Abstract
Identifying gene mutations in individual tumors is critical to improve the efficacy of cancer therapy by matching targeted drugs to specific mutations. Gastrointestinal stromal tumors (GIST) are stromal or mesenchymal subepithelial neoplasms affecting the gastrointestinal tract and frequently contain activating gene mutations in either KIT or platelet-derived growth factor A (PDGFRA). Although GIST is highly responsive to several selective tyrosine kinase inhibitors, combined use of inhibitors targeting other mutations is needed to further prolong survival in patients with GIST. In this study, we aim to screen and identify genetic mutations in GIST for targeted therapy using the new Ion Torrent next-generation sequencing platform. Utilizing the Ion Ampliseq Cancer Panel, we sequenced 737 loci from 45 cancer-related genes using DNA extracted from formalin-fixed and paraffin-embedded (FFPE) samples of 121 human gastrointestinal stromal tumors, set up stringent parameters for reliable variant calling by filtering out potential raw base calling errors, and identified frequent mutations in the KIT gene. This study demonstrates the utility of using Ion Torrent sequencing to efficiently identify human cancer mutations. This may provide a molecular basis for clinically developing new drugs targeting these gene mutations for GIST therapy.
Gastrointestinal stromal tumor (GIST) is a stromal or mesenchymal subepithelial neoplasm affecting the gastrointestinal (GI) tract. Population-based studies showed an annual incidence of 14.5 per million in Swedish, 11 per million in Icelandic1, 11.1 per million in French, 19.6 per million in Swiss2, and 14.2 per million in Italian populations. Epidemiologic studies show the annual incidence of GIST in United States to be at least 4,000 to 6,000 new cases per year, or roughly 7 to 20 cases per million people3,4. While GISTs may occur at any age, they are rare in children. GISTs commonly originate from the stomach (55%), small intestine (35%), and rectum (5%). Esophageal and colonic GISTs are rare, and these tumors also rarely occur outside the alimentary tract such as in the omentum, mesentery, and peritoneum, and are called extragastrointestinal GISTs or “E-GISTs.” GIST may originate from interstitial cells of Cajal (ICC) or from their stem cell-like precursors, although this is not certain5,6. Because of their relatively broad morphologic spectrum, GISTs were formerly called leiomyomas, leiomyosarcomas, and leiomyoblastomas of the gastrointestinal tract, until they were found to have clinical, histopathological, and molecular biological features that differentiated them from other soft tissue tumors.
GIST frequently contains oncogenic mutations in one of two receptor tyrosine kinases: KIT or PDGFRA (platelet-derived growth factor receptor alpha)7,8. KIT and PDFGRA proteins are growth factor receptors, which are activated by ligands such as PDGF-AA and stem cell factor, respectively triggering cell pathways that up-regulate proliferation, down-regulate apoptosis, and control cell differentiation, adhesion, and motility in normal conditions. Approximately 95% of GISTs express the CD117 antigen, an epitope of the KIT receptor tyrosine kinase7,9, hence the most commonly used marker for GIST is CD117. Mutations of KIT and PDGFRA lead to constitutive activation of these cell pathways leading to spontaneous proliferation and uncontrolled growth of a tumor. The downstream events following activation of mutant KIT or mutant PDGFRA are very similar10,11. Different mutations can be found in different exons or in different regions of a single exon occurring as point mutations, deletions and insertions in the KIT (exon: 9, 11, 13 and 17) and PGFRA (exon: 12, 14 and 18) genes. However, some GISTs have no detectable KIT or PDGFRA mutations and fewer than 5% of GIST occur as symptoms of syndromic diseases, such as neurofibromatosis type 1 (NF1), Carney triad syndrome, and other familial diseases8,12.
Accurate detection of mutations in GIST is critical for targeted therapy with drugs, such as KIT/PDGFRA tyrosine kinase inhibitors (TKI)8,13,14. Next-generation sequencing technologies have revolutionized cancer genomics research by providing an unbiased and comprehensive method of detecting somatic cancer genome alterations15. These technologies have several advantages over Sanger sequencing by capillary electrophoresis, including the ability to sequence gigabases of nucleotides to detect individually unique mutations16. However, routine usage of these technologies leaves us with several limitations such as the cost of entry, long processing time, and sample scalability. A new sequencing technology, Ion Torrent (Life Technologies, Carlsbad, CA, USA), has substantially circumvented many of these issues. The Ion Torrent method relies on standard DNA polymerase sequencing with unmodified dNTPs and uses semiconductor-based detection of hydrogen ions released during every cycle of DNA polymerization17. Each nucleotide incorporation into the growing complementary DNA strand causes the release of a hydrogen ion that is sensed by a hypersensitive ion sensor17. Ion Torrent Personal Genome Machine (PGM) can currently generate 10–100 Mb pairs (Mbp) of sequence data on various chips within several hours of instrument's run time. In this study, we sequenced 737 loci of 45 cancer-related genes from 121 human gastrointestinal stromal tumors using the Ion Ampliseq Cancer Panel, set up stringent parameters for reliable variant calling by filtering out potential raw base calling errors, and found frequent missense mutations in KIT gene consistent to that of other reports.
Results
Mutation analysis of human gastrointestinal stromal tumors (GISTs) with Ion Ampliseq Cancer Panel
The sample set for this study was GISTs from 121 Chinese patients, and 76 samples were of gastric origin and 45 samples had an intestinal origin (Table 1). Ion Ampliseq technology was used to identify mutations in 189 amplicons covering 737 loci of 45 tumor suppressor genes and oncogenes in these tumor samples. The sequenced data were processed and mutations identified using Torrent Suite Software v3.0 with a plug-in “variant caller.” To generate reliable variant calling, three filtering steps were used to eliminate erroneous base calling (Supplementary figure 1), as described in the Materials and Methods. The mean read length was 74 bp, and the average sequence per sample was approximately 27 Mb. With normalization to 300,000 reads per specimen, there was an average of 1,630 reads per amplicon (range: 174 to 4024) (Supplemental figure 2A), and 183/189 (96.8%) amplicons averaged at least 100 reads, and 177/189 (93.7%) amplicons averaged at least 300 reads (Supplemental figure 2B).
Table 1. Mutations (including Missense point mutations/deletion/insertion) frequencies in 45 genes (737 loci) in female and male GISTs. The p-value of Fisher exact test result of KIT mutations in female and male is 0.429.
| Genes | Number of samples with mutations (Mutation frequency in 121 samples) | Number of female samples with mutations (Mutation frequency in 59 female samples) | Number of male samples with mutations (Mutation frequency in 62 male samples) |
|---|---|---|---|
| ABL1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| AKT1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ALK | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| APC | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ATM | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| BRAF | 1(0.8%) | 0(0.0%) | 1(1.6%) |
| CDH1 | 1(0.8%) | 0(0.0%) | 1(1.6%) |
| CDKN2A | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| CSF1R | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| CTNNB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| EGFR | 1(0.8%) | 1(1.7%) | 0(0.0%) |
| ERBB2 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ERBB4 | 1(0.8%) | 1(1.7%) | 0(0.0%) |
| FBXW7 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR2 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR3 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FLT3 | 3(2.5%) | 2(3.4%) | 1(1.6%) |
| GNAS | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| HNF1A | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| HRAS | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| IDH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| JAK3 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| KDR | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| KIT | 60(49.6%) | 25(42.4%) | 35(56.5%) |
| KRAS | 1(0.8%) | 1(1.7%) | 0(0.0%) |
| MET | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MLH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MPL | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NOTCH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NPM1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NRAS | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PDGFRA | 1(0.8%) | 1(1.7%) | 0(0.0%) |
| PIK3CA | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PTEN | 1(0.8%) | 1(1.7%) | 0(0.0%) |
| PTPN11 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| RB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| RET | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMAD4 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMARCB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMO | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SRC | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| STK11 | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| TP53 | 1(0.8%) | 0(0.0%) | 1(1.6%) |
| VHL | 1(0.8%) | 0(0.0%) | 1(1.6%) |
Identification of frequent mutations in a set of cancer-related genes in GISTs
The incurred mutations were of both gastric and intestinal origin (Supplementary Table 1). Of the 45 oncogenes and tumor suppressor genes sequenced in the 121 GISTs from Chinese patients, mutations were detected in the genes BRAF, CDH1, EGFR, ERBB4, FLT3, KRAS, PDGFRA, PTEN, TP53, and VHL, and the most common mutations occurred in the KIT gene (Table 1). A detailed list of missense, point mutations, insertions, and deletions identified in this study is provided in Supplementary Table 2.
KIT mutations in GISTs
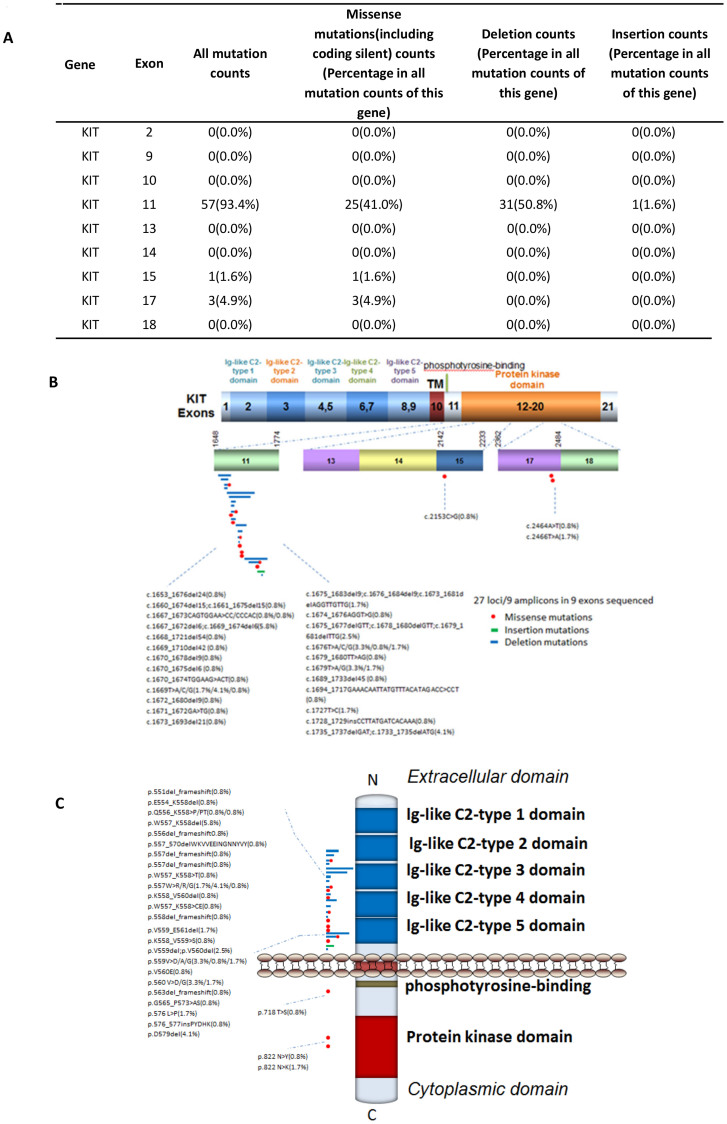
Accumulating evidence indicates that activating mutations of tyrosine kinases KIT or PDGFRA are the initiating event in GISTs. Activation of KIT or PDGFR leads to downstream signaling in the PI3K, Ras, and Jak/Stat pathways resulting in increased cell proliferation and inhibition of apoptosis18. Accordingly, 60 of the 121 GIST tumors sequenced in this study had KIT gene mutations: exons 2, 9, 10, 11, 13, 14, 15, 17, and 18 were found to be mutated along its transmembrane and cytoplasmic domain but not along its extracellular domain (Fig. 1A–C). Mutations of exon 11 (94.4%), consisting of the regulatory domain of the enzyme included nucleotide deletions in 31 cases (50.80%), missense mutations in 25 cases (41.0%) and insertion in 1 case. Different size deletions in Kit can affect KIT intron 10- exon-11 splice-acceptor sites. Exon 17 encoding the kinase activation loop was mutated in only 3 cases (4.9%) with a substitution of A2466 for T2466 or a substitution of T2464 for A2464. These data support the critical role of KIT mutations in the development and progression of GISTs.
Figure 1. Mutation distribution in the exons and functional domains of KIT.
(A). Frequencies of detected mutations in different exons. (B). Mutation distribution in exons. (C). Mutation distribution in functional domains.
Multiple mutations and mutation hotspots in GISTs
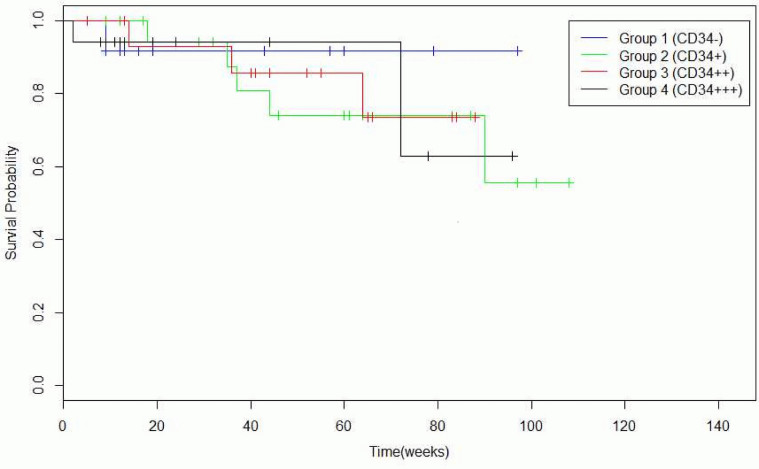
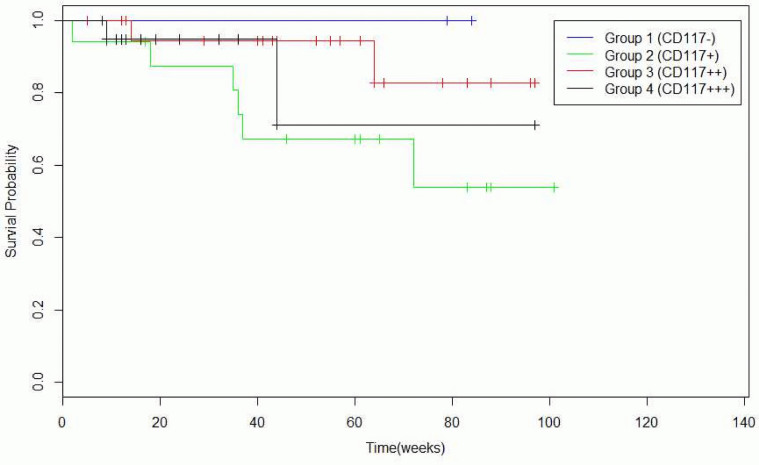
Identifying single or combination of mutations for delivering individualized treatment with a single or combination of target agents against the detected mutational combinations has been an effective cancer therapy in the recent years. 54.5% of samples had at least one or more missense mutations, 5.8% had at least two or more missense mutations, and 45.5% of samples incurred no deleterious mutations (Table 2). It is known that functional SNPs may contribute to the development of GISTs, and Table 3 and Table 4 illustrate the incurrence of SNPs in combination with other mutations in our sample set. CD34 and CD117 expression levels can help to predict the prognosis of GIST patients, and mutations found in CD117+ and CD117− GISTs and CD34+ and CD34− GISTs are summarized in Table 5 and Table 6, respectively. There was a significant reduction in the survival rate between patients with one CD34 mutation compared to patients with no CD34 mutations (Table 7 and Fig. 2), and between patients with triple CD117 mutations compared to those with double CD117 mutations (Table 8 and Fig. 3).
Table 2. Single and multiple mutations in 121 GISTs.
| Mutations combination (including missense point mutations/deletion/insertion) | Number of samples with mutation combination | Percentage in all sequenced samples |
|---|---|---|
| Single and more | 66 | 54.50% |
| Double and more | 7 | 5.80% |
| Three and more | 0 | 0.00% |
| No missense, deletion, insert or substitution-nonsense | 55 | 45.50% |
Table 3. Single and multiple mutations and SNPs in 121 GISTs.
| Mutations and SNPs combination (including missense point mutation/deletion/insertion/SNP) | Number of samples with mutation/snp combination | Percentage in all sequenced samples |
|---|---|---|
| Single and more | 88 | 72.70% |
| Double and more | 30 | 24.80% |
| Three and more | 4 | 3.30% |
| No missense, deletion, insert or substitution-nonsense | 33 | 27.30% |
Table 4. List of multiple mutations and SNPs within more than one GIST patients.
| Genes with Mutations combination | CDS and amino acid change | Numbers of patients with this mutation combination |
|---|---|---|
| MLH1 SNP and KIT mutation | c.1151T > A p.V384D;c.1667_1672del6/c.1669_1674del6 p.W557_K558del | 3 |
| MLH1 SNP and KIT SNP | c.1151T > A p.V384D;c.1621A > C p.M541L | 2 |
| KIT SNP and KIT mutation | c.1621A > C p.M541L;c.1735_1737delGAT/c.1733_1735delATG p.D579del | 2 |
| MLH1 SNP and MET SNP | c.1151T > A p.V384D;c.1124A > G p.N375S | 2 |
Table 5. Mutations (including Missense point mutations/deletion/insertion) frequencies in 45 genes (737 loci) in CD117+ and CD117− GISTs.
| Genes | Number of samples with mutations (Mutation frequency in 121 samples) | Number of unknown samples with mutations (Mutation frequency in 57 unknown samples) | Number of CD117(+) samples with mutations (Mutation frequency in 17 CD117(+) samples) | Number of CD117(++) samples with mutations (Mutation frequency in 23 CD117(++) samples) | Number of CD117(+++) samples with mutations (Mutation frequency in 21 CD117(+++) samples) | Number of CD117(−) samples with mutations (Mutation frequency in 3 CD117(−) samples) |
|---|---|---|---|---|---|---|
| ABL1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| AKT1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ALK | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| APC | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ATM | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| BRAF | 1(0.8%) | 0(0.0%) | 1(5.9%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| CDH1 | 1(0.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 1(4.8%) | 0(0.0%) |
| CDKN2A | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| CSF1R | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| CTNNB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| EGFR | 1(0.8%) | 0(0.0%) | 1(5.9%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ERBB2 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ERBB4 | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FBXW7 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR2 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR3 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FLT3 | 3(2.5%) | 0(0.0%) | 1(5.9%) | 0(0.0%) | 2(9.5%) | 0(0.0%) |
| GNAS | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| HNF1A | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| HRAS | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| IDH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| JAK3 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| KDR | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| KIT | 60(49.6%) | 27(47.4%) | 8(47.1%) | 12(52.2%) | 12(57.1%) | 1(33.3%) |
| KRAS | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MET | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MLH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MPL | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NOTCH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NPM1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NRAS | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PDGFRA | 1(0.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 1(4.8%) | 0(0.0%) |
| PIK3CA | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PTEN | 1(0.8%) | 0(0.0%) | 1(5.9%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PTPN11 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| RB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| RET | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMAD4 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMARCB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMO | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SRC | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| STK11 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| TP53 | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| VHL | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
Table 6. Mutations (including Missense point mutations/deletion/insertion) frequencies in 45 genes (737 loci) in CD34+ and CD34− GISTs.
| Genes | Number of samples with mutations in 121 samples (Mutation frequency) | Number of unknown samples with mutations (Mutation frequency in 56 unknown samples) | Number of CD34(+) samples with mutations (Mutation frequency in 20 CD34(+) samples) | Number of CD34(++) samples with mutations (Mutation frequency in 16 CD34(++) samples) | Number of CD34(+++) samples with mutations (Mutation frequency in 17 CD34(+++) samples) | Number of CD34(−) samples with mutations (Mutation frequency in 12 CD34(−) samples) |
|---|---|---|---|---|---|---|
| ABL1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| AKT1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ALK | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| APC | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ATM | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| BRAF | 1(0.8%) | 0(0.0%) | 0(0.0%) | 1(6.2%) | 0(0.0%) | 0(0.0%) |
| CDH1 | 1(0.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 1(10.0%) |
| CDKN2A | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| CSF1R | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| CTNNB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| EGFR | 1(0.8%) | 0(0.0%) | 1(5.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ERBB2 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| ERBB4 | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FBXW7 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR2 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FGFR3 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| FLT3 | 3(2.5%) | 0(0.0%) | 1(5.0%) | 1(6.2%) | 1(5.9%) | 0(0.0%) |
| GNAS | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| HNF1A | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| HRAS | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| IDH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| JAK3 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| KDR | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| KIT | 60(49.6%) | 26(46.4%) | 11(55.0%) | 9(56.2%) | 9(52.9%) | 5(41.7%) |
| KRAS | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MET | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MLH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| MPL | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NOTCH1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NPM1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| NRAS | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PDGFRA | 1(0.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 1(5.9%) | 0(0.0%) |
| PIK3CA | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PTEN | 1(0.8%) | 0(0.0%) | 1(5.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| PTPN11 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| RB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| RET | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMAD4 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMARCB1 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SMO | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| SRC | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| STK11 | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| TP53 | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| VHL | 1(0.8%) | 1(1.8%) | 0(0.0%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
Table 7. Survival time of patients with CD34+ and CD34− GISTs.
| Number of samples | Survival time (average, range) | P-value (log-rank test) | |
|---|---|---|---|
| CD34(−) | 12 | 38 (9–97+ months) | CD34− and CD34+: 0.526 |
| CD34(+) | 20 | 55 (9+–108+ months) | |
| CD34(++) | 16 | 52 (5+–88+ months) | CD34++ and CD34+++: 0.935 |
| CD34(+++) | 17 | 29 (2–96+ months) |
Figure 2. Survival probability in weeks of patients with CD34+ and CD34− GISTs.
Table 8. Survival time of patients with CD117+ and CD117− GISTs.
| Number of samples | Survival time (average, range) | P-value(log-rank test) | |
|---|---|---|---|
| CD117(−) | 3 | 58 (12+–84+ months) | |
| CD117(+) | 17 | 52 (2–101+ months) | |
| CD117(++) | 23 | 49 (5+–97+ months) | CD117++ and CD117+++: 0.367 |
| CD117(+++) | 21 | 24 (8+–97+ months) |
Figure 3. Survival probabiliy in weeks of patients with CD117+ and CD117− GISTs.
Discussion
Cancer is an accumulation of many genetic changes within cells, and mutation patterns vary widely among different types of cancers and among patients with the same cancer. These mutations can be used to direct targeted drug therapy, an approach has significantly changed the treatment of cancer over the past 10 years. Drugs like small-molecule inhibitors or monoclonal antibodies that target these genetic alterations specifically have been developed, and such drugs act as a potential therapy for many common malignancies and are generally better tolerated with fewer side effects over the traditional cytotoxic chemotherapy.
Since the development of targeted therapy in the last decade, screening the genetic mutations of cancer cells has been of keen interest to direct the use of targeted anticancer drugs. Because of this, identifying mutations in each individual tumor can be highly beneficial to the patient, but has yet to be a feasible option. The first near-complete human genome sequence assembly required more than 12 years of sequencing and more than $3 billion19. The urgent need for alternative sequencing approaches employing a lower cost and faster pace led to the development of Next generation sequencing (NGS) methods such as Illumina HiSeq and Miseq, 454 Pyrosequencing, Helicos Heliscope, SOLiD Sequencing, and Ion Torrent Sequencing20,21,22. These new next-generation sequencing strategies are believed to enable high-throughput screening and detection of cancer cell genomes and epigenomes at much faster rates and reduced sequencing costs per base23, facilitating the advancement of personalized medicine for cancer patients. In addition, the cost and complexity associated with the 4-color optical detection used in all other NGS platforms is circumvented in Ion Torrent sequencing through the employment of Post Light sequencing technology. This study aimed to identify mutation patterns in human GISTs to demonstrate the possibility of using Ion Torrent sequencing to direct personalized cancer treatment in the near future.
In our sample group of 121 GISTs, 189 amplicons covering 737 loci of 45 oncogenes and tumor suppressor genes from 121 human GISTs were sequenced using the Ion Torrent PGM platform. Analysis revealed a unique pattern of distribution and co-occurrence (Tables 2–4 and Tables 7 and 8): KIT was the most frequently mutated gene, and MLH1 and MET SNPS frequently occurred in combination with the KIT mutations. While KIT, PDFRA, and PIK3CA mutations have previously been reported, mutations in the MET and MLH1 genes in GISTs are previously not well known. Interestingly, our tumor group harbored mutations in the exon 11 of KIT but mutations along the PDGFRA gene were rare. Defects in genes that control mismatch repair (MMR) such as MLH1, MSH2, and PMS2 generally exhibit microsatellite instability, leading to the formation of tumors such as endometrial and colon cancer24. MET signaling is affected by mutations in this gene, and dysregulation of the MET signaling pathway occurs in a wide range of human cancers25.
Recent data indicate that GIST patients respond differently to tyrosine kinase inhibitors (drugs like Gleevec and Sutent), depending on the specific mutations displayed by their tumors. Gleevec (Imatinib) is considered as the standard treatment of metastatic GIST, for approximately 65–70% of patients being treated with this drug achieve a partial response, but 15–20% maintain stable disease26,27,28,29. Reports indicate that GIST patients respond better to Gleevec if their tumors harbor KIT mutations in exon 1110,30 versus those with mutations in exon 931, or if they have GIST negative for KIT expression. The response to Gleevac for PDGFR mutations also depends on the specific mutations involved, but data indicates that 35% of patients with PDGFR mutations benefit from the drug32. Hence, Gleevac is shown to benefit virtually all patients with metastatic GIST regardless of the mutational status of the tumor; however, patients with particular GIST mutations on exon 18 (D842V) are resistant to Gleevac. In addition, newly acquired secondary mutations can also confer drug resistance to Gleevec. Such patients who had grown resistant to Gleevec and with KIT mutations in exon 9 responded well to Sutent, another tyrosine kinase inhibitor33. Sunitinib (SU11248), Masitinib, Nilotinib, and Dasatinib are other oral small molecule inhibitors of multiple receptor tyrosine kinases including KIT, PDGFRA, VEGFR FLT3, CSF-1R, and RET5,34,35. The metastases resistant to drugs are often with new mutations that were not detected in primary tumors36,37,38. For example, drug resistant metastases may lose KIT expression, potentially mimicking other types of tumors39. This demands the constant need for the evaluation of GISTs for new mutations for appropriate drug-selection against the specific mutations harboring a tumor.
Frequent SNPs were detected along MLH1 and MET genes in our sample set and are often associated with the development of GISTs when they co-occur with other mutations. While drugs against tyrosine kinases have either entered clinical trials or are being tested massively today, drug targets against MLH1 and MET mutations are still in early stages. Martin et al. suggest that MSH2 and MLH1 deficiencies are associated with increased expression of POLB and POLG, respectively, and therefore DNA sequencing-based detection and IHC detection of these latter proteins in tumor biopsies may serve as a predictive biomarker for the application of POLG or POLB inhibitors40. Several MET inhibitors have been tested to date; they are broadly divided into those inhibiting either the ligand or receptor of HGF, and they occur as monoclonal antibodies or small-molecule kinase inhibitors25. Tivantinib (ARQ197), a selective, non-adenosine triphosphate-competitive inhibitor targeting MET tyrosine kinase, and abozantinib (XL184), a dual c-Met/VEGFR2 inhibitor, are under early clinical evaluation41. However, validated biomarkers for proper evaluation of MET mutations such as copy number and amplification are not yet available25, and high throughput DNA sequencing techniques may be highly beneficial to investigate along this line for developing personalized drug-targets against MET mutations.
Our current study identifies several mutations and mutation combinations in GISTs from Chinese patients using the Ion Torrent sequencing platform. As more experience and information is gained from the next generation technologies, it is necessary to expand our understanding in the sensitivity of individualized therapies to specific mutations. So, gathering a complete profile of mutations in GISTs or any specific tumors for the application of personalized and tailored targeted therapy is critical for the development of improved cancer treatments. We believe a faster and more cost effective genotyping tool such as Ion Torrent sequencing will be greatly beneficial for the assignment of such specific therapeutics in the near future.
Methods
Ethics statement
The study has been approved by the Human Research Ethics Committee of Nanjing First Hospital, China. For Formalin fixed and paraffin embedded (FFPE) tumor samples from the tumor tissue bank at the Department of Pathology of the hospital, the institutional ethics committee waived the need for consent. All samples and medical data used in this study have been irreversibly anonymized.
Patient information
Tumor samples used in the study were collected from Nanjing First Hospital, China. A total of 121 FFPE tumor samples from GIST patients were analyzed (Supplementary Table 4). DNA was isolated from FFPE samples after deparaffinization and extraction of 3–5 μm thick paraffin sections in xylene and by using the QIAamp DNA Mini Kit (Qiagen) per the manufacturer's instruction.
Variant calling
Data from the PGM runs were processed initially using the Ion Torrent platform-specific pipeline software Torrent Suite to generate sequence reads, trim adapter sequences, filter, and remove poor signal-profile reads. The Torrent Suite Software v3.0 with a plug-in “variant caller v3.0” program was used to generate initial variant calling from the Ion AmpliSeq sequencing data. In order to eliminate erroneous base calling, four filtering steps were used to generate final variant calling (Supplementary Fig. 1). The first filter was set at an average total coverage depth of >100, each variant coverage of >20, a variant frequency of each sample > 10%, and P-value < 0.01. The second filter was set for base calling < 4 bases homopolymer tracts and >3 bases from the terminus of amplicons, due to an observation of false positive mutations at the ends of some reads. The third filtering step was employed by visually examining mutations using Integrative Genomics Viewer (IGV) software (http//www.broadinstitute.org/igv) or Samtools software SAMtools software (http://samtools.sourceforge.net), as well as by filtering out possible strand-specific errors, like a mutation that was only detected in the “+” or “−” DNA strand, but not in both strands. The last filter step eliminated variants in amplicon AMPL339432 (PIK3CA, exon13, chr3:178938822–178938906), for it is not uniquely matched in human genome. False deletion data were generated from the JAK2 gene locus from our sequencing runs with the Ion Ampliseq Cancer Panel; thus the sequencing data from this locus were excluded from further analysis.
Variant calling is a subjective process and considerably influenced by the particular software used and the specific parameters employed to filter out raw error base calling. Various next-generation sequencing platforms, including the Ion Torrent sequencing, can generate wrong base calling. The data generated using the Ion Torrent PGM platform were reported to have a raw error rate of ~1.8%42, largely due to the inability of calling the correct number of bases in homopolymers > 8 bases long.
Somatic mutations
To distinguish between somatic and germline mutations, detected mutations were compared to variants in the 1000 Genomes Project43 and 6500 exomes of the National Heart, Lung, and Blood Institute Exome Sequencing Project44.
Bioinformatical and experimental validation
To assess reappearing mutations in GIST, we used the COSMIC (version 64)45, MyCancerGenome database (http://www.mycancergenome.org/), and other publications (Supplementary Table 1). Additionally, Sanger's sequencing was used to confirm some detected missense mutations (Supplementary Table 3).
Statistical analysis
We selected reappearing somatic missense/in-del mutations in the GIST to perform the statistical analysis. Some categorical data were compared using Fisher's exact tests. Log-rank statistics were used to determine whether patients' survival was influenced by CD34 and CD117. Survival analyses were also performed using Kaplan-Meier survival curves. All statistical analyses were performed using R version 3.0.2.
Author Contributions
S.C. and J.C. designed the study. Z.X., X.H., C.T., F.L., D.Z., S.J., H.S., H.D., G.Z., Z.L., Z.D., B.G., H.Y., C.Y., L.W., Z.S., Y.L., D.G., X.Z., X.W., X.W., L.H. Y.Z., J.Y., Y.G. and C.T. performed experiments and data analysis. C.T., V.N. and L.J. prepared the figure and tables. V.N., L.J., X.H. and S.C. wrote the main manuscript text. All authors reviewed the manuscript.
Supplementary Material
Supplemental Information
Acknowledgments
We would like to thank Rong Shi at the Wu Jieping Foundation, and Haibo Wang, Ying Li, and other members of San Valley Biotechnology Inc. Beijing for their assistance in sample and data collection. We would also like to thank the staffs at Nanjing First Hospital, Nanjing Medical University, and Beijing Military Hospital for their generous support for DNA sequencing and data collection. This research was supported by the grants from National Natural Science Foundation of China (Grant No. 81272469 to JC and Grant No. 81000880 to ZX); Special Foundation for Clinical Medicine of the Department of Jiangsu Province Science and Technology (Grant No. BL2012016 to JC); Jiangsu Provincial 12th Five-Year Program on Developing Health by Technology and Education Project (JC), the Wu Jieping Foundation, and the National Institute of Health (R01 CA90427 & R01 AI084811 to SY Chen).
Footnotes
Authors Chuanning Tang, Hua Ye, Feng Lou, Dandan Zhang, Shouwen Jiang, Hong Sun, Haichao Dong, Guangchun Zhang, Zhiyuan Liu, Zhishou Dong, Baishuai Guo, He Yan, Chaowei Yan, Lu Wang, Ziyi Su, and Yangyang Li are paid employees of San Valley Biotechnology, Inc. Authors Zhi Xu, Xinying Huo, Vijayalakshmi Nandakumar, Dongying Gu, Xiaojing Zhang, Xiaomin Wu, Xiaowei Wei, Lingzhi Hong, Yangmei Zhang, Jinsong Yang, Yonglin Gong, Cuiju Tang, Lindsey Jones, Xue F. Huang, Si-Yi Chen, and Jinfei Chen declare no potential conflict of interest.
References
- Tryggvason G., Gíslason H. G., Magnússon M. K. & Jónasson J. G. Gastrointestinal stromal tumors in Iceland, 1990–2003: The Icelandic GIST study, a population-based incidence and pathologic risk stratification study. Int. J. Cancer 117, 289–293; 10.1002/ijc.21167 (2005). [DOI] [PubMed] [Google Scholar]
- Nilsson B. et al. Gastrointestinal stromal tumors: The incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era. Cancer 103, 821–829; 10.1002/cncr.20862 (2005). [DOI] [PubMed] [Google Scholar]
- Tran T., Davila J. A. & El-Serag H. B. The Epidemiology of Malignant Gastrointestinal Stromal Tumors: An Analysis of 1,458 Cases from 1992 to 2000. Am. J. Gastroenterol. 100, 162–168; 10.1111/j.1572-0241.2005.40709.x (2005). [DOI] [PubMed] [Google Scholar]
- Fletcher C. D. M. et al. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum. Pathol. 33, 459–465; 10.1053/hupa.2002.123545 (2002). [DOI] [PubMed] [Google Scholar]
- Hirota S. et al. Gain-of-Function Mutations of c-kit in Human Gastrointestinal Stromal Tumors. Science 279, 577–580; 10.1126/science.279.5350.577 (1998). [DOI] [PubMed] [Google Scholar]
- Kindblom L. G., Aldenborg H. E. R. F. & Meis-Kindblom J. M. Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am. J. Pathol. 152, 1259–1269 (1998). [PMC free article] [PubMed] [Google Scholar]
- Corless C. L. & Heinrich M. C. Molecular Pathobiology of Gastrointestinal Stromal Sarcomas. Annu. Rev. Pathol.-Mech. 3, 557–586; 10.1146/annurev.pathmechdis.3.121806.151538 (2008). [DOI] [PubMed] [Google Scholar]
- Miettinen M. & Lasota J. Gastrointestinal Stromal Tumors: Review on Morphology, Molecular Pathology, Prognosis, and Differential Diagnosis. Arch. Pathol. Lab. Med. 130, 1466–1478; 10.1043/1543-2165(2006)130[1466:GSTROM]2.0.CO;2 (2006). [DOI] [PubMed] [Google Scholar]
- Joensuu H. Gastrointestinal stromal tumor (GIST). Ann. Oncol. 17, x280–x286; 10.1093/annonc/mdl274 (2006). [DOI] [PubMed] [Google Scholar]
- Heinrich M. C. et al. PDGFRA Activating Mutations in Gastrointestinal Stromal Tumors. Science 299, 708–710; 10.1126/science.1079666 (2003). [DOI] [PubMed] [Google Scholar]
- Duensing A. et al. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumors (GISTs). Oncogene 23, 3999–4006; 10.1038/sj.onc.1207525 (2004). [DOI] [PubMed] [Google Scholar]
- Carney J. A. Gastric Stromal Sarcoma, Pulmonary Chondroma, and Extra-adrenal Paraganglioma (Carney Triad): Natural History, Adrenocortical Component, and Possible Familial Occurrence. Mayo Clin. Proc. 74, 543–552; 10.4065/74.6.543 (1999). [DOI] [PubMed] [Google Scholar]
- Judson I. & Demetri G. Advances in the treatment of gastrointestinal stromal tumours. Ann. Onc. 18, x20–x24; 10.1093/annonc/mdm410 (2007). [DOI] [PubMed] [Google Scholar]
- Antonescu C. R. Targeted therapy of cancer: new roles for pathologists in identifying GISTs and other sarcomas. Mod. Pathol. 21, S31–S36; 10.1038/modpathol.2008.9 (2008). [DOI] [PubMed] [Google Scholar]
- Metzker M. L. Sequencing technologies [mdash] the next generation. Nat. Rev. Genet. 11, 31–46; 10.1038/nrg2626 (2010). [DOI] [PubMed] [Google Scholar]
- Suzuki S., Ono N., Furusawa C., Ying B.-W. & Yomo T. Comparison of Sequence Reads Obtained from Three Next-Generation Sequencing Platforms. PLoS ONE 6, e19534; 10.1371/journal.pone.0019534 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmand N. et al. Direct electrical detection of DNA synthesis. Proc. Natl. Acad. Sci. 103, 6466–6470; 10.1073/pnas.0601184103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu H. & DeMatteo R. P. The Management of Gastrointestinal Stromal Tumors: A Model for Targeted and Multidisciplinary Therapy of Malignancy. Ann. Rev. Med. 63, 247–258; 10.1146/annurev-med-043010-091813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945; 10.1038/nature03001 (2004). [DOI] [PubMed] [Google Scholar]
- Schuster S. C. Next-generation sequencing transforms today's biology. Nat. Meth. 5, 16–18; 10.1038/nmeth1156 (2008). [DOI] [PubMed] [Google Scholar]
- Voelkerding K. V., Dames S. A. & Durtschi J. D. Next-Generation Sequencing: From Basic Research to Diagnostics. Clin. Chem. 55, 641–658; 10.1373/clinchem.2008.112789 (2009). [DOI] [PubMed] [Google Scholar]
- Meyerson M., Gabriel S. & Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696; 10.1038/nrg2841 (2010). [DOI] [PubMed] [Google Scholar]
- Roukos D. H. Trastuzumab and beyond: sequencing cancer genomes and predicting molecular networks. Pharmacogenomics J. 11, 81–92; 10.1038/tpj.2010.81 (2011). [DOI] [PubMed] [Google Scholar]
- Basil J. B., Goodfellow P. J., Rader J. S., Mutch D. G. & Herzog T. J. Clinical significance of microsatellite instability in endometrial carcinoma. Cancer 89, 1758–1764; 10.1002/1097-0142(20001015)89:81758::AID-CNCR16>3.0.CO;2-A (2000). [DOI] [PubMed] [Google Scholar]
- Peters S. & Adjei A. A. MET: a promising anticancer therapeutic target. Nat. Rev. Clin. Oncol. 9, 314–326; 10.1038/nrclinonc.2012.71 (2012). [DOI] [PubMed] [Google Scholar]
- Demetri G. D. et al. Efficacy and Safety of Imatinib Mesylate in Advanced Gastrointestinal Stromal Tumors. New Engl. J. Med. 347, 472–480; 10.1056/NEJMoa020461 (2002). [DOI] [PubMed] [Google Scholar]
- van Oosterom A. T. et al. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet 358, 1421–1423; 10.1016/s0140-6736(01)06535-7 (2001). [DOI] [PubMed] [Google Scholar]
- Demetri G. D. et al. Imatinib Plasma Levels Are Correlated With Clinical Benefit in Patients With Unresectable/Metastatic Gastrointestinal Stromal Tumors. J. Clin. Oncol. 27, 3141–3147; 10.1200/JCO.2008.20.4818 (2009). [DOI] [PubMed] [Google Scholar]
- Joensuu H. & DeMatteo R. P. The management of gastrointestinal stromal tumors: a model for targeted and multidisciplinary therapy of malignancy. Ann. Rev. Med. 63, 247–258; 10.1146/annurev-med-043010-091813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debiec-Rychter M. et al. Gastrointestinal stromal tumours (GISTs) negative for KIT (CD117 antigen) immunoreactivity. J. Pathol. 202, 430–438; 10.1002/path.1546 (2004). [DOI] [PubMed] [Google Scholar]
- Heinrich M. C. & Corless C. L. Gastric GI stromal tumors (GISTs): The role of surgery in the era of targeted therapy. J. Surg. Oncol. 90, 195–207; 10.1002/jso.20230 (2005). [DOI] [PubMed] [Google Scholar]
- Blanke C. D. & Corless C. L. State-of-the Art Therapy for Gastrointestinal Stromal Tumors. Cancer Invest. 23, 274–280; 10.1081/CNV-200055972 (2005). [DOI] [PubMed] [Google Scholar]
- Raut C. P. et al. Surgical Management of Advanced Gastrointestinal Stromal Tumors After Treatment With Targeted Systemic Therapy Using Kinase Inhibitors. J. Clin. Oncol. 24, 2325–2331; 10.1200/JCO.2005.05.3439 (2006). [DOI] [PubMed] [Google Scholar]
- Blanke C. D. et al. Phase III Randomized, Intergroup Trial Assessing Imatinib Mesylate At Two Dose Levels in Patients With Unresectable or Metastatic Gastrointestinal Stromal Tumors Expressing the Kit Receptor Tyrosine Kinase: S0033. J. Clin. Oncol. 26, 626–632; 10.1200/JCO.2007.13.4452 (2008). [DOI] [PubMed] [Google Scholar]
- Verweij J. et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet 364, 1127–1134; 10.1016/s0140-6736(04)17098-0 (2004). [DOI] [PubMed] [Google Scholar]
- Willett C. G. et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat. Med. 10, 145–147; 10.1038/nm988 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu C. R. et al. Acquired Resistance to Imatinib in Gastrointestinal Stromal Tumor Occurs Through Secondary Gene Mutation. Clin. Cancer Res. 11, 4182–4190; 10.1158/1078-0432.CCR-04-2245 (2005). [DOI] [PubMed] [Google Scholar]
- Wardelmann E. et al. Acquired resistance to imatinib in gastrointestinal stromal tumours caused by multiple KIT mutations. Lancet Oncol. 6, 249–251; 10.1016/s1470-2045(05)70097-8 (2005). [DOI] [PubMed] [Google Scholar]
- Van Putte B. P. et al. Combination chemotherapy with gemcitabine with isolated lung perfusion for the treatment of pulmonary metastases. J. Thorac. Cardiovasc. Surg. 130, 125–130; 10.1016/j.jtcvs.2004.07.045 (2005). [DOI] [PubMed] [Google Scholar]
- Martin S. A. et al. DNA Polymerases as Potential Therapeutic Targets for Cancers Deficient in the DNA Mismatch Repair Proteins MSH2 or MLH1. Cancer Cell 17, 235–248; 10.1016/j.ccr.2009.12.046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes F. M. et al. Cabozantinib (XL184), a Novel MET and VEGFR2 Inhibitor, Simultaneously Suppresses Metastasis, Angiogenesis, and Tumor Growth. Mol. Cancer Ther. 10, 2298–2308; 10.1158/1535-7163.MCT-11-0264 (2011). [DOI] [PubMed] [Google Scholar]
- Quail M. et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341; 10.1186/1471-2164-13-341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G. P. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073; 10.1038/nature09534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/), (Accessed March, 2013).
- Bamford S. et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 91, 355–358; 10.1038/sj.bjc.6601894 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information