Abstract
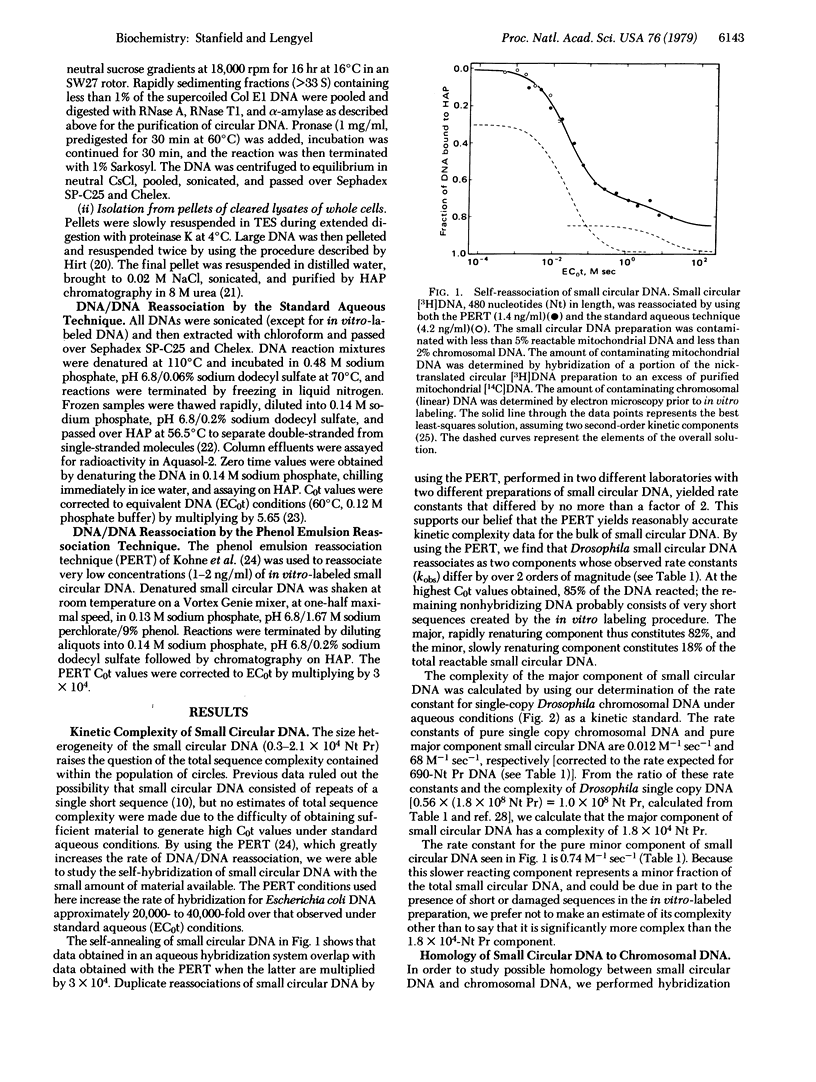
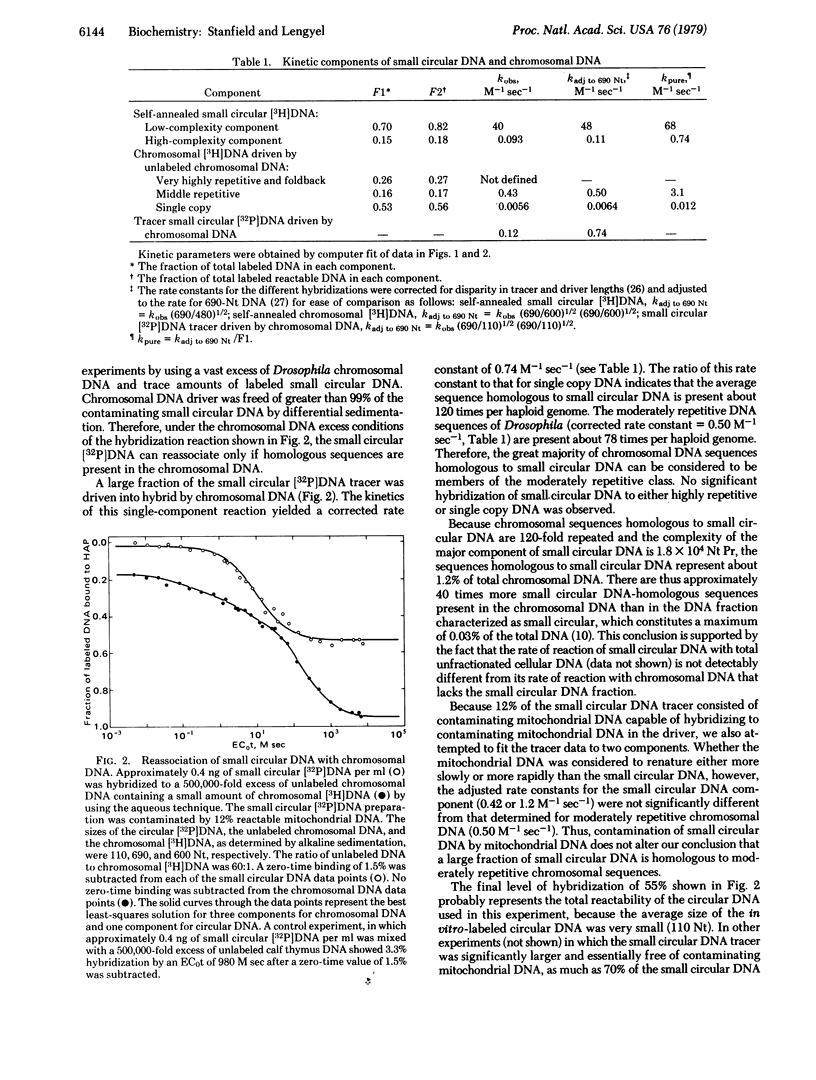
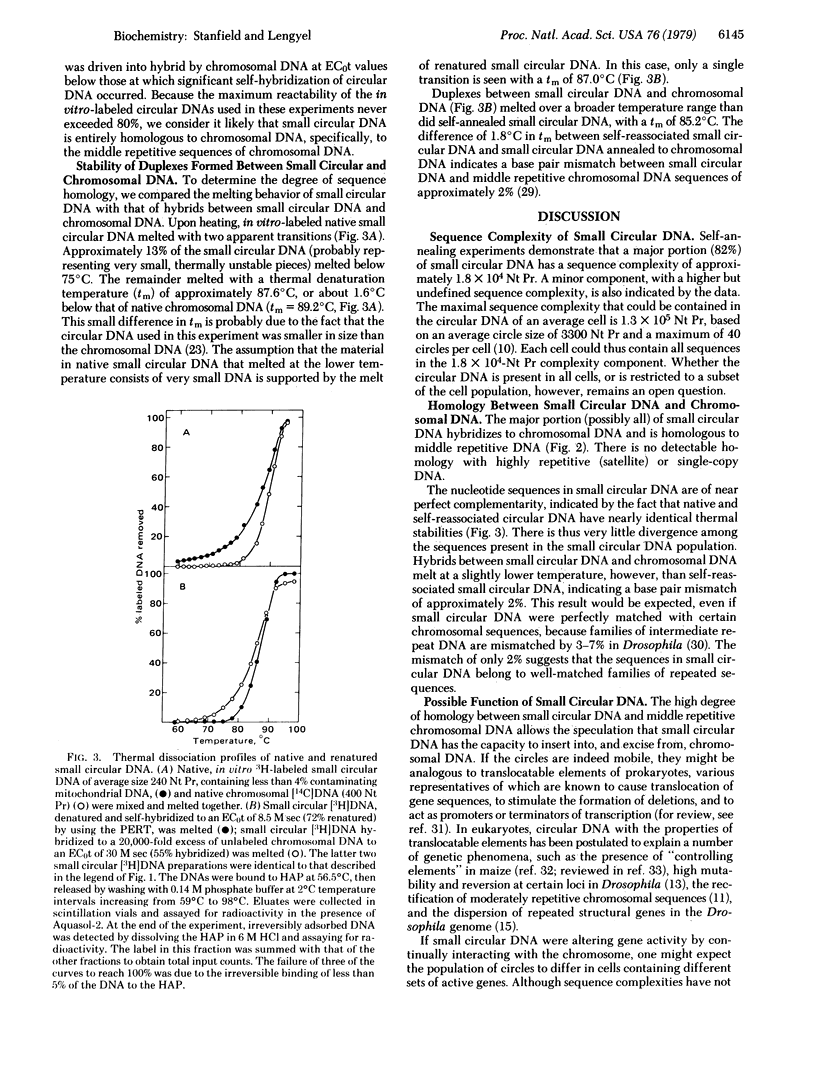
Nucleic acid reassociation techniques were used to determine the kinetic complexity of small circular DNA in cultured cells of Drosophila melanogaster. Two kinetic components are present. One of these constitutes 82% of the mass of the circular DNA and has a complexity of 1.8 x 10(4) nucleotide pairs; the other constitutes 18% of the mass and the a significantly higher but undefined sequence complexity. We have demonstrated that these circular molecules hybridize to middle repetitive chromosomal sequences by hybridization of in vitro-labeled circular DNA tracer with a vast excess of Drosophila chromosomal DNA. Thermal stability measurements indicate that base-pair mismatch between small circular DNA and middle repetitive chromosomal DNA does not exceed 2%. We discuss possible functions of these small circular DNAs in light of the above findings.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agsteribbe E., Kroon A. M., van Bruggen E. F. Circular DNA from mitochondria of Neurospora crassa. Biochim Biophys Acta. 1972 May 10;269(2):299–303. doi: 10.1016/0005-2787(72)90439-x. [DOI] [PubMed] [Google Scholar]
- Billheimer F. E., Avers C. J. Nuclear and mitochondrial DNA from wild-type and petite yeast: circularity, length, and buoyant density. Proc Natl Acad Sci U S A. 1969 Oct;64(2):739–746. doi: 10.1073/pnas.64.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner T. I., Brenner D. J., Neufeld B. R., Britten R. J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973 Dec 5;81(2):123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M., Amaldi F., Lava-Sanchez P. A. Electron microscope analysis of amplifying ribosomal DNA from Xenopus laevis. Exp Cell Res. 1976 Mar 1;98(1):95–103. doi: 10.1016/0014-4827(76)90467-5. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. E., Galau G. A., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: V. Effects of disparity in tracer and driver fragment lengths. Nucleic Acids Res. 1978 Jun;5(6):2073–2094. doi: 10.1093/nar/5.6.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Botchan P. Sequences homologous to ribosomal insertions occur in the Drosophila genome outside the nucleolus organizer. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4233–4237. doi: 10.1073/pnas.74.10.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLap R. J., Rush M. G. Change in quantity and size distribution of small circular DNAs during development of chicken bursa. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5855–5859. doi: 10.1073/pnas.75.12.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLap R. J., Rush M. G., Zouzias D., Khan S. Isolation and preliminary characterization of the small circular DNA present in African green monkey kidney (BSC-1) cells. Plasmid. 1978 Sep;1(4):508–521. doi: 10.1016/0147-619x(78)90008-2. [DOI] [PubMed] [Google Scholar]
- Fincham J. R., Sastry G. R. Controlling elements in maize. Annu Rev Genet. 1974;8:15–50. doi: 10.1146/annurev.ge.08.120174.000311. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Glover D. M. Cloned segment of Drosophila melanogaster rDNA containing new types of sequence insertion. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4932–4936. doi: 10.1073/pnas.74.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovsky M. D., Ivano Y. N., Green M. M. Genetic instability in Drosophila melanogaster: putative multiple insertion mutants at the singed bristle locus. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2973–2975. doi: 10.1073/pnas.74.7.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani F., Caizzi R., Gargano S. Circular ribosomal DNA during ribosomal magnification in Drosophila melanogaster. J Mol Biol. 1977 May 5;112(1):49–63. doi: 10.1016/s0022-2836(77)80155-1. [DOI] [PubMed] [Google Scholar]
- Green M. M. Genetic instability in Drosophila melanogaster: De novo induction of putative insertion mutations. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3490–3493. doi: 10.1073/pnas.74.8.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOTTA Y., BASSEL A. MOLECULAR SIZE AND CIRCULARITY OF DNA IN CELLS OF MAMMALS AND HIGHER PLANTS. Proc Natl Acad Sci U S A. 1965 Feb;53:356–362. doi: 10.1073/pnas.53.2.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Kohne D. E., Levison S. A., Byers M. J. Room temperature method for increasing the rate of DNA reassociation by many thousandfold: the phenol emulsion reassociation technique. Biochemistry. 1977 Nov 29;16(24):5329–5341. doi: 10.1021/bi00643a026. [DOI] [PubMed] [Google Scholar]
- Meinke W., Goldstein D. A., Hall M. R. Rapid isolation of mouse DNA from cells in tissue culture. Anal Biochem. 1974 Mar;58(1):82–88. doi: 10.1016/0003-2697(74)90444-8. [DOI] [PubMed] [Google Scholar]
- Nass M. M., Ben-Shaul Y. A novel closed circular duplex DNA in bleached mutant and green strains of Euglena gracilis. Biochim Biophys Acta. 1972 Jun 22;272(1):130–136. doi: 10.1016/0005-2787(72)90041-x. [DOI] [PubMed] [Google Scholar]
- Ono T., Ozeki Y., Okubo S., Inoki S. Characterization of nuclear and satellite DNA from trypanosomes. Biken J. 1971 Sep;14(3):203–215. [PubMed] [Google Scholar]
- Pearson W. R., Davidson E. H., Britten R. J. A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 1977 Jun;4(6):1727–1737. doi: 10.1093/nar/4.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasch E. M., Barr H. J., Rasch R. W. The DNA content of sperm of Drosophila melanogaster. Chromosoma. 1971;33(1):1–18. doi: 10.1007/BF00326379. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972 Apr;27(2):353–365. [PubMed] [Google Scholar]
- Smith C. A., Vinograd J. Small polydisperse circular DNA of HeLa cells. J Mol Biol. 1972 Aug 21;69(2):163–178. doi: 10.1016/0022-2836(72)90222-7. [DOI] [PubMed] [Google Scholar]
- Stanfield S., Helinski D. R. Small circular DNA in Drosophila melanogaster. Cell. 1976 Oct;9(2):333–345. doi: 10.1016/0092-8674(76)90123-9. [DOI] [PubMed] [Google Scholar]
- Wensink P. C. Sequence homology within families of Drosophila melanogaster middle repetitive DNA. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1033–1039. doi: 10.1101/sqb.1978.042.01.103. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wong F. Y., Wildman S. G. Simple procedure for isolation of satellite DNA's from tobacco leaves in high yield and demonstration of minicircles. Biochim Biophys Acta. 1972 Jan 18;259(1):5–12. doi: 10.1016/0005-2787(72)90468-6. [DOI] [PubMed] [Google Scholar]


