Background: The Sec translocon is a universally conserved protein transport channel that interacts with many auxiliary proteins.
Results: PpiD binds to the lateral gate of SecY and is detached by nascent membrane proteins but not by SecA.
Conclusion: PpiD is a novel transient subunit of the Sec translocon.
Significance: This is the first description of a direct SecY-chaperone interaction.
Keywords: Chaperone, Escherichia coli (E. coli), Protein Cross-linking, Protein Sorting, Protein Translocation, Periplasm, Protein Quality Control
Abstract
The Sec translocon constitutes a ubiquitous protein transport channel that consists in bacteria of the three core components: SecY, SecE, and SecG. Additional proteins interact with SecYEG during different stages of protein transport. During targeting, SecYEG interacts with SecA, the SRP receptor, or the ribosome. Protein transport into or across the membrane is then facilitated by the interaction of SecYEG with YidC and the SecDFYajC complex. During protein transport, SecYEG is likely to interact also with the protein quality control machinery, but details about this interaction are missing. By in vivo and in vitro site-directed cross-linking, we show here that the periplasmic chaperone PpiD is located in front of the lateral gate of SecY, through which transmembrane domains exit the SecY channel. The strongest contacts were found to helix 2b of SecY. Blue native PAGE analyses verify the presence of a SecYEG-PpiD complex in native Escherichia coli membranes. The PpiD-SecY interaction was not influenced by the addition of SecA and only weakly influenced by binding of nontranslating ribosomes to SecYEG. In contrast, PpiD lost contact to the lateral gate of SecY during membrane protein insertion. These data identify PpiD as an additional and transient subunit of the bacterial SecYEG translocon. The data furthermore demonstrate the highly modular and versatile composition of the Sec translocon, which is probably essential for its ability to transport a wide range of substrates across membranes in bacteria and eukaryotes.
Introduction
Protein transport via the Sec translocon represents an evolutionarily conserved mechanism for transporting newly synthesized proteins from the cytosol into or across membranes (1, 2). The Sec translocon is embedded in the endoplasmic reticulum membrane of eukaryotes and in the cytoplasmic membrane of bacteria (3, 4). The three-subunit core of the bacterial Sec translocon, termed SecYEG, constitutes the minimal membrane-embedded unit required for protein transport (5–7) and provides the platform for multiple interacting partner proteins (8). These partner proteins determine the mechanism of substrate movement across the channel and facilitate substrate folding and release from the Sec translocon. For signal recognition particle (SRP)-dependent2 co-translational targeting of bacterial membrane proteins (9, 10), the Sec translocon associates with the bacterial SRP receptor FtsY (11–14), which allows SRP to deliver ribosome-associated nascent chains (RNCs) directly to the Sec translocon. After RNC binding to SecY, the translational activity of the ribosome threads the nascent protein directly into the Sec channel (15). For post-translational translocation of secretory proteins across the Sec translocon, the ATPase SecA associates with SecY, and substrate translocation is probably achieved by an ATP-dependent pushing mechanism (16, 17). This mechanism is also required for translocating large periplasmic loops of membrane proteins (18, 19). The cytosolic loops of SecY provide the binding sites for these cytosolic ligands, and SecA, FtsY, and ribosomes engage a partially overlapping binding site on the fifth cytosolic loop of SecY (13).
In addition to these cytosolic contacts, SecYEG interacts with the membrane integral SecDFYajC complex and YidC. SecDFYajC is a nonessential and very low abundant partner protein of the Escherichia coli SecYEG, which is thought to be involved in energizing protein transport, possibly by utilizing the proton-motive force (20–22). However, details on how SecYEG and SecDFYajC interact are currently unknown. YidC is a conserved and essential membrane protein that cooperates with SecYEG during membrane protein insertion but can also insert membrane proteins independently of SecYEG (23–25). YidC is located in front of a lateral opening (lateral gate) of SecY, through which transmembrane substrates are thought to exit the channel for entering the lipid phase (26). The position of YidC in front of the lateral gate is in line with a sequential transfer of substrates from SecY to YidC (27, 28) and also with the proposed function of YidC in helping transmembrane domains to exit the SecY channel and in facilitating their subsequent folding (29). Although protein transport across the eukaryotic Sec complex requires several proteins on the trans-side of the membrane (1, 8), it is largely unknown how the bacterial SecYEG complex interacts with proteins on the trans-side of the membrane, i.e. periplasmic proteins. The E. coli periplasm contains several chaperones and proteases that assist in the maturation of β-barrel proteins (30) after their transport via the Sec translocon. A localization in close proximity to the Sec translocon has been suggested for the small chaperone Skp and the peptidyl-prolyl isomerase PpiD (31, 32). Skp is a trimeric chaperone that was shown to interact with the outer membrane proteins OmpA (32) and PhoE (33). It is thought that Skp influences the release of fully translocated substrates from the cytoplasmic membrane into the periplasm (32). PpiD is single-spanning membrane protein with a large periplasmic peptidyl-prolylisomerase (PPIase) domain (34) and one of several PPIases (SurA, PpiA, and FkpA) found in the E. coli periplasm (35). This probably explains why a ΔppiD strain shows only a weak phenotype (36). Like Skp, PpiD might be required for the release of a substrate from the membrane into the periplasm, but different from Skp, PpiD probably interacts with substrates while they are translocated through SecY (31). This is deduced from the observation that PpiD cross-links to a translocation intermediate of a single-spanning membrane protein (31), which furthermore suggests that PpiD does not exclusively act on outer membrane proteins but also on periplasmic domains of inner membrane proteins. The interaction of PpiD with nascent membrane proteins indicates that PpiD is located in close vicinity to SecYEG.
For gaining insight into the interaction between PpiD and the Sec translocon, we performed an in vivo site-directed cross-linking approach and found that PpiD is located at the lateral gate of SecY. Our data furthermore show that PpiD is detached from the lateral gate when SecY is engaged in membrane protein insertion. These data support the emerging concept that the Sec translocon in bacteria and eukaryotes exhibits a modular composition, which not only involves the direct contact to targeting modules but also contacts to the cellular protein quality machinery.
EXPERIMENTAL PROCEDURES
Plasmids, Strains, and Growth Conditions
The following E. coli strains were used: MC4100, DH5α (37), BL21 pSup-BpaRS-6TRN (26, 38), C43 pSup-BpaRS-6TRN (26), KC6(DE3) pftsQ-tnaC (a gift from R. Beckmann, Munich), SecY39 (39), K12 ΔppiD (a gift from Dan Daley, University of Stockholm), and MC4100 ΔppiD (a gift from Michaela Fürst and Matthias Müller, University Freiburg). Cells were grown in LB medium at either 30 or 37 °C. TAG stop codons were incorporated at the indicated positions of pTRc99aSecY(His)EG (13) using the Phusion PCR Kit (NE Biolabs, Frankfurt, Germany) with 5′-phosphorylated oligonucleotides (26).
In Vivo and in Vitro pBpa Cross-linking
For in vivo cross-linking, E. coli BL21 and K12 ΔppiD harboring the plasmids pSup-BpaRS-6TRN and pTrc99a-SecY(His)EG were grown at 30 °C in minimal medium in the presence of 1 mm pBpa as previously described (13, 41). Only the C43 strain carrying the same plasmids but with TAG amber stop codons in TMs 7 and 8 of SecY was grown at 37 °C in LB medium in the presence of 1 mm pBpa. In this case, cells were induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside after reaching the log phase and were grown for another 5 h at 25 °C. After harvesting, the cells were washed once with 50 mm triethanolamine acetate, pH 7.5, and incubated on ice for 30 min in a UV box (Vilbert-Lourmat BLX-365) in PBS buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.76 mm NaH2HPO4, pH 7.6). Cells were lysed in a French pressure cell, and membranes were prepared and solubilized with 1% n-dodecyl β-d-maltoside (Thermo Scientific). SecY was further purified via Talon® affinity resin (Clontech), and its cross-linking partners were identified by immune detection.
For in vitro cross-linking, inner membrane vesicles (INVs) were prepared from E. coli cells expressing SecYpBpa in the presence of 1 mm pBpa according to the procedure described previously (26, 42). INV (4 μg/μl) were incubated on ice with ribosomes or ribosome-associated nascent chains in INV buffer (100 mm triethanolamine acetate, pH 8, 250 mm sucrose, 5 mm Mg(Ac)2) and UV-irradiated for 20 min. The reaction mixture was then solubilized with 1% n-dodecyl-β-d-maltoside, and SecY cross-linking products were purified via Talon affinity resin and visualized by Western blotting. The N-terminally His-tagged RNCs containing the first 102 amino acids of FtsQ followed by an HA tag and a TnaC stalling sequence were expressed in vivo and purified essentially as described (43). The purification of SecA and ribosomes followed previously published protocols (42).
In Vitro Protein Synthesis and Transport
Proteins were in vitro synthesized using a CTF cell extract (42) and radioactively labeled with [35S]methionine/[35S]cysteine (PerkinElmer Life Sciences). Transport of in vitro synthesized proteins was carried out for 30 min at 37 °C, and the reaction mixture was further divided in two parts. One part was directly TCA-precipitated, whereas the other part was digested with 0.5 mg/ml proteinase K and incubated for 25 min at 25 °C. The proteins were afterward TCA-precipitated and separated on SDS-PAGE.
BN-PAGE Analysis
Purified INV (100 μg of protein) were dissolved in buffer containing 50 mm imidazole/HCl, pH 7.0, 5 mm 6-aminocaproic acid, 50 mm NaCl, solubilized with 1% final concentration of n-dodecyl β-d-maltoside (Roche Applied Science) and incubated for 5 min at 25 °C. Nonsolubilized material was pelleted by centrifugation for 30 min at 45,000 rpm at 4 °C (TLA-45 Beckman rotor). The solubilized proteins were separated on 4–15% BN gels and analyzed by immune detection.
Affinity Purification of α-SecY Antibodies
Affinity purification of SecY antibodies was performed by using purified recombinant protein. His-tagged SecYEG was purified by metal affinity purification as previously described (7). Purified SecY was separated on SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane. The membrane was blocked by incubation with 3% milk powder in TBS. After washing with TBS, α-SecY serum was added to the membrane and incubated for 90 min at room temperature. After washing with TBS, the bound SecY antibody was eluted from the membrane by incubation with 100 mm glycine/HCl, pH 2.5. The affinity-purified antibody was stored in 100 mm Tris/HCl, pH 8.0, 2 mg/ml BSA.
Quantification of SecY, YidC, and PpiD
Cells were grown on minimal medium at 30 °C to an A600 of approximately 1.0 and subsequently directly precipitated with 10% TCA (final concentration). After 30 min on ice, samples were centrifuged for 10 min, and the pellet was directly resuspended in SDS loading dye and denatured. Samples were then separated on 5–15% SDS-PAGE and after Western transfer decorated with polyclonal antibodies against SecY, YidC, and PpiD. As controls, purified proteins were loaded. Quantification was performed with the ImageJ software (National Institutes of Health). For calculating the molar concentration, the E. coli cell volume was considered to be 1 × 10−15 liter/cell (44).
RESULTS
The Lack of PpiD Does Not Significantly Impair in Vitro Protein Transport
A ΔppiD strain shows enhanced sensitivity toward SDS and EDTA, which is commonly associated with defects in outer membrane and cell envelope assembly (45). We therefore analyzed the influence of PpiD on outer membrane protein transport across the SecYEG translocon in an in vitro transcription/translation system employing purified INV of a ΔppiD strain. Western blotting confirmed the absence of PpiD in these INV and also demonstrated that the lack of PpiD did not influence the steady state amounts of SecY or YidC (Fig. 1A). We then measured the translocation of the SecA-dependent outer membrane protein pOmpA into INV of the ΔppiD strain. In comparison to wild type INV, neither signal sequence cleavage nor translocation was significantly impaired in the absence of PpiD (Fig. 1B). As a second substrate, we analyzed the integration of YidC into ΔppiD INV. The SRP-dependent, multispanning membrane protein YidC was selected as substrate because it contains a large periplasmic loop of 320 amino acids. Translocation of this loop depends on the ATPase activity of SecA, and its folding could require folding catalysts like PpiD. Thus, by using YidC as a substrate for in vitro insertion into INV, we can simultaneously detect any defects in lateral gate opening, i.e. TM insertion and in the translocation of large periplasmic loops. Comparing the insertion of YidC into wild type INV with the insertion into ΔppiD INV did not reveal any insertion defect (Fig. 1B). Although PpiD was shown to cross-link to a nascent chain exiting the SecY translocon (31), our in vitro data do not reveal a significant protein transport defect in the absence of PpiD.
FIGURE 1.
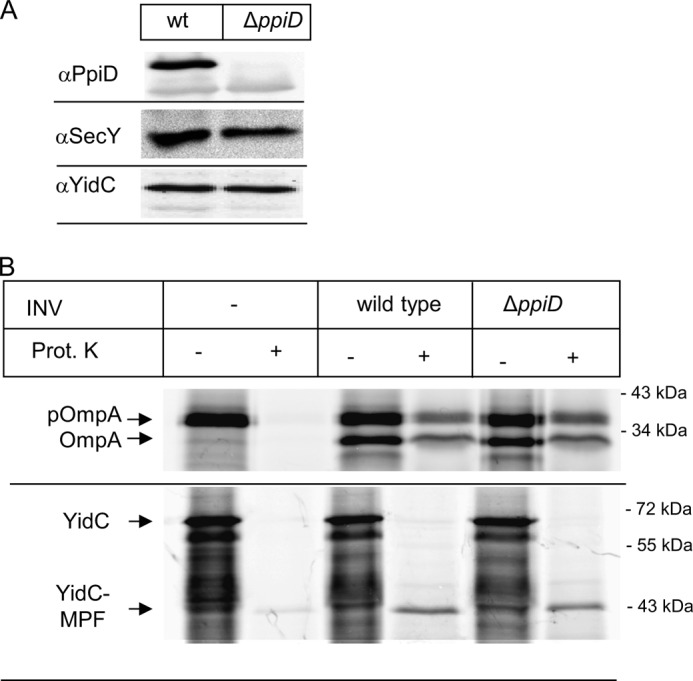
The absence of PpiD does not cause a significant protein transport defect in vitro. A, INV of wild type and a ΔppiD strain were separated on SDS-PAGE and after Western transfer decorated with the indicated antibodies. B, YidC and OmpA were in vitro synthesized using a coupled transcription/translation system in the presence of inner membrane vesicles (INV), derived from either a wild type E. coli strain or a ΔppiD strain. After 30 min of synthesis at 37 °C, the samples were split in half, and one part was directly precipitated with TCA (−). The other half was first treated for 20 min at 25 °C with 0.5 mg/ml proteinase K (Prot. K) and only then TCA-precipitated. pOmpA corresponds to the signal sequence containing pro-OmpA and OmpA to the mature version. YidC and its membrane-protected fragment (YidC-MPF) are indicated. The YidC-MPF corresponds to the first two transmembrane domains of YidC plus the connecting 325-amino acid-long periplasmic loop (66, 67).
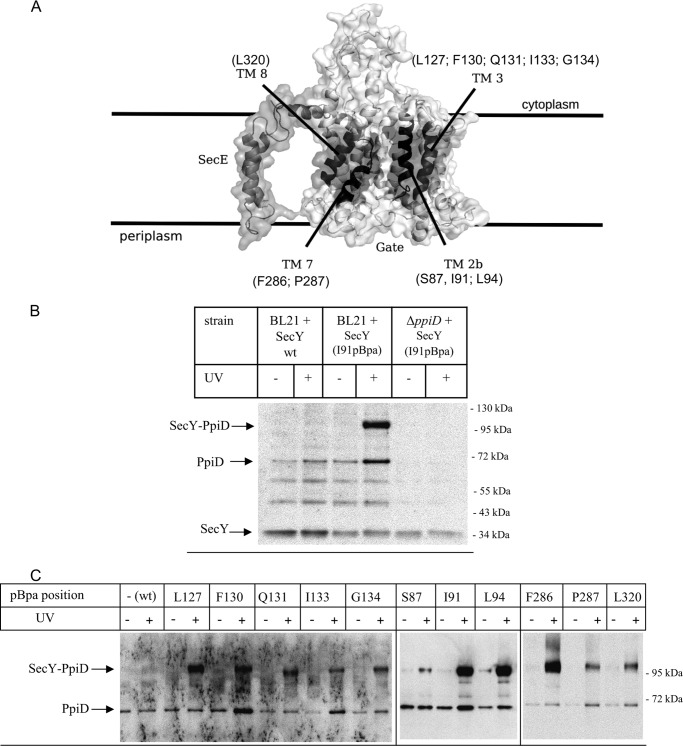
PpiD Is Located Close to the Lateral Gate of SecY
In a recent cross-linking approach, we had determined the interaction of SecY with the Sec translocon associated protein YidC and found that YidC makes contact to all four helices that constitute the lateral gate of SecY (26) (Fig. 2A). Analyzing potential cross-linking products by MS also revealed cross-links between SecY and PpiD (26). This was further analyzed in the current study by in vivo site-directed cross-linking employing the UV-activated phenylalanine derivative para-benzoyl-l-phenylalanine (pBpa). A TAG amber stop codon was incorporated at position 91 of a plasmid borne secY copy, and this construct was expressed in E. coli cells carrying a plasmid-borne orthogonal aminoacyl tRNA synthetase/tRNACUA pair, resulting in the in vivo incorporation of pBpa at the stop codon position when the growth media were supplemented with pBpa (38). Position 91 within helix 2b of the lateral gate of SecY (Fig. 2A) was selected because incorporation of pBpa at this position gave strong cross-links to YidC and MS-detectable cross-links to PpiD (26).
FIGURE 2.
The chaperone PpiD is located at the lateral gate of SecY. A, cryo-EM structure of the E. coli SecYEG complex based on the study by Frauenfeld et al. (15) (Protein Data Bank codes 3J00 and 3J01) with the lateral gate consisting of the four transmembrane domains highlighted in dark. pBpa was incorporated at the indicated residues. B, E. coli BL21 expressing either wild type SecY or SecY(I91pBpa) were treated as described under “Experimental Procedures.” As control, SecY(I91pBpa) was expressed in a ΔppiD strain. After SecY purification, samples were probed with antibodies against PpiD. Indicated are the SecY-PpiD cross-link, PpiD that co-purified with SecY, and the SecY band itself that cross-reacted with the PpiD antibody. The two bands at approximately 50 and 60 kDa probably correspond to proteolysis products of PpiD, because they were not detected in the ΔppiD strain, but this was not further analyzed. C, E. coli cells expressing either wild type SecY or SecY containing pBpa at the indicated positions were kept in the dark or were UV-exposed. Subsequently, SecY was purified and after Western transfer decorated with α-PpiD antibodies.
E. coli BL21 cells expressing the tRNA synthetase/tRNACUA pair and SecY(I91pBpa) were grown in the presence of pBpa and exposed to UV light. After cell fractionation, SecY and its cross-linked partner proteins were purified via an N-terminal His6 tag on SecY and analyzed by immune detection with α-PpiD antibodies. SecY(I91pBpa) gave a strong UV-specific band at approximately 105 kDa, which was recognized by α-PpiD antibodies (Fig. 2B). The mass of the cross-linking product is consistent with the predicted mass of a cross-link between SecY (migrating at approximately 34 kDa on SDS-PAGE) and PpiD (migrating at approximately 68 kDa) and is in agreement with our previous MS analyses (26). The cross-linking experiment was also repeated in an E. coli ΔppiD strain and with E. coli cells expressing wild type SecY lacking pBpa. The 105-kDa cross-linking product was neither observed in cells expressing SecY lacking pBpa nor in the ΔppiD strain, which further verifies that the 105-kDa band corresponds to a specific SecY-PpiD complex. We observed in all samples, with the exception of the ΔppiD strain, three additional, UV-independent bands that were recognized by α-PpiD antibodies. The band at approximately 66 kDa most likely corresponds to PpiD that co-purifies with SecY, whereas the two smaller bands at approximately 60 and 50 kDa could be PpiD degradation products, but this was not further analyzed. We also observed that the α-PpiD antibodies cross-reacted with purified SecY (Fig. 2B), but this did not interfere with our analyses, because our previous MS analyses established the SecY-PpiD cross-linking product at 105 kDa (26).
The SecY-PpiD interaction was further analyzed by incorporating pBpa into different positions within all four helices of the lateral gate (Fig. 2A). The functionality of the pBpa-containing SecY derivatives was previously confirmed by testing their ability to suppress the cold-sensitive phenotype of the conditional SecY39 mutant strain (26). After UV exposure and SecY purification, we found strong SecY-PpiD cross-links for positions Leu-127 and Phe-130 and weaker cross-links for positions Gln-131, Ile-133, and Gly-134 (Fig. 2C). A strong SecY-PpiD cross-linking product was confirmed for position Ile-91 and also found for position Leu-94, whereas position Ser-87 gave only a weaker cross-linking product. These data demonstrate that PpiD is located in close proximity to helices 2b and 3 on one side of the SecY lateral gate. Helices 7 and 8 form the other side of the lateral gate and SecY-PpiD cross-links were observed for positions Phe-286 and Pro-287 in helix 7 and for position Leu-320 in helix 8. However, the SecY-PpiD cross-linking products for Pro-287 and Leu-320 were weaker than the cross-linking product with Phe-286.
For further controlling that PpiD specifically contacts the lateral gate of SecY, we incorporated pBpa into position Val-225, which is located in a loop connecting TM5 and TM6 at the back of SecY. Here we did not observe any SecY-PpiD cross-linking product (Fig. 3). We also did not observe any PpiD cross-linking product when pBpa was incorporated into the N terminus of SecG (Fig. 3), which is located close to helix 3 of the lateral gate (16, 46). In summary, these data show that PpiD is positioned in immediate vicinity to the lateral gate of SecY where it contacts all four helices of the lateral gate.
FIGURE 3.

PpiD does not interact with the back of the SecYEG translocon or SecG. pBpa was incorporated into the indicated positions within SecY and SecG. E. coli cells expressing either wild type SecY or SecY/SecG containing pBpa at the indicated positions were kept in the dark or were UV-exposed. Subsequently, SecY was purified and after Western transfer decorated with α-PpiD antibodies.
PpiD Is a Component of the SecYEG Translocon
The in vivo cross-linking was performed with cells that moderately overexpressed SecYEG. The PpiD-SecY interaction could therefore reflect the increased need for chaperone activity during SecYEG overexpression. For excluding this possibility, we employed blue native PAGE of INV derived from either wild type cells or the ΔppiD strain. Blue native PAGE allows the detection of membrane protein complexes and has been used before for analyzing the composition of the Sec translocon (47, 48). Affinity-purified SecY antibodies recognized two complexes migrating at ∼200 and 300 kDa, respectively (Fig. 4A). The 200-kDa complex has been detected before and likely reflects a SecYEG dimer (47, 49), whereas the 300-kDa band was only detected with affinity-purified SecY antibodies. In ΔppiD INV, the 200-kDa band was still detected, but the 300-kDa band was absent. α-PpiD antibodies recognized the 300-kDa band and a band at approximately 70 kDa in wild type INV. Both bands were absent in ΔppiD INV (Fig. 4A). In this experimental setup, the α-PpiD antibody did not cross-react with SecY, indicating that cross-reactivity is only observed with purified SecY (Fig. 2B). These data indicate that PpiD is present in at least two pools in the E. coli membrane. The slightly diffuse band at approximately 70 kDa probably contains the PpiD monomer but could also contain the PpiD-YfgM complex that was recently described (48, 50). The second pool corresponds to a PpiD-SecYEG complex that migrates at approximately 300 kDa. These data demonstrate that the PpiD-SecY interaction is observed in membranes containing only the endogenous SecYEG levels and thus not the result of SecYEG overexpression. The data also show that it is not the incorporation of pBpa into SecY that causes the PpiD-SecY interaction, because the SecYEG-PpiD interaction was detectable on BN-PAGE in the absence of pBpa.
FIGURE 4.

PpiD forms a complex with the SecYEG translocon in native E. coli membranes. A, blue native PAGE analysis containing wild type (E. coli MC4100) INV or INV derived from the ΔppiD strain. After Western transfer, the blot was decorated with α-SecY or α-PpiD antibodies. B, E. coli BL21 cells were grown on minimal medium at 30 °C up to an A600 of 1 and were then directly TCA-precipitated. TCA pellets corresponding to the indicated cell number were separated on SDS-PAGE and decorated with polyclonal antibodies against SecY, YidC, or PpiD. Purified proteins served as reference. C, quantification of the Western blot using the ImageJ software. As reference, the concentrations of SecY and YidC in wild type E. coli MC4100 cells as estimated previously (2, 40) are indicated.
The BN-PAGE data show that the greater part of the cellular PpiD pool is not in complex with SecYEG but exists as monomer or as a PpiD-YfgM complex. For determining whether the cellular PpiD concentration is higher than the SecYEG concentration, we determined their concentration in E. coli membranes by quantitative Western blotting. By quantifying Western blots of TCA-precipitated E. coli BL21 cells using purified proteins as reference, we determined a cellular PpiD concentration of approximately 4 μm (Fig. 4, B and C). As reference, we verified also the concentration of SecY and YidC, for which the cellular concentrations have been determined before in the E. coli wild type strain MC4100 (2). The SecY concentration was determined to be 0.8 μm, which is very close to the 1 μm concentration determined previously (2). YidC was present at a cellular concentration of 5 μm, also very similar to the 4.2 μm determined in previous studies (2). Thus, PpiD is in surplus over SecYEG, which explains the presence of a PpiD pool not in complex with SecYEG.
Ribosomes and SecA Do Not Influence the SecY-PpiD Interaction
The membrane localization and topology of PpiD makes it a very likely substrate for SecYEG-dependent insertion. Therefore, the in vivo SecY-PpiD cross-linking product could correspond to PpiD that is in the process of being co-translationally membrane inserted via SecYEG. We therefore switched to an in vitro approach using sucrose-gradient purified inner membrane vesicles (INVs), derived from cells expressing the different SecY(pBpa) derivatives. After UV exposure of INV, SecY(pBpa) and its cross-linking adducts were purified via the N-terminal His tag on SecY. This in vitro approach has the intrinsic advantage that translating ribosomes and mRNA are largely absent from INV (26, 42, 47), and thus the large majority of the SecYEG translocons are in their resting state but can be activated by the addition of ligands like SecA, ribosomes, or substrates.
When INVs were purified from E. coli expressing SecY(I91pBpa) and subsequently UV-exposed, the 105-kDa SecY-PpiD cross-linking product was detected in the UV-exposed sample but absent in the UV control (Fig. 5A). SecY-PpiD cross-links were also observed for INVs derived from cells expressing SecY(F130pBpa) or SecY(L320pBpa) (Fig. 5A). This demonstrates that the SecY-PpiD interaction persists even for the resting translocon, which excludes the possibility that these cross-links represent insertion intermediates of PpiD. This is also supported by the observation that full-length PpiD co-purifies with His-tagged SecY (Fig. 5A), confirming the co-purification already observed after in vivo cross-linking (Fig. 2). Co-purification of PpiD was also observed for wild type SecY lacking pBpa (Fig. 2), confirming that the SecY-PpiD interaction is not caused by enhanced folding requirements of pBpa containing SecY derivatives. In this in vitro approach, we observed some UV-independent cross-link formation, which was not observed in vivo. This is probably the result of extended light exposure during INV preparation.
FIGURE 5.
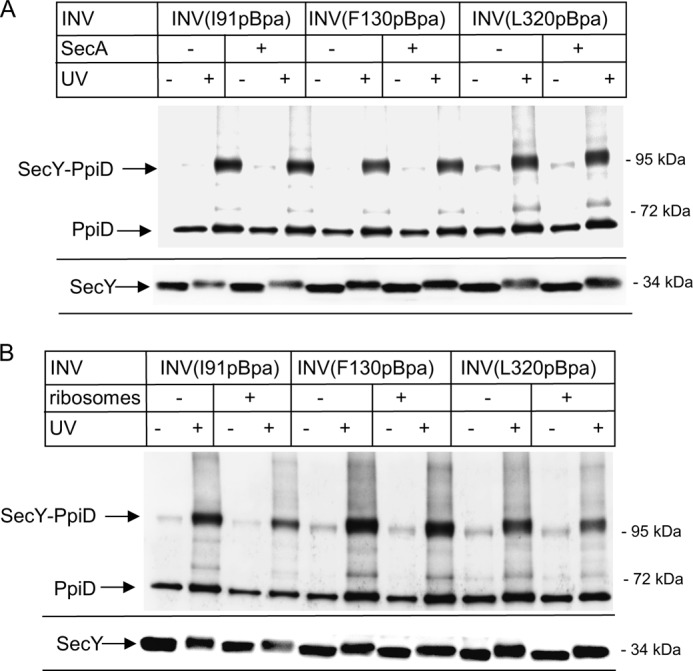
The SecY-PpiD interaction is not significantly influenced by SecA or by nontranslating ribosomes. INVs from BL21 cells with pBpa incorporated at the indicated positions were isolated by sucrose-gradient centrifugation. SecYpBpa INVs (0.6 μm SecY) were incubated with SecA (A) or ribosomes (B) in a 1:1 molar ratio, and the reaction mixture was UV-exposed or kept in the dark. Subsequently, SecY and its cross-linking products were purified via metal affinity chromatography separated on SDS-PAGE and analyzed by immune detection using α-PpiD antibodies. For monitoring the SecY concentration in these experiments, the lower part of the blot was also decorated with α-SecY antibodies.
SecA has been shown to activate the SecYEG translocon and to induce a partial opening of the lateral gate. However, the SecY-PpiD interaction was not influenced by the addition of SecA. For all three SecY(pBpa) derivatives tested, the addition of SecA did not change the efficiency of SecY-PpiD cross-linking (Fig. 5A). The amount of PpiD co-purifying with the SecY derivatives was also not influenced by the addition of SecA (Fig. 5A), suggesting that a partial opening of the lateral gate does not change the SecY-PpiD interaction. Western blotting using α-SecY antibodies confirmed that all samples contained similar amounts of SecY (Fig. 5A, lower panel). The slight UV-induced shift in the migration pattern of SecY is probably due to intramolecular cross-linking.
Like SecA, the addition of ribosomes induces a preactivation of the SecY translocon. Although Cryo-EM structures did not reveal significant changes at the SecY lateral gate upon ribosome binding (51, 52), electrophysiological experiments revealed that ribosomes induce an opening of the SecY channel, resulting in ion flux (53). The addition of sucrose gradient-purified 70 S ribosomes slightly weakened the SecY-PpiD interaction (Fig. 5B), but for all three pBpa positions, the SecY-PpiD cross-linking product was still detectable. In conclusion, neither SecA nor ribosome binding to SecY significantly influenced the interaction of PpiD with the lateral gate of SecY.
PpiD Is Displaced from the Lateral Gate of SecY in the Presence of Substrates
Complete lateral gate opening is probably required during protein transport for intercalating cleavable signal sequences of secretory proteins (46) and to allow transmembrane domains to enter the lipid phase of the membrane (52, 54). Recent Cryo-EM structures have visualized significant movements at the lateral gate after docking of RNCs onto SecY (52, 54). We therefore monitored the SecY-PpiD interaction in the presence of RNCs of the single spanning membrane protein FtsQ, which is targeted by SRP to the SecYEG translocon (28). In the presence of FtsQ-RNCs, the PpiD contact to helix 2b of SecY (SecY(I91pBpa)) almost completely disappeared, and a similar observation was also made for SecY(F130pBpa) (Fig. 6). Western blotting verified that comparable amounts of SecY were present in these experiments (Fig. 6, lower panel). This demonstrates that in the presence of a nascent membrane protein, PpiD loses contact to the lateral gate of SecY.
FIGURE 6.
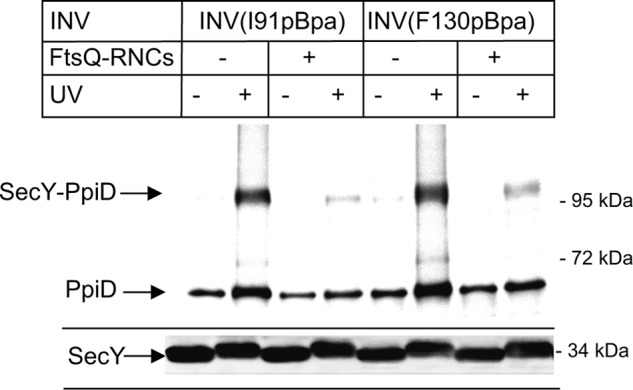
PpiD is displaced from the lateral gate of SecY in the presence FtsQ RNCs. SecYpBpa INVs (0.6 μm SecY) with pBpa incorporated at positions Ile-91 and Phe-130 were incubated with 0.6 μm affinity-purified FtsQ-RNCs and treated as described in the legend to Fig. 4.
DISCUSSION
The SecYEG translocon is a highly dynamic protein complex that associates with multiple cytosolic and membrane-bound partner proteins during protein transport. This is probably a prerequisite for its ability to handle the vast array of different protein substrates that have to be inserted into or translocated across the cytoplasmic membrane of bacteria. Although interactions with the SecA- and SRP-dependent targeting machineries are largely established, the interactions with partner proteins that facilitate protein folding or are involved in protein quality control are so far only ill-defined. Our study now presents four important details on the interaction of SecYEG with the periplasmic chaperone PpiD: 1) a fraction of PpiD is stably associated with SecYEG under native conditions; 2) PpiD is located in immediate vicinity to the lateral gate of SecY; 3) the SecY-PpiD interaction is only weakly influenced by a partial opening of the lateral gate, induced by either SecA or nontranslating ribosomes; and 4) complete opening of the lateral gate by the addition of FtsQ-RNCs displaces PpiD from the lateral gate.
PpiD is one of several chaperones in the periplasmic space of E. coli (30). It contains a periplasmic parvulin-like PPIase domain that is membrane-anchored via a single transmembrane helix. PpiD has been shown to bind peptides via its PPIase domain, but no catalytic activity has been detected (55). Instead, it was suggested that PpiD functions as a gatekeeper on the periplasmic side of the SecYEG translocon (31, 55). This was deduced from the observation that the periplasmic loop of a nascent membrane protein exiting the SecYEG translocon was cross-linked to PpiD (31). Our results now provide definite proof for a physical interaction between SecY and PpiD and localize PpiD close to the lateral gate of SecY. Contacts to PpiD were detected for several SecY residues that are deeply buried within the membrane, suggesting that the SecY-PpiD contact is mainly established between the single TM of PpiD and the lateral gate helices. The importance of the single TM of PpiD is underscored by the observation that a PpiD derivative lacking it is unable to suppress the synthetic lethality of a surA skp double knock-out (36).
The contacts between PpiD and SecY appear to be stronger for the two N-terminal lateral gate helices 2b and 3 of SecY. Although the intensity of cross-linking bands is not necessarily directly proportional to the strength of interaction, the N-terminal helices 2b and 3 were also preferentially cross-linked to YidC (26), and a stronger interaction of the N-terminal SecY helices with the trimeric SecDFYajC complex was also proposed (56). This probably indicates that the N-terminal half of SecY comprising TMs 1–5 provides the preferred interface for membrane-integral partner proteins. Binding of the transmembrane domain of PpiD to the lateral gate of SecY would localize the periplasmic PPIase domain in close proximity to the periplasmic vestibule of the SecY channel. PpiD could then facilitate the rapid release of secretory proteins or soluble domains of membrane proteins from the Sec channel, which would be in line with the observation that the release of the outer membrane OmpA from the periplasmic side of the cytoplasmic membrane was delayed in the ΔppiD strain (31).
The periplasmic domain of PpiD could also help sealing the SecYEG channel and thus preventing uncontrolled ion flux. In eukaryotic cells, uncontrolled ion flux via the Sec61 complex is restricted by binding of the Hsp70 homologue Bip to the lumenal side of Sec61 (57) and by binding of calmodulin to the cytoplasmic side of the Sec61 channel (58). Such a gating function could also be important in bacteria because it was recently shown in planar lipid bilayer studies that ion flow across SecYEG was induced by binding of nontranslating ribosomes to SecYEG (53). Nontranslating ribosomes have a significant affinity for the bacterial SecYEG (59–61), and thus uncontrolled ion flux needs to be controlled in vivo.
These SecY-associated functions of PpiD would argue against a specific role of PpiD in the assembly of outer membrane proteins, which is in agreement with a recent study (36). In E. coli, PpiD is not essential, indicating that other periplasmic proteins can functionally substitute PpiD (36, 45). PpiD could be replaced by Skp, which was also suggested to facilitate release of newly translocated substrates from the SecYEG channel (32, 33) or by one of the other three PPIases of the E. coli periplasm: SurA, PpiA, or FkpA (35). Functional redundancy appears to be a general concept in the bacterial chaperone network and has been shown, for example, for the trigger factor, which chaperones proteins exiting the ribosomal tunnel and which can be replaced by DnaK (62).
We have recently shown that the SecYEG-associated membrane protein YidC is located in front of the lateral gate of SecY (26). Our current data now show that PpiD contacts the same SecY residues as YidC, and it therefore appears rather unlikely that PpiD and YidC are simultaneously in contact with SecY. PpiD and YidC could compete for access to SecYEG, which is difficult to analyze in our experimental setting, because PpiD is up-regulated upon YidC depletion (63). The SecYEG translocon could also exist in two pools: one in contact with YidC and the other in contact with PpiD. However, the 5-fold cellular excess of YidC and PpiD over SecYEG probably rather favors a dynamic and substrate-dependent exchange between both proteins at the SecY lateral gate. This conclusion is also supported by recent BN-PAGE analyses, which indicate that SecYEG is in contact either with YfgM/PpiD or with YidC (50). The SecY-YidC contact is probably stabilized if transmembrane domains need to be lipid inserted, whereas the contact between SecY and PpiD is important for the folding of periplasmic proteins and periplasmic loops of membrane proteins. This concept concurs with the observation that on blue native PAGE, the SecYEG-YidC interaction is stabilized by the presence of transmembrane domains (47). It is also in line with our observation that the PpiD interaction with the SecY lateral gate is weakened by short (102 amino acids) RNCs of the membrane protein FtsQ, whereas the same RNCs fail to dissociate the SecY-YidC interaction (26). It is important to emphasize that RNCs dissociate PpiD from the lateral gate of SecY, but our data do not reveal whether PpiD loses contact to SecY completely. This needs to be analyzed in future studies. Finally, a substrate-dependent contact of SecY with either YidC or PpiD is supported by data showing a length-dependent cross-linking of the single-spanning membrane protein Momp2 to PpiD (31). Cross-links were observed for Momp2-RNCs of 220 amino acids in length, but not for Momp2-RNCs of 146 amino acids. In the longer construct, a large portion of the periplasmic domain of Momp2 is already translocated to the periplasm and therefore accessible to PpiD. Importantly, a dynamic exchange of interacting proteins is not only seen at the lateral gate of SecY, but also at its cytosolic loops, where FtsY, SecA, and the ribosome compete for overlapping binding sites during the targeting step of protein transport (13, 61). This supports the concept of a need-based association of the SecYEG channel with accessory subunits.
The presence of PpiD close to the lateral gate has strong implications for the definition of the holo-translocon. So far, the holo-translocon was defined as a complex comprising SecYEG, SecDFYajC, and YidC, and recent biochemical data have demonstrated that the SecYEG-SecDFYajC-YidC complex is more efficient in membrane protein insertion than the SecYEG core complex (56). Protein translocation across this holo-translocon also differs from translocation across the SecYEG core by being more dependent on the protein motive force (56). This indicates that although in vitro SecYEG is sufficient for protein transport of probably most SecA- and SRP-dependent proteins, the interaction with additional proteins enhances transport efficiency and allows the Sec translocon to adapt to the specific needs of a protein substrate. It is therefore probably not surprising that SecDFYajC and YidC are not the only proteins that associate with the Sec translocon. As a consequence, the organization of the holo translocon is probably highly dynamic.
A dynamic and need-based organization of protein transport complexes is also observed for the eukaryotic Sec61 complex (1), the mitochondrial TIM machinery (64), or the chloroplast TIC (translocase of the inner chloroplast envelope) import machinery (65). This probably provides the means to adjust protein transport to the specific folding and modification requirements of a given substrate.
Acknowledgments
We thank Diana Boy (Universität Freiburg) for performing the BN-PAGE, Michaela Fürst and Matthias Müller (Universität Freiburg) for α-PpiD antibodies, and Dan Daley (University of Stockholm) for the ΔppiD strain.
This work was supported by Deutsche Forschungsgemeinschaft Grants GRK1478, DFG-FOR967, and DFG-FOR929 (to H.-G. K.), funds from the Excellence Initiative of the German Federal and State Governments GSC-4 Spemann Graduate School of Biology and Medicine (to H.-G. K.), and a Ph.D. fellowship from the Deutscher Akademischer Austauschdienst (to N.-A. P.).
- SRP
- signal recognition particle
- INV
- inverted inner membrane vesicle
- pBpa
- para-benzoyl-l-phenylalanine
- RNC
- ribosome-associated nascent chain
- TM
- transmembrane domain
- PPIase
- peptidyl-prolylisomerase
- BN-PAGE
- blue native PAGE.
REFERENCES
- 1. Zimmermann R., Eyrisch S., Ahmad M., Helms V. (2011) Protein translocation across the ER membrane. Biochim. Biophys. Acta 1808, 912–924 [DOI] [PubMed] [Google Scholar]
- 2. Kudva R., Denks K., Kuhn P., Vogt A., Müller M., Koch H. G. (2013) Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534 [DOI] [PubMed] [Google Scholar]
- 3. du Plessis D. J., Nouwen N., Driessen A. J. (2011) The Sec translocase. Biochim. Biophys. Acta 1808, 851–865 [DOI] [PubMed] [Google Scholar]
- 4. Park E., Rapoport T. A. (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 [DOI] [PubMed] [Google Scholar]
- 5. Park E., Rapoport T. A. (2012) Bacterial protein translocation requires only one copy of the SecY complex in vivo. J. Cell Biol. 198, 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kedrov A., Kusters I., Krasnikov V. V., Driessen A. J. (2011) A single copy of SecYEG is sufficient for preprotein translocation. EMBO J. 30, 4387–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braig D., Mircheva M., Sachelaru I., van der Sluis E. O., Sturm L., Beckmann R., Koch H. G. (2011) Signal sequence-independent SRP-SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle. Mol. Biol. Cell 22, 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denks K., Vogt A., Sachelaru I., Petriman N. A., Kudva R., Koch H. G. (2014) The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31, 58–84 [DOI] [PubMed] [Google Scholar]
- 9. Akopian D., Shen K., Zhang X., Shan S. O. (2013) Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bange G., Sinning I. (2013) SIMIBI twins in protein targeting and localization. Nat. Struct. Mol. Biol. 20, 776–780 [DOI] [PubMed] [Google Scholar]
- 11. Angelini S., Boy D., Schiltz E., Koch H. G. (2006) Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J. Cell Biol. 174, 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Angelini S., Deitermann S., Koch H. G. (2005) FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 6, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuhn P., Weiche B., Sturm L., Sommer E., Drepper F., Warscheid B., Sourjik V., Koch H. G. (2011) The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic 12, 563–578 [DOI] [PubMed] [Google Scholar]
- 14. Akopian D., Dalal K., Shen K., Duong F., Shan S. O. (2013) SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J. Cell Biol. 200, 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frauenfeld J., Gumbart J., Sluis E. O., Funes S., Gartmann M., Beatrix B., Mielke T., Berninghausen O., Becker T., Schulten K., Beckmann R. (2011) Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 18, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zimmer J., Nam Y., Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manting E. H., van der Does C., Driessen A. J. (1997) In vivo cross-linking of the SecA and SecY subunits of the Escherichia coli preprotein translocase. J. Bacteriol. 179, 5699–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deitermann S., Sprie G. S., Koch H. G. (2005) A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J. Biol. Chem. 280, 39077–39085 [DOI] [PubMed] [Google Scholar]
- 19. Neumann-Haefelin C., Schäfer U., Müller M., Koch H. G. (2000) SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 19, 6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duong F., Wickner W. (1997) Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 16, 2756–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Echizen Y., Tsukazaki T., Dohmae N., Ishitani R., Nureki O. (2011) Crystallization and preliminary X-ray diffraction of the first periplasmic domain of SecDF, a translocon-associated membrane protein, from Thermus thermophilus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1367–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsukazaki T., Mori H., Echizen Y., Ishitani R., Fukai S., Tanaka T., Perederina A., Vassylyev D. G., Kohno T., Maturana A. D., Ito K., Nureki O. (2011) Structure and function of a membrane component SecDF that enhances protein export. Nature 474, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luirink J., von Heijne G., Houben E., de Gier J. W. (2005) Biogenesis of inner membrane proteins in Escherichia coli. Annu. Rev. Microbiol. 59, 329–355 [DOI] [PubMed] [Google Scholar]
- 24. Scotti P. A., Urbanus M. L., Brunner J., de Gier J. W., von Heijne G., van der Does C., Driessen A. J., Oudega B., Luirink J. (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., Dalbey R. E. (2000) YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 [DOI] [PubMed] [Google Scholar]
- 26. Sachelaru I., Petriman N. A., Kudva R., Kuhn P., Welte T., Knapp B., Drepper F., Warscheid B., Koch H. G. (2013) YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J. Biol. Chem. 288, 16295–16307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beck K., Eisner G., Trescher D., Dalbey R. E., Brunner J., Müller M. (2001) YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Houben E. N., Urbanus M. L., Van Der Laan M., Ten Hagen-Jongman C. M., Driessen A. J., Brunner J., Oudega B., Luirink J. (2002) YidC and SecY mediate membrane insertion of a Type I transmembrane domain. J. Biol. Chem. 277, 35880–35886 [DOI] [PubMed] [Google Scholar]
- 29. Dalbey R. E., Wang P., Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 [DOI] [PubMed] [Google Scholar]
- 30. Merdanovic M., Clausen T., Kaiser M., Huber R., Ehrmann M. (2011) Protein quality control in the bacterial periplasm. Annu. Rev. Microbiol. 65, 149–168 [DOI] [PubMed] [Google Scholar]
- 31. Antonoaea R., Fürst M., Nishiyama K., Müller M. (2008) The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47, 5649–5656 [DOI] [PubMed] [Google Scholar]
- 32. Schäfer U., Beck K., Müller M. (1999) Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274, 24567–24574 [DOI] [PubMed] [Google Scholar]
- 33. Harms N., Koningstein G., Dontje W., Muller M., Oudega B., Luirink J., de Cock H. (2001) The early interaction of the outer membrane protein PhoE with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J. Biol. Chem. 276, 18804–18811 [DOI] [PubMed] [Google Scholar]
- 34. Dartigalongue C., Raina S. (1998) A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17, 3968–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raivio T. L., Silhavy T. J. (2001) Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55, 591–624 [DOI] [PubMed] [Google Scholar]
- 36. Matern Y., Barion B., Behrens-Kneip S. (2010) PpiD is a player in the network of periplasmic chaperones in Escherichia coli. BMC Microbiol. 10, 251–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanahan D. (1983) Studies on transfromation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 38. Ryu Y., Schultz P. G. (2006) Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3, 263–265 [DOI] [PubMed] [Google Scholar]
- 39. Baba T., Jacq A., Brickman E., Beckwith J., Taura T., Ueguchi C., Akiyama Y., Ito K. (1990) Characterization of cold-sensitive secY mutants of Escherichia coli. J. Bacteriol. 172, 7005–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Drew D., Fröderberg L., Baars L., de Gier J. W. (2003) Assembly and overexpression of membrane proteins in Escherichia coli. Biochim. Biophys. Acta 1610, 3–10 [DOI] [PubMed] [Google Scholar]
- 41. Braig D., Bär C., Thumfart J. O., Koch H. G. (2009) Two cooperating helices constitute the lipid-binding domain of the bacterial SRP receptor. J. Mol. Biol. 390, 401–413 [DOI] [PubMed] [Google Scholar]
- 42. Koch H. G., Hengelage T., Neumann-Haefelin C., MacFarlane J., Hoffschulte H. K., Schimz K. L., Mechler B., Müller M. (1999) In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10, 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seidelt B., Innis C. A., Wilson D. N., Gartmann M., Armache J. P., Villa E., Trabuco L. G., Becker T., Mielke T., Schulten K., Steitz T. A., Beckmann R. (2009) Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326, 1412–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moran U., Phillips R., Milo R. (2010) SnapShot: Key Numbers in Biology. Cell 141, 1262. [DOI] [PubMed] [Google Scholar]
- 45. Weski J., Ehrmann M. (2012) Genetic analysis of 15 protein folding factors and proteases of the Escherichia coli Cell envelope. J. Bacteriol. 194, 3225–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hizlan D., Robson A., Whitehouse S., Gold V. A., Vonck J., Mills D., Kühlbrandt W., Collinson I. (2012) Structure of the SecY complex unlocked by a preprotein mimic. Cell Rep. 1, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boy D., Koch H. G. (2009) Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol. Biol. Cell 20, 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maddalo G., Stenberg-Bruzell F., Götzke H., Toddo S., Björkholm P., Eriksson H., Chovanec P., Genevaux P., Lehtiö J., Ilag L. L., Daley D. O. (2011) Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome Res. 10, 1848–1859 [DOI] [PubMed] [Google Scholar]
- 49. Duong F. (2003) Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 22, 4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Götzke H., Palombo I., Muheim C., Perrody E., Genevaux P., Kudva R., Müller M., Daley D. O. (2014) YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. J. Biol. Chem. 27, 19089–19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Becker T., Bhushan S., Jarasch A., Armache J. P., Funes S., Jossinet F., Gumbart J., Mielke T., Berninghausen O., Schulten K., Westhof E., Gilmore R., Mandon E. C., Beckmann R. (2009) Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park E., Ménétret J. F., Gumbart J. C., Ludtke S. J., Li W., Whynot A., Rapoport T. A., Akey C. W. (2014) Structure of the SecY channel during initiation of protein translocation. Nature 506, 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Knyazev D. G., Lents A., Krause E., Ollinger N., Siligan C., Papinski D., Winter L., Horner A., Pohl P. (2013) The bacterial translocon SecYEG opens upon ribosome binding. J. Biol. Chem. 288, 17941–17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gogala M., Becker T., Beatrix B., Armache J. P., Barrio-Garcia C., Berninghausen O., Beckmann R. (2014) Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107–110 [DOI] [PubMed] [Google Scholar]
- 55. Weininger U., Jakob R. P., Kovermann M., Balbach J., Schmid F. X. (2010) The prolyl isomerase domain of PpiD from Escherichia coli shows a parvulin fold but is devoid of catalytic activity. Protein Sci. 19, 6–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schulze R. J., Komar J., Botte M., Allen W. J., Whitehouse S., Gold V. A., M., Lycklama A., Nijeholt J. A., Huard K., Berger I., Schaffitzel C., Collinson I. (2014) Membrane protein insertion and proton-motive-force dependent aecretion through the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Proc. Natl. Acad. Sci. 111, 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schäuble N., Lang S., Jung M., Cappel S., Schorr S., Ulucan Ö., Linxweiler J., Dudek J., Blum R., Helms V., Paton A. W., Paton J. C., Cavalié A., Zimmermann R. (2012) BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 31, 3282–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Erdmann F., Schäuble N., Lang S., Jung M., Honigmann A., Ahmad M., Dudek J., Benedix J., Harsman A., Kopp A., Helms V., Cavalié A., Wagner R., Zimmermann R. (2011) Interaction of calmodulin with Sec61a limits Ca2+ leakage from the endoplasmic reticulum. EMBO J. 30, 17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Prinz A., Behrens C., Rapoport T. A., Hartmann E., Kalies K. U. (2000) Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 19, 1900–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schaletzky J., Rapoport T. A. (2006) Ribosome binding to and dissociation from translocation sites of the endoplasmic reticulum membrane. Mol. Biol. Cell 17, 3860–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu Z. C., de Keyzer J., Kedrov A., Driessen A. J. (2012) Competitive binding of the SecA ATPase and ribosomes to the SecYEG translocon. J. Biol. Chem. 287, 7885–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Deuerling E., Patzelt H., Vorderwülbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B. (2003) Trigger factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 47, 1317–1328 [DOI] [PubMed] [Google Scholar]
- 63. Wickström D., Wagner S., Simonsson P., Pop O., Baars L., Ytterberg A. J., van Wijk K. J., Luirink J., de Gier J. W. (2011) Characterization of the consequences of YidC depletion on the inner membrane proteome of E. coli using 2D blue native/SDS-PAGE. J. Mol. Biol. 409, 124–135 [DOI] [PubMed] [Google Scholar]
- 64. Becker T., Böttinger L., Pfanner N. (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci. 37, 85–91 [DOI] [PubMed] [Google Scholar]
- 65. Di Cola A., Klostermann E., Robinson C. (2005) The complexity of pathways for protein import into thylakoids: it's not easy being green. Biochem. Soc. Trans. 33, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 66. Koch H. G., Moser M., Schimz K. L., Muller M. (2002) The integration of YidC into the cytoplasmic membrane of Escherichia coli requires the signal recognition particle, SecA and SecYEG. J. Biol. Chem. 277, 5715–5718 [DOI] [PubMed] [Google Scholar]
- 67. Welte T., Kudva R., Kuhn P., Sturm L., Braig D., Müller M., Warscheid B., Drepper F., Koch H. G. (2012) Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol. Biol. Cell 23, 464–479 [DOI] [PMC free article] [PubMed] [Google Scholar]