Abstract
The lone star tick Amblyomma americanum is host to a wide diversity of endosymbiotic bacteria. We identified a novel Wolbachia symbiont infecting A. americanum. Multilocus sequence typing phylogenetically placed the endosymbiont in the increasingly diverse F supergroup. We assayed a total of 1031 ticks (119 females, 78 males and 834 nymphs in 89 pools) from 16 Maryland populations for infection. Infection frequencies in the natural populations were approximately 5% in females and <2% (minimum infection rate) in nymphs; infection was not detected in males. Infected populations were only observed in southern Maryland, suggesting the possibility that Wolbachia is currently invading Maryland A. americanum populations. Because F supergroup Wolbachia have been detected previously in filarial nematodes, tick samples were assayed for nematodes by PCR. Filarial nematodes were detected in 70% and 9% of Wolbachia-positive and Wolbachia-negative tick samples, respectively. While nematodes were more common in Wolbachia-positive tick samples, the lack of a strict infection concordance (Wolbachia-positive, nematode-negative and Wolbachia-negative, nematode-positive ticks) suggests that Wolbachia prevalence in ticks is not due to nematode infection. Supporting this hypothesis, phylogenetic analysis indicated that the nematodes were likely a novel species within the genus Acanthocheilonema, which has been previously shown to be Wolbachia-free.
Keywords: Wolbachia, Amblyomma americanum, tick, nematode, Acanthocheilonema, MLST
Introduction
Amblyomma americanum (the lone star tick) is broadly distributed, with a host range spanning the Midwest to the eastern United States (Childs & Paddock, 2003). Amblyomma americanum is a vector of pathogens that cause diseases of humans and domestic animals such as granulocytic ehrlichiosis, tularemia and borreliosis, and is associated with Southern tick-associated rash illness (Goddard & Varela-Stokes, 2009). Several endosymbiotic bacterial species (including Coxiella, Rickettsia and Arsenophonus) have been detected previously in A. americanum (Clay et al., 2008). Wolbachia is a common endosymbiotic associate of arthropods and filarial nematodes (Rowley et al., 2004; Baldo et al., 2007; Hilgenboecker et al., 2008; Werren et al., 2008) and has been described previously in the ticks Ixodes ricinus (Hartelt et al., 2004) and Ixodes scapularis (Benson et al., 2004). In this report, we identified a novel Wolbachia symbiont in A. americanum. The infection was found only in southern Maryland at a low frequency in female ticks and pooled nymphs, but was never observed in males. Multilocus sequence typing (MLST) placed the infection into the F supergroup. Filarial nematode infection was observed in some tick samples, but concordance between Wolbachia infection and nematode infection was not observed. Nematodes were phylogenetically placed into the genus Acanthocheilonema, members of which were previously shown to be Wolbachia-free.
Materials and methods
Tick collection information and sample sizes are listed in Table 1 and Fig. 1. All ticks were collected in 2008 using tick drags. Individual adult ticks and pooled nymphs (10 per pool) were homogenized using a TissueLyser II bead mill (Qiagen, Valencia, CA) with 5-mm stainless-steel bead. Genomic DNA was isolated using the MasterPure DNA purification kit (Epicentre Biotechnologies, Madison, WI) and eluted in 30 µL water. Samples were screened for Wolbachia infection using Wolbachia-specific 16S primers WSpecF and WSpecR (Sakamoto et al., 2006; Werren & Windsor, 2000) using a PTC thermocycler (Bio-Rad, Hercules, CA) with a program of 95 °C for 5 min; 40 cycles of 95 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min; and a final extension of 72 °C for 5 min. Each 40 µL reaction consisted of 2.0 µL template DNA, 1 µM concentrations of all forward and reverse primers, 0.4mM dNTPs and 2.0U Taq polymerase. Infections were confirmed using wsp gene amplification using primers wspF1, wspR1 (Wolbachia MLST website: http://pubmlst.org/wolbachia/). The PCR protocol used was 94 °C for 2 min, followed by 37 cycles of 94 °C for 30 s, 59 °C for 45 s and 72 °C for 1.5 min and a final extension of 72 °C for 10 min (http://pubmlst.org/wolbachia/). Template DNA from a Wolbachia-infected mosquito cell line was used as a positive control; a sample containing deionized water instead of template DNA was used as a negative control. For Wolbachia MLST, the four MLST genes (coxA, gatB, hcpA and fbpA) were amplified using nested PCR. Primary PCR amplification was performed using F3/R3 ~64-fold degenerate primer sets (http://pubmlst.org/wolbachia/). Each 40 µL reaction consisted of 2 µL template DNA, 1 µM concentrations of all forward and reverse primers, 0.4mM dNTPs and 1.0U HotStar Taq polymerase (Qiagen). Primary PCR cycling conditions followed the ‘alternative protocols’ listed on the MLST website. Secondary PCR amplification was performed using the MLST website ‘standard protocols’ and F1/R1 primer sets for these four MLST genes (Baldo et al., 2006). The ftsZuniF/ftsZuniR (Lo et al., 2002) primer pair was used for the ftsZ gene PCR. PCR products were directly sequenced in both directions using an Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). Sequences were deposited in GenBank under accession numbers HM061157–HM061163.
Table 1.
Collection site, tick stage and infection rate
| Location | County | Stage | N | # Positive |
% Infected |
|---|---|---|---|---|---|
| Idylwild WMA | Caroline | Female | 19 | 2 | 10.5% |
| Male | 10 | 0 | 0 | ||
| Nymph | 5 pools (44) | 0 | 0 | ||
| Linkwood WMA | Dorchester | Female | 29 | 1 | 3.5% |
| Male | 19 | 0 | 0 | ||
| Nymph | 3 pools (27) | 0 | 0 | ||
| Assateague SP | Worchester | Female | 9 | 0 | 0 |
| Male | 7 | 0 | 0 | ||
| Nymph | 7 pools (68) | 0 | 0 | ||
| Pocomoke SP | Worchester | Nymph | 7 pools (63) | 3 | 4.76%* |
| Cedarville SF | PG and Charles | Female | 2 | 0 | 0 |
| Male | 1 | 0 | 0 | ||
| Nymph | 6 pools (51) | 3 | 5.88%* | ||
| Chapel Point SP | Charles | Female | 3 | 0 | 0 |
| Nymph | 6 pools (56) | 4 | 7.14%* | ||
| Chapman SP | Charles | Female | 17 | 2 | 11.8% |
| Male | 9 | 0 | 0 | ||
| Nymph | 11 pools (103) | 1 | 0.97%* | ||
| Purse SP | Charles | Nymph | 5 pools (46) | 1 | 2.17%* |
| Serpentine | Montgomery | Female | 15 | 0 | 0 |
| Male | 17 | 0 | 0 | ||
| Nymph | 4 pools (40) | 0 | 0 | ||
| St Mary’s SP | St Mary’s | Female | 4 | 1 | 25% |
| Male | 1 | 0 | 0 | ||
| Nymph | 3 pools (29) | 0 | 0 | ||
| Greenwell SP | St Mary’s | Female | 3 | 0 | 0 |
| Male | 1 | 0 | 0 | ||
| Sandy Point SP | Anne Arundel | Female | 8 | 0 | 0 |
| Male | 9 | 0 | 0 | ||
| Nymph | 4 pools (38) | 0 | 0 | ||
| Millington WMA | Kent | Female | 2 | 0 | 0 |
| Male | 3 | 0 | 0 | ||
| Nymph | 1 pool (11) | 0 | 0 | ||
| Calvert Cliffs SP | Calvert | Female | 7 | 0 | 0 |
| Nymph | 19 pools (183) | 0 | 0 | ||
| Elk Neck SP | Cecil | Female | 1 | 0 | 0 |
| Nymph | 3 pools (28) | 0 | 0 | ||
| Tuckahoe SP | Queen Anne’s | Male | 1 | 0 | 0 |
| Nymph | 5 pools (47) | 0 | 0 | ||
| Total | Female | 119 | 6 | 5.04% | |
| Male | 78 | 0 | 0 | ||
| Nymph | 89 pools (834) | 12 | 1.44%* |
Infection rate of nymphs is reported as the MIR; the number of infected pools divided by the total number of ticks tested.
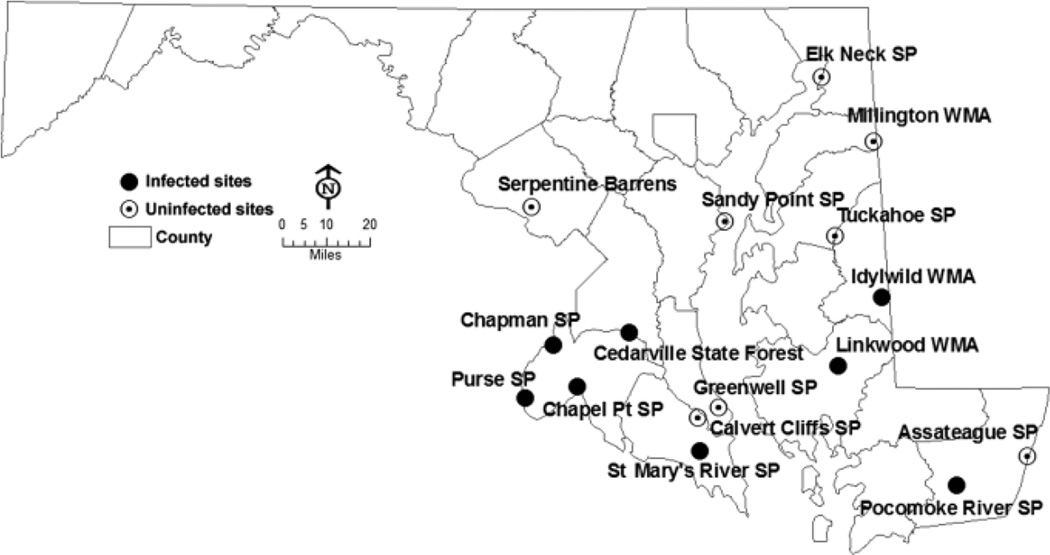
Fig. 1.
Map of Maryland Amblyomma americanum collection sites. Closed circles indicate populations with infected ticks; open circles indicate populations where no infected ticks were collected.
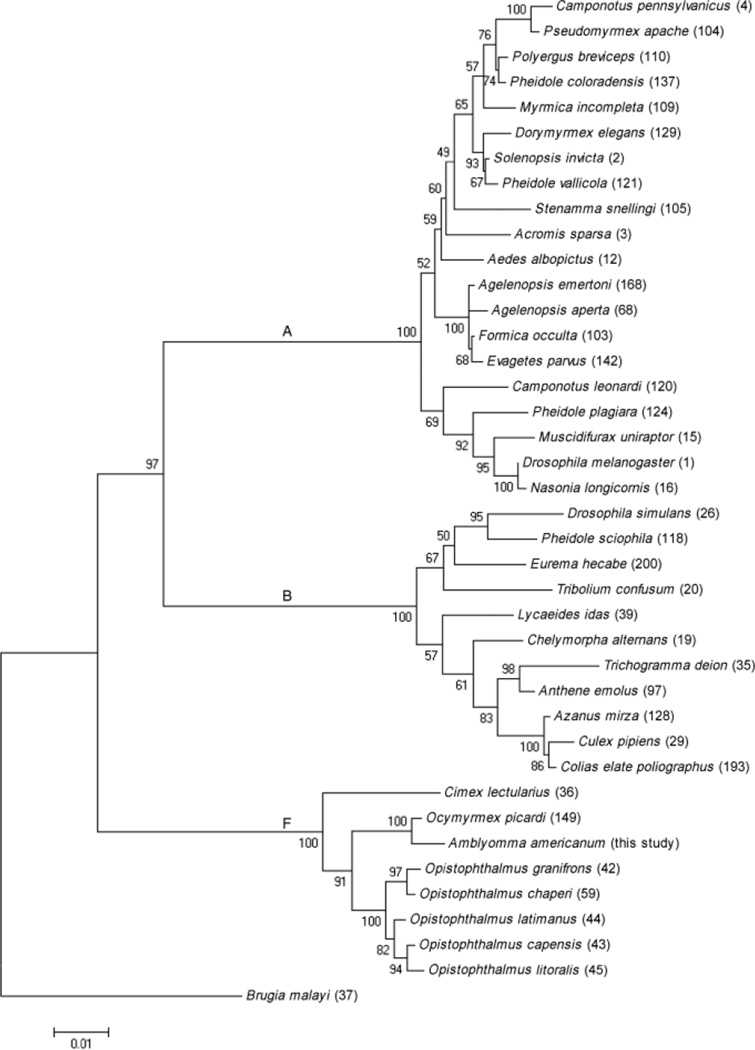
Thirty-nine concatenated MLST sequences were retrieved from the Wolbachia MLST database. The Wolbachia strains are listed by host species name and concatenated MLST sequence ID number in Fig. 2. Sequence alignment and neighbor-joining phylogenetic analyses were conducted using MEGA version 4 (Tamura et al., 2007). Tree support was evaluated by bootstrapping with 1000 replications. Phylogenies were produced for concatenated and gene-specific data sets.
Fig. 2.
Neighbor-joining phylogenetic analysis of a 2079 bp alignment of five Wolbachia MLST concatenated gene sequences (coxA, gatB, hcpA, ftsZ and fbpA). Numbers at nodes indicate bootstrap support values (1000 replicates). Taxon names are host species. Values in parentheses represent MLST sequence ID numbers (searchable at http://pubmlst.org/wolbachia/). Letters represent Wolbachia supergroup designations. The tree is rooted by the supergroup D strain from Brugia malayi.
Tick samples were tested for the presence of filarial nematodes using specific 12S primers as described (Casiraghi et al., 2004). Template DNA from a Wuchereria bancrofti-infected Culex pipiens mosquito was used as a positive control; a sample containing deionized water instead of template DNA was used as a negative control. The positive amplicons obtained were directly sequenced. Phylogenetic placement of nematode sequences was conduced as described above.
Results and discussion
Out of 16 sampled Maryland populations, Wolbachia-infected ticks were found in eight populations. Infection rates within populations were low, ranging from 3.5% to 25% in females, and a minimum infection rate (MIR) from 1% to 7% in pooled nymphs. Wolbachia was not detected in males. The overall statewide infection rate was approximately 5% in females and 1.4% (MIR) in pooled nymphs (Table 1). Wolbachia infection was spatially structured; all infected populations were located in southern Maryland, while infection was not detected in central and northern populations (Fig. 1). This distribution suggests the possibility that Wolbachia is currently invading A. americanum populations in Maryland. Future sampling efforts will be required to confirm or refute this hypothesis.
The coxA, gatB, hcpA, ftsZ and fbpA gene amplicons were produced by PCR from sample #366 from Dorchester County. Additionally, the fbpA gene was amplified and sequenced from all positive samples, while the coxA, gatB, hcpA and ftsZ genes were amplified and sequenced from selected samples (Table 2). All sequences were identical to those obtained from sample #366. According to the phylogenetic analysis of these concatenated sequences, the Wolbachia strain found in A. americanum was placed into supergroup F and was 99% identical to Wolbachia from the ant Ocymyrmex picardi (Russell et al., 2009). Gene-specific phylogenetics analyses showed the same result (Supporting Information). This is the first report of supergroup F Wolbachia in ticks. Up to this point, F supergroupWolbachia infections have been detected in nematodes, scorpions and several insect hosts (termites, weevils, bush crickets, bedbugs, lice, louse flies, cockroaches and antlions) (Lo et al., 2002; Casiraghi et al., 2005; Dunn & Stabb, 2005; Sakamoto et al., 2006; Baldo et al., 2007; Covacin & Barker, 2007; Panaram & Marshall, 2007; Vaishampayan et al., 2007).
Table 2.
Sequences obtained from assayed ticks
| Specimen | Stage assayed | Wolbachia | FbpA | CoxA | GatB | HcpA | ftsZ | Nematode |
|---|---|---|---|---|---|---|---|---|
| 366 | Adult female | + | × | × | × | × | × | Not tested |
| 294 | Adult female | + | × | × | × | − | ||
| 295 | Adult female | + | × | × | + | |||
| 995 | Adult female | + | × | × | + | |||
| 998 | Adult female | + | × | + | ||||
| 1178 | Adult female | + | × | + | ||||
| 445–454 | Pooled nymphs | + | × | × | − | |||
| 465–474 | Pooled nymphs | + | × | + | ||||
| 485–494 | Pooled nymphs | + | × | + | ||||
| 790–799 | Pooled nymphs | + | × | − | ||||
| 817–826 | Pooled nymphs | + | × | − | ||||
| 827–836 | Pooled nymphs | + | × | + | ||||
| 868–877 | Pooled nymphs | + | × | + | ||||
| 927–932 | Pooled nymphs | + | × | + | ||||
| 1016–1025 | Pooled nymphs | + | × | + | ||||
| 1032–1041 | Pooled nymphs | + | × | + | ||||
| 1042–1051 | Pooled nymphs | + | × | + | ||||
| 1062–1071 | Pooled nymphs | + | × | − | ||||
| 1337 | Adult female | − | − | |||||
| 1338 | Adult female | − | − | |||||
| 255–258 | Pooled nymphs | − | − | |||||
| 475–484 | Pooled nymphs | − | + | |||||
| 837–840 | Pooled nymphs | − | − | |||||
| 907–916 | Pooled nymphs | − | − | |||||
| 917–926 | Pooled nymphs | − | − | |||||
| 997 | Adult female | − | − | |||||
| 999 | Adult female | − | − | |||||
| 1000 | Adult female | − | − | |||||
| 1001 | Adult female | − | − |
F supergroup Wolbachia have been identified in filarial nematodes (genus Mansonella) in addition to arthropods. Although ticks collected for this study were collected by dragging, and were unlikely to be blood fed, we investigated the hypothesis that Wolbachia infection in Amblyomma was due to filarial nematode infection. All Wolbachia-positive tick samples and a random selection of Wolbachia-negative tick samples were assayed for filarial nematode infection by specific 12S PCR. Filarial nematodes were detected in both Wolbachia-positive and Wolbachia-negative adult female and nymphal ticks. Nematode infection was more common in Wolbachia-infected ticks (70% vs. 9%), but strict concordance of infection types was not observed (both Wolbachia-positive, nematode-negative ticks and Wolbachia-negative, nematode-positive ticks were identified) (Table 2). Phylogenetic analysis indicated that the filarial nematode infecting A. americanum was most closely related to nematodes in the genus Acanthocheilonema (formerly Dipetalonema) (Fig. 3). Acanthocheilonema nematodes have not been described previously from Amblyomma, but their transmission by both soft ticks (Ornithodoros) and hard ticks (Rhipicephalus) has been demonstrated (Londoño, 1976; Olmeda-García et al., 1993). Acanthocheilonema nematodes have also been shown previously to lack Wolbachia infection (Casiraghi et al., 2004). In total, these results indicate that Wolbachia prevalence in A. americanum is not explained by nematode infection.
Fig. 3.
Unrooted neighbor-joining phylogenetic analysis of of nematode 12S gene sequences. Amblyomma americanum nematode sequence has been deposited in GenBank under accession number JF732757. Alphanumeric codes represent GenBank ID numbers. Numbers at nodes indicate bootstrap support values (1000 replicates). Bold indicates nematode sequence identified in this study.
The frequency of Wolbachia infection is low in A. americanum. Some Wolbachia reproductive phenotypes, such as cytoplasmic incompatibility, tend to equilibrate at high frequencies in populations (Turelli & Hoffmann, 1999). In contrast, phenotypes such as male killing tend to equilibrate at low frequencies (Hurst & Jiggins, 2000).We did not detect any infected males in our study – this could reflect sex-specific infection differences (possibly related to male killing) or could simply be due to sampling. The reproductive phenotype of Wolbachia in A. americanum is unknown and needs further study.
Supplementary Material
Acknowledgements
This research was funded by NIH/NIAID grants R21AI067386 and R03AI079297 to D.E.N., and R21AI070178 to J.L.R. X.Z. was partially supported by the JHSPH Ralph and Sylvia Barr fellowship. We thank Timothy Shields and the Johns Hopkins Environmental Surveillance Core for assistance in producing Fig. 1.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Phylogenetic analysis of Wolbachia gatB sequences.
Fig. S2. Phylogenetic analysis of Wolbachia coxA sequences.
Fig. S3. Phylogenetic analysis of Wolbachia hcpA sequences.
Fig. S4. Phylogenetic analysis of Wolbachia fbpA sequences.
Fig. S5. Phylogenetic analysis of Wolbachia ftsZ sequences.
References
- Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MCJ, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis . Appl Environ Microb. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Prendini L, Corthals A, Werren JH. Wolbachia are present in southern African scorpions and cluster with supergroup F. Curr Microbiol. 2007;55:367–373. doi: 10.1007/s00284-007-9009-4. [DOI] [PubMed] [Google Scholar]
- Benson MJ, Gawronski JD, Eveleigh DE, Benson DR. Intracellular symbionts and other bacteria associated with deer ticks (Ixodes scapularis) from Nantucket and Wellfleet, Cape Cod, Massachusetts. Appl Environ Microb. 2004;70:616–620. doi: 10.1128/AEM.70.1.616-620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiraghi M, Bain O, Guerrero R, Martin C, Pocacqua V, Gardner SL, Franceschi A, Bandi C. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: evidence for symbiont loss during evolution. Int J Parasitol. 2004;34:191–203. doi: 10.1016/j.ijpara.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Casiraghi M, Bordenstein SR, Baldo L, Lo N, Beninati T, Wernegreen JJ, Werren JH, Bandi C. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology. 2005;151:4015–4022. doi: 10.1099/mic.0.28313-0. [DOI] [PubMed] [Google Scholar]
- Childs JE, Paddock CD. The ascendancy of Amblyomma americanum as a vector of pathogens affecting humans in the United States. Annu Rev Entomol. 2003;48:307–337. doi: 10.1146/annurev.ento.48.091801.112728. [DOI] [PubMed] [Google Scholar]
- Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C. Microbial communities and interactions in the lone star tick, Amblyomma americanum . Mol Ecol. 2008;17:4371–4381. doi: 10.1111/j.1365-294x.2008.03914.x. [DOI] [PubMed] [Google Scholar]
- Covacin C, Barker SC. Supergroup F Wolbachia bacteria parasitise lice (Insecta: Phthiraptera) Parasitol Res. 2007;100:479–485. doi: 10.1007/s00436-006-0309-6. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Stabb EV. Culture-independent characterization of the microbiota of the ant lion Myrmeleon mobilis (Neuroptera: Myrmeleontidae) Appl Environ Microb. 2005;71:8784–8794. doi: 10.1128/AEM.71.12.8784-8794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J, Varela-Stokes AS. Role of the lone star tick, Amblyomma americanum (L.), in human and animal diseases. Vet Parasitol. 2009;160:1–12. doi: 10.1016/j.vetpar.2008.10.089. [DOI] [PubMed] [Google Scholar]
- Hartelt K, Oehme R, Frank H, Brockmann SO, Hassler D, Kimmig P. Pathogens and symbionts in ticks: prevalence of Anaplasma phagocytophilum (Ehrlichia sp.), Wolbachia sp., Rickettsia sp., Babesia. sp. in Southern Germany. Int J Med Microbiol. 2004;293(suppl 37):86–92. doi: 10.1016/s1433-1128(04)80013-5. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? – A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GD, Jiggins FM. Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg Infect Dis. 2000;6:329–336. doi: 10.3201/eid0604.000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. How many Wolbachia supergroups exist? Mol Biol Evol. 2002;19:341–346. doi: 10.1093/oxfordjournals.molbev.a004087. [DOI] [PubMed] [Google Scholar]
- Londoño I. Behavior of Dipetalonema viteae (Filarioidea) during escape from the vector tick, Ornithodoros tartakowskyi (Argasidae) J Parasitol. 1976;62:596–603. [PubMed] [Google Scholar]
- Olmeda-García AS, Rodríguez-Rodríguez JA, Rojo-Vázquez FA. Experimental transmission of Dipetalonema dracunculoides (Cobbold 1870) by Rhipicephalus sanguineus (Latreille 1806) Vet Parasitol. 1993;47:339–342. doi: 10.1016/0304-4017(93)90034-k. [DOI] [PubMed] [Google Scholar]
- Panaram K, Marshall JL. F supergroup Wolbachia in bush crickets: what do patterns of sequence variation reveal about this supergroup and horizontal transfer between nematodes and arthropods? Genetica. 2007;130:53–60. doi: 10.1007/s10709-006-0020-7. [DOI] [PubMed] [Google Scholar]
- Rowley SM, Raven RJ, McGraw EA. Wolbachia pipientis in Australian spiders. Curr Microbiol. 2004;49:208–214. doi: 10.1007/s00284-004-4346-z. [DOI] [PubMed] [Google Scholar]
- Russell JA, Goldman-Huertas B, Moreau CS, Baldo L, Stahlhut JK, Werren JH, Pierce NE. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution. 2009;63:624–640. doi: 10.1111/j.1558-5646.2008.00579.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto JM, Feinstein J, Rasgon JL. Wolbachia infections in the Cimicidae: museum specimens as an untapped resource for endosymbiont surveys. Appl Environ Microb. 2006;72:3161–3167. doi: 10.1128/AEM.72.5.3161-3167.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Turelli M, Hoffmann AA. Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol Biol. 1999;8:243–255. doi: 10.1046/j.1365-2583.1999.820243.x. [DOI] [PubMed] [Google Scholar]
- Vaishampayan PA, Dhotre DP, Gupta RP, Lalwani P, Ghate H, Patole MS, Shouche YS. Molecular evidence and phylogenetic affiliations of Wolbachia in cockroaches. Mol Phylogenet Evol. 2007;44:1346–1351. doi: 10.1016/j.ympev.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267:1277–1285. doi: 10.1098/rspb.2000.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.