Abstract
MicroRNAs (miRNAs) constitute a robust regulatory network with post-transcriptional regulatory efficiency for almost one half of human coding genes, including oncogenes and tumour suppressors. We determined the expression profile of 667 miRNAs in colorectal cancer (CRC) tissues and paired non-tumoural tissues and identified 42 differentially expressed miRNAs. We chose miR-215, miR-375, miR-378, miR-422a and miR-135b for further validation on an independent cohort of 125 clinically characterized CRC patients and for in vitro analyses. MiR-215, miR-375, miR-378 and miR-422a were significantly decreased, whereas miR-135b was increased in CRC tumour tissues. Levels of miR-215 and miR-422a correlated with clinical stage. MiR-135b was associated with higher pre-operative serum levels of CEA and CA19-9. In vitro analyses showed that ectopic expression of miR-215 decreases viability and migration, increases apoptosis and promotes cell cycle arrest in DLD-1 and HCT-116 colon cancer cell lines. Similarly, overexpression of miR-375 and inhibition of miR-135b led to decreased viability. Finally, restoration of miR-378, miR-422a and miR-375 inhibited G1/S transition. These findings indicate that miR-378, miR-375, miR-422a and miR-215 play an important role in CRC as tumour suppressors, whereas miR-135b functions as an oncogene; both groups of miRNA contribute to CRC pathogenesis.
Keywords: colorectal cancer, microRNA, expression profiling, apoptosis, migration
Introduction
Colorectal cancer (CRC) is the third most common cancer in western countries and the third leading cause of cancer-related death worldwide [1]. CRC carcinogenesis is associated with multiple alterations of tumour suppressor genes and oncogenes. However, clinical utility of these genes as markers for early diagnosis and determination of prognosis and prediction is limited. Therefore, several efforts have been made to find new biomarkers for early detection of asymptomatic disease, accurate differentiation between particular clinical stages of CRC and better individualization of therapy.
MicroRNAs (miRNAs) are small, non-coding, single-stranded RNAs, 18–25 nucleotides in length. They are endogenously expressed and post-transcriptionally regulate gene expression by binding to 3′ untranslated region (3′ UTR) of target mRNAs [2]. There is increasing evidence that miRNAs can function as tumour suppressors as well as oncogenes [3] and therefore are important in regulation of many biological processes, such as cell cycle, proliferation, differentiation, apoptosis or invasiveness [4].
A growing number of studies have addressed miRNA expression in CRC [5–9]. Nevertheless, different profiling platforms, methodology and lacking clinicopathological data resulted in discrepancies across miRNAs identified in these studies. Therefore, only a small fraction of miRNAs identified this way may be further used as diagnostic and prognostic markers or therapeutics targets. miR-143, miR-145, miR-21, miR-31, miR-183 and miR-17-92 cluster are among miRNAs most consistently reported to have deregulated expression in CRC [5,9–11]. While miR-143 and miR-145 function as tumour suppressors, miR-21, miR-31, miR-183 and miR-17-92 are reported to be oncogenes. Moreover, association between miRNA expression, prognosis and therapy response prediction in CRC patients was repeatedly described [5,8,12]. Recently, several studies have indicated that circulating miRNAs could be used as new non-invasive biomarkers for CRC [13–15]. Although miRNA profiling significantly contributes to a better understanding of CRC pathogenesis, only little is known about miRNAs target molecules and signalling pathways in which they are involved.
The aim of our study was to identify and validate new miRNAs that are deregulated in CRC using highly standardized qRT-PCR-based TaqMan Low Density miRNA arrays (TLDAs). Furthermore, we screened effects of selected miRNAs on cell viability, migration, cell cycle and apoptosis of DLD-1 and HCT-116 colon cancer cell lines.
Materials and methods
Patients and tissue samples
Eight tumour tissues from patients diagnosed with colorectal carcinoma at Masaryk Memorial Cancer Institute (MMCI, Brno, Czech Republic) between 2003 and 2008 (four men, four women; one stage II, four stage III, three stage IV) and eight paired adjacent non-tumoural tissues were used for profiling analyses. Patients’ age ranged from 45 to 76 years with a median of 63.5 years. Validation was performed on a cohort of 125 patients (78 men, 47 women) with histologically confirmed colorectal adenocarcinoma undergoing surgery at MMCI between 2009 and 2011; their age ranged from 30 to 92 with median 68 years. Native tumour and paired non-tumoural colon tissues were obtained after surgical resection and immediately placed in liquid nitrogen. Informed consent was obtained from all patients, and the local Ethical Board approved the study protocol. Clinicopathological features of the validation cohort are summarized in Table 1.
Table 1.
Clinicopathological features of CRC patients in the validation cohort and summary of results in validation phase of the study
| n | miR-215 | miR-375 | miR-378 | miR-422a* | miR-135b | |
|---|---|---|---|---|---|---|
| Tumour versus mucosa | ||||||
| Normal mucosa | 125 | 0.47 (0.29–0.65) | 0.95 (0.59–1.94) | 1.32 (0.88–1.95) | 0.69 (0.40–1.00) | 0.02 (0.01–0.04) |
| Colorectal tumour | 125 | 0.07 (0.02–0.15) | 0.23 (0.07–0.46) | 0.26 (0.14–0.38) | 0.11 (0.05–0.19) | 0.33 (0.13–0.59) |
| P-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| Clinical stage | 125 | |||||
| I | 26 | 0.13 (0.07–0.32) | 0.29 (0.14–0.73) | 0.31 (0.15–0.36) | 0.23 (0.14–0.32) | 0.53 (0.37–0.88) |
| II | 39 | 0.08 (0.05–0.23) | 0.22 (0.06–0.42) | 0.30 (0.21–0.51) | 0.12 (0.06–0.22) | 0.36 (0.21–0.64) |
| III | 31 | 0.03 (0.02–0.07) | 0.20 (0.06–0.37) | 0.16 (0.12–0.32) | 0.06 (0.04–0.14) | 0.16 (0.06–0.41) |
| IV | 29 | 0.05 (0.02–0.12) | 0.33 (0.16–0.69) | 0.26 (0.13–0.39) | 0.12 (0.04–0.17) | 0.39 (0.20–0.55) |
| P-value | < 0.0001 | 0.2115 | 0.1476 | 0.0001 | 0.0003 | |
| pT category | 125 | |||||
| pT1 | 2 | 0.35 (–) | 0.55 (–) | 1.43 (–) | 0.52 (–) | 0.77 (–) |
| pT2 | 28 | 0.12 (0.08–0.30) | 0.33 (0.14–0.50) | 0.32 (0.16–0.37) | 0.20 (0.14–0.29) | 0.40 (0.23–0.74) |
| pT3 | 82 | 0.06 (0.02–0.12) | 0.21 (0.06–0.41) | 0.24 (0.14–0.38) | 0.10 (0.05–0.17) | 0.31 (0.16–0.56) |
| pT4 | 13 | 0.04 (0.02–0.08) | 0.34 (0.21–0.89) | 0.19 (0.09–0.44) | 0.10 (0.04–0.17) | 0.19 (0.06–0.55) |
| P-value | 0.0006 | 0.0580 | 0.8633 | 0.0001 | 0.3004 | |
| Lymph nodes | 125 | |||||
| Negative | 68 | 0.11 (0.07–0.26) | 0.25 (0.13–0.49) | 0.30 (0.19–0.44) | 0.17 (0.09–0.26) | 0.41 (0.23–0.73) |
| Positive | 57 | 0.03 (0.02–0.07) | 0.20 (0.06–0.41) | 0.19 (0.11–0.35) | 0.09 (0.04–0.14) | 0.19 (0.07–0.46) |
| P-value | <0.0001 | 0.2536 | 0.0415 | 0.0002 | 0.0013 | |
| Distant metastases | 125 | |||||
| No | 96 | 0.07 (0.03–0.17) | 0.23 (0.08–0.43) | 0.27 (0.15–0.38) | 0.13 (0.06–0.22) | 0.34 (0.16–0.62) |
| Yes | 29 | 0.04 (0.02–0.11) | 0.29 (0.16–0.66) | 0.20 (0.12–0.38) | 0.12 (0.04–0.16) | 0.39 (0.20–0.55) |
| P-value | 0.1824 | 0.3670 | 0.5862 | 0.2468 | 0.6976 | |
| Grading | 121 | |||||
| G1 | 29 | 0.09 (0.04–0.33) | 0.22 (0.06–0.51) | 0.25 (0.14–0.45) | 0.13 (0.05–0.27) | 0.29 (0.06–0.54) |
| G2 | 67 | 0.07 (0.03–0.12) | 0.26 (0.10–0.41) | 0.27 (0.15–0.38) | 0.13 (0.06–0.21) | 0.38 (0.20–0.55) |
| G3 | 25 | 0.02 (0.02–0.13) | 0.22 (0.06–0.69) | 0.23 (0.12–0.37) | 0.11 (0.04–0.18) | 0.38 (0.17–0.93) |
| P-value | 0.0388 | 0.8646 | 0.3211 | 0.2300 | 0.1304 | |
| Localization | 124 | |||||
| Proximal colon | 57 | 0.07 (0.02–0.18) | 0.30 (0.09–0.67) | 0.27 (0.15–0.35) | 0.11 (0.08–0.21) | 0.38 (0.18–0.68) |
| Distal colon | 67 | 0.07 (0.03–0.14) | 0.22 (0.07–0.43) | 0.25 (0.13–0.44) | 0.12 (0.05–0.19) | 0.32 (0.13–0.53) |
| P-value | 0.6036 | 0.2986 | 0.6212 | 0.9042 | 0.3593 | |
| Tumour diameter | 111 | |||||
| ≤50 mm | 25 | 0.07 (0.02–0.14) | 0.24 (0.06–0.51) | 0.24 (0.14–0.36) | 0.11 (0.05–0.21) | 0.33 (0.16–0.60) |
| >50 mm | 86 | 0.07 (0.03–0.20) | 0.22 (0.18–0.33) | 0.32 (0.14–0.44) | 0.14 (0.07–0.17) | 0.38 (0.15–0.59) |
| P-value | 0.4972 | 0.9877 | 0.4211 | 0.6258 | 0.9789 | |
| Pre-s-CEA levels | 74 | |||||
| ≤4.6 μg/l | 39 | 0.07 (0.02–0.13) | 0.18 (0.05–0.41) | 0.28 (0.15–0.39) | 0.12 (0.08–0.24) | 0.23 (0.15–0.39) |
| >4.6 μg/l | 35 | 0.07 (0.02–0.13) | 0.21 (0.07–0.49) | 0.20 (0.10–0.36) | 0.11 (0.04–0.18) | 0.42 (0.19–0.76) |
| P-value | 0.9827 | 0.3704 | 0.2617 | 0.1658 | 0.0338 | |
| Pre-s-CA19-9 levels | 75 | |||||
| ≤40 kU/l | 64 | 0.07 (0.02–0.13) | 0.21 (0.06–0.45) | 0.24 (0.14–0.37) | 0.12 (0.06–0.22) | 0.31 (0.13–0.55) |
| >40 kU/l | 11 | 0.07 (0.02–0.23) | 0.18 (0.05–0.37) | 0.33 (0.09–0.44) | 0.16 (0.05–0.18) | 0.41 (0.39–1.18) |
| P-value | 0.8869 | 0.4846 | 0.8411 | 0.8456 | 0.0360 |
The statistically significant values are bolded.
All values for miR-422a are multiplied by 10.
Tissue sample preparation and miRNA isolation
All analysed tissues were homogenized and total RNA enriched for small RNAs was isolated using mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). Concentration and purity of RNA were determined spectrophotometrically by measuring optical density (A260/280 >2.0; A260/230 >1.8) using Nanodrop ND-1000 (Thermo Scientific, Rockford, IL, USA).
Large-scale miRNA profiling
Expression profiling of miRNAs was performed using TLDAs. A set of two cards (TaqMan® Array Human MicroRNA Card Set v2.0; Applied Biosystems, Foster City, CA., USA) enabling quantification of 667 human miRNAs and 3 endogenous controls for data normalization was used. Two sets of megaplex miRNA RT primers with special stem-loop structure allowed synthesis of all cDNAs in two separate reactions. Each reverse transcriptase mixture consisted of 700 ng of RNA sample, 0.67 mM of each dNTPs, 6.67 U/μl of MultiScribe reverse transcriptase, 1 × Megaplex RT primers, 1 × RT buffer, 0.27 U/μl RNase inhibitor and 3 mM MgCl2 (all part of TaqMan MicroRNA Reverse Transcription kit; Applied Biosystems). Reaction mixtures were incubated for 2 min at 16°C, 1 min. at 42 °C and 1 sec. at 50°C for 40 cycles, then 5 min. at 85°C and finally held at 4°C (TGradient thermal cycler; Biotherma, Biometra, Goettingen, Germany). Real-time PCR was performed using the Applied Biosystems 7900 HT Version 2.3 Sequence Detection System. Each 800 μl PCR reaction mixture consisted of 5.3 μl of RT product, 1 × TaqMan (NoUmpErase UNG) Universal PCR Master Mix and 394.7 μl of nuclease free water. 100 μl of each PCR reaction mix was dispensed into each port of the TaqMan MicroRNA Array. Reactions were run at 50°C for 2 min., 95°C for 10 min., followed by 40 cycles at 95°C for 30 sec. and 60°C for 1 min.
Reverse transcription and qRT-PCR for validation assays
cDNA was synthesized using gene-specific primers according to the TaqMan MicroRNA Assay protocol (Applied Biosystems). For reverse transcription, 10 ng of RNA sample, 0.25 mM of each dNTPs, 3.33 U/μl of MultiScribe reverse transcriptase, 50 nM of stem–loop RT primer, 1 × RT buffer and 0.25 U/μl of RNase inhibitor (all from TaqMan MicroRNA Reverse Transcription kit; Applied Biosystems) were used. Reaction mixtures were incubated for 30 min. at 16°C, 30 min. at 42°C, 5 min. at 85°C and then held at 4°C (TGradient thermal cycler; Biotherma). Real-time PCR was performed using the Applied Biosystems 7500 Sequence Detection System. The 20-μl PCR reaction mixture consisted of 1.33 μl of RT product, 1 × TaqMan (NoUmpErase UNG) Universal PCR Master Mix and 1 μl of primer and probe mix of the TaqMan MicroRNA Assay kit (Applied Biosystems). Reactions were run in a 96-well optical plate at 95°C for 10 min., followed by 40 cycles at 95°C for 15 sec. and 60°C for 10 min.
Data normalization and statistical analysis
The CT values (CT) were calculated by SDS 2.0.1 software (Applied Biosystems) using the manual threshold settings (threshold = 0.2). All real-time PCR reactions were run in triplicates, and average threshold cycles and S.D. values were calculated. The average expression levels of all analysed miRNAs were normalized using RNU48 (Assay No. 4427975; Applied Biosystems) as a reference gene and subsequently the 2−ΔCT method was applied. Acquired ΔCT values were analysed in R using the Bioconductor package LIMMA concerning miRNA profiling [16]. RNU48 was selected as reference gene through combination of standard geneNorm and NormFinder algorithms from six reference genes on the TLDAs. In case of validation cohort, statistical differences between miRNAs levels in CRCs and non-tumoural tissues were evaluated by the two-tailed non-parametric Wilcoxon test for 125 paired samples. Furthermore, two-tailed Mann-Whitney U-test and Kruskal-Wallis test were used to analyse the correlation between the miRNA expression levels and clinical–pathological features of the patients. All calculations were performed using MedCalc software version 11.2.1. P-values lower than 0.05 were considered statistically significant.
Cell lines and growth conditions
Two human colon carcinoma cell lines were used in this study: HCT-116 (Dukes’ type D colon cancer, CCL-247™) and DLD-1 (Dukes’ type C colon cancer, CCL–221™). Both cell lines were obtained from the American Type Culture Collection (ATCC). Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS, 100 μg/ml of penicillin, 100 μg/ml of streptomycin, 0.1 mM non-essential amino acids, 2 mM l–glutamin, 1 mM sodium pyruvate (all purchased from Invitrogen, Carlsbad, CA, USA, Gibco) in 10% CO2 at 37°C.
Cell transfection
Both cell lines were transfected with 10 nM of pre-miR-215, pre-miR-375, pre-miR-378, pre-miR-422a or pre-miRNA negative control precursors, resp. or with 30 nM of anti-miR-135b and anti-miRNA negative control inhibitors, resp. (Ambion) using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's protocol. Transfection efficiency was evaluated by qRT-PCR as described above.
Detection of apoptosis
Cells were seeded in 60 mm plates at a density of 1 × 106 cells per plate, incubated for 24 hrs and transfected with 10 nM precursors or 30 nM inhibitors of analysed miRNAs. Forty-eight hours after transfection, the cells were trypsinized and apoptosis was measured using Annexin V–FITC kit (Miltenyi Biotec Inc., Cambridge, MA, USA) according to the manufacturer's protocol. All measurements were repeated three times.
Cell cycle analysis
Cells were seeded in 60 mm plates at a density of 1 × 106 cells per plate, incubated for 24 hrs and transfected with 10 nM precursors or 30 nM inhibitors of analysed miRNAs, resp. After 48 hrs, the cells were trypsinized and fixed in 70% ethanol. Subsequently, cells were washed in PBS and treated with 0.1 mg/ml of RNase for 30 min. at 37°C. Finally, 1 mg/ml of propidium iodide was added and another 10 min. incubation at room temperature was performed. Cell cycle analysis was measured using BD Facs Canto II (BD, San Jose, CA, USA). The data were analysed by FlowJo v 7.6.5. All measurements were repeated three times.
MTT assay
Cell viability was measured using the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay (MTT; Sigma Aldrich, St. Louis, MO, USA). Cells were seeded in 96-well plate at a density of 7.5 × 103 cells per well, 24 hrs before transfection. Subsequently, the cells were transfected with 10 nM precursors or 30 nM inhibitors of analysed miRNAs. To each well, 20 μl of 5 mg/ml MTT solution in PBS was added 24 and 48 hrs after transfection. The plates were incubated for 3 hrs at 37°C. The precipitate was solubilized in 100% DMSO (200 μl per well), and absorbance was measured on Elisa reader Multi–Detection Microplate Reader (BIO-TEK, USA) at wave lengths of 490 and 650 nm (background). Each measurement was performed in six plicates, and all experiments were repeated three times.
Scratch wound migration assay
Cells were seeded in 6-well plates at a density of 4 × 105 cells per well 24 hrs before they were transfected with 10 nM precursors or 30 nM inhibitors of analysed miRNAs. The cell monolayer was wounded with a sterile pipette tip 24 hrs after transfection and then rinsed twice with PBS to remove cellular debris. Subsequently, warm fresh medium was added. Migration was measured at time 0 and 24 hrs after wounding using microscope Nikon Diaphod 300 INV (10×) and camera Canon Power shot A95. Images were analysed by the Tscratch software (CSE, Switzerland). All measurements were repeated three times.
Results
MiRNAs differentially expressed in CRC tissue
To identify miRNAs that are differentially expressed between CRC tissues and normal adjacent tissues, we analysed expression profiles of 667 miRNAs in 8 tissues of patients with CRC and 8 paired non-tumoural tissues. Unsupervised hierarchical clustering was carried out, and only miRNAs with P < 0.0005 were considered significant. Using this criterion, we gained a set of 42 miRNAs that were differentially expressed between tumoural and non-tumoural colon tissue by LIMMA method (Table 2). All these miRNAs showed decreased expression in CRC tissues except for miR-135b, miR-766, miR-183* and miR-135b* that were significantly overexpressed in tumour tissues.
Table 2.
MiRNAs differentially expressed between CRC tissues and normal colon tissue
| miRNA | Fold change | P | miRNA | Fold change | P |
|---|---|---|---|---|---|
| hsa-miR-215 | 0.04 | 2.39 × 10−5 | hsa-miR-100 | 0.28 | 1.52 × 10−4 |
| hsa-miR-190 | 0.09 | 1.24 × 10−4 | hsa-miR-411 | 0.28 | 5.64 × 10−5 |
| hsa-miR-139-5p | 0.09 | 2.47 × 10−6 | hsa-miR-30c | 0.29 | 1.89 × 10−5 |
| hsa-miR-138 | 0.09 | 1.85 × 10−4 | hsa-miR-30a* | 0.29 | 3.47 × 10−5 |
| hsa-miR-451 | 0.11 | 6.93 × 10−5 | hsa-miR-192* | 0.29 | 1.59 × 10−4 |
| hsa-miR-422a | 0.12 | 4.67 × 10−7 | hsa-miR-30e* | 0.30 | 3.88 × 10−6 |
| hsa-miR-378* | 0.14 | 6.06 × 10−5 | hsa-miR-342-3p | 0.30 | 2.33 × 10−5 |
| hsa-miR-375 | 0.15 | 7.70 × 10−6 | hsa-miR-26b | 0.31 | 3.85 × 10−5 |
| hsa-miR-133b | 0.16 | 3.00 × 10−4 | hsa-miR-101 | 0.32 | 4.20 × 10−4 |
| hsa-miR-145 | 0.17 | 4.88 × 10−5 | hsa-miR-127-3p | 0.34 | 3.16 × 10−4 |
| hsa-miR-376c | 0.17 | 4.36 × 10−7 | hsa-miR-200b | 0.35 | 1.96 × 10−4 |
| hsa-miR-378 | 0.17 | 1.99 × 10−7 | hsa-miR-140-5p | 0.36 | 1.15 × 10−5 |
| hsa-miR-144* | 0.18 | 1.06 × 10−4 | hsa-miR-30b | 0.36 | 3.58 × 10−5 |
| hsa-miR-195 | 0.19 | 9.33 × 10−7 | hsa-miR-186 | 0.36 | 2.78 × 10−4 |
| hsa-miR-486-5p | 0.19 | 6.69 × 10−5 | hsa-miR-26a | 0.38 | 2.31 × 10−4 |
| hsa-miR-598 | 0.20 | 3.80 × 10−5 | hsa-miR-30e | 0.38 | 5.78 × 10−5 |
| hsa-miR-99a | 0.24 | 5.52 × 10−5 | hsa-miR-16 | 0.41 | 4.40 × 10−4 |
| hsa-miR-143 | 0.26 | 2.73 × 10−4 | hsa-miR-135b | 6.06 | 4.01 × 10−4 |
| hsa-miR-194 | 0.27 | 1.43 × 10−5 | hsa-miR-766 | 10.56 | 9.33 × 10−5 |
| hsa-miR-636 | 0.27 | 1.42 × 10−4 | hsa-miR-183* | 29.04 | 2.69 × 10−4 |
| hsa-miR-192 | 0.27 | 3.41 × 10−5 | hsa-miR-135b* | 31.12 | 2.59 × 10−4 |
Validation of selected miRNAs by qRT-PCR
Based on the significance of the difference (fold change, P-value), previous observations and biological plausibility (according to putative miRNA targets and/or Pubmed hits when particular miRNA is combined with keyword ‘cancer’), and favourable expression levels (Ct < 30), we chose five miRNAs for further validation. MiR-215, miR-375, miR-378 and miR-422a were down-regulated, whereas miR-135b was up-regulated. For validation, we used independent collection of 125 paired samples of tumoural tissue and adjacent non-tumoural mucosa. To determine the differences between these two groups, we used two-tailed non-parametric Wilcoxon test. Furthermore, two-tailed Mann–Whitney U-test and Kruskal–Wallis test were used to analyse the correlation between the miRNA expression levels and clinical–pathological features of the patients.
We confirmed significantly decreased expression of miR-215, miR-375, miR-378 and miR-422a and increased expression of miR-135b in colon cancer tissues (Fig. 1, P < 0.0001 for all evaluated miRNAs). Furthermore, levels of miR-215 and miR-422a correlated with CRC clinical stages, except stage IV (Fig. 2A and B). Interestingly, miR-135b showed similar trend, with the highest expression in stage I, which is in accordance with the previous studies that described the role of this miRNA in early stages of CRC (Fig. 2C). MiR-375 and miR-378 indicated no correlation with CRC clinical stage. Additionally, tumours with higher grade were characterized by lower expression of miR-215. Moreover, we observed the relationship between lymph node positivity and expression of miR-215, miR-378, miR-422a and miR-135b (Fig. 3A–D). Finally, we detected an increased expression of miR-135b in samples of patients with higher pre-operative serum levels of CEA and CA19-9 (Fig. 3E and F). The complete results of the validation phase of study are summarized in Table 1.
Fig. 1.
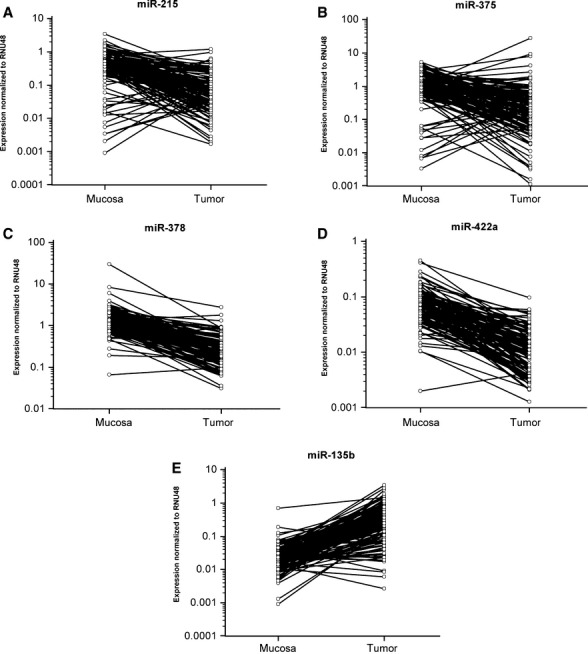
Different expressions of miR-215, miR-375, miR-378, miR-422a and miR-135b in 125 paired samples of CRC and adjacent mucosa. MiR-215 (A), miR-375 (B), miR-378 (C) and miR-422a (D) showed significantly lower expression in tumour tissue, whereas miR-135b (E) was overexpressed in tumour tissue (all miRNAs reached P < 0.0001, Wilcoxon paired test).
Fig. 2.
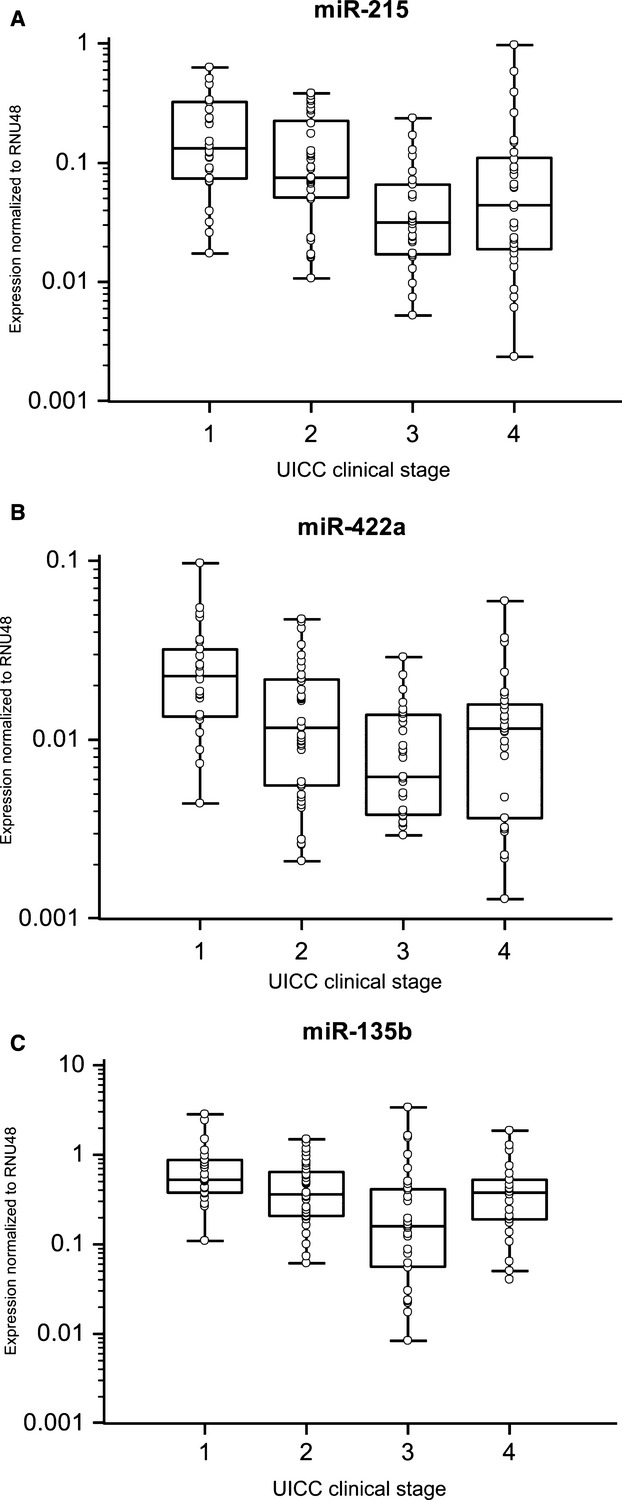
Correlation of miRNA expression with clinical stage of CRC patients. (A) Expression of miR-215 correlates with stage of CRC (P < 0.0001). (B) Expression of miR-422a correlates with a stage of CRC (P = 0.0001). (C) Expression of miR-135b negatively correlates with a stage of CRC (P = 0.0003).
Fig. 3.
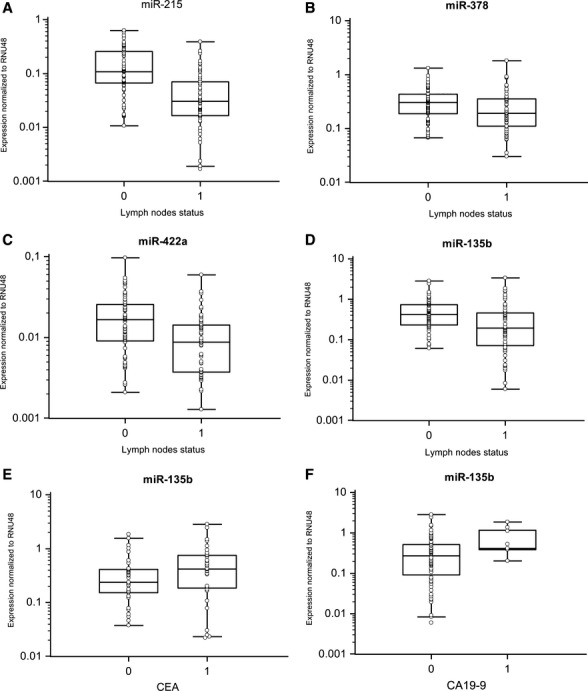
MiRNAs expression based on lymph node positivity and pre-operative serum levels of CEA and CA19-9. (A) MiR-215 is underexpressed in lymph node positive samples (P < 0.0001). (B) MiR-378 is underexpressed in lymph node positive samples (P = 0.0415). (C) MiR-422a is underexpressed in lymph node positive samples (P = 0.0002). (D) MiR-135b is underexpressed in lymph node positive samples (P = 0.0008). (E, F) Higher levels of miR-135b are associated with elevated pre-operative serum levels of CEA (cut-off 4.6 μg/l); P = 0.0338) and CA19-9 (cut-off 40 kU/l; P = 0.0360).
Determination of transfection efficiency
To analyse the effects of validated miRNA on cell viability, migration, cell cycle and apoptosis, we first optimized the transfection process of corresponding miRNA precursors or inhibitors. HCT-116 and DLD-1 cells were transfected using Lipofectamine RNAiMAX, and efficiency was subsequently determined by qRT-PCR 24 and 48 hrs after transfection. We significantly increased levels of tumour suppressive miRNAs, miR-215, miR-375, miR-378 and miR-422a (for all miRNAs, more than 10 000-fold increase was achieved, P < 0.0001, n = 3) and also inhibited the expression of oncogenic miR-135b by 70.5 ± 16.1% (P = 0.01, n = 3) in HCT-116 cells and by 60.0 ± 17.3% (P = 0.01, n = 3) in DLD-1 cells (Fig. 4A) using 10 nM precursors and 30 nM inhibitors, respectively. The effect of transfection was observed 24 hrs after transfection.
Fig. 4.
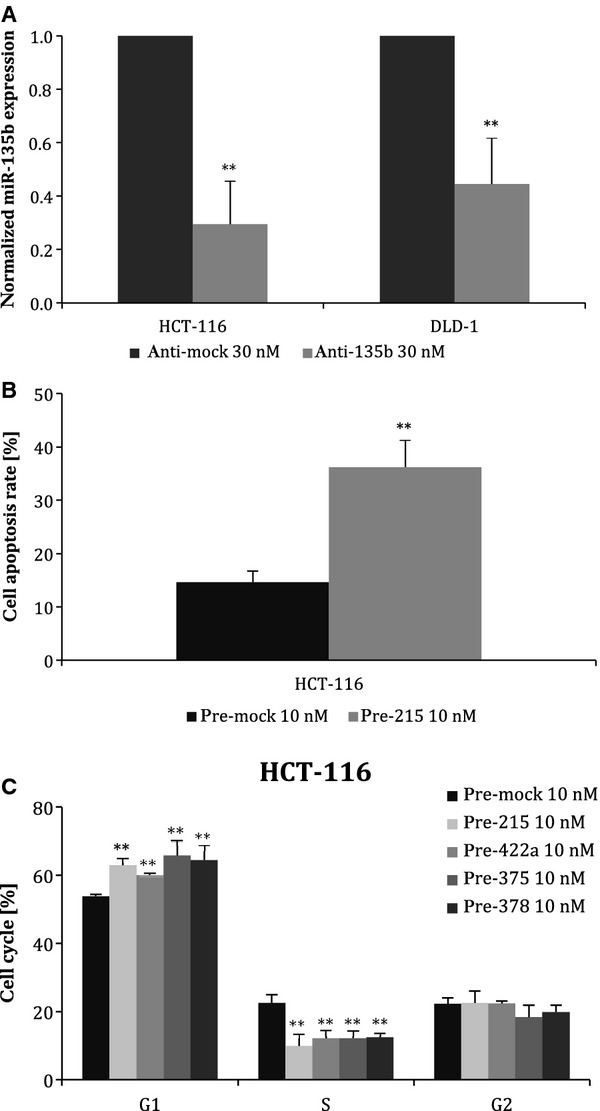
Efficiency of transfection and effects of anti- and pre-miR oligonucleotides on apoptosis and cell cycle. (A) Suppression of miR-135b expression by anti-miR-135b as detected by TaqMan qRT-PCR. Compared with negative control, miR-135b was reduced by 70.5 ± 16.1% in HCT-116 cells and by 60.0 ± 17.3% in DLD-1 cells. (B) Effect of miR-215 on apoptosis of HCT-116 cells. Transfection of 10 nM pre-miR-215 significantly increased the number of apoptotic cells from 14.6 ± 2.1% to 36.2 ± 5.1%. C. Staining with propidium iodide for cell cycle analysis. miR-215, miR-375, miR-378 and miR-422a significantly increased the number of cells in G0–G1 phase and reduced S-phase cells in HCT-116 (wt-p53) cell line. Results from three independent experiments (*t-test significant at P < 0.05, **P < 0.01).
MiR-215 affects apoptosis of HCT-116 cells
To evaluate the effect of particular miRNA on the rate of apoptosis, we used the Annexin V assay. We ascertained that higher levels of miR-215 resulted in an increase in the number of apoptotic cells from 14.6 ± 2.1% to 36.2 ± 5.1% (P = 0.005, n = 3) (Fig. 4B). Nevertheless, this effect was observed only in the case of HCT-116 cells. The rest of analysed miRNAs do not induce apoptosis, at least in the tested cell lines. All results are summarized in Table 3.
Table 3.
Summarized results of in vitro analyses (observed effects of particular pre- and anti-miR oligonucleotides)
| pre-miR-378 | pre-miR-422a | pre-miR-375 | pre-miR-215 | anti-miR-135b | ||
|---|---|---|---|---|---|---|
| HCT-116 cells | ||||||
| Apoptosis | – | – | – | Increased apoptosis P = 0.005 | – | |
| Cell cycle | G1/S increased P = 0.005 | G1/S increased P = 0.01 | G1/S increased P = 0.01 | G1/S increased P = 0.01 | – | |
| MTT | Trend (P > 0.05) | – | Decreased viability P = 0.005 | Decreased viability P = 0.05 | Decreased viability P = 0.05 | |
| SWA* | – | Trend (P > 0.05) | – | Trend (P > 0.05) | – | |
| DLD-1 cells | ||||||
| Apoptosis | – | – | – | – | – | |
| Cell cycle | – | – | – | – | – | |
| MTT | Trend (P > 0.05) | – | Trend (P > 0.05) | Decreased viability P = 0.05 | Decreased viability P = 0.005 | |
| SWA | – | Trend (P > 0.05) | – | Decreased viability P = 0.05 | – | |
SWA, scratch wound assay.
MiR-215, miR-375, miR-378 and miR-422a affect G1/S transition of HCT-116 cells
To investigate the role of selected miRNAs on cell cycle regulation, we used propidium iodide staining, and cells were subsequently analysed by flow cytometry. Our results show that miR-215, miR-375, miR-378 and miR-422a significantly increased the number of cells in G0–G1 phase and reduced S-phase cells (P = 0.01) in HCT-116 (wt-p53) cell line (Fig. 4C). However, in the DLD-1 (mut-p53) cell line, we did not observe any significant change in distribution of cell cycle phases. All results are summarized in Table 3.
MiR-135b, miR-215, miR-375 and miR-378 affect viability of HCT-116 and DLD-1 cells
To determine whether any of analysed miRNAs affect cell viability, the HCT-116 and DLD-1 cells were transfected with particular miRNA precursor or inhibitor, respectively, and subsequently MTT assay was used and absorbance was measured 48 hrs after transfection. We observed that up-regulated expression of miR-215 decreased viability of HCT-116 cells by 21.7 ± 0.3% (P = 0.05, n = 3) and DLD-1 cells by 19.7 ± 7.1% (P = 0.05, n = 3). Similarly, miR-375 decreased viability of HCT-116 cells by 32.9 ± 6.9% (P = 0.005, n = 3). In case of miR-135b, viability of DLD–1 cells was reduced by 27.4 ± 19.2% (P = 0.005, n = 3) and viability of HCT-116 by 16.0 ± 11.7 % (P = 0.05, n = 3) 48 hrs after transfection when corresponding inhibitor of this miRNA was used (Fig. 5A–C). These results are in accordance with our previous assumption that miR-215 and miR-375 function as tumour suppressors, while miR-135b functions as an oncogene. All results are summarized in Table 3.
Fig. 5.
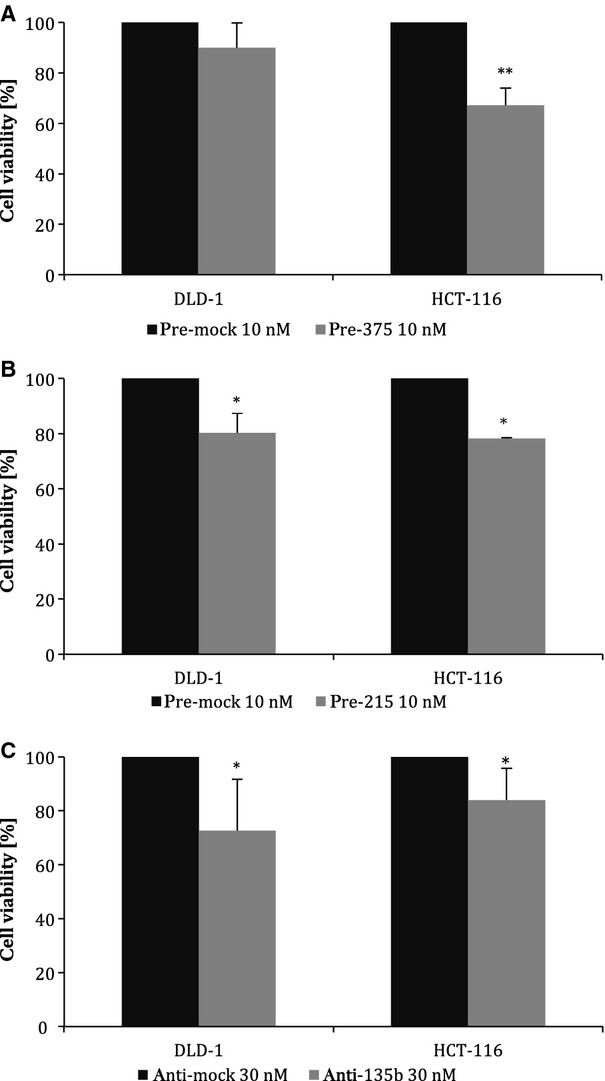
Viability of DLD-1 and HCT-116 cells determined with MTT assay. Cells were transfected with 10 nM precursors or 30 nM inhibitor of particular miRNAs, resp., and cell viability was measured by MTT test 48 hrs after transfection. (A) miR-375 decreased viability of DLD-1 cells by 10.0 ± 9.7% and HCT-116 cells by 32.9 ± 6.9%. (B) miR-215 decreased viability of DLD-1 cells by 19.7 ± 7.1% and HCT-116 cells by 21.7 ± 0.3%. (C) Inhibition of miR-135b expression led to decrease in viability of DLD-1 cells by 27.4 ± 19.2 % and HCT-116 by 16.0 ± 11.7%. Results from three independent experiments (*t-test significant at P < 0.05, **P < 0.01).
MiR-215 significantly suppresses migration of DLD-1 and HCT-116 cells
As our previous results showed that miR-215, miR-378, miR-422a and miR-135b are differentially expressed depending on lymph node involvement, we wanted to find out whether they play any role in cell migration. Therefore, DLD-1 and HCT-116 cells were transfected with particular miRNA precursors or inhibitors; subsequently, in vitro scratch wound migration assay was performed. We observed that migration of DLD-1 cells was reduced by 46.1 ± 20.5% (P = 0.05, n = 3, see Fig. 6A–B) and migration of HCT-116 cells decreased by 26.6 ± 35.2% (P > 0.05, n = 3) when 10 nM pre-miR-215 was used. Similar trend was also detected using 10 nM pre-miR-422a; however, in this case, the decrease was not statistically significant. Concerning miR-135b and miR-378, there was no difference in cell migration. All results are summarized in Table 3.
Fig. 6.
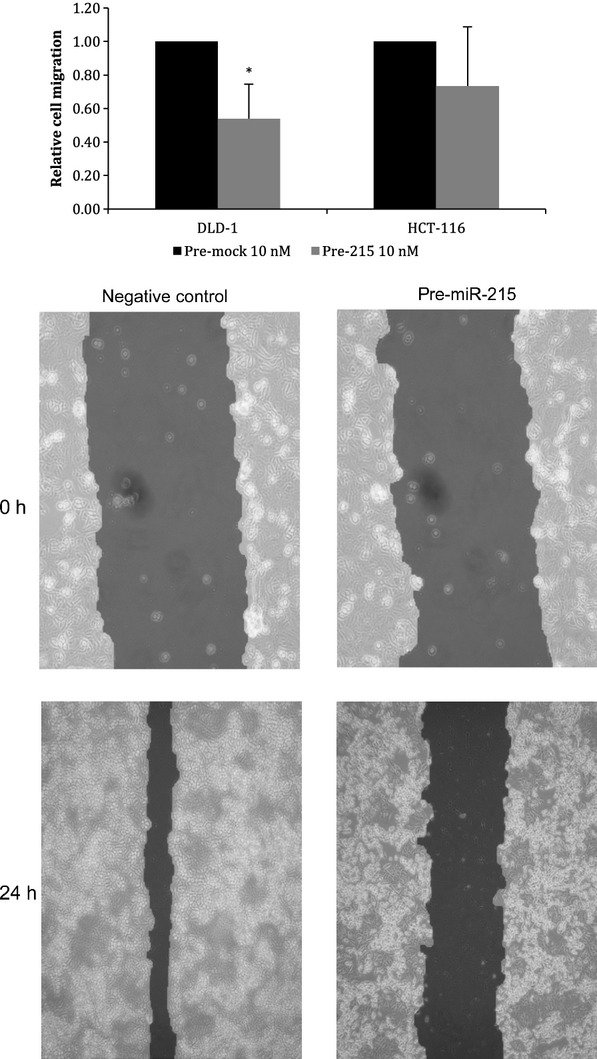
Effect of transfection of pre-miR-215 on cell migration of DLD-1 cells. Scratch wound assay proved that higher levels of miR-215 decreased migration ability of DLD-1 cells by 46.1 ± 20.5% 24 hrs after transfection. Results from three independent experiments (*t-test significant at P < 0.05).
Discussion
It is becoming clear that deregulated expression of miRNAs is connected to pathogenesis of many cancer types. In this study, we analysed expression profile of 667 miRNAs in 8 tissues of patients with CRC and paired non-tumoural tissues. Using unsupervised hierarchical clustering, we gained a set of 42 miRNAs that showed significantly different expression between tumoural tissue and adjacent normal mucosa. Four of them (miR-135b, miR-766, miR-183* and miR-135b*) were significantly overexpressed in tumour tissues, and they are supposed to function as oncogenes, although their specific role in pathogenesis of CRC is not known. Nevertheless, miR-183 is one in a cluster of three related miRNAs; this cluster also includes miR-182 and miR-96. It is situated on chromosome 7q32.2 [9] and was described to be overexpressed in many other types of cancer, including lung cancer [17], breast cancer [18], colon cancer [9] and prostate cancer [19]. These miRNAs were also up-regulated in our study, but they did not meet the criterion P < 0.0005. Furthermore, Nagel et al.[20] proved that miR-135b binds 3′ UTR of the APC gene, which leads to accumulation of free β-catenin in cells. The other identified miRNAs were down-regulated in CRC tissues, which is in accordance with previous observations that miRNA levels are globally decreased in human cancers [4]. Several of them were already described in connection with CRC in previous studies (miR-192, miR-215, miR-26b, miR-143, miR-145, miR-16 [10], miR-139-5p [21], miR-138 [22], miR-451 [23], miR-378, miR-378* [24], miR-133b [9], miR-144* [25], miR-195 [26], miR-194 [27], miR-30c, miR-26a [28], miR-342-3p [24] and miR-101 [29]), whereas remaining were identified in this study for the first time (miR-190, miR-422a, miR-375, miR-376, miR-486-5p, miR-598, miR-99a, miR-636, miR-100, miR-411, miR-30b, miR-30e, miR-30e*, miR-30a*, miR-192a*, miR-127-3p, miR-200b, miR-140-5p and miR-186). It seems that miR-30 family is important in CRC, because most of its members were down-regulated in tumour tissue. Xi et al. [28] described decreased expression of these miRNAs in CRC cell lines with TP53 gene deletion. MiR-143 and miR-145 are tumour suppressive miRNAs that are very repeatedly described as associated with CRC. Among their target molecules, there are oncogenes APC [9], ERK, RAF [30], EGFR [31], MYC [32], MAPK, CCND2 and transcription factors FOS, YES and FLI [5]. Boominathan [33] showed that expression of miR-145 depends on correct function of p53. This important molecule is also connected with other miRNAs – miR-192, miR-194 and miR-215- that belong to the same family and affect cell cycle arrest through p21 accumulation [34]. Lower expression of miR-195 is associated with lymph node positivity and shorter survival of patients with CRC. It is supposed that this miRNA can bind anti-apoptotic protein BLC-2 and thus promote apoptosis [26]. Similarly, miR-133b is able to regulate some proteins from the BCL-2 family [35]. Moreover, among important targets of this miRNA, there are oncogenes KRAS and MAP3K1 [9] supporting miR-133b function as an important tumour suppressor. Decreased levels of miR-101 are connected with enhanced function of COX-2 (cyclooxygenase 2), oversized oxidation of arachidonic acid and subsequently deregulated activation of prostaglandins, which leads to increased proliferation of cancer cells [29]. Although the number of research groups interested in miRNAs functioning in cancer in last years dramatically increased, new pathogenic miRNAs are still being discovered and further studies are necessary to understand the modus operandi of this complex regulatory network in CRC pathogenesis.
Therefore, we chose five miRNAs (miR-215, miR-375, miR-378, miR-422a and miR-135b) for further validation and in vitro analyses to find out what is their role in CRC. We used new collection of 125 paired-samples of tumour tissue and adjacent non-tumoural mucosa to confirm our results from miRNA profiling. We proved that miR-215, miR-375, miR-378 and miR-422a are significantly down-regulated, whereas miR-135b is up-regulated in tumour tissue (P < 0.0001). Furthermore, we analysed the correlation between the miRNA expression levels and clinical–pathological features of the CRC patients (see Table 1). It was found that lower expression of miR-215 and miR-422a is associated with advanced stages of the disease. Interestingly, expression of miR-135b was the highest in stage I, which is in accordance with previous studies describing the role of this miRNA in early stages of CRC by targeting APC and activating Wnt signalling [36]. Importantly, we also observed the relationship between lymph node positivity and the expression of miR-215, miR-378, miR-422a and miR-135b. This fact indicates a possible role of these miRNAs in migration of colon cancer cells, which was also analysed in this study. Moreover, expression of these miRNAs could contribute to better differentiation between clinical stages II and III, which is currently based on examination of lymph node positivity. Unfortunately, this approach is not precise enough, because about 25% of patients with lymph node involvement are not detected due to insufficient number of examined lymph nodes. Subsequently, these patients do not receive adjuvant systemic therapy increasing probability of relapse. Finally, we detected increased expression of miR-135b in samples of patients with higher pre-operative serum levels of CAE and CA19-9. This is in agreement with previous observations describing higher levels of miR-135b also in plasma of CRC patients, indicating its potential usage as circulating biomarker in CRC [13].
To explore the function of validated miRNAs in pathogenesis of CRC, DLD-1 and HCT-116 colon cancer cells were transfected with particular miRNA precursors or inhibitors. The functional analyses of miR-215 showed that overexpression of this miRNA leads to cell cycle arrest and enhanced apoptosis of HCT–116 cells carrying wt-p53, but no effect was observed in the case of DLD–1 cells containing mut-p53. These results are in accordance with previous studies that describe direct connection between miR-215 function and p53 status. Braun et al.[34] found out not only p53-responsive induction of miR-215 but also direct feedback of this miRNA on the activity of p53, which results in activation of apoptosis and cell cycle arrest in cells with wild-type p53, but not in cells with mutated p53. Moreover, we detected significantly decreased migration of cells transfected with pre-miR-215, supporting our observation that expression of this miRNA is lower in primary tumours with lymph node positivity. Karaayvaz et al. ([35]) observed prognostic potential of miR-215 in the small cohort of 34 CRC patients of II and III clinical stages. Expression levels of miR-215 were decreased in tumours, but, interestingly, higher levels of miR-215 were associated with worse survival (P = 0.025). Differences in miR-215 levels between tumour of clinical stages II and III were not evaluated [35]. These results together indicate that miR-215 is important in pathogenesis of CRC and could be used not only as a new biomarker of the disease but also as a potential therapeutic target for prevention of metastases.
The analyses of other miRNAs revealed that higher expression of miR-378, miR-375 and miR-422a is associated with accumulation of cells in G1 phase in HCT-116 cells. Concerning DLD-1 cells, we did not observe any significant effect on cell cycle and apoptosis. Therefore, we suppose that these miRNAs function in a similar way as miR-215, although the precise mechanism is not known and further identification of target molecules will be necessary. Using MTT assay, we observed a decreased viability of both DLD-1 and HCT-116 cells 48 hrs after transfection with pre-miR-375 or anti-miR-135b. Tsukamoto et al. [37] described that miR-375 inhibits expression of PDK1 (phosphoinositide-dependent kinase-1) and anti-apoptotic protein 14–3–3zeta by binding to its 3′ UTR, and its ectopic expression markedly reduced viability of gastric cancer cells.
In conclusion, we proved that miR-215, miR-375, miR-378 and miR-422a evince significant tumour suppressive properties, whereas miR-135b functions as an oncogene. Our observations from analysis of clinical CRC samples indicate potential usage of validated miRNAs as biomarkers, and our functional screening suggests that some of them, mainly miR-215, represent potentially important targets for novel therapeutic strategies in CRC. However, exact molecular mechanisms of these miRNAs functioning could not be fully understood as their target molecules have not been experimentally validated.
Acknowledgments
This study was supported by Internal Grant Agency of the Czech Ministry of Health (IGA MZ CR) NS 9814-4/2008 and by project MZ0MOU2005, by The Ministry of Education, Youth and Sports for the project BBMRI CZ (LM2010004), and by the project “CEITEC – Central European Institute of Technology” (CZ.1.05/1.1.00/02.0068).
Conflicts of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Siegel R, Ward E, Brawley O, et al. Cancer statistics, 2011. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Janga SC, Vallabhaneni S. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv Exp Med Biol. 2011;722:59–74. doi: 10.1007/978-1-4614-0332-6_4. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Lee CG. MicroRNA and cancer – focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slaby O, Svoboda M, Michalek J, et al. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 6.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–24. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 7.Sarver AL, French AJ, Borralho PM, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandrés E, Cubedo E, Agirre X, et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earle JSL, Luthra R, Romans A, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433–40. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koga Y, Yasunaga M, Takahashi A, et al. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res. 2010;3:1435–42. doi: 10.1158/1940-6207.CAPR-10-0036. [DOI] [PubMed] [Google Scholar]
- 12.Díaz R, Silva J, García JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosom Cancer. 2008;47:794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 13.Ng EKO, Chong WWS, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 14.Pu X-xiang, Huang G-liang, Guo H-qiang, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674–80. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang L-G, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2011 doi: 10.1016/j.canep.2011.05.002. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Reimers M, Carey VJ. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 2006;411:119–134. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- 17.Miko E, Czimmerer Z, Csánky E, et al. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35:646–64. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 18.Hannafon BN, Sebastiani P, de las Morenas A, et al. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13:R24. doi: 10.1186/bcr2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer A, Jung M, Mollenkopf H-J, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–76. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 20.Nagel R, le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 21.Chang KH, Miller N, Kheirelseid EAH, et al. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26:1415–22. doi: 10.1007/s00384-011-1279-4. [DOI] [PubMed] [Google Scholar]
- 22.Paun BC, Cheng Y, Leggett BA, et al. Screening for microsatellite instability identifies frequent 3′-untranslated region mutation of the RB1-inducible coiled-coil 1 gene in colon tumors. PLoS One. 2009;4:e7715. doi: 10.1371/journal.pone.0007715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bitarte N, Bandres E, Boni V, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells. 2011;29:1661–71. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 24.Wang YX, Zhang XY, Zhang BF, et al. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–4. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 25.Kalimutho M, Del Vecchio Blanco G, Di Cecilia S, et al. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011;46:1391–402. doi: 10.1007/s00535-011-0456-0. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Chen L, Xu Y, et al. MicroRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–40. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 27.Sundaram P, Hultine S, Smith LM, et al. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res. 2011;71:7490–501. doi: 10.1158/0008-5472.CAN-11-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi Y, Formentini A, Chien M, et al. Prognostic values of microRNAs in colorectal cancer. Biomark Insights. 2006;2:113–21. [PMC free article] [PubMed] [Google Scholar]
- 29.Strillacci A, Griffoni C, Sansone P, et al. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–47. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 31.Cho WCS, Chow ASC, Au JSK. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011;8:125–31. doi: 10.4161/rna.8.1.14259. [DOI] [PubMed] [Google Scholar]
- 32.Sachdeva M, Mo Y-Y. miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res. 2010;2:170–80. [PMC free article] [PubMed] [Google Scholar]
- 33.Boominathan L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 2010;29:613–39. doi: 10.1007/s10555-010-9257-9. [DOI] [PubMed] [Google Scholar]
- 34.Braun CJ, Zhang X, Savelyeva I, et al. p53-responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karaayvaz M, Pal T, Song B, et al. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. 2011;10:340–7. doi: 10.1016/j.clcc.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Necela BM, Carr JM, Asmann YW, Thompson EA. Differential expression of microRNAs in tumours from chronically inflamed or genetic (APC(Min/+)) models of colon cancer. PLoS One. 2011;6:e18501. doi: 10.1371/journal.pone.0018501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–49. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]


