 11. Suzuki N., Ohashi E., Kolbanovskiy A., Geacintov N. E., Grollman A. P., Ohmori H., Shibutani S. (2002) Translesion synthesis by human DNA polymerase κ on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-N(2)-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene). Biochemistry 41, 6100–6106 [DOI] [PubMed] [Google Scholar]
- 12. Bi X., Slater D. M., Ohmori H., Vaziri C. (2005) DNA polymerase κ is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J. Biol. Chem. 280, 22343–22355 [DOI] [PubMed] [Google Scholar]
- 13. Sale J. E., Lehmann A. R., Woodgate R. (2012) Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nelson J. R., Lawrence C. W., Hinkle D. C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382, 729–731 [DOI] [PubMed] [Google Scholar]
- 15. Edmunds C. E., Simpson L. J., Sale J. E. (2008) PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell 30, 519–529 [DOI] [PubMed] [Google Scholar]
- 16. Salehan M. R., Morse H. R. (2013) DNA damage repair and tolerance: a role in chemotherapeutic drug resistance. Br. J. Biomed. Sci. 70, 31–40 [DOI] [PubMed] [Google Scholar]
- 17. O-Wang J., Kawamura K., Tada Y., Ohmori H., Kimura H., Sakiyama S., Tagawa M. (2001) DNA polymerase κ, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res. 61, 5366–5369 [PubMed] [Google Scholar]
- 18. Yang J., Chen Z., Liu Y., Hickey R. J., Malkas L. H. (2004) Altered DNA polymerase ι expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res. 64, 5597–5607 [DOI] [PubMed] [Google Scholar]
- 19. Ceppi P., Novello S., Cambieri A., Longo M., Monica V., Lo Iacono M., Giaj-Levra M., Saviozzi S., Volante M., Papotti M., Scagliotti G. (2009) Polymerase η mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin. Cancer Res. 15, 1039–1045 [DOI] [PubMed] [Google Scholar]
- 20. Teng K. Y., Qiu M. Z., Li Z. H., Luo H. Y., Zeng Z. L., Luo R. Z., Zhang H. Z., Wang Z. Q., Li Y. H., Xu R. H. (2010) DNA polymerase η protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy. J. Transl. Med. 8, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Albertella M. R., Lau A., O'Connor M. J. (2005) The overexpression of specialized DNA polymerases in cancer. DNA Repair 4, 583–593 [DOI] [PubMed] [Google Scholar]
- 22. Doles J., Oliver T. G., Cameron E. R., Hsu G., Jacks T., Walker G. C., Hemann M. T. (2010) Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc. Natl. Acad. Sci. U.S.A. 107, 20786–20791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie K., Doles J., Hemann M. T., Walker G. C. (2010) Error-prone translesion synthesis mediates acquired chemoresistance. Proc. Natl. Acad. Sci. U.S.A. 107, 20792–20797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmitt M. W., Matsumoto Y., Loeb L. A. (2009) High fidelity and lesion bypass capability of human DNA polymerase δ. Biochimie 91, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamath-Loeb A. S., Hizi A., Kasai H., Loeb L. A. (1997) Incorporation of the guanosine triphosphate analogs 8-oxo-dGTP and 8-NH2-dGTP by reverse transcriptases and mammalian DNA polymerases. J. Biol. Chem. 272, 5892–5898 [DOI] [PubMed] [Google Scholar]
- 26. Akagi J., Masutani C., Kataoka Y., Kan T., Ohashi E., Mori T., Ohmori H., Hanaoka F. (2009) Interaction with DNA polymerase η is required for nuclear accumulation of REV1 and suppression of spontaneous mutations in human cells. DNA Repair 8, 585–599 [DOI] [PubMed] [Google Scholar]
- 27. Kamath-Loeb A. S., Shen J. C., Schmitt M. W., Loeb L. A. (2012) The Werner syndrome exonuclease facilitates DNA degradation and high fidelity DNA polymerization by human DNA polymerase δ. J. Biol. Chem. 287, 12480–12490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen J. C., Fox E. J., Ahn E. H., Loeb L. A. (2014) A rapid assay for measuring nucleotide excision repair by oligonucleotide retrieval. Sci. Rep. 4, 4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seber G. A. (2003) Linear Regression Analysis, 2nd Ed., Wiley, Boston [Google Scholar]
- 30. Sasaki H., Arai H., Cocco M. J., White S. H. (2009) pH dependence of sphingosine aggregation. Biophys. J. 96, 2727–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morad S. A., Cabot M. C. (2013) Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 13, 51–65 [DOI] [PubMed] [Google Scholar]
- 32. Simbulan C. M., Tamiya-Koizumi K., Suzuki M., Shoji M., Taki T., Yoshida S. (1994) Sphingosine inhibits the synthesis of RNA primers by primase in vitro. Biochemistry 33, 9007–9012 [DOI] [PubMed] [Google Scholar]
- 33. Zhang H., Buckley N. E., Gibson K., Spiegel S. (1990) Sphingosine stimulates cellular proliferation via a protein kinase C-independent pathway. J. Biol. Chem. 265, 76–81 [PubMed] [Google Scholar]
- 34. Zorn J. A., Wells J. A. (2010) Turning enzymes ON with small molecules. Nat. Chem. Biol. 6, 179–188 [DOI] [PubMed] [Google Scholar]
- 35. Ohanian J., Ohanian V. (2001) Sphingolipids in mammalian cell signalling. Cell Mol. Life Sci. 58, 2053–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ponnusamy S., Meyers-Needham M., Senkal C. E., Saddoughi S. A., Sentelle D., Selvam S. P., Salas A., Ogretmen B. (2010) Sphingolipids and cancer: ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol. 6, 1603–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pyne N. J., Pyne S. (2010) Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer 10, 489–503 [DOI] [PubMed] [Google Scholar]
- 38. Verheij M., Bose R., Lin X. H., Yao B., Jarvis W. D., Grant S., Birrer M. J., Szabo E., Zon L. I., Kyriakis J. M., Haimovitz-Friedman A., Fuks Z., Kolesnick R. N. (1996) Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature 380, 75–79 [DOI] [PubMed] [Google Scholar]
- 39. Mao C., Obeid L. M. (2008) Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim. Biophys. Acta 1781, 424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Urs A. N., Dammer E., Sewer M. B. (2006) Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology 147, 5249–5258 [DOI] [PubMed] [Google Scholar]
- 41. Velasco-Miguel S., Richardson J. A., Gerlach V. L., Lai W. C., Gao T., Russell L. D., Hladik C. L., White C. L., Friedberg E. C. (2003) Constitutive and regulated expression of the mouse Dinb (Polκ) gene encoding DNA polymerase κ. DNA Repair 2, 91–106 [DOI] [PubMed] [Google Scholar]
- 42. Singer W. D., Osimiri L. C., Friedberg E. C. (2013) Increased dietary cholesterol promotes enhanced mutagenesis in DNA polymerase κ-deficient mice. DNA Repair 12, 817–823 [DOI] [PubMed] [Google Scholar]
- 43. Goodman M. F., Tippin B. (2000) Sloppier copier DNA polymerases involved in genome repair. Curr. Opin. Genet. Dev. 10, 162–168 [DOI] [PubMed] [Google Scholar]
- 44. Lemée F., Bavoux C., Pillaire M. J., Bieth A., Machado C. R., Pena S. D., Guimbaud R., Selves J., Hoffmann J. S., Cazaux C. (2007) Characterization of promoter regulatory elements involved in downexpression of the DNA polymerase κ in colorectal cancer. Oncogene 26, 3387–3394 [DOI] [PubMed] [Google Scholar]
- 45. Brauze D., Rawluszko A. A. (2012) The effect of aryl hydrocarbon receptor ligands on the expression of polymerase (DNA directed) κ (Polκ), polymerase RNA II (DNA directed) polypeptide A (PolR2a), CYP1B1 and CYP1A1 genes in rat liver. Environ. Toxicol. Pharmacol. 34, 819–825 [DOI] [PubMed] [Google Scholar]
- 46. Chen Y. W., Cleaver J. E., Hatahet Z., Honkanen R. E., Chang J. Y., Yen Y., Chou K. M. (2008) Human DNA polymerase η activity and translocation is regulated by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 16578–16583 [DOI] [PMC free article] [PubMed] [Google Scholar]