Synopsis
Survivin, a member of the Inhibitor of Apoptosis Protein (IAP) family, plays important roles in maintaining cellular homeostasis and regulating cell cycle progression. This IAP is overexpressed in esophageal cancer cells leading to uncontrolled cell growth and resistance to apoptosis. CUG-binding protein 1 (CUG-BP1) is an RNA-binding protein that regulates the stability and translational efficiency of target mRNAs. In this paper, we report that CUG-BP1 is overexpressed in esophageal cancer cell lines and human esophageal cancer specimens. CUG-BP1 associates with the 3′ untranslated region of survivin mRNA, thereby stabilizing the transcript and elevating its expression in esophageal cancer cells. Our results show that overexpression of CUG-BP1 in esophageal epithelial cells results in increased survivin mRNA stability and consequently survivin protein expression. Conversely, silencing CUG-BP1 in esophageal cancer cells destabilizes survivin mRNA, lowering survivin protein levels. In addition, we have found that altering CUG-BP1 expression modulates susceptibility to chemotherapy-induced apoptosis. Overexpression of CUG-BP1 in esophageal epithelial cells increases resistance to apoptosis, while silencing CUG-BP1 makes esophageal cancer cells more susceptible to chemotherapy-induced apoptosis. Cotransfection experiments with siRNA directed against survivin suggest that the anti-apoptotic role of CUG-BP1 is not entirely dependent on its effect on survivin expression.
Keywords: CUG-BP1, survivin, mRNA stability, apoptosis, 3′-untranslated region, post-transcriptional regulation
Introduction
The prevalence of esophageal cancer in the United States has been increasing over the last 4 decades. The American Cancer Society estimates that there will be 16,980 new cases of esophageal cancer and 14,710 deaths from this disease in 2011. Despite improvements in early detection and the improved efficacy of multimodality therapy in the treatment of this disease, overall 5-year survival rates remain only 20% [1].
Esophageal cancer cells demonstrate a marked resistance to apoptosis that is partly mediated by the overexpression of survivin, a member of the Inhibitor of Apoptosis Protein (IAP) family [2]. Survivin is expressed during the G2/M phase of the cell cycle and associates with microtubules of the mitotic spindle. Overexpression of survivin functions to overcome this apoptotic checkpoint and favors the aberrant progression of transformed cells through mitosis [3]. In two separate studies examining squamous esophageal cancer specimens, overexpression of survivin has been associated with poor prognosis [4–5]. Survivin overexpression has also been correlated with resistance to chemotherapy-induced apoptosis in esophageal cancer specimens [6]. More recently, overexpression of survivin mRNA and protein have been observed in Barrett’s esophagus compared to normal esophageal epithelium, suggesting that survivin overexpression likely occurs relatively early in the metaplasia-dysplasia-invasive carcinoma sequence [7–8]. Importantly, a recent study of 59 patients found decreased overall survival in patients with elevated survivin expression in their resected esophageal cancer specimens following induction of chemoradiation [9].
Current understanding of the mechanisms responsible for the regulation of survivin expression is incomplete. Transcription of the survivin gene is activated by a number of transcriptional factors including the β-catenin/TCF complex, c-Myc, and Stat-3 [10–12], and conversely inhibited by p53 [13–14]. However, little is known about the post-transcriptional regulation of survivin. CUG-BP1 is part of the CELF family of RNA-binding proteins and its role in cancer is beginning to be elucidated. It was first discovered in myotonic dystrophy and was found to bind to abnormally extended CUG mRNA repeats [15]. The binding of CUG-BP1 to its mRNA targets has most often been associated with decreased translation and target mRNA degradation. Recent reports demonstrate that the binding of CUG-BP1 to the mRNA of both tumor necrosis factor-α and p21 results in enhanced target mRNA expression, through increased mRNA stability and translation, respectively [16–17].
We have observed that CUG-BP1 is overexpressed in esophageal cancer cells compared to esophageal epithelial cells. Because survivin mRNA contains multiple potential CUG-BP1 binding sites, we hypothesized that CUG-BP1 bound to survivin mRNA and enhanced survivin protein expression. The experiments presented in this manuscript demonstrate for the first time that 1) overexpression of CUG-BP1 is strongly correlated with the overexpression of survivin observed in esophageal cancer cell lines and human specimens; 2) CUG-BP1 binds to the 3′untranslated region (UTR) of survivin mRNA; 3) this interaction increases the half-life of survivin mRNA in esophageal epithelial cells; and 4) silencing CUG-BP1 in esophageal cancer cells increases their susceptibility to chemotherapy-induced apoptosis.
Experimental
Cells and cell culture
The human esophageal cancer cell lines TE7 and TE10 were received as gifts from Dr. Nishihira. They were derived from an esophageal adenocarcinoma and squamous cell carcinoma, respectively, and characterized in the Cell Resource Center for Biomedical Research, Tohoku University. These cell lines were cultured in Roswell Park Memorial Institute (Mediatech Inc, Herndon, VA) supplemented with 10% heat-inactivated fetal bovine serum, and 1% L-glutamine (from Mediatech Inc), and maintained in a 37°C incubator with 5% CO2 humidified air.
Human esophageal (nhESO) epithelial cells were derived from esophageal specimens harvested at the time of donor lung procurement. Esophageal specimens were opened and washed with sterile medium. The mucosa was then removed by careful dissection along the submucosal plane separating the mucosal and muscular layers of the esophagus. This mucosal layer was then cut into small pieces and digested in a collagenase medium to create a single cell suspension. These esophageal mucosal cells were then maintained in BEBM media (Lonza Corporation, Walkersville, MD) supplemented with 20% heat-inactivated fetal bovine serum and the BEGM BulletKit. Passages 5 to 20 were used for the experiments. Stock cells were maintained in T-150 flasks and incubated at 37°C with 5% CO2 humidified air.
Reagents and antibodies
Anti-human survivin was purchased from R&D Systems (Minneapolis, MN). Anti-CUG-BP1, anti-caspase-3, anti-procaspase-3, anti-Lsm4, anti-Ago2, and anti-CDK4 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-actin mouse monoclonal antibody was obtained from EMD Biosciences (La Jolla, CA). (S)-(+)-Camptothecin was purchased from Sigma-Aldrich (St. Louis, MO).
Histologic analysis of human esophageal specimens
Representative sections were taken from both tumor and normal epithelium distant from the tumor from resected esophagectomy specimens. The sections were deparaffinized in xylene and rehydrated in graded series of ethanol. Sections were pre-treated with heat-induced epitope retrieval using a pressure cooker and Target Retrieval Solution pH 6.1 (Dako, Carpinteria, CA), followed by endogenous peroxidase blocking for 5 minutes with 0.3% hydrogen peroxide. The sections were then incubated overnight with rabbit anti-survivin or murine anti-CUG-BP1 antibodies at 1:250 dilution at 4°C in a hydration chamber. Antibody detection was performed by incubation with biotinylated goat anti-rabbit or anti-mouse secondary antibody (Dako), respectively, for 30 minutes at room temperature. Slides were developed for 5 minutes using diaminobenzidine as the chromagen (Dako) and were counterstained with hematoxylin. As a negative control, tissue sections were reacted with non-immune rabbit IgG plus the secondary antibody.
Western blot analysis
Whole cell lysates were prepared in buffer containing 2% sodium dodecyl sulfate sample buffer (250 nM Tris·HCl, pH 6.8, 2% sodium dodecyl sulfate, 20% glycerol, and 5% mercaptoethanol), and protein concentrations were determined using the BCA Protein Assay kit (Pierce Biotechnology, Rockford, IL). Twenty-five micrograms of protein were denatured in 5× SDS loading buffer at 95°C for 5 minutes, separated on 10% or 15% SDS-PAGE gels (Bio-Rad, Hercules, CA), and transferred onto nitrocellulose membranes. After 1 hour of blocking with 5% non-fat milk, membranes were incubated with specific antibodies followed by horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin, and signal was detected by Chemiluminescence Reagent (NEN, Boston, MA) and visualized by autoradiography. Densitometry analysis was performed via ImageJ software (NIH, Bethesda, MD).
RNA interference
Cells were transfected with oligonucleotides targeting CUG-BP1, Lsm4 (Santa Cruz Biotechnology, Santa Cruz, CA), or survivin (Cell Signaling Technologies, Danvers, MA) or a control scrambled sequence (Qiagen, Valencia, CA) at a concentration of 300pmol with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) for 6 hours.
CUG-BP1 overexpression
Cells were transfected at 60–70% confluency in 60-mm plates with Homo sapiens CUG-BP, Elav-like family member 1 cDNA or the empty pCMV6-XL5 control vector (OriGene, Rockville, MD). Two micrograms of cDNA were transfected into nhESO or TE7 cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for 6 hours.
Preparation of synthetic RNA transcripts
Complementary DNA (cDNA) from nhESO and TE7 cells were used as PCR templates for in vitro synthesis of biotinylated survivin coding region (CR) and 3′-UTR transcripts. The T7 RNA polymerase promoter sequence (5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′) was appended to the 5′-end of all fragments. Primers used for the preparation of biotinylated transcripts spanning the CR and 3′-UTR are listed in Table 1.
Table 1.
Primers used for preparation of biotinylated transcripts
| Sequence | Region | |
|---|---|---|
| CR | (T7) 5′-CCATTAACCGCCAGATTTGA-3′ 5′-TCAATCCATGGCAGCCAGCT-3′ |
66–619 |
| FL | (T7) 5′-TGTCTTGAAAGTGGCACCAG-3′ 5′-ACTTTCCAGGATGGCAGTTG-3′ |
773–2576 |
| F1 | (T7) 5′-CATGGATTGAGGCCTCTGG-3′ 5′-CCTCCCTCACTTCTCACCTG-3′ |
610–918 |
| F2 | (T7) 5′-AGGGAGGAAGAAGGCAGTGT-3′ 5′-AGAGGAGCCAGGGACTCTGT-3′ |
912–1199 |
| F3 | (T7) 5′-CAGAGTCCCTGGCTCCTCTA-3′ 5′-TAGGCCACTGCCTTTTTCTG-3′ |
1181–1458 |
| F4 | (T7) 5′-AGGCAGTGGCCTAAATCCTT-3′ 5′-TGACCTCCAGAGGTTTCCAG-3′ |
1446–1723 |
| F5 | (T7) 5′-ATCTCGGCTGTTCCTGAGAA-3′ 5′-CAAGCGCCCAGAGACAACT-3′ |
1723–1903 |
| F6 | (T7) 5′-CTTGCCAGAGCCACGAAC-3′ 5′-CCATTTACAGACTGACACAAATATCA-3′ |
1900–2136 |
| F7 | (T7) 5′-GTCAGTCTGTAAATGGATACTTCACT-3′ 5′-ATGTCGAGGAAGCTTTCAGGT-3′ |
2121–2380 |
| F8 | (T7) 5′-CCTGAAAGCTTCCTCGACAT-3′ 5′-AAAACCACATGAGACTTTATTGG-3′ |
2361–2695 |
RNA-protein binding assays
Six micrograms of biotinylated transcripts were incubated with 120μg of cytoplasmic lysates for 1 hour at room temperature. Paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) was utilized to isolate bound complexes, and Western blot analysis using antibodies recognizing CUGBP1 was subsequently performed. To assess the association of both CUG-BP1 and Ago2 with endogenous survivin mRNA, mRNP IP (immunoprecipitation) assays were performed. Whole cell lysates were used for IP in the presence of anti-CUG-BP1, anti-Ago2, or non-specific IgG. RNA in the IP materials was used in qRT-PCR reactions to detect the presence of survivin mRNA.
RT (reverse transcription)-PCR and qRT-PCR (quantitative real-time PCR) analysis
Total RNA isolation was carried out with the RNeasy mini kit (QIAGEN, Valencia, CA) and reverse transcribed with the Reverse Transcription System (Promega Corporation, Madison, WI). The resulting complimentary DNA (cDNA) was amplified with primers specific for survivin and GAPDH. The forward primer for survivin was 5′-TTTCTGCCACATCTGAGTCG-3′ and the reverse primer was 5′-TGTCGAGGAAGC TTTCAGGT-3′. The forward GAPDH primer was 5′-GTCAGTGGTGGACCTGACCT-3′ and the reverse primer was 5′-AGGGGTCTACATGGCAACTG-3′. The levels of GAPDH PCR product were assessed to monitor the even input of RNA in RT-PCR samples. qRT-PCR was performed using the 7500-Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) with specific primers, probes and software (Applied Biosystems). The levels of survivin mRNA were quantified by qRT-PCR analysis and normalized with to GAPDH levels.
Apoptosis assays
Cells were incubated with camptothecin and cell apoptosis was assessed after 6 hours. Cells were photographed with a Nikon inverted microscope and quantified by morphologic analysis. Approximately 75 and 300 cells were counted for nhESO and TE7, respectively, and the numbers of apoptotic cells were expressed as a fraction of the total number of cells counted. For caspase activation assays, cells were prepared in a similar manner and caspase-3 activity was assessed using the Caspase-3 Colorimetric Assay (R&D Systems, Minneapolis, MN) according to manufacturer’s instructions.
Messenger RNA stability
Messenger RNA stability assays were performed to measure the differences in mRNA half-lives. Briefly, pretreated cells were incubated with actinomycin D (Sigma-Aldrich, St. Louis, MO) for specified time points at a final concentration of 5μg/μl and total RNA was extracted for PCR analyses.
Statistics
Analyses were performed using Prism 5.0 software (GraphPad, La Jolla, CA). Data derived from multiple determinations were subjected to two-sided t tests and Two-way ANOVA analyses. P values ≤0.05 were considered statistically significant.
Results
CUG-BP1 and survivin levels are increased in human esophageal cancer specimens and esophageal cancer cell lines
Esophagectomy specimens from patients with esophageal adenocarcinoma containing both normal and malignant tissue were examined for both CUG-BP1 and survivin expression using immunohistochemical (IHC) staining. Figure 1A-a shows hematoxylin and eosin (H&E) staining of a representative adjacent benign squamous mucosa (top) and esophageal cancer specimen (bottom) from the same patient. IHC staining for CUG-BP1 (Figure 1A-b) depicts expression in the basal zone of the benign squamous mucosa with a loss of expression in the luminal surface. The adjacent malignant glandular tissue shows diffuse staining of CUG-BP1. Similarly, IHC staining for survivin (Figure 1A-c) reveals high expression in the basal zone of the benign squamous mucosa, which is also reduced in the luminal surface. The adjacent malignant glandular tissue exhibits strong, diffuse staining for survivin.
Figure 1.
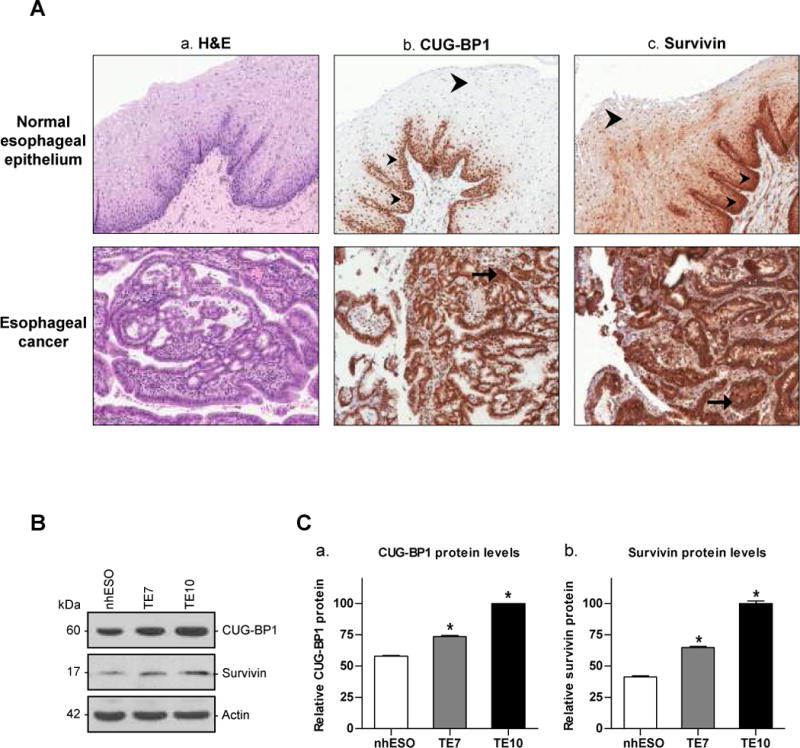
A: A representative esophagectomy specimen containing both normal and malignant tissue was examined after H&E staining and IHC staining for CUG-BP1 and survivin. a. Representative photomicrograph (100×) of H&E stained benign squamous mucosa (top) and malignant and invasive glandular tissue (bottom). b. IHC staining for CUB-GP1 (100×). The benign squamous mucosa shows expression in the basal zone (small arrowheads), with loss of expression at the luminal surface (large arrowhead). The adjacent malignant glandular tissue shows diffuse staining (arrow) of CUG-BP1. c. IHC staining for survivin (100×). The benign squamous mucosa shows marked staining for survivin in the basal zone (small arrowheads), which is reduced at the luminal surface (large arrowhead). The adjacent malignant glandular tissue shows strong diffuse staining for survivin (arrow). B: Baseline levels of CUG-BP1 and survivin protein in nhESO cells, and two esophageal cancer cell lines, TE7 and TE10. Twenty-five micrograms of total protein were loaded into each lane, and immunoblots were probed with either a CUG-BP1 or survivin-specific antibody. Actin hybridization was performed as an internal control to ensure equal loading. The approximate sizes of CUG-BP1, survivin, and actin are 60kDa, 17kDa and 42kDa, respectively. C: Western blot analysis from (B) was quantified by densitometry and graphed relative to TE10 cells. *p < 0.05 compared with nhESO.
Expression patterns of CUG-BP1 and survivin were also examined in nhESO cells and the TE7 and TE10 esophageal cancer cell lines. Western blot analysis reveals that CUG-BP1 and survivin levels are significantly increased in both TE7 and TE10 cells in comparison to nhESO (Figure 1B). Densitometry analysis shows an approximate 1.3- and 1.75-fold increase in CUG-BP1 and a 1.6- and 2.2-fold increase in survivin in TE7 and TE10 cells, respectively (Figure 1C). These findings indicate that the up-regulated expression of survivin is correlated with increased levels of CUG-BP1 in both human esophageal cancer tissue samples and cell lines. This primary observation led us to speculate that the increased levels of CUG-BP1 in esophageal cancer cells may play an important role in regulating survivin expression.
CUG-BP1 binds to the survivin mRNA 3′ untranslated region (UTR)
We first examined the interaction of intracellular survivin mRNA with CUG-BP1 by immunoprecipitating CUG-BP1 under conditions which preserve its association with target mRNAs in ribonucleoprotein (RNP) complexes using an anti-CUG-BP1 antibody. Following isolation of RNA, the presence of survivin mRNA in these RNP complexes was confirmed by qRT-PCR analysis (Figure 2A). The association of survivin mRNA with CUG-BP1 was 8-fold higher in TE7 cells compared to nhESO cells in keeping with the increased levels of survivin in these cells. This interaction was specific because survivin mRNA was almost undetectable in complexes isolated with control non-specific IgG antibodies.
Figure 2.
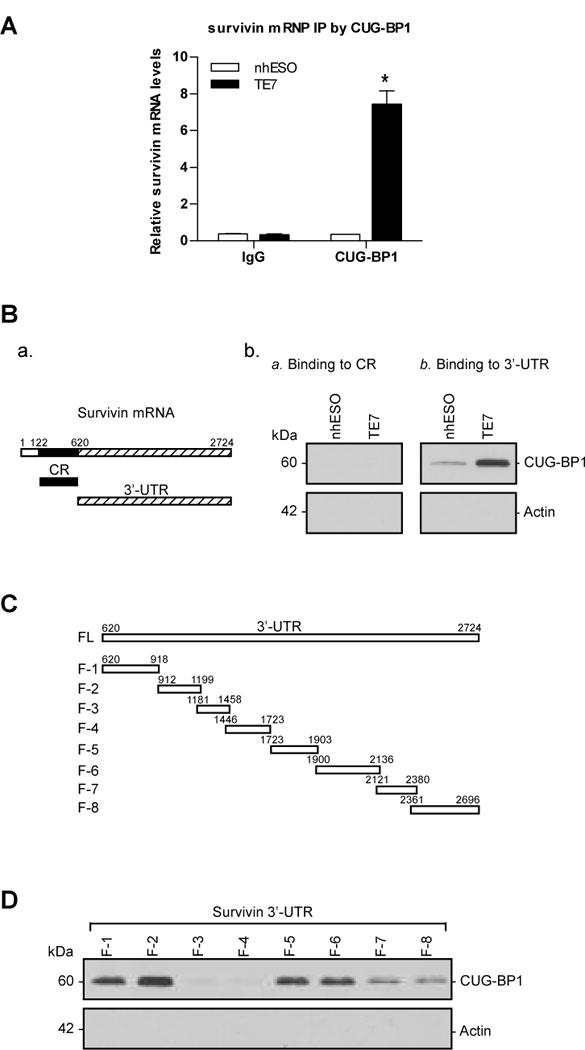
A: Levels of association of endogenous CUG-BP1 with survivin mRNA in nhESO and TE7 cell lines. Whole cell lysates from nhESO and TE7 cells were used for immunoprecipitation (IP) in the presence of anti-CUG-BP1 antibody, or non-specific IgG. RNA in the IP materials was used in qRT-PCR reactions to detect the presence of survivin mRNA. *p < 0.05 compared nhESO cells immunoprecipitated with CUG-BP1. B: a. Schematic representation of survivin mRNA coding region (CR) and 3′ untranslated region (UTR). b. Binding of CUG-BP1 to the CR (a) and 3′ UTR (b) of survivin mRNA in biotin pull-down experiments. Cytoplasmic lysates from nhESO and TE7 cell lines were isolated and incubated with 6μg of biotinylated survivin CR or 3′-UTR. Streptavidin-coated beads were used to pull-down the resultant RNP complexes. The presence of CUG-BP1 in the pull-down material was assayed by Western blotting. Actin in the pull-down material was also examined and served as a negative control. c. Levels of input CUG-BP1 and actin proteins in each cell line are shown by Western blot. C: Schematic representation of the fragmented survivin 3′-UTR biotinylated transcripts used in this series of experiments. D: Representative CUG-BP1 immunoblots of the material pulled down by the different biotinylated fragments of the survivin mRNA 3′-UTR (F-1 to F-8), using the same methods as described in (B).
Based on sequence analysis of survivin mRNA, we found multiple potential CUG-BP1 binding sites in both the coding region (CR) and 3′-UTR. The formation of CUG-BP1-survivin mRNA complexes was tested by using biotinylated transcripts spanning either the CR or 3′-UTR regions of survivin mRNA. As shown in Figure 2B-b, Western blot analyses of pull-down materials reveal that the survivin 3′-UTR transcript readily associates with CUG-BP1 in both nhESO and TE7 cells. These associations could be seen more prominently in TE7 cells, as expected because of previously observed increases in CUG-BP1 and survivin expression in TE7 cells. However, no complexes could be observed with the CR transcript, despite the presence of potential CUG-BP1 binding sites.
The survivin 3′-UTR was further subdivided into smaller transcripts in order to determine the specific binding location of CUG-BP1 (Figure 2C). Biotinylated transcripts spanning eight fragments of the survivin 3′-UTR (each containing several potential CUG-BP1 binding sites) were constructed and analyses of pull-down materials were performed. CUG-BP1 was found to associate most strongly to F-2, moderately to F-1, F-5 and F-6, and less strongly to F-7 and F-8. CUG-BP1-survivin mRNA complexes were not observed in F-3 and F-4 even though several potential CUG-BP1 binding motifs were located in these regions (Figure 2D). Binding of survivin mRNA was specific to CUG-BP1 because no complexes were formed when transcripts were pulled down with actin. These findings clearly indicate that CUG-BP1 is able to bind and associate specifically with survivin 3′-UTR, and that its strongest affinity is to the region spanning the 912–1199 bp region.
CUG-BP1 manipulation leads to altered survivin mRNA stability and protein expression
Because basal levels of CUG-BP1 in nhESO cells are low, transfection of CUG-BP1 cDNA into these cells was performed in order to transiently elevate CUG-BP1 levels and assess the effects on survivin mRNA stability. As shown in Figure 3A, a CUG-BP1-expressing vector was successfully transfected into nhESO cells, resulting in an approximately 3-fold increase in CUG-BP1 protein levels compared with control vector. Importantly, this increase in CUG-BP1 protein levels was associated with an approximately 5-fold increase in survivin protein levels. Concurrently, overexpression of CUG-BP1 increased survivin mRNA levels (Figure 3B). This finding was due, in part, to stabilization of survivin mRNA as revealed by an increase in its half-life from approximately 3 hours to 16 hours (Figure 3C and D). These experiments reveal that overexpression of CUG-BP1 stabilizes survivin mRNA resulting in elevated survivin protein levels in esophageal epithelial cells.
Figure 3.
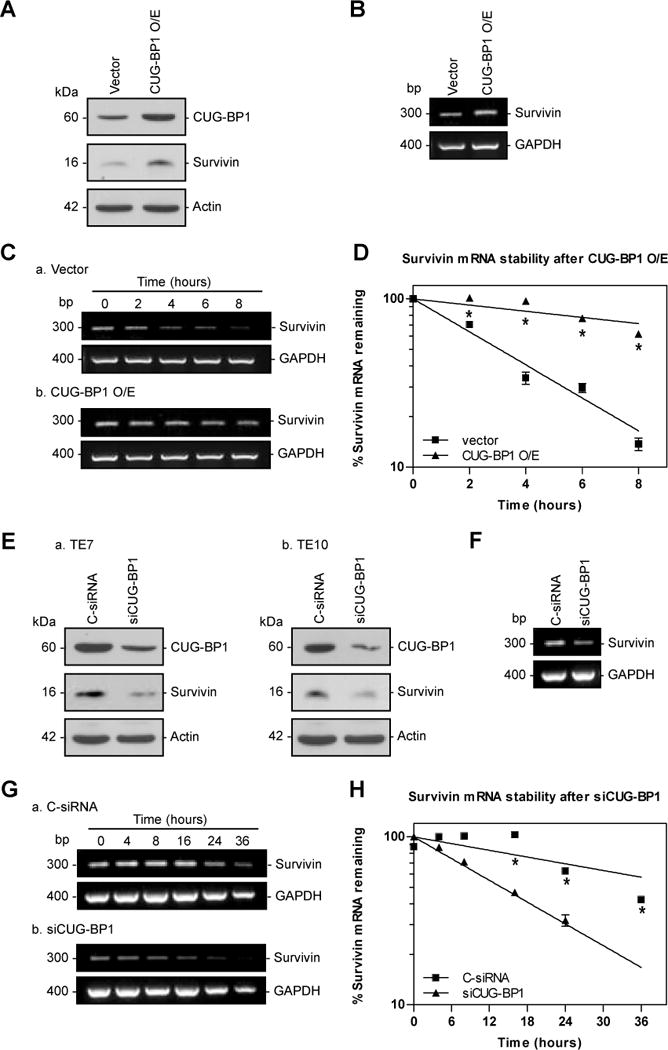
Changes in survivin mRNA stability following modulation of CUG-BP1. A: Representative immunoblots of CUG-BP1 and survivin after overexpression of CUG-BP1 in nhESO cells. Following transfection with control vector or a CUG-BP1 overexpressing vector whole cell lysates were harvested for Western blot analysis. Actin was used as a loading control. B: Levels of survivin mRNA in cells that were processed as described in (A). Total cellular RNA was extracted and levels of survivin mRNA were measured by RT-PCR. GAPDH was concurrently amplified to serve as a control for equal loading. Survivin and GAPDH mRNA sizes are as indicated on the left. C: Half-life of survivin mRNA in cells described in (A). Total cellular RNA was isolated at indicated times after administration of actinomycin D (5μg/μl), and the remaining levels of survivin and GAPDH mRNA were measured by qRT-PCR analysis. D: Percentage of survivin mRNA remaining in cells that were processed as described in (C). Relative levels of survivin mRNA were normalized to the amount of GAPDH mRNA (optical density of survivin mRNA/optical density of GAPDH mRNA). *p < 0.001 compared with nhESO cells transfected with vector control. E: Representative immunoblots of CUG-BP1 and survivin protein in CUG-BP1-silenced cells. After TE7 and TE10 cells were transfected with either siRNA targeting CUG-BP1 (siCUG-BP1) or C-siRNA for 48 hours, whole cell lysates were isolated for Western blot analysis. Actin was used as an internal loading control. CDK4 levels were also determined after silencing CUG-BP1 in TE7 cells. F: Levels of survivin mRNA in TE7 cells that were processed as described in (E). G: Half-life of survivin mRNA in TE7 cells described in (E), following procedure described in (C). H: Percentage of survivin mRNA remaining in TE7 cells that were processed as described in (G). *p < 0.001 compared with TE7 cells transfected with C-siRNA.
In reciprocal experiments, siRNA directed against CUG-BP1 (siCUG-BP1) was employed in order to reduce CUG-BP1 levels in TE7 and TE10 cells because basal CUG-BP1 levels are elevated in these cell lines. Forty-eight hours post-transfection, CUG-BP1 protein was effectively reduced by approximately 3-fold compared to control siRNA (C-siRNA) transfection (Figure 3E). This reduction in CUG-BP1 expression resulted in decreased levels of survivin protein in TE7 and TE10 cells. To demonstrate that the down-regulation of survivin expression following CUG-BP1 silencing was not merely a reflection of global changes in cell growth, we examined the effect of silencing CUG-BP1 on Cyclin-Dependent Kinase 4 (CDK4). Western blot analysis shows that silencing CUG-BP1 increases CDK4 protein expression (Figure 3E-c). Survivin mRNA was also decreased after silencing CUG-BP1 (Figure 3F). Messenger RNA stability analysis of survivin was performed in TE7 cells which reveal a decrease in the survivin mRNA half-life from approximately 45 hours to 14 hours (Figure 3G and H). These studies show that silencing CUG-BP1 destabilizes survivin mRNA leading to decreased survivin protein expression in esophageal cancer cells.
Binding of CUG-BP1 to survivin mRNA prevents its association with P-bodies
To further determine the underlying mechanism by which CUG-BP1 stabilizes survivin mRNA, we examined the role of P-bodies in this system. P-bodies are intracellular organelles in which mRNA degradation occurs. We postulate that in the presence of CUG-BP1, survivin mRNA is prevented from being transported to P-bodies, leading to its increased expression. To test this hypothesis, we measured the amount of survivin mRNA associated with Ago2, an important component of the P-body, following CUG-BP1 silencing. After silencing CUG-BP1 in TE7 cells, the detected levels of survivin mRNA were increased in the Ago2-immunoprecipitated materials compared with cells treated with C-siRNA. Survivin mRNA levels were minimal when immunoprecipitated with non-specific anti-IgG antibody (Figure 4A-b). In separate experiments, we simultaneously silenced both CUG-BP1 and Lsm4, another important P-body component. As seen in Figure 4B, silencing both CUG-BP1 and Lsm4 abrogates the reduction in survivin protein levels observed when CUG-BP1 alone is silenced. Together, these studies suggest that CUG-BP1 prevents transport of survivin mRNA to the P-body, thus maintaining high survivin expression levels in esophageal cancer cells.
Figure 4.
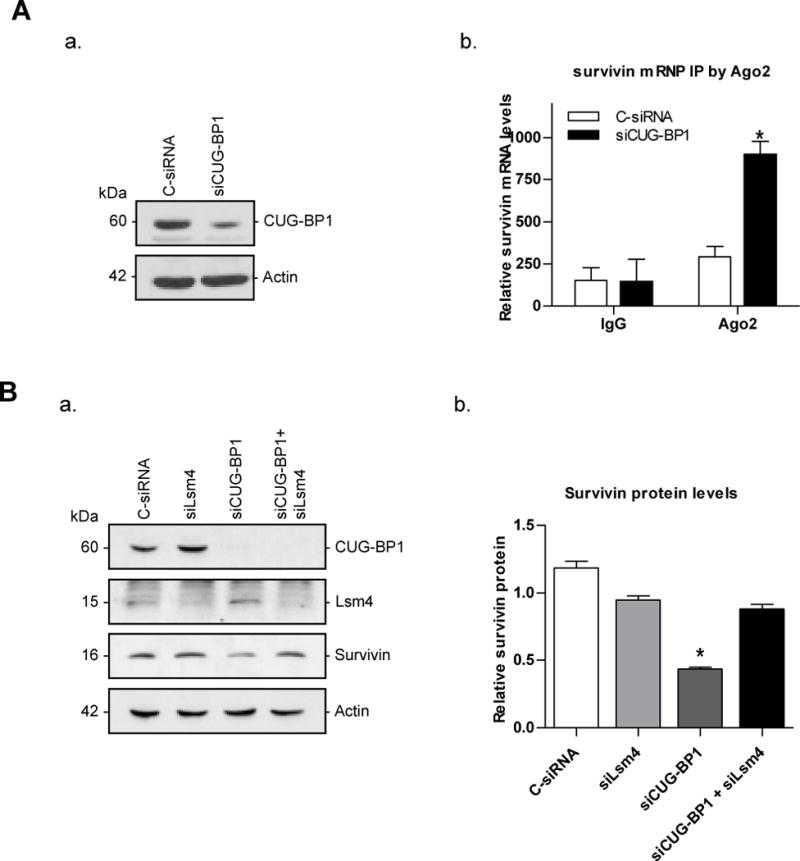
Silencing CUG-BP1 in esophageal cancer cells increases association of survivin mRNA with P-body components. A: a. Western blots confirming the successful silencing of CUG-BP1 by siRNA in TE7 cells. b. Levels of association of endogenous Ago2 with survivin mRNA in CUG-BP1-silenced TE7 cell lines. Whole cell lysates from TE7 cells treated with either C-siRNA or siCUG-BP1 were used for immunoprecipitation (IP) in the presence of anti-Ago2 antibody, or non-specific IgG. RNA in the IP materials was used in qRT-PCR reactions to detect the presence of survivin mRNA. B: Effect on survivin protein level after silencing both CUG-BP1 and Lsm4 in TE7 cells. a. Western blots confirming the silencing of CUG-BP1 and Lsm4, as well as the change in survivin protein levels. b. Densitometry analysis of survivin protein levels. *p < 0.05 compared with TE7 cells transfected with siLsm4 alone or in combination with siCUG-BP1.
Overexpression of CUG-BP1 desensitizes esophageal epithelial cells to apoptosis
As we have previously shown, the responsiveness of nhESO and TE7 cells to camptothecin-induced apoptosis correlates with survivin expression [2]. Based on our findings that CUG-BP1 overexpression leads to increased survivin expression in nhESO cells, we hypothesized that overexpression of CUG-BP1 in nhESO cells would result in increased resistance to apoptosis. CUG-BP1-overexpressing nhESO cells were incubated with 1μM camptothecin for 6 hours followed by measurement of caspase-3 levels. Western blot analysis confirmed the successful overexpression of CUG-BP1 and correspondingly revealed a decrease in caspase-3 protein levels following exposure to camptothecin (Figure 5A). Apoptosis was also quantified by morphometry and the percentage of apoptotic cells was significantly decreased in cells overexpressing CUG-BP1 (Figure 5B). Furthermore, a caspase-3 colorimetric analysis was done as another method to quantify apoptosis. Consistent with our previous findings, apoptosis was significantly reduced after the overexpression of CUG-BP1 compared to vector control (Figure 5C). This series of experiments indicates that the overexpression of CUG-BP1 significantly enhances resistance of nhESO cells to camptothecin-induced apoptosis, possibly through increased survivin protein expression.
Figure 5.
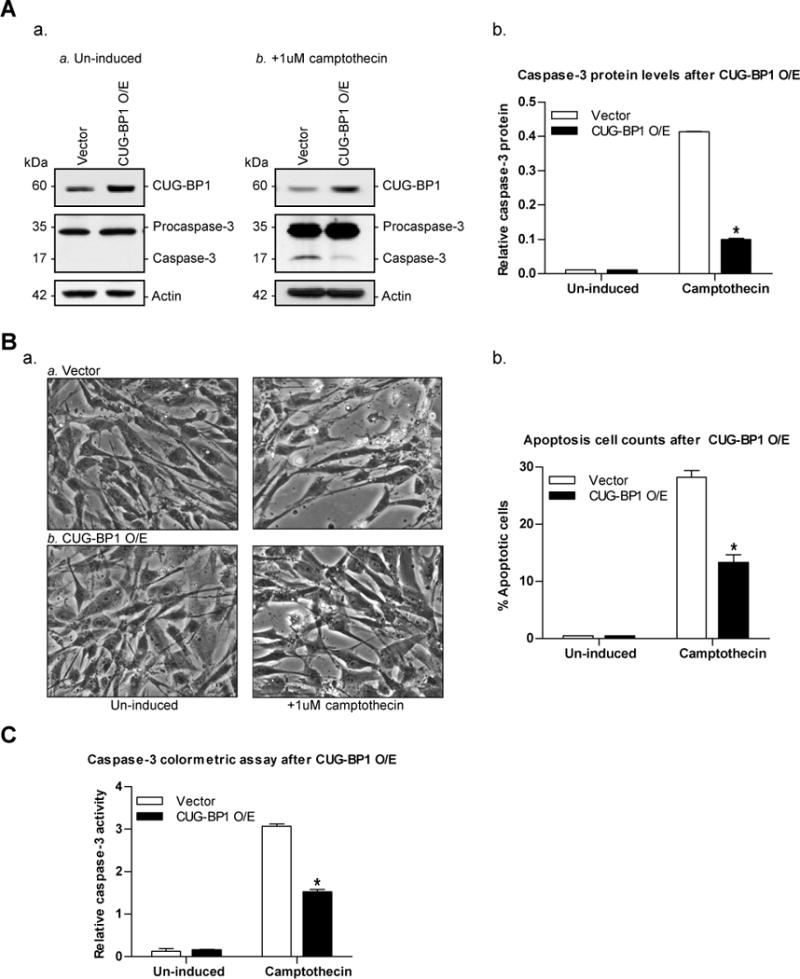
Overexpression of CUG-BP1 enhances resistance to apoptosis in nhESO cells. A: nhESO cells were transfected with either control vector (Vector) or the CUG-BP1 cDNA (CUG-BP1 O/E). Following transfection, cells were either not induced or exposed to camptothecin (1μM) for 6 hours and whole cell lysates were obtained. a. Western blot analysis of un-induced (Panel a) or camptothecin-induced (Panel b) levels of CUG-BP1, procaspase-3, caspase-3, and actin. b. Densitometry analysis quantifying caspase-3 protein levels relative to levels in un-induced samples. *p < 0.0001 compared to vector control. B: Apoptosis was measured in transfected cells as described in (A). a. Representative morphological analysis of treated cells. b. Percentage of apoptotic cells. *p < 0.01 compared to vector control. C: Early apoptosis was detected by caspase-3 colorimetric assay in cells prepared as described in (A). Whole cell lysates were collected and lysed by Lysis Buffer, then incubated with appropriate reaction buffers, and the caspase-3 colorimetric substrate (DEVD-pNA) at 37°C. Enzymatic activities for each condition were then read and recorded on a microplate reader using a 405 nm wavelength light after 1 hour. *p < 0.05 compared to vector control.
Silencing CUG-BP1 enhances susceptibility of TE7 cells to apoptosis
Based on these findings, we further hypothesized that silencing CUG-BP1 would increase the susceptibility of TE7 cells to apoptosis. Following successful CUG-BP1 silencing, TE7 cells were induced with 10μM camptothecin for 6 hours and apoptosis was measured. Western blot analysis confirmed the successful silencing of CUG-BP1 and correspondingly revealed an increase in caspase-3 protein levels following exposure to camptothecin (Figure 6A). Apoptotic cell counts, quantified by morphological changes associated with cell death, showed a 2-fold increase following CUG-BP1 silencing compared to C-siRNA transfection (Figure 6B). A caspase-3 colorimetric assay reveals an increased number of cells undergoing apoptotic cell death in siCUG-BP1-treated TE7 cells (Figure 6C). These findings indicate that silencing CUG-BP1 leads to increased susceptibility of esophageal cancer cells to apoptosis, possibly through decreased survivin protein levels.
Figure 6.
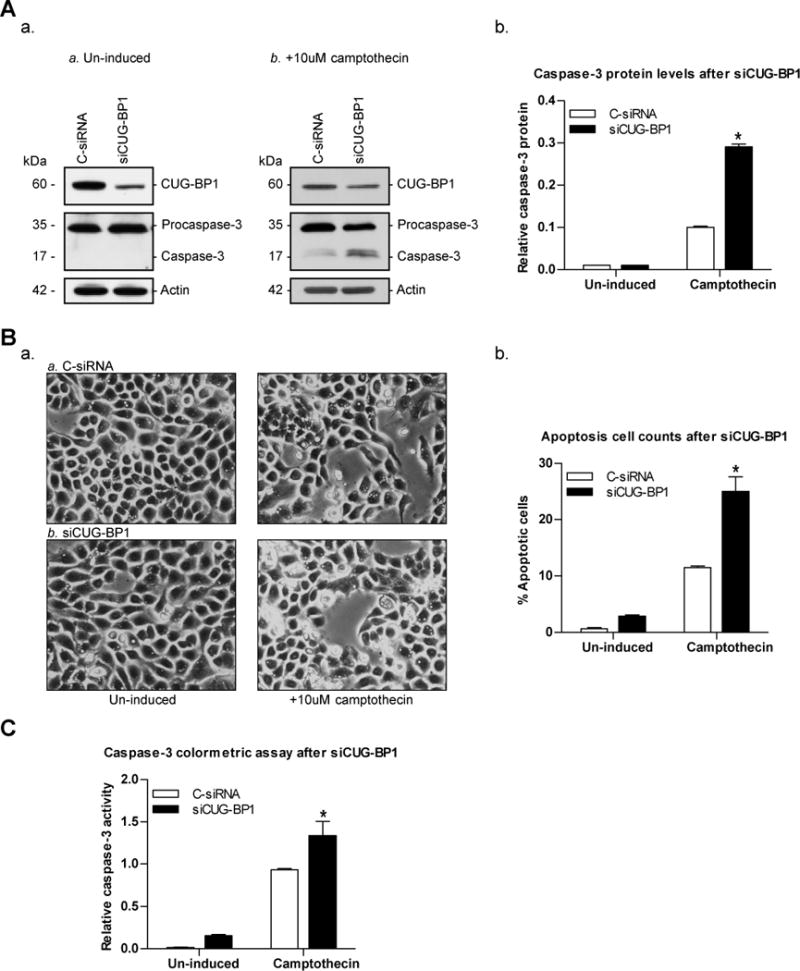
Down-regulation of CUG-BP1 in esophageal cancer cells sensitizes cells to apoptosis. A: a. Representative immunoblots of CUG-BP1, procaspase-3, caspase-3, and actin in CUG-BP1-silenced TE7 cells following no induction (Panel a) or exposure to camptothecin (10μM) for 6 hours (Panel b). b. Densitometry analysis of caspase-3 protein levels. *p < 0.0001 compared to C-siRNA. B: a. Representative morphologic analysis of treated cells. Cells were prepared as described in (A). b. Percentage of apoptotic cells. *p < 0.01 compared to C-siRNA. C: Caspase-3 colorimetric assay was performed as described in Figure 5D. *p < 0.05 compared to C-siRNA.
To test whether the increased susceptibility of TE7 cells to camptothecin-induced apoptosis observed following CUG-BP1 silencing was directly related to reduced survivin levels, co-transfection experiment was performed. TE7 cells were transfected with both siRNA directed against survivin (siSurvivin) and the CUG-BP1 overexpression vector, or appropriate controls, and then subjected to camptothecin for 6 hours. Western blot and densitometry analyses reveal successful silencing and overexpression of both survivin and CUG-BP1, respectively (Figure 7A). Despite the robust expression of CUG-BP1 in TE7 cells, CUG-BP1 levels approaching twice the baseline levels were achieved following overexpression. Apoptosis of un-induced and camptothecin-treated cells was visualized by pictomicrographs (Figure 7B) and quantified by cell counts (Figure 7C). In keeping with our previous results, TE7 cells treated with siSurvivin and control vector display a 3-fold increase in susceptibility to camptothecin-induced apoptosis compared to the double negative control (C-siRNA + vector). Interestingly, when CUG-BP1 was overexpressed in TE7 cells co-transfected with siSurvivin, these cells displayed a marked resistance to camptothecin-induced apoptosis, returning to baseline levels seen in the double negative control group. These important experiments suggest the anti-apoptotic effect exerted by CUG-BP1 is not entirely dependent on its effect on survivin expression.
Figure 7.
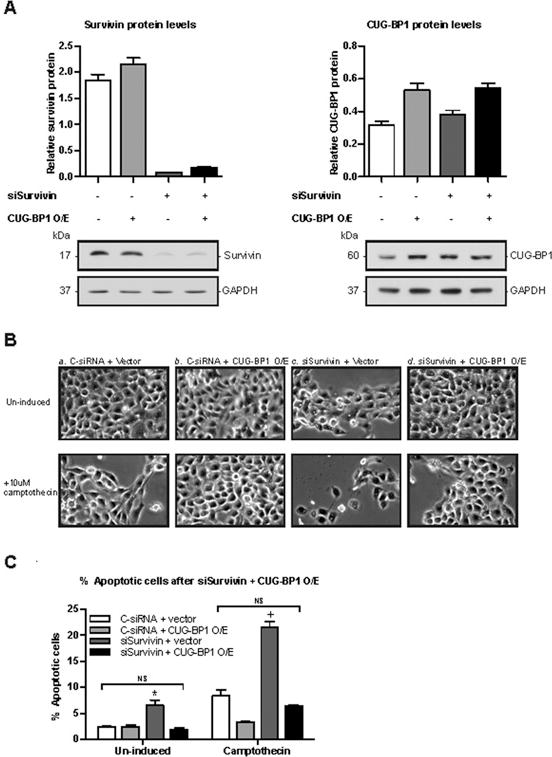
Overexpression of CUG-BP1 in esophageal cancer cells abrogates increased sensitivity to apoptosis following down-regulation of survivin. A: Representative immunoblots and densitometry analyses of survivin and CUG-BP1 after the co-transfection of siSurvivin plus CUG-BP1 cDNA in TE7 cells. B: Representative morphologic analyses of un-induced (top panels) and camptothecin-induced (bottom panels) TE7 cells after a. C-siRNA + vector, b. C-siRNA + CUG-BP1 O/E, c. siSurvivin + vector, and d. siSurvivin + CUG-BP1 O/E. Cells were transfected with appropriate siRNA and cDNA for 48 hours and then exposed to camptothecin for 6 hours. C: Percentage of apoptotic cells after co-transfection. Cells were prepared as described in (B) and apoptotic cells were counted and graphed as a percentage of total cells. *p < 0.05 compared to C-siRNA + CUG-BP1 O/E and siSurvivin + CUG-BP1 O/E; +p < 0.001 compared to C-siRNA + CUG-BP1 O/E and siSurvivin + CUG-BP1 O/E.
Discussion
Despite accumulating evidence detailing the importance of survivin overexpression in esophageal cancer, the intracellular mechanisms regulating survivin expression are not clearly known [4–6, 9]. The survivin gene has not been shown to be mutated in esophageal cancer, suggesting critical roles for its transcriptional and post-transcriptional regulation. In this study we describe the novel role of the RBP CUG-BP1 in survivin overexpression in esophageal cancer cells. CUG-BP1 is overexpressed in esophageal cancer cells and interacts specifically with the survivin mRNA 3′UTR. This interaction results in enhanced survivin mRNA stability, leading to increased survivin protein expression.
To date, data on the post-transcriptional regulation of survivin has been scarce. We have previously shown that reducing survivin transcription in p53-null esophageal cancer cells by overexpressing p53 does not decrease survivin protein levels, suggesting an important role for post-transcriptional mechanisms in regulating survivin expression [2]. Vaira and colleagues have described how activation of the IGF-1/mTOR signaling pathway markedly increased survivin protein levels in serum-deprived prostate cancer cells [18]. This increase in survivin protein was independent of increased survivin gene transcription or changes in protein stability. Instead, IGF-1 was found to modulate improved stability and enhanced translation of a pre-existing pool of survivin mRNA, although the mechanism by which this occurred was not elucidated. Finally, we have recently shown that the RBP HuR bound to the 3′UTR of survivin mRNA and prolonged its half-life in esophageal epithelial cells [19]. However, this interaction proved to be quite complex, as HuR overexpression in these cells also increased levels of p53, leading to decreased survivin transcription.
Our current results indicate that CUG-BP1 is overexpressed in both human esophageal cancer specimens and esophageal cancer cell lines compared to esophageal epithelial cells. In human specimens, the expression patterns of CUG-BP1 and survivin are well correlated. In the normal squamous mucosa of the esophagus, expression of both CUG-BP1 and survivin was localized to the regenerative zone of the submucosa with minimal expression in the epithelial layer. However, both CUG-BP1 and survivin expression were diffusely expressed throughout the esophageal cancer tissue. Further studies are needed to elucidate the mechanisms whereby CUG-BP1 levels are increased in malignant tissue.
We found that CUG-BP1 binds to multiple sites in the 3′-UTR of survivin mRNA. Survivin mRNA contains several GU-rich and CUG repeats in both the 3′ UTR and CR. These sites have been recognized as target sequences for CUG-BP1 [20–21]. Interestingly, despite the presence of such sites in the CR of survivin mRNA, we were not able to demonstrate binding between the CR and CUG-BP1. Furthermore, experiments mapping CUG-BP1 binding activity in the survivin 3′UTR demonstrated significant differences in binding affinity among the different fragments, although each fragment contained potential binding sites. One of the most interesting aspects of CUG-BP1 biology is that it can lead to accelerated decay of some targets while prolonging the half-life and enhancing translation of others. It is tempting to speculate that differences in the particular binding characteristics of CUG-BP1 to its target mRNA may account for these divergent effects.
The results reported here further show that overexpression of CUG-BP1 in esophageal epithelial cells resulted in an increase in survivin mRNA half-life and protein expression. In reciprocal experiments, down-regulation of CUG-BP1 in esophageal cancer cells was associated with a decrease in survivin mRNA half-life and decreased survivin protein expression. To determine if the relationship between CUG-BP1 and surviving was specific or related to a more global role of CUG-BP1 in regulating cell division, we examined the relationship between CUG-BP1 and CDK4 in TE7 cells. Previous data from our group showed that overexpressing CUG-BP1 leads to decreased translation of CDK4 mRNA and ultimately decreased CDK4 protein levels in rat intestinal crypt cells [22]. Similarly, in TE7 cells, silencing CUG-BP1 in TE7 cells was associated with an increase in CDK4 levels.
The precise mechanism by which CUG-BP1 contributes to increased survivin mRNA stability remains unknown. P-bodies function to repress expression of mRNAs delivered to these cytoplasmic structures [23–24]. Argonaute-2 protein (Ago2), a core component of the RNA-induced silencing complex, has been demonstrated to be an important component of the P-body and is involved in the repression of a number of mRNA transcripts [25–27]. We found that down-regulation of CUG-BP1 in esophageal cancer cells is associated with an increase in the association of survivin mRNA with Ago2 protein. In addition, we found that silencing Lsm4, an important activator of decapping in P-bodies, abrogates the reduction of survivin levels in TE7 cells following silencing of CUG-BP1 [28–29]. Taken together, these findings provide preliminary evidence to suggest that CUG-BP1 prevents the trafficking of survivin mRNA to the P-body. Further studies will be required to more fully characterize the mechanisms by which the binding of CUG-BP1 to survivin mRNA prevents the association of survivin mRNA with P-body proteins.
A key finding of our study was the ability to modulate cellular responsiveness to chemotherapy-induced apoptosis by altering levels of CUG-BP1. CUG-BP1 overexpression in esophageal epithelial cells was associated with an increased resistance to apoptosis. Conversely, silencing CUG-BP1 in esophageal cancer cells increased the susceptibility of these cells to apoptosis. We have previously shown that the degree of responsiveness of nhESO cells and TE7 cells to camptothecin-induced apoptosis largely depends on the level of survivin expression [2]. The finding that altering CUG-BP1 expression directly correlated with survivin expression and cellular sensitivity to apoptosis suggested that CUG-BP1 mediates sensitivity to apoptosis by affecting survivin levels. However, by simultaneously overexpressing CUG-BP1 and silencing surviving in TE7 cells, we were able to completely abrogate the increased sensitivity to camptothecin-induced apoptosis seen only with survivin silencing. This finding raises the possibility that CUG-BP1 can enhance the expression of other anti-apoptotic proteins in addition to survivin. A recent report from Gareau and colleagues describes the ability of CUG-BP1 to mediate resistance to the proteasome inhibitor bortezomid by stabilizing p21 mRNA and thereby increasing p21 levels [30]. It is also possible that CUG-BP1 may destabilize transcripts of pro-apoptotic genes. We are currently exploring the broader role of CUG-BP1 in in modulating apoptosis in esophageal cancer cells.
Acknowledgments
Funding
This work was supported by the Department of Veterans Affairs, U.S.A. [Merit Review Grant (to J.-Y.W.), Career Development Award (to J.D.)]; the National Institutes of Health [grant numbers DK-57819, DK-61972 and DK-68491 (to J.-Y. W.)], and departmental funds from the Department of Surgery, University of Maryland (to J.D. and R.B.).
Abbreviations used
- 3′-UTR
3′-untranslated region
- Ago2
Argonaute-2 protein
- C-siRNA
control siRNA
- CDK4
Cyclin-dependent kinase 4
- CR
coding region
- CUG-BP1
CUG-binding protein 1
- GRE
GU-rich element
- H&E
hematoxylin
- HuR
Hu-antigen R
- IAP
Inhibitor of Apoptosis Protein
- IGF-1
Insulin-like growth factor-1
- IHC
immunohistochemistry
- IP
immunoprecipitation
- mTOR
mammalian target of rapamycin
- nhESO
normal human esophageal
- P-body
processing body
- PCR
polymerase chain reaction
- qRT-PCR
quantitative real-time PCR
- RBP
RNA-binding protein
- RNP
ribonucleoprotein
- RT
reverse transcription
- siCUG-BP1
siRNA directed against CUG-BP1
- siLsm4
siRNA directed against Lsm4
- siSurvivin
siRNA directed against surviving
- TE7
adenocarcinoma esophageal cell line
- TE10
esophageal squamous cell carcinoma cell line
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Chang E, Donahue J, Smith A, Hornick J, Rao JN, Wang JY, Battafarano RJ. Loss of p53, rather than beta-catenin overexpression, induces survivin-mediated resistance to apoptosis in an esophageal cancer cell line. J Thorac Cardiovasc Surg. 2010;140:225–232. doi: 10.1016/j.jtcvs.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 3.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 4.Grabowski P, Kuhnel T, Muhr-Wilkenshoff F, Heine B, Stein H, Hopfner M, Germer CT, Scherubl H. Prognostic value of nuclear survivin expression in oesophageal squamous cell carcinoma. Br J Cancer. 2003;88:115–119. doi: 10.1038/sj.bjc.6600696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeguchi M, Yamaguchi K, Kaibara N. Survivin gene expression positively correlates with proliferative activity of cancer cells in esophageal cancer. Tumour Biol. 2003;24:40–45. doi: 10.1159/000070659. [DOI] [PubMed] [Google Scholar]
- 6.Kato J, Kuwabara Y, Mitani M, Shinoda N, Sato A, Toyama T, Mitsui A, Nishiwaki T, Moriyama S, Kudo J, Fujii Y. Expression of survivin in esophageal cancer: correlation with the prognosis and response to chemotherapy. Int J Cancer. 2001;95:92–95. doi: 10.1002/1097-0215(20010320)95:2<92::aid-ijc1016>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Vallböhmer D, Peters JH, Oh D, Kuramochi H, Shimizu D, Demeester SR, Hagen JA, Chandrasoma PT, Danenberg KD, DeMeester TR, Danenberg P. Survivin, a potential biomarker in the development of Barrett’s adenocarcinoma. Surgery. 2005;138:701–706. doi: 10.1016/j.surg.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 8.Parenti A, Leo G, Porzionato A, Zaninotto G, Rosato A, Ninfo V. Expression of survivin, p53, and caspase 3 in Barrett’s esophagus carcinogenesis. Hum Pathol. 2006;37:16–22. doi: 10.1016/j.humpath.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Vallböhmer D, Kuhn E, Warnecke-Eberz U, Brabender J, Hoffmann AC, Metzger R, Baldus SE, Drebber U, Hoelscher AH, Schneider PM. Failure of downregulation of intratumoral survivin expression following neoadjuvant chemoradiation in esophageal cancer. Pharmacogenomics. 2008;9:681–690. doi: 10.2217/14622416.9.6.681. [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, Otevrel T, Gao Z, Ehrlich SM, Fields JZ, Boman BM. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001;61:8664–8667. [PubMed] [Google Scholar]
- 11.Cosgrave N, Hill AD, Young LS. Growth factor-dependent regulation of survivin by c-myc in human breast cancer. J Mol Endocrinol. 2006;37:377–390. doi: 10.1677/jme.1.02118. [DOI] [PubMed] [Google Scholar]
- 12.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, Yoder S, Enkemann S, Eschrich S, Lee JH, Beam CA, Cheng J, Minton S, Muro-Cacho CA, Jove R. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 14.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, Nielsen LL, Pickett CB, Liu S. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 15.Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUG-BP1 and alter protein levels and activity of CUG-BP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUG-BP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUG-BP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaira V, Lee CW, Goel HL, Basari S, Languino LR, Altieri DC. Regulation of survivin expression by IGF-1/mTOR signaling. Oncogene. 2007;26:2678–2684. doi: 10.1038/sj.onc.1210094. [DOI] [PubMed] [Google Scholar]
- 19.Donahue JM, Chang ET, Xiao L, Wang PY, Rao JN, Turner DJ, Wang JY, Battafarano RJ. The RNA binding protein HuR stabilizes survivin mRNA in human esophageal epithelial cells. Biochem J. 2011;437:89–96. doi: 10.1042/BJ20110028. [DOI] [PubMed] [Google Scholar]
- 20.Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, Sugano S, Güntert P, Muto Y, Yokoyama S. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao L, Cui YH, Rao JN, Zou T, Liu L, Smith A, Turner DJ, Gorospe M, wang JY. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22:3055–3069. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, Yang L, Li H, Li L, Chen J. Residues that affect human Argonaute2 concentration in cytoplasmic processing bodies. Biochem. Biophys. Res Commun. 2009;378:620–624. doi: 10.1016/j.bbrc.2008.11.098. [DOI] [PubMed] [Google Scholar]
- 26.Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009;15:1078–1089. doi: 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jagannath A, Wood MJ. Localization of small interfering RNA to cytoplasmic processing bodies is Ago2 dependent and results in up-regulation of GW182 and Argonaute-2. Mol Biol Cell. 2009;20:521–529. doi: 10.1091/mbc.E08-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Bio. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gareau C, Fournier MJ, Filion C, Coudert L, Martel D, Labelle Y, Mazroui R. p21 upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. Plos One. 2011;6:1–14. doi: 10.1371/journal.pone.0020254. [DOI] [PMC free article] [PubMed] [Google Scholar]


