Abstract
Strain RB38 was recovered from a former dumping area in Malaysia. MALDI-TOF mass spectrometry and genomic analysis identified strain RB-38 as Pandoraea pnomenusa. Various biosensors confirmed its quorum sensing properties. High resolution triple quadrupole liquid chromatography–mass spectrometry analysis was subsequently used to characterize the N-acyl homoserine lactone production profile of P. pnomenusa strain RB38, which validated that this isolate produced N-octanoyl homoserine lactone as a quorum sensing molecule. This is the first report of the production of N-octanoyl homoserine lactone by P. pnomenusa strain RB38.
Keywords: matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF), mass spectrometry (MS), cell-to cell communication, triple quodruopole liquid chromatography mass spectrometry (LC-MS/MS), N-octanoyl homoserine lactone (C8-HSL)
1. Introduction
Pandoraea is a Gram-negative rod shape proteobacteria of the Burkholderiaceae family that is often isolated from sputa of cystic fibrosis patients and soil [1]. Initially documented by Coenye and co-workers [1], this genus is primarily represented by four cystic fibrosis (CF) isolates, namely: Pandoraea pnomenusa, Pandoraea pulmonicola, Pandoraea apista and Pandoraea sputorum [2] which are often misclassified as their neighboring organisms (Burkholderia and Ralstonia) or even described as non-fermentative Gram-negative bacteria [3–5]. P. pnomenusa in particular, is recognized as a causing agent for bacteremia, multiple organ failure and it also causes mortality [6]. P. pnomenusa has been reported as a multi-drug resistant pathogen [4].
Quorum sensing (QS) bacteria produce diffusible signaling molecules to communicate with one another as the bacterial population increases in order to regulate gene expression in unison [7–9]. Gram-negative bacteria have the ability to secrete N-acylhomoserine lactones (AHLs) as their autoinducers. QS modulates a battery of bacterial physiological activities which include swimming, swarming, sporulation, motility, bioluminescence, biofilm, antibiotic biosynthesis and plasmid conjugal transfer by coordinating the gene expression of a population [7,10,11]. The latest discovery is the documentation of QS properties in Pandoraea sp. [12]. Because of the importance of AHLs [13], this study aims to characterize the AHL secreted by P. pnomenusa RB-38 using triple quadrupole mass spectrometry (LC-MS/MS) in order to gain a better understanding of the QS activity of this bacterium.
Here, we present the QS properties of P. pnomenusa RB38. Its identity was confirmed via MALDI-TOF MS analysis and 16S rDNA sequencing. Different AHL biosensors were used for preliminary screening of AHLs and subsequently the P. pnomenusa RB38 AHL profile was characterized by LC-MS/MS analysis. To our best knowledge, this is the first documentation of the QS profile in the species P. pnomenusa.
2. Experimental Section
2.1. Bacterial Growth Conditions
All strains (Table 1) were cultured aerobically at 28 °C in Luria-Bertani (LB) broth or LB agar with exception for Escherichia coli [pSB401], which was incubated at 37 °C. The antibiotic tetracycline (20 μg/mL) was supplemented into the growth media when necessary.
Table 1.
Bacteria strains used in this study.
| Bacterial Strains Used | Descriptions | Source/Reference |
|---|---|---|
| Pandoraea pnomenusa RB38 | Isolated from ex-landfill site | This study |
| Chromobacterium violaceum CV026 | AHL biosensor that synthesizes purple pigment in detection of short chain N-acyl-side chain length of four to eight carbons. | [14] |
| Erwinia carotovora GS101 | Positive control of the CV026 AHL detection | [15] |
| Erwinia carotovora PNP22 | Negative control of the CV026 AHL detection | [15] |
| Escherichia coli [pSB401] | Short chain AHLs biosensor used for the bioluminescence assay, TetR | [16] |
2.2. Strain Isolation
In the search for QS bacteria from a dumping ground (subsurface of 10 cm from the top soil) in Malaysia (GPS coordinate of N03'00'12.1, E101' 39'33'1), strain RB38 was isolated for further studies. The sampling site was 61 m above sea level. To isolate soil bacteria, we used KGm [12,17]. The soil sample was inoculated into KGm and underwent four enrichment cycles using a reported method [12,17]. Single pure colonies of soil bacteria were obtained by serial dilution streaking on LB agars.
2.3. Bacterial Identification
MALDI-TOF MS analysis and 16S rDNA nucleotide sequencing was performed as described previously [12,18–20]. Nucleotide comparison was performed using the BLASTN program in the NCBI database and the phylogenetic tree was constructed using Molecular Evolutionary Genetic Analysis (MEGA).
2.4. Detection of Short Chain AHLs Productions
AHL screening was performed using the CV026 biosensor. P. pnomenusa RB38 and the experimental control strains were cross-streaked with CV026. Purple pigmentation after 24 h indicates QS activity.
2.5. AHL Extractions
P. pnomenusa RB38 was cultured in 100 mL LB broth supplemented with MOPS buffer. Growth media were buffered to acidic condition at pH 5.5 to prevent lactonolysis because AHLs are unstable in alkaline conditions [21,22]. Triplicate AHL extractions were performed twice with 150 mL of acidified ethyl acetate. The upper non-polar immiscible solvent layer was then transferred into a sterile beaker and the other layer was discarded. AHL extracts were dried under sterile conditions. Extracts from LB broth (with no inoculum) served as negative control.
2.6. Bioluminescence Assay
Planktonic culture of E. coli biosensor was adjusted to OD600 nm of 0.1 using sterile LB broth as diluent. The diluted biosensor cells were used to dissolve AHL extracts and the mixtures transferred to microtitre wells in a 96-well microtitre plate. The luminescence intensity and OD495 nm were determined and recorded simultaneously at 60 min intervals for the duration of 24 h in Infinite M200 luminometer (Tecan, Männerdorf, Switzerland). The bioluminescence assay result was determined as relative light unit per OD495 nm (RLU/OD495 nm) vs. time [12,23,24].
2.7. Determination of AHL by LC-MS/MS
AHL extracts were resuspended with 100 µL acetonitrile and analyzed as described previously [12,17–19] using an Agilent 1290 Infinity LC system (Agilent Technologies Inc., Santa Clara, CA, USA) coupled with an Agilent ZORBAX Rapid Resolution High Definition SB-C18 Threaded column (2.1 mm × 50 mm, 1.8 µm particle size). Electrospray ionization (ESI) with jet-stream positive mode was used as the ion source and detection of m/z 102 product ion was performed using precursor ion scan mode. Data analysis was conducted using the Agilent Mass Hunter software.
2.8. Nucleotide Sequence Accession Number
We have deposited the 16S rDNA nucleotides sequences of P. pnomenusa RB38 into NCBI and was assigned GenBank accession No. KJ507404. For molecular analysis, we obtained the other 16S rDNA nucleotides sequences from GenBank database.
3. Results and Discussion
3.1. Isolation and Identity Classification of Soil Bacterium Strain RB38
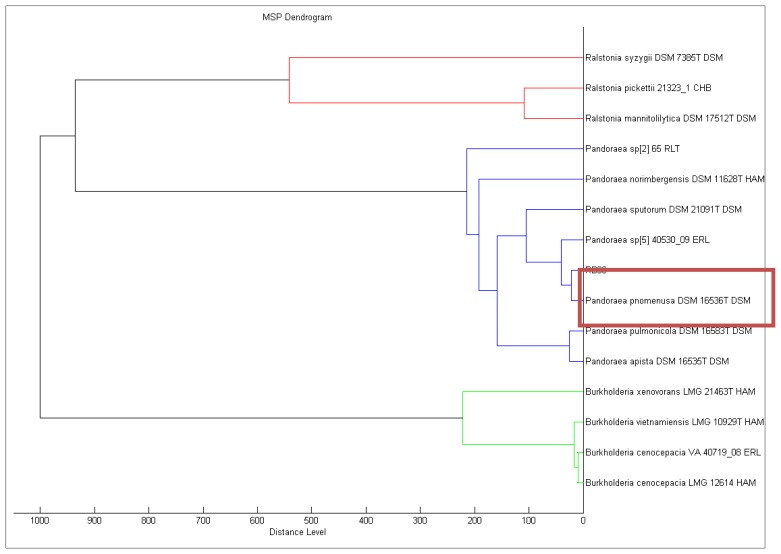
KGm medium supplemented with 3-oxo-C6-HSL (50 mM final concentration, Sigma-Aldrich, St Loius, MO, USA) was used for the soil bacteria enrichment process [12,17]. Within 48 h post-incubation, viable cell count showed that microbial growth occurred in KGm medium. Several distinct morphological colonies of bacteria were selected and a succession of streaking was conducted to obtained pure colonies. A strain labeled as RB38 which was identified as P. pnomenusa MALDI-TOF (Figure 1) was selected for further analysis.
Figure 1.
Dendrogram of isolate RB38. MALDI-TOF analysis confirmed isolate RB38 clusters with Pandoraea pnomenusa.
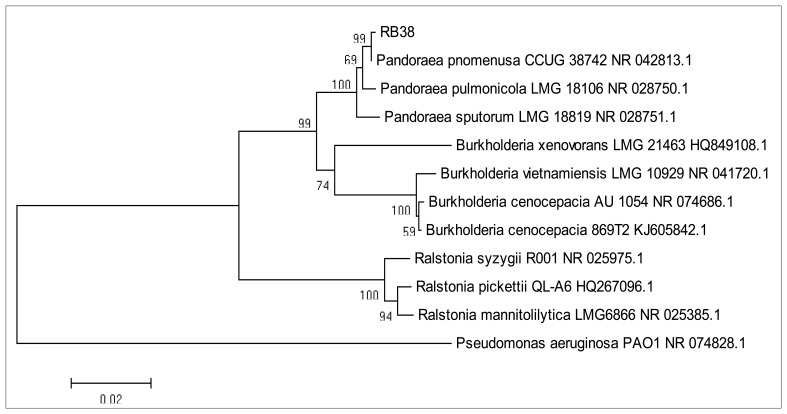
Biochemical identification using the Microbial Identification System has often misclassified Pandoraea as Burkholderia and Ralstonia species due to the similarity of biochemical activities between these two genera. Thus, 16S rDNA sequence analysis was performed to further confirm the identity of isolate RB38 (Figure 2). Phylogeny of 16S rDNA nucleotide sequences of isolate RB38 with its closest neighbours was constructed using Neighbour-Joining algorithm [25]. The evolutionary distances were computed using the Maximum Likelihood method [26] and expressed as the number of base changes per site. The analysis involved seven nucleotide sequences datasets. There were a total of 1,394 positions in the final dataset. For evolutionary analyses, we used MEGA 5 as reported elsewhere [27].
Figure 2.
Evolutionary tree of 16S rDNA nucleotide sequences of isolate RB-38 with its closest neighbours using the Neighbour-Joining method.
16S rDNA analysis has been reported as a better option if there is difficulty in identifying a pathogen as B. cepacia complex or Ralstonia species [6]. Molecular analysis also confirmed the data from MALDI-TOF for microorganism identification [17]. In 2002, Coenye has documented a better method of detection of Pandoraea using restriction fragment length polymorphism (RFLP) analysis and nucleotide sequencing of a house-keeping gene (gyr B) for better resolution to distinguish closely-related species in the genus of Pandoraea [28].
3.2. AHL Screening
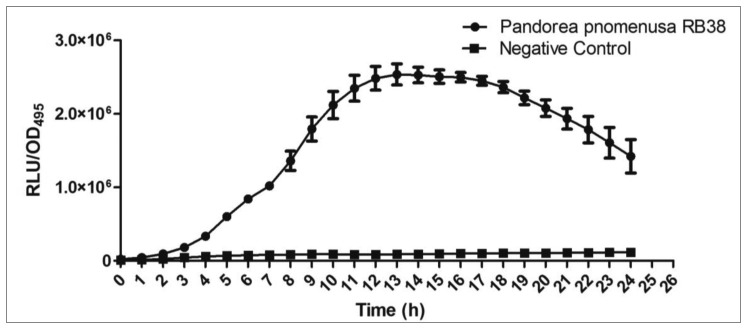
Rapid screening of AHL production by soil isolates was conducted using the C. violaceum CV026 biosensor [14]. Formation of purple pigmentation in C. violaceum CV026 after a 1-day incubation with P. pnomenusa RB38 indicated detection of short chain AHLs produced by P. pnomenusa RB38 (Figure 3). The C. violaceum CV026 biosensor is capable of detecting short chain AHLs in the range of N-butanoyl homoserine lactone (C4-HSL) to N-octanoyl homoserine lactone (C8-HSL). Further investigation was performed in bioluminescence assay using lux-based E. coli [pSB401] biosensor with a detection range of C4-HSL to C8-HSL [16]. Luminescence bioassay involving E. coli [pSB401] suggested production of short chain AHLs by P. pnomenusa RB38 (Figure 4).
Figure 3.
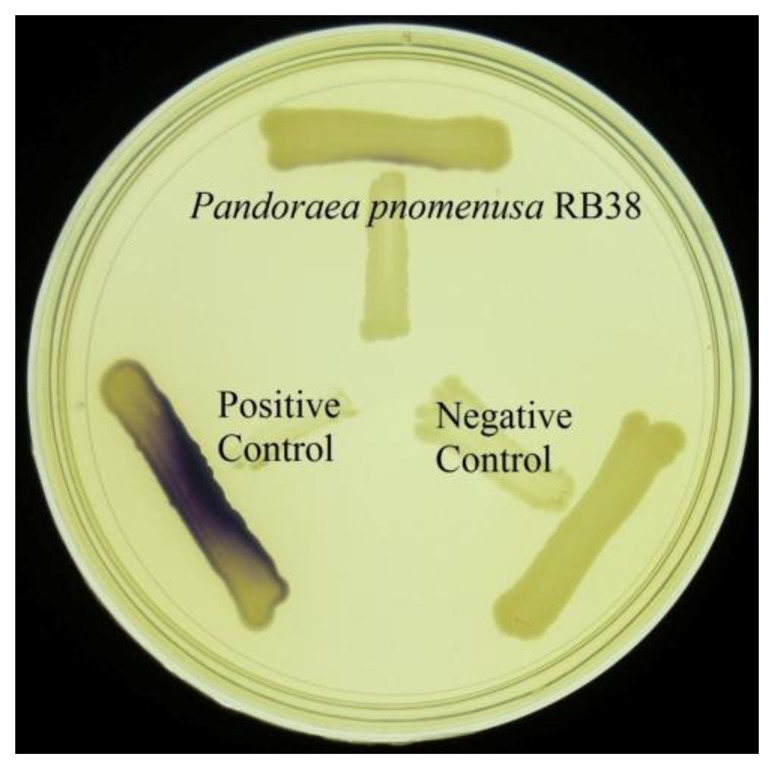
AHL screening of P. pnomenusa RB38. Biosensors C. violaceum CV026 was streaked adjacent to the test strains. Note that purple pigment shows quorum sensing activity. Erwinia carotovora GS101 (Positive control) and Erwinia carotovora PNP22 (Negative control) were included as controls.
Figure 4.
Bioluminescence assay for production of AHLs by P. pnomenusa RB38. Each dot symbolizes the mean results of triplicate experiments. Negative control was included which is an extract from uninoculated LB broth.
3.3. LC-MS/MS Analysis of AHLs
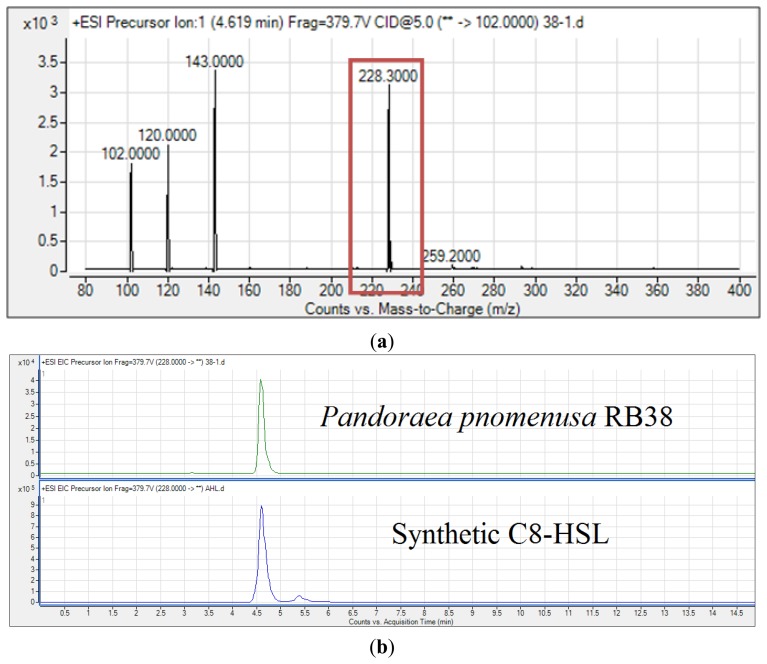
There are many reports on the regulation of virulence factors by QS mechanisms in CF pathogens such as Pseudomonas aeruginosa and Burkholderia spp. [29–32]. Thus, it is important to ensure P. pnomenusa produces AHLs. To identify the AHL profile of P. pnomenusa RB38 in the spent supernatant of P. pnomenusa RB38, our MS data validated the presence of C8-HSL (m/z value of 228.3, retention time: 4.602 min; abundance: 3342.7; abundance %: 95.71) in the supernatant of this strain (Figure 5). The presence of three product fragment ions with m/z values of 102, 120 and 143 was also detected, thus confirming unequivocal presence of this AHL.
Figure 5.
Mass spectrometry analysis of AHLs produced by P. pnomenusa RB38. Mass spectrum shows that P. pnomenusa RB38 produced C8-HSL (a); Retention time of C8-HSL from spent supernatant of P. pnomenusa RB38 is the same as the retention time of synthetic C8-HSL (b).
4. Conclusions
Here, we report P. pnomenusa RB38 produced C8-HSL. To our best knowledge, this is the first documentation of short chain AHL production in the species P. pnomenusa.
Acknowledgments
This work was supported by the University of Malaya for High Impact Research Grant (UM-MOHE HIR Grant UM.C/625/1/HIR/MOHE/CHAN/01, No. A000001-50001) awarded to Kok-Gan Chan which is gratefully acknowledged.
Author Contributions
Robson Ee, Yan-Lue Lim, Lin-Xin Kin, Wai-Fong Yin conducted the experiments and data analysis. Kok-Gan Chan designed, supervised, and guided the experiments. All authors contributed in the preparation and revision of this paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Coenye T., Falsen E., Hoste B., Ohlen M., Goris J., Govan J.R., Gillis M., Vandamme P. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 2000;50:887–899. doi: 10.1099/00207713-50-2-887. [DOI] [PubMed] [Google Scholar]
- 2.Daneshvar M.I., Hollis D.G., Steigerwalt A.G., Whitney A.M., Spangler L., Douglas M.P., Jordan J.G., MacGregor J.P., Hill B.C., Tenover F.C., et al. Assignment of CDC weak oxidizer group 2 (WO-2) to the genus Pandoraea and characterization of three new Pandoraea genomo species. J. Clin. Microbiol. 2001;39:1819–1826. doi: 10.1128/JCM.39.5.1819-1826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coenye T., Liu L., Vandamme P., LiPuma J.J. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J. Clin. Microbiol. 2001;39:4452–4455. doi: 10.1128/JCM.39.12.4452-4455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caraher E., Collins J., Herbert G., Murphy P.G., Gallagher C.G., Crowe M.J., Callaghan M., McClean S. Evaluation of in vitro virulence characteristics of the genus Pandoraea in lung epithelial cells. J. Med. Microbiol. 2008;57:15–20. doi: 10.1099/jmm.0.47544-0. [DOI] [PubMed] [Google Scholar]
- 5.Aravena-Roman M. Cellular fatty acid-deficient Pandoraea isolated from a patient with cystic fibrosis. J. Med. Microbiol. 2008;57:252. doi: 10.1099/jmm.0.47671-0. [DOI] [PubMed] [Google Scholar]
- 6.Stryjewski M.E., LiPuma J.J., Messier R.H., Jr., Reller L.B., Alexander B.D. Sepsis, multiple organ failure, and death due to Pandoraea pnomenusa infection after lung transplantation. J. Clin. Microbiol. 2003;41:2255–2257. doi: 10.1128/JCM.41.5.2255-2257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua C., Winans S.C., Greenberg E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 8.Fuqua C., Greenberg E.P. Self perception in bacteria: Quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 9.Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 10.Salmond G.P., Bycroft B.W., Stewart G.S., Williams P. The bacterial “enigma”: Cracking the code of cell-cell communication. Mol. Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 11.Hardman A.M., Stewart G.S., Williams P. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and non-pathogenic bacteria. Antonie Van Leeuwenhoek. 1998;74:199–210. doi: 10.1023/a:1001178702503. [DOI] [PubMed] [Google Scholar]
- 12.Han-Jen R.E., Wai-Fong Y., Kok-Gan C. Pandoraea sp. RB-44, a novel quorum sensing soil bacterium. Sensors. 2013;13:14121–14132. doi: 10.3390/s131014121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong K.W., Koh C.L., Sam C.K., Yin W.F., Chan K.G. Quorum quenching revisited—From signal decays to signalling confusion. Sensors. 2012;12:4661–4696. doi: 10.3390/s120404661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClean K.H., Winson M.K., Fish L., Taylor A., Chhabra S.R., Camara M., Daykin M., Lamb J.H., Swift S., Bycroft B.W., et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 15.McGowan S., Sebaihia M., Jones S., Yu B., Bainton N., Chan P., Bycroft B., Stewart G.S.A.B., Williams P., Salmond G.P.C. Carbapenem antibiotic production in Erwinia carotovora is regulated by carR, a homologue of the luxR transcriptional activator. Microbiology. 1995;141:541–550. doi: 10.1099/13500872-141-3-541. [DOI] [PubMed] [Google Scholar]
- 16.Winson M.K., Swift S., Fish L., Throup J.P., Jørgensen F., Chhabra S.R., Bycroft B.W., Williams P., Stewart G.S. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen J.W., Koh C.L., Sam C.K., Yin W.F., Chan K.G. Short chain n-acyl homoserine lactone production by soil isolate Burkholderia sp. strain A9. Sensors. 2013;13:13217–13227. doi: 10.3390/s131013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngeow Y.F., Cheng H.J., Chen J.W., Yin W.F., Chan K.G. Short chain N-acylhomoserine lactone production by clinical multidrug resistant Klebsiella pneumoniae strain CSG20. Sensors. 2013;13:15242–15251. doi: 10.3390/s131115242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau Y.Y., Sulaiman J., Chen J.W., Yin W.F., Chan K.G. Quorum sensing activity of Enterobacter asburiae isolated from lettuce leaves. Sensors. 2013;13:14189–14199. doi: 10.3390/s131014189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong T.M., Koh C.L., Sam C.K., Choo Y.M., Yin W.F., Chan K.G. Characterization of quorum sensing and quorum quenching soil bacteria isolated from Malaysian tropical montane forest. Sensors. 2012;12:4846–4859. doi: 10.3390/s120404846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates E.A., Philipp B., Buckley C., Atkinson S., Chhabra S.R., Sockett R.E., Goldner M., Dessaux Y., Camara M., Smith H., et al. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortori C.A., Dubern J.F., Chhabra S.R., Camara M., Hardie K., Williams P., Barrett D.A. Simultaneous quantitative profiling of N-acyl-L-homoserine lactone and 2-alkyl-4(1H)-quinolone families of quorum-sensing signaling molecules using LC-MS/MS. Anal. Bioanal. Chem. 2011;399:839–850. doi: 10.1007/s00216-010-4341-0. [DOI] [PubMed] [Google Scholar]
- 23.Tan L.Y., Yin W.F., Chan K.G. Silencing quorum sensing through extracts of Melicope lunu-ankenda. Sensors. 2012;12:4339–4351. doi: 10.3390/s120404339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Priya K., Yin W.F., Chan K.G. Anti-quorum sensing activity of the traditional Chinese herb, Phyllanthus amarus. Sensors. 2013;13:14558–14569. doi: 10.3390/s131114558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coenye T., LiPuma J.J. Use of the gyrB gene for the identification of Pandoraea species. FEMS Microbiol. Lett. 2002;208:15–19. doi: 10.1111/j.1574-6968.2002.tb11053.x. [DOI] [PubMed] [Google Scholar]
- 29.Koch G., Nadal-Jimenez P., Reis C.R., Muntendam R., Bokhove M., Melillo E., Dijkstra B.W., Cool R.H., Quax W.J. Reducing virulence of the human pathogen Burkholderia by altering the substrate specificity of the quorum-quenching acylase PvdQ. Proc. Natl. Acad. Sci USA. 2014;111:1568–1573. doi: 10.1073/pnas.1311263111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suppiger A., Schmid N., Aguilar C., Pessi G., Eberl L. Two quorum sensing systems control biofilm formation and virulence in members of the Burkholderia cepacia complex. Virulence. 2013;4:400–409. doi: 10.4161/viru.25338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabeen M.T. Stationary phase-specific virulence factor overproduction by a LasR mutant of Pseudomonas aeruginosa. PLoS One. 2014;9:e88743. doi: 10.1371/journal.pone.0088743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiricny N., Molin S., Foster K., Diggle S.P., Scanlan P.D., Ghoul M., Johansen H.K., Santorelli L.A., Popat R., West S.A., et al. Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS One. 2014;9:e83124. doi: 10.1371/journal.pone.0083124. [DOI] [PMC free article] [PubMed] [Google Scholar]