Abstract
Human Campylobacter jejuni infection can result in an asymptomatic carrier state, watery or bloody diarrhea, bacteremia, meningitis, or autoimmune neurological sequelae. Infection outcomes of C57BL/6 IL-10−/− mice orally infected with twenty-two phylogenetically diverse C. jejuni sstrains were evaluated to correlate colonization and disease phenotypes with genetic composition of the strains. Variation between strains was observed in colonization, timing of development of clinical signs, and occurrence of enteric lesions. Five pathotypes of C. jejuni in C57BL/6 IL-10−/− mice were delineated: little or no colonization, colonization without disease, colonization with enteritis, colonization with hemorrhagic enteritis, and colonization with neurological signs with or without enteritis. Virulence gene content of ten sequenced strains was compared in silico; virulence gene content of twelve additional strains was compared using a C. jejuni pan-genome microarray. Neither total nor virulence gene content predicted pathotype; nor was pathotype correlated with multilocus sequence type. Each strain was unique with regard to absences of known virulence-related loci and/or possession of point mutations and indels, including phase variation, in virulence-related genes. An experiment in C. jejuni 11168-infected germ-free mice showed that expression levels of ninety open reading frames (ORFs) were significantly up- or down-regulated in the mouse cecum at least two-fold compared to in vitro growth. Genomic content of these ninety C. jejuni 11168 ORFs was significantly correlated with the capacity to colonize and cause enteritis in C57BL/6 IL-10−/− mice. Differences in gene expression levels and patterns are thus an important determinant of pathotype in C. jejuni strains in this mouse model.
Keywords: Campylobacter jejuni, mouse, colonization, enteritis, genome, gene expression
1. INTRODUCTION
Campylobacter jejuni is a leading cause of sporadic food-borne enteritis throughout the world. Some individuals are refractory to primary C. jejuni infection or develop an asymptomatic carrier state [1]. In most instances, C. jejuni infection in humans causes acute enteritis with watery or bloody diarrhea that resolves within 7–10 days. Reactive inflammatory disorders can also occur after C. jejuni infection. In the gastrointestinal tract, transmural inflammatory changes in colon or small intestines may occur that resemble inflammatory bowel disease (IBD)[2–3], while joint disease can also result. In addition, peripheral neuropathies (Miller Fisher and Guillain Barré Syndromes) are known sequelae of C. jejuni infection where autoimmune mechanisms are triggered [4–5]. It is known that immunocompromised patients have both a higher incidence of infection with Campylobacter and more severe disease manifestations with profuse diarrhea and persistence of the organism [6–7]. Furthermore, those at the extremes of age or with underlying immune compromise are predisposed to bacteremia and subsequent spread to other organs [7–8]. In this study we focused on examining clinical phenotypes potentially arising from strain-specific attributes.
Attempts have been made to associate particular disease outcomes with particular pathogen genetic and/or phenotypic characteristics with mixed success [9–10]. The most compelling such association discovered to date has been that of strains expressing particular lipooligosaccharides with autoimmune neurological sequelae [11]. The most likely reason for this general lack of correspondence between pathogen genotype and clinical presentation is that C. jejuni is known to have extensive genetic variation driven by multiple mutagenic mechanisms [12] and lateral gene transfer within C. jejuni and between C. jejuni and its closest relative, C. coli [13–14].
C57BL/6 IL-10−/−, NOD IL-10−/−, and C3H/HeJ IL-10−/− murine models of C. jejuni 11168 infection [15–16] were recently developed and characterized in our laboratory; the C57BL/6 IL-10−/− model was used to explore the capacity of C. jejuni 11168 and six other C. jejuni strains to colonize and undergo genetic adaptation to a new host during serial passage [17]. Some data from the latter study are reproduced here under open license agreement with BioMed Central and BMC Microbiology; the relevant citation is given at each occurrence.
The initial stage of the serial passage experiment revealed that an array of C. jejuni strains from humans and animals produced a spectrum of clinical outcomes that reflected that seen in human campylobacteriosis. In addition, the results showed that some C. jejuni strains, but not others, were able to evolve increased pathogenicity during serial passage in mice. This ability—or lack of ability—to respond to selection indicates that the genomic content of each strain is crucial for its virulence phenotype. The term “pathotype” will be used below to denote the virulence phenotype of C. jejuni strains in C57BL/6 IL-10−/− mice, that is, the constellation of clinical, gross pathological, and histopathological outcomes of infection with a particular C. jejuni strain in C57BL/6 IL-10−/− mice.
The experiments reported here were undertaken to correlate disease phenotypes in the murine model with the genetic composition of C. jejuni isolates tested and to extend our understanding of the ability of the murine model to replicate the spectrum of C. jejuni-associated disease seen in humans. Similar approaches using combinations of in vivo and in silico methods have been used to define the “virulome,” or the set of genes necessary for pathogenicity, of Brucella suis and Burkholderia mallei [18–19].
We therefore infected C57BL/6 IL-10−/− mice with fifteen additional C. jejuni strains from a variety of clinical and non-clinical sources. This strain set included six minimally passaged clinical isolates from the Centers for Disease Control and nine genome-sequenced strains. The genome-sequenced strains include nine C. jejuni subsp. jejuni strains: one chicken isolate, four isolates from human cases of enteritis, one isolate from a case of meningitis, two isolates from cases of Guillain Barré syndrome, one isolate from a case of Miller Fisher syndrome, and one C. jejuni subsp. doylei isolate from a case of bacteremia. We were able to delineate five pathotypes of C. jejuni in this murine model: no colonization, colonization with little or no disease, colonization with moderate or severe enteritis, colonization with hemorrhagic enteritis, and colonization with neurological sequelae with or without enteritis. A detailed description of the neurological sequelae will be published separately. In order to identify additional genes involved in host colonization and disease induction, we compared the gene expression profiles of a C. jejuni 11168 culture grown in broth to that of C. jejuni 11168 cells recovered from the ceca of germ-free mice. Ninety open reading frames (ORFs) were significantly (P ≤ 0.05) up-or down-regulated more than two-fold in C. jejuni 11168 cells from mouse ceca, including thirty-nine up-regulated ORFs not previously shown to be involved in colonization or virulence.
Publically available sequence data and genomic analysis tools were used to compare the genome-sequenced strains. Among the genome-sequenced strains, neither total genomic content nor the composition of putative experimentally identified virulence factors correlated with pathotype. However, these results should be interpreted cautiously, since many of the genome-sequenced strains, especially strains 81–176 and 33560 (type strain), have been used in multiple laboratory studies and may have lost virulence during passage on laboratory media. The genomic content of each of the minimally passaged strains and of each of the previously studied strains [17] was compared to that of C. jejuni 11168 using a pan-genomic microarray developed for this study using data from the ten genome-sequenced C. jejuni strains tested in mice. Again, no correlation was found between pathotypes of C. jejuni strains in C57BL/6 IL-10−/− mice and either clonal complexes defined by multilocus sequence typing or total genome content. We therefore concluded that particular constellations of virulence genes, point mutations or indels (including phase variation in contingency genes) in individual loci, or differences in gene expression levels are likely to determine the pathotypes of individual strains.
A statistically significant correlation was found between colonization and disease phenotypes of both the set of genome-sequenced strains and the set of strains analyzed by microarray and genomic content of ninety ORFs that were up- and down-regulated at least two-fold in the ceca of germ-free C57BL/6 IL-10−/− mice. Finally, the results of strain comparisons based on documented virulence factors and the results of the gene expression study were used to establish a first approximation of the C. jejuni C57BL/6 IL-10−/− mouse virulome.
2. MATERIALS AND METHODS
2.1. Campylobacter jejuni strains, media, and growth conditions
Campylobacter jejuni strains used in these experiments are listed in Table 1. (Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology). Strains D6844–D6849 were kindly supplied as blinded cultures by Drs. Collette Fitzgerald and Linda Demma of the Centers for Disease Control (CDC), Atlanta, GA, USA; strains CG8486 and CG8421 were gifts of Dr. Patricia Guerry; strain 84-25 was the gift of Drs. Martin Blazer and Nicole Iovine, and strain 81–176 was the gift of Dr. William Miller. Cultures to be inoculated into mice were grown on tryptic soy agar (Accumedia, Neogen, Lansing, MI, USA) containing 5% defibrinated sheep’s blood (Cleveland Scientific, Bath, Ohio, USA) under an atmosphere of 10% H2, 10% CO2, and 80% N2 at 37°C; harvested; and suspended in tryptic soy broth as previously described [15]. Purity and motility of cultures were verified by light microscopy and Gram stain and colony-forming units (cfu) per milliliter determined by serial dilution. Inoculum sizes are given in (Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology). C. jejuni was isolated from mouse tissues and feces by streaking on selective tryptose soya agar containing 5% defibrinated sheep’s blood, 2 µg/ml amphotericin B, 10 µg/ml vancomycin, and 20 µg/ml cefaperazone (all antibiotics from Sigma Aldrich, St. Louis, MO, USA). Plates were incubated in closed containers at 37°C with CampyGen packs (Oxoid, Basingstoke, Hampshire, England, UK) for 48 hours before scoring semi-quantitatively as previously described; cultures were verified using a C. jejuni-specific gyrA PCR assay [15]. Results for C. jejuni strains 33560T, D0121, D0835, D2586, D2600, and NW derived from passage 1 data in a genetic adaptation study were reported by Bell et al. [17]. (These data are reproduced under an open licence agreement with BioMed Central and BMC Microbiology.)
Table 1. Campylobacter jejuni strains: MLST typing, inoculum size, colonization, and disease phenotypes.
(Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology).
| Straina | Disease status and geographic origin |
MLST sequence type (clonal complex) |
Inoculum (cfu per mouse) |
C. jejuni detectable by culture |
C. jejuni detectable only by PCR |
Early euthanasia requiredb |
Moderate to severe enteritisc |
|---|---|---|---|---|---|---|---|
| (number of mice colonized or showing disease/total number infected) |
|||||||
| Experiment 1 | |||||||
| D6844 | mild enteritis/USAd | 22 (22) | 2.5 × 1010 | 8/9 | 1/9 | 1/9 | 2/9 |
| D6845 | watery enteritis/USAd | 353 (353) | 4.2 × 109 | 10/10 | -- | 1/10 | 1/10 |
| D6846 | bloody enteritis/USAd | 824 (257) | 1.3 × 1010 | 10/10 | -- | 4/10 | 6/10 |
| D6847 | bloody enteritis/USAd | 353 (353) | 1.1 × 1010 | 8/10 | 2/10 | 1/10 | 1/10 |
| D6848 | watery enteritis/USAd | 50 (21) | 1.4 × 1010 | 10/10 | -- | 1/10 | 2/10 |
| D6849 | mild enteritis/USAd | 48 (48) | 1.2 × 1010 | 10/10 | -- | 5/10 | 5/10 |
| Experiment 2 | |||||||
| RM1221 | chicken/USAe | 353 (354) | 7.8 × 109 | 8/10 | 0/10 | 2/10 | 3/10 |
| 269.97 | bacteremia/South Africae | 1846 (403) | 3.6 × 109 | 1/10 | 0/10 | 0/10 | 0/10 |
| HB93-13 | Guillain Barré Syndrome/Chinae | new (22) | 9.0 × 109 | 6/10 | 1/10 | 1/10 | 2/10 |
| 260.94 | Guillain Barré Syndrome/South Africae | 362 (362) | 9.6 × 109 | 10/10 | -- | 0/10 | 0/10 |
| Experiment 3 | |||||||
| 84-25 | meningitis/USAf | 21 (21) | 1.9 × 1010 | 6/10 | 0/10 | 0/10 | 0/10 |
| CF93-6 | Miller Fisher Syndrome/Japane | 883 (21) | 2.3 × 1010 | 9/10 | 0/10 | 5/10 | 5/10 |
| CG8486 | enteritis/Thailandg | 2943 (574) | 1.9 × 1010 | 9/10 | 0/10 | 1/10 | 1/10 |
| CG8421 | enteritis/Thailandg | 1919 (52) | 5.2 × 109 | 10/10 | -- | 9/10 | 9/10 |
| 81–176 | enteritis/USAh | 913 (42) | 5.4 × 109 | 4/9 | 0/9 | 0/9 | 0/9 |
| Experiments 1, 2, and 3 combined | |||||||
| 11168 | enteritis/UKd | 43 (21) | 1.7 × 1010, 7.8 × 109, 2.4 × 109 | 27/27 | -- | 21/27 | 22/27 |
| Control | NA | NA | NA | 0/29 | 0/29 | 0/29 | 0/29 |
| Previously studied strains (data from Bell et al., 2009)i | |||||||
| D0121 | unknown/Canadaj | 45 (45) | 0/5 | 1/5 | 0/5 | 0/5 | |
| D0835 | chicken/USAj | 429 (48) | 5/5 | -- | 0/5 | 2/5 | |
| D2586 | enteritis/USAj | 43 (21) | 5/5 | -- | 0/5 | 1/5 | |
| D2600 | enteritis/USAj | 452 (353) | 5/5 | -- | 0/5 | 0/5 | |
| NW | enteritis/Kenyak | 354 (354) | 5/5 | -- | 0/5 | 1/5 | |
| 33560 | bovine/USAe | 403 (403) | 0/5 | 5/5 | 0/5 | 0/5 | |
All strains are of human origin except RM1221 (chicken), D0835 (chicken), and 33560T (bovine).
All mice requiring early euthanasia exhibited moderate to severe enteritis.
Gross pathology: mild, enlarged ileocecocolic lymph node only; moderate, thickened colon wall with or without enlarged ileocecocolic lymph node; severe, thickened colon wall, enlarged ileocecocolic lymph node, and bloody lumen contents.
Gifts of Linda Demma, and Collette Fitzgerald, Centers for Disease Control, Atlanta, GA.
American Type Culture Collection, Manassas, VA, USA. ATCC designations are as follows: strain 11168, ATCC 700819; RM1221, ATCC BAA-1062; 269.97, ATCC BAA-1458; HB93-13, ATCC 700297; 260.94, ATCC BAA-1234; CF93-6, ATCC BAA-1456.
Gift of Martin Blaser, Department of Microbiology, New York University School of Medicine, New York, NY.
Gift of Patricia Guerry, Department of Enteric Diseases, Naval Medical Research Center, Uniformed Services University of the Health Sciences
Gift of William Miller, U.S. Department of Agriculture, Agricultural Research Service, Western Regional Research Center, Albany, CA.
Data given are for the initial passage in a serial mouse passage experiment; these mice were on a different dietary regime from mice in Experiments 1, 2 and 3 [17]. These data are reproduced under open licence agreement with BioMed Central and BMC Microbiology.
Gift of Patricia Fields, Centers for Disease Control, Atlanta, GA. MLST results as reported by Sails et al. [47]
Sparrow Hospital, Lansing, MI, USA
For gene expression studies, C. jejuni NCTC11168 was grown to mid-exponential phase (approximate OD600 = 0.7; approximately 18 h) in Bolton broth at 37°C with agitation in a 10% CO2 incubator, before cultures were centrifuged (9260 rcf, 4°C, 8 minutes), cell pellets were re-suspended in RNAlater (Ambion), and RNA extraction performed exactly as described for cecal content samples [20].
2.2. Animals
All procedures involving animals were performed according to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and under protocols approved by the Michigan State University institutional Animal Use and Care Committee (approvals 04/07-030-00 and 06/09-092-00) and the University of Michigan Committee on Use and Care of Animals (Application Number: 09116).
Three separate screening experiments in conventional mice were conducted at Michigan State University using identical protocols. All animal breeding, husbandry, randomization of mice to different treatments, and oral gavage and other procedures are described in detail in Mansfield et al. [15]. C57BL/6 IL-10−/− mice were produced in our specific pathogen free breeding colony, genotyped, and tested for Campylobacter spp. and Helicobacter spp. as previously described [15]; mice were housed under filtered air, given sterile water, and fed a sterile modified NIH 31 diet (approximately 6% fat; Harlan Teklad 7913; Harlan Teklad, Indianapolis, IN, USA). At 8 weeks of age, mice were transferred to the University Research Containment facility prior to inoculation; inoculated mice were housed individually in sterile filter-topped cages. Inoculation per os with approximately 1010 cfu of each C. jejuni strain was carried out using 3.5 Fr red rubber feeding tubes as previously described [15]. Five male and five female mice were inoculated with each C. jejuni strain except as noted in (Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology). Mice were monitored daily for clinical signs of enteric disease (lethargy, ruffled hair coat, hunched posture, diarrhea, dehydration) according to an established scoring system; mice exceeding a predetermined score were euthanized immediately to prevent suffering [15]. Mice not developing clinical signs were monitored for 35 days and then euthanized and necropsied. Euthanasia was carried out by CO2 overdose, blood samples were obtained by cardiac puncture, and the diaphragm cut to prevent resuscitation during necropsy. Three samples each of stomach, jejunum, cecum and colon tissue were obtained; one sample was spread on selective agar and two samples snap frozen for DNA isolation or other studies as previously described [15]. In three separate experiments, ten mice were inoculated with each of a panel of test strains, C. jejuni 11168 (positive control), and tryptose soya broth (negative vehicle control); results are presented separately for each experiment.
The C. jejuni gene expression experiment in germ-free mice was performed as described in Jerome et al. [20]. Germ-free, male, six week old C57BL/6 IL-10−/− mice were inoculated with approximately 1×1010 CFU of Campylobacter jejuni NCTC11168. Mice were euthanized and necropsied 96 hours after infection, and RNA was extracted from cecal contents as previously described [20].
2.3. Gross pathology and histopathological changes
The gastrointestinal (GI) tract was removed in its entirety and placed on clean absorbent bench paper. Gross pathological changes were noted in all portions of the GI tract by trained individuals. The ileocecocolic junction was gently injected at necropsy with 10% phosphate buffered formalin (Fisher Scientific, Pittsburgh, PA, USA) and placed in a histological cassette; the cassette was submerged in 10% phosphate buffered formalin. After 24 hours, the cassette was drained and transferred to 60% ethanol. Tissue processing, paraffin embedment, sectioning, and hematoxylin and eosin staining were carried out at the Investigative Histopathology Laboratory, Division of Human Pathology, Department of Physiology, Michigan State University). Slides were randomized and coded so that their identities were concealed; histopathological features were evaluated by a single investigator (LSM) according to a previously described scoring system [15].
2.4. Enzyme-linked immunosorbent assays (ELISA)
Assays for anti-C. jejuni IgG2b and Bradford assays for total plasma protein were carried out as previously described [15].
2.5. Multilocus sequence typing (MLST) and phylogenetic analyses
DNA was extracted from strains to be typed as previously described [17]. Multilocus sequence typing was carried out and the clonal complexes and sequence types of newly typed strains identified using genes, primers, PCR reaction conditions, and software at the Campylobacter jejuni Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford [21] or downloaded from the Campylobacter jejuni Multi Locus Sequence Typing website [22]. PCR products required for sequencing of the housekeeping genes used for Campylobacter jejuni MLST were produced in our laboratory for strains D6844–D6849; sequencing was done on an ABI 3730 Genetic Analyzer at the Michigan State University Research Technology Support Facility. Sequences were obtained for genome-sequenced strains by BLAST from the respective genome sequences at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Concatenated sequences were aligned with MUSCLE and GBlocks and maximum likelihood phylogenetic trees generated using PhyML utilities at the Phylogeny.fr website (http://www.phylogeny.fr/; [23]; default parameters were used throughout the analysis. The thirteen reference MLST types designated by Wareing et al. [24] were included to provide phylogenetic context; the tree was rooted using C. coli 12526.
2.6. Development of a pan-genome whole-open reading frame (ORF) C. jejuni microarray
Campylobacter jejuni genes not present in strain 11168 [25] were identified using procedures described in Lefebure and Stanhope [26]. Fifteen Campylobacter genomes were used; these genomes were available at NCBI in June, 2007, and included ten C. jejuni subsp. jejuni and subsp. doylei genomes. OrthoMCL was used to define orthologous clusters. All strain-specific genes were checked using BLAST. Three hundred eight putatively strain-specific genes had strong BLAST hits (P>1e-10) with other Campylobacter jejuni genes and were excluded. Then clusters that were absent from C. jejuni 11168 strain but present in one or more other C. jejuni strains were identified; 683 clusters corresponding to that criterion were identified. Sequences that were incomplete because they occurred at the beginning or end of a contig were not exported. The resulting 683 ORFs are listed in Supplementary Table 1. Primers were developed for the newly identified ORFs using the web-based Primer3 program (http://jura.wi.mit.edu/rozen/papers/rozen-and-skaletsky-2000-primer3.pdf)[27]. As much of the sequence as possible of the longest representative of each ORF was included in the PCR product. Primers were synthesized at the Macromolecular Structure, Sequencing and Synthesis Facility, Department of Biochemistry, Michigan State University. PCR products were synthesized and verified by Sanger sequencing at the Research Technology Support Facility, Michigan State University. These products were added to our existing C. jejuni 11168 whole-ORF microarray (NCBI GEO ID: SE1379400), which contains all C. jejuni 11168 and pVir ORFs; new arrays were printed at the Research Technology Support Facility, Michigan State University as previously described [17]. All spots were printed in duplicate. Details of the construction of the new array are at NCBI GEO ID: GPL8954. Strain NW, which was previously analyzed using our C. jejuni 11168 whole-ORF microarray [17], was reanalyzed for this study using the expanded microarray.
2.7. Microarray analyses
DNA for microarray analysis of genomic content was isolated as follows. C. jejuni strains were spread on tryptose soya agar (Difco, Becton Dickinson, Sparks, MD) plates containing 5% defibrinated sheep’s blood (Cleveland Scientific, Bath, OH) and incubated 48 hours at 37 C in a sealed jar in the atmosphere generated by a CampyGen sachet (Oxoid, Cambridge, UK). Single isolated colonies were selected and spread heavily on tryptose soya agar plates containing 5% defibrinated sheep’s blood; these cultures were incubated 48 hours at 37 C in a sealed jar in the atmosphere generated by a CampyGen sachet. Growth was harvested using a sterile cotton swab and suspended in 900 µL tryptose soya broth (Difco, Becton Dickinson, Sparks, MD) in a microcentrifuge tube. Cells were pelleted by centrifugation at 16,000 rcf in an Eppendorf 5415D centrifuge (Hamburg, Germany) for 30 seconds at room temperature. The supernatant was removed and DNA extracted from the pelleted cells using a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA) according the manufacturer’s instructions for Gram-negative bacteria, including RNase A (Roche Diagnostics, Indianapolis, IN, USA) treatment after cell lysis and proteinase K digestion. DNA was eluted once from the columns using 200 µL elution buffer. DNA preparations were kept at room temperature for three hours, heated at 37 C for 30 minutes, and centrifuged at 16,000 rcf in an Eppendorf 5415D centrifuge at room temperature for 30 minutes. A volume of 150 µL was removed to a fresh microcentrifuge tube, taking care not to touch the bottom or sides of the tube with the pipette tip, and stored frozen at minus 20 C.
DNA was sheared to a molecular size of 1.5–3.0 kilobase pairs using a Covaris S2 Adaptive Focused Acoustic Disruptor (Covaris, Woburn, MA) and quantified using a QUBIT spectrophotometer (Invitrogen, Carlsbad, CA) prior to labeling with amino-allyl dUTP and coupling to Cy3 and Cy5 dyes (Amersham, GE Healthcare UK, Little Chalfont, Buckinghamshire, England, UK) as previously described; all slide blocking, hybridization, and washing procedures were also as previously described [17]. Four biological replicate hybridizations, two of which were dye swaps, were performed, yielding a total of eight comparisons for each ORF in the microarray. Scanning was done using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA) and GenePixPro6 software. Spots were edited using GenPixPro3 software (Molecular Devices, Sunnyvale, CA). Data analysis is described below. Data are available in NCBI GEO ID: GSE24764 and summarized in Supplementary Table 2.
For gene expression studies, dual color microarray analysis and data analysis were performed as described previously [20]. cDNAs derived from RNA obtained during C. jejuni growth in mice and in broth were compared using whole open-reading frame arrays (NCBI GEO ID: GPL8707). RNA obtained from each of four mice was compared to RNA obtained from four replicate broth cultures. Reports from microarray experiments are deposited in NCBI GEO ID: GSE28578 and conform to MIAME guidelines.
2.8. Genomic and virulence gene content analyses
Data for sequenced strains and strains analyzed by microarray were analyzed separately. Whole genome comparison of gene content of sequenced strains was performed using abundance profiles of clusters of orthologous groups of proteins (COGS) and pfam domains downloaded from the Joint Genome Institute Integrated Microbial Genomes website (http://img.jgi.doe.gov/cgi-bin/w/main.cgi)[28]. The array of numbers of COGS in each genome was used to calculate Hamming distances between strains and the resulting matrix was clustered by the unweighted pair groups with arithmetic averaging (UPGMA) method with 1000 bootstrap runs using PAST software [29].
Microarray data were analyzed as previously described [17] using the GACK method of Kim et al. [30] at http://falkow.stanford.edu/whatwedo/software/software.html for genes found in C. jejuni 11168 (reference DNA present in the hybridization) and analysis by method 4 of Carpaij et al. [31] for ORFs not found in C. jejuni 11168 (reference DNA not present in the hybridization). For GACK analysis of all strains except D6848, ORFs were counted as present (P) if 5 of 8 spots had GACK values of 0.45 or 0.5 and as absent (A) if 5 of 8 spots had GACK values of −0.45 or −0.5. For strain D6848, only three comparisons were obtained that could be processed by the GACK program, and ORFs were counted as present (P) if 4 of 6 spots had GACK values of 0.45 or 0.5 and as absent (A) if 4 of 6 spots had GACK values of -0.45 or -0.5. All ORFs that did not fall into these “present” or “absent” categories were counted as divergent (D). Method 4 of Carpaij et al. [31] was used for ORFs not present in strain 11168 (no reference DNA present in the hybridization). For ORFs not found in C. jejuni 11168, the signal at an individual microarray spot was designated as positive if the signal attributable to the non-C. jejuni 11168 DNA was both more than twice the background signal attributable to that DNA and greater than the signal attributable to the C. jejuni 11168 DNA; otherwise signals are designated by the method as negative. For the data presented here, ORFs were counted as present (P) if the criteria required in Method 4 of Carpaij et al. [31] were met for five of the eight spots. ORFs were counted as absent (A) if the criteria were not met in five of the eight spots. All ORFs that did not fall into these “present” or “absent” categories were counted as ambiguous (?). Results for strain D6848 were treated as described above.
Comparison of virulence-associated gene content of sequenced strains was performed using presence, absence or ambiguous scoring as described above for all ORFs except those belonging to Campylobacter Insertion Elements (CJIEs) and the plasmid pVir. UPGMA clustering with 1000 bootstrap runs was performed using the Hamming matching coefficient and PAST software as described above [29].
Putative virulence genes were identified in a literature search for C. jejuni genes in the following categories: (1) genes which were mutated in a parent strain either by random mutagenesis or by targeted mutation and shown to affect adherence and or invasion of tissue culture cells or ability to colonize a host, (2) genes not demonstrated experimentally to be involved in adherence, invasion, or colonization but known or hypothesized to be involved in production of virulence determinants such as flagella, capsule, or lipo-oligosaccharide (LOS), and (3) C. jejuni 11168 genes shown to be upregulated in more adherent, more invasive, and better colonizing minimally passaged variants of C. jejuni 11168 compared to highly laboratory passaged variants [32–33]. Genes involved in energy generation or intermediary metabolism (e.g., aspA and aspB [34–35]; oorB and nuoD [36]; and fdrA [37] were included; it is recognized that impairment in these functions would indirectly affect colonization and virulence [38]. The 314 genes identified, including virulence-related phenotypes and citations, appear in Supplementary Table 3; the literature search was concluded in March, 2012. Protein sequences for the alleles of these genes found in C. jejuni 11168 were obtained from GenBank accession AL111168 and protein vs translated nucleotide (tBLASTn) searches of all genome-sequenced C. jejuni strains performed using the batch BLAST utility at the NCBI genome website (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi; [39]). Assignments of gene function are those given in the reannotation of the C. jejuni 11168 genome by Gundogdu et al. [40].
Alleles in strains other than 11168 were classified as matches to the 11168 allele, divergent from the 11168 allele, or missing. Classification as a match required full or nearly full length alignment with an alignment score of ≥200 throughout the length of the sequence. Sequences too short to generate an alignment score >200 of the 11168 allele with itself were classified as matches if alignment scores were ≥80 and ≤200 throughout the length of the sequence. Classification as divergent from the 11168 allele was made when either the alignment scores were ≤200 for all or part of the sequence or when alignments exhibited internal deletions or truncations at the N or C termini. Classification as absent was made when no alignment was obtained, when only a small portion of the sequence was aligned, and/or when alignment scores were ≤80 throughout the length of the sequence. For each strain, alleles that matched the 11168 allele were given a value of 2 (present); alleles divergent from the 11168 allele were given a value of 1(divergent); and 11168 alleles not found were given a value of 0 (absent) [41]. For strain 81–176, a gene was recorded as present or divergent if it was present or divergent in one or both of the two sequenced isolates. Scoring of presence, absence or divergence of colonization- and/or virulence-associated ORFs in strains analyzed by microarray was performed as described above. For both sets of strains, cluster analyses of the resulting virulence gene profiles were performed and dendrograms generated as described above.
Predicted protein sequences were derived using ExPASy software (http://web.expasy.org/translate/) [42].
2.9. Statistical analyses
Statistical analysis of histopathology and ELISA data was performed as previously described [15]. Briefly, Kruskal Wallis one-way ANOVA on ranks (SigmaStat 3.1, Systat Software, Port Richmond, CA) was used to detect the presence of differences; if a significant result was obtained, post hoc tests were performed using Fisher’s exact tests on categorized data for histopathology scores (VassarStats, http://vassarstats.net/ [15, 43] and t-tests (two-tailed, unequal variances) for ELISA data. P values from post hoc tests were adjusted by the Holm-Šidák step-down procedure to correct for multiple comparisons [44]. Kaplan Meier log rank survival analysis with post hoc tests by the Holm-Šidák procedure was done using SigmaStat 3.1. Corrected P values for all pairwise post hoc comparisons appear in the appropriate figure legends.
The Mantel test [45] correlating colonization and disease phenotypes with the presence, absence, and sequence divergence or ambiguity of microarray result of differentially regulated open reading frames was conducted using PAST software [29]. Data for sequenced strains and strains analyzed by microarray were analyzed separately. For this analysis, the colonization/disease matrix contained the percent of mice colonized and the percent of mice exhibiting enteritis (watery and hemorrhagic enteritis combined); the genomic composition matrix contained the absence, sequence divergence or ambiguity of microarray result, and presence of ORFs coded as 1, 2, and 3, respectively. The Bray-Curtis distance measure was used for the colonization disease matrix and the Hamming matching coefficient was used for the genomic data.
3. RESULTS
3.1. Study design
The three experiments to evaluate C. jejuni disease in C57BL/6 IL-10−/− mice had identical designs and were conducted using the same protocols throughout. In each experiment, eight to ten mice were orally inoculated with each test strain. Details concerning the strains used are given in Table 1. (Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology). In each experiment, eight to ten mice were inoculated with C. jejuni 11168 (positive control), and eight to ten mice were inoculated with tryptose soya broth (TSB; negative control). Colonization, clinical disease, survival, antibody responses, gross pathology findings, and histopathological scoring of the ileocecocolic junction were conducted as previously described [15]. In these experiments, strain 11168 colonized 100% of the mice and produced enteritis in 81% of them; 78% of the C. jejuni 11168-inoculated mice had to be euthanized to prevent suffering due to severe clinical signs of disease; and 51% of them experienced hemorrhagic enteritis. No mice inoculated with TSB were colonized by C. jejuni or developed enteritis.
Phylogenetic (MLST) and total and virulence gene content (BLAST and microarray) comparisons of C. jejuni strains were made as described in Materials and Methods. Data were then examined for correlations between total and virulence gene content and disease phenotype. (Data for strains examined in a previous serial passage experiment [17] are given for easy reference; these data are reproduced under open license agreement with BioMed Central and BMC Microbiology; the citation is given at each occurrence of reproduced data.) Comparison of gene expression profiles in C. jejuni 11168 grown in broth and recovered from the ceca of germ-free mice four days after inoculation were performed as previously described [20].
The results of these studies are discussed below in four main sections: Section 3.2, disease phenotype and immune responses; Section 3.3, phylogenetic relationships among strains; Section 3.4, genomic and virulence gene content comparisons; and Section 3.5, gene expression profiles and the relationship of the genomic content of significantly up- and down-regulated genes to colonization and disease in C57BL/6 IL-10−/− mice.
3.2. Disease phenotype and immune responses in C57BL/6 IL-10−/− mice
3.2.1. Colonization
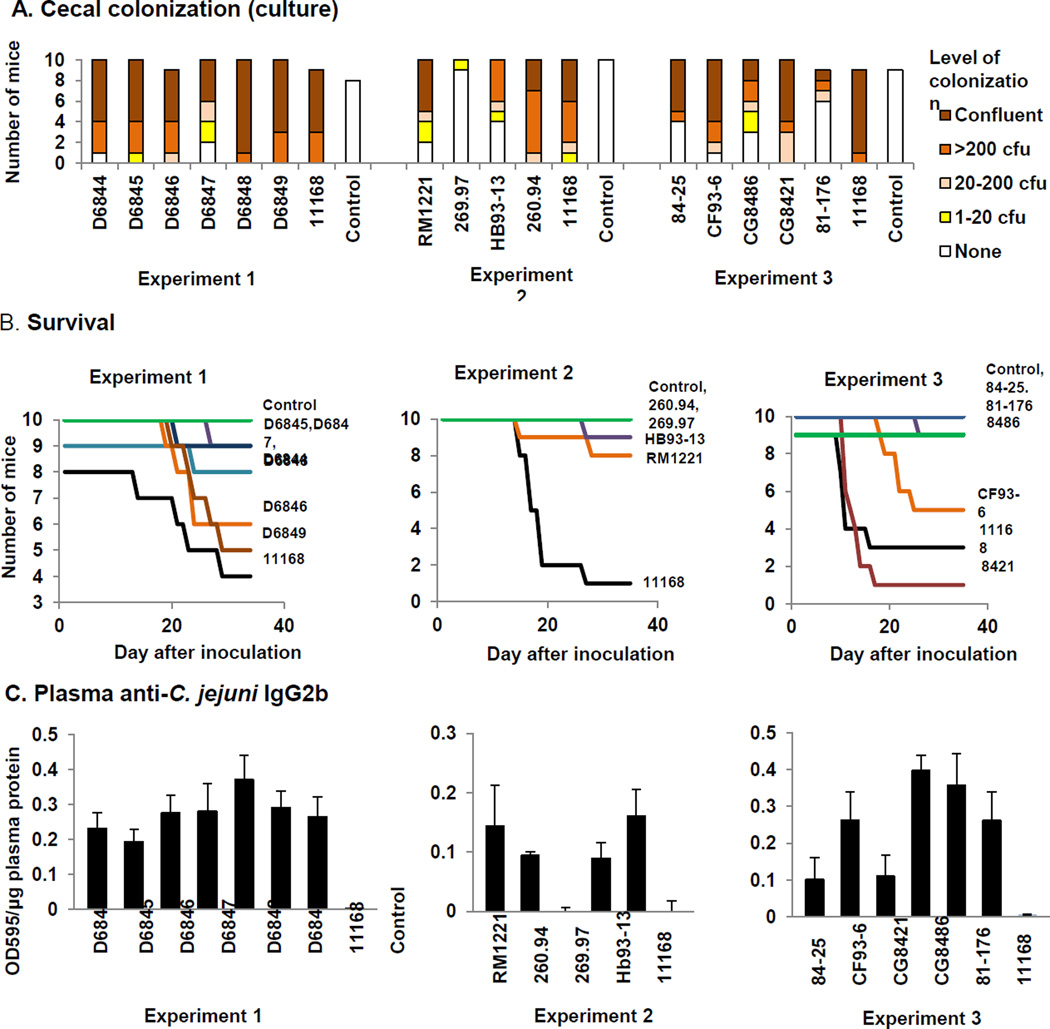
Colonization was assessed by culture of cecal tissue obtained at necropsy 35 days after inoculation as previously described; if C. jejuni was not obtained by culture, bulk DNA was isolated from a snap-frozen replicate sample and assayed by C. jejuni-specific PCR [15]. Of the sixteen strains newly tested here, only strain 269.97 was unable to establish persistent populations in C57BL/6 IL-10−/− mice, since it was recovered by culture from only one of ten inoculated mice and could not be detected by PCR in the remaining mice (Table 1; Figure 1A). (Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology.) Strain 269.97 belongs to C. jejuni subsp. doylei, while all of the other strains tested belong to C. jejuni subsp. jejuni. In previously reported work, strains 33560T and D0121 were unable to establish persistent populations in C57BL/6 IL-10−/− mice [17] (data reproduced under open license agreement with BioMed Central and BMC Microbiology). Strains HB93-13, 84-25, and 81–176 colonized 44–60% of inoculated mice; all other strains colonized 80–100% of inoculated mice. No broth-inoculated mice were colonized as assessed by culture and C. jejuni-specific PCR assay of DNA isolated from cecal tissue.
Figure 1. Cecal colonization, survival, gross pathology, histopathology, and antibody responses in C57BL/6 IL-10−/− mice.
A. Colonization. Level of colonization in the cecum was scored semi-quantitatively as indicated in the legend. B. Survival. Timing of appearance of clinical signs that required euthanasia prior to 35 days after inoculation is shown. In experiment 2, differences in survival were significant for comparisons of mice infected with C. jejuni 11168 vs mice infected with strains 269.97 (Pcorrected =4.96 × 10−5), 260.94 (Pcorrected ==, 0.00015), HB93-13 (Pcorrected =0.00054), RM1221 (Pcorrected =0.00613), and control mice (Pcorrected = 9.92 × 10−5). For experiment 3, differences in survival were significant for pairwise comparisons of mice infected with C. jejuni 11168 vs mice infected with strains 84-25 (Pcorrected = 0.01218) and 81–176 (Pcorrected = 0.02610) and control mice (Pcorrected = 0.02287). In addition, comparisons were significant for pairwise comparisons of mice infected with C. jejuni CG8421 vs mice infected with C. jejuni 84-25 (Pcorrected =3.74 × 10−5), CG8486 (Pcorrected = 0.00015), 81–176 (Pcorrected = 0.00033), and CF93-6 (Pcorrected = 0.00468) and control mice (Pcorrected = 0.00025). C. Plasma anti-C. jejuni IgG2b levels. Determined by ELISA as previously described; OD595/µg plasma protein; statistically significant differences are as follows: in experiment 1, all strains vs control (for D6844, Pcorrected = 0.0045; D6845, 0.0023; D6846, 0.0029; D6847, 0.0406; D6848, 0.0014; D6849, 0.0002; 11168, 0.0133); in experiment 2, strains 11168, HB93-13, and RM1221 vs control (Pcorrected = 0.0077, 0.0162, and 0.0309, respectively) and strain 269.97 vs 11168, HB93-13, and RM1221 (Pcorrected = 0.0038, 0.0120, and 0.0256, respectively), and in experiment 3, strain CG8486 vs control, 11168, CG8421, and 84-25 (Pcorrected = 1.16 × 10−6, 0.0007, 0.0014, and 0.0034, respectively), strain 81–176 vs control (Pcorrected = 0.0094), and strain CF93-6 vs control (Pcorrected =0.0395).
3.2.2. Clinical signs and timing of disease development
The percentages of mice exhibiting five clinical signs of enteric disease (ruffled hair coat, hunched posture, soft or mucoid feces, bloody feces, inactivity) are given in Table 2. If severe clinical signs developed in inoculated mice prior to 35 days after infection, the animals were monitored twice daily to ensure timely euthanasia to prevent suffering. The day when the first mouse inoculated with a particular strain exhibited clinical signs varied between strains, from day 10 to day 23 after inoculation. Some mice developed clinical signs requiring euthanasia within the 12 hour period between health checks; others exhibited milder signs for 1 to 5 days before signs became severe enough to trigger euthanasia. A few mice, notably four mice inoculated with strain D6845, exhibited clinical signs for up to 6 days without developing signs severe enough to trigger euthanasia.
Table 2.
Clinical signs of enteritis.
| Strain | Number of days after infection before clinical signs appeared |
Number of days clinical signs present before euthansia required |
Percent of mice exhibiting clinical sign | ||||
|---|---|---|---|---|---|---|---|
| Rough hair coat |
Hunched posture |
Soft or mucoid feces |
Bloody feces |
Reduced activity |
|||
| Experiment 1 | |||||||
| D6844 (9) | 18–22 | 1–5 | 20 | 30 | 10 | 10 | 10 |
| D6845 (10) | 17–29 | 0 (1 mouse)a | 0 | 0 | 0 | 10 | 0 |
| D6846 (10) | 17–23 | 1–4 | 40 | 30 | 20 | 20 | 40 |
| D6847 (10) | 20–21 | 1 (1 mouse)b | 10 | 10 | 0 | 10 | 10 |
| D6848 (10) | 23 | 0 | 20 | 20 | 10 | 10 | 10 |
| D6849 (10) | 17–27 | 0–3 | 20 | 30 | 10 | 30 | 30 |
| 11168 (8) | 12–28 | 0–2 | 44 | 55 | 11 | 44 | 55 |
| Control (10) | no clinical signs | no clinical signs | 0 | 0 | 0 | 0 | 0 |
| Experiment 2 | |||||||
| RM1221 (10) | 10–25 | 1–3 | 0 | 20 | 0 | 20 | 10 |
| 260.94 (10) | no clinical signs | no clinical signs | 0 | 0 | 0 | 0 | 0 |
| 269.97 (10) | no clinical signs | no clinical signs | 0 | 0 | 0 | 0 | 0 |
| HB93-13 (10) | 23 | 4 | 0 | 0 | 0 | 10 | 0 |
| 11168 (10) | 12–18 | 0–2 | 0 | 100 | 10 | 70 | 30 |
| Control (10) | no clinical signs | no clinical signs | 0 | 0 | 0 | 0 | 0 |
| Experiment 3 | |||||||
| 8421 (10) | 11–17 | 0–1 | 60 | 90 | 30 | 10 | 0 |
| 8425 (10) | no clinical signs | no clinical signs | 0 | 0 | 0 | 0 | 0 |
| 8486 (10) | 21 | 5 | 0 | 10 | 20 | 0 | 10 |
| 81–176 (9) | no clinical signs | no clinical signs | 0 | 0 | 0 | 0 | 0 |
| CF93-6 (10) | 19–26 | 0 | 30 | 50 | 30 | 0 | 0 |
| 11168 (9) | 10–13 | 0–3 | 55 | 67 | 33 | 22 | 11 |
| Control (9) | no clinical signs | no clinical signs | 0 | 0 | 0 | 0 | 0 |
Four mice exhibited clinical signs for 1–6 days without requiring euthanasia.
One mouse exhibited clinical signs for 6 days without requiring euthanasia.
Only strains 260.94, 269.97, 84-25, and 81–176 did not cause severe enough clinical signs to trigger early euthanasia in at least one mouse (Tables 1 and 2; Figure 1B). Strains D6844, D6845, D6847, D6848, HB93-13, RM1221, and CG8486 caused severe enough clinical signs to trigger early euthanasia in 1–3 mice; strains 11168, D6846, D6849, CF93-6, and CG8421 caused severe enough clinical signs to trigger early euthanasia in 4–9 mice. Mice infected with the latter four strains also became ill earlier than mice infected with other strains. Six of ten C57BL/6 IL-10−/− mice infected with strain CF93-6 apparently developed neurological signs, although this observation was complicated by the concurrent development of severe enteritis; we regard this finding as suggestive but not definitive at the time of writing. Further work is currently underway to examine this finding using a full panel of neurological tests in mice infected with strain CF93-6. C57BL/6 IL-10−/− mice infected with strains HB93-13 and 260.94 did not exhibit neurological signs but mice of the NOD genetic background infected with strains HB93-13 and 260.94 did develop neurological signs in experiments to be reported elsewhere [46]. No clinical signs consistent with central nervous system inflammation were observed in mice infected with strain 84-25 from a patient with meningitis. Since strain 269.97 colonized the mice so poorly, it was not possible to determine whether it could produce bacteremia.
It should be noted that the genome-sequenced strains may have lost virulence during laboratory passage [33]. All the minimally passaged strains from the CDC, which were obtained from humans with diarrhea, did cause enteritis in at least one mouse; however, the severity of the enteritis in mice did not exactly correspond to the severity of the diarrhea in the human of origin. IL-10 deficient mice tended to have more severe disease than reported in the human case from which the strain was isolated. At least one mouse infected with each of the six minimally passaged strains produced bloody feces (observed either on the perineum of the animal or on the cage or bedding), although these strains were associated with a spectrum of clinical presentations in the humans from which they were isolated (Table 1). No broth-inoculated mice developed clinical signs of either enteric or neurological disease or required early euthanasia.
Kaplan Meier log rank survival analysis revealed significant differences in survival of mice receiving different inocula in all three experiments (P = 0.026 for experiment 1; P ≤ 0.001 for experiment 2, and P ≤ 0.001 for experiment 3). No pairwise strain comparisons were significant in experiment 1 indicating that differences in development of disease were not pronounced enough to produce different survival patterns. Strains D6844, D6846, and D6849 produced survival patterns intermediate between those of uninfected control mice and mice infected with C. jejuni 11168; whereas strains D6845, D6847, and D6848 each only caused disease severe enough to trigger euthanasia in a single mouse. In experiment 2, differences in survival were significant for comparisons of 11168-infected mice with 269.97-, 260.94-, HB93-13-, and RM1221-infected mice and control mice; thus strain 11168 produced severe disease in mice earlier than the other strains in experiment 1. For experiment 3, differences in survival were significant for comparisons of strain 11168-infected mice and 84-25- and 81–176-infected mice and control mice. Differences were also significant for comparisons of CG8421-infected mice and 84-25-, CG8486-, 81–176-, and CF93-6-infected mice and control mice.
3.2.3. Antibody responses
Mice inoculated with all strains except C. jejuni subsp. doylei 269.97 developed IgG2b anti-C. jejuni plasma antibody responses (Figure 1C), which comprise the main humoral adaptive response to C. jejuni infection [15–17]. Thus all strains except C. jejuni subsp. doylei 269.97 were able to interact strongly enough with the host to produce an adaptive immune response. No control mice developed IgG2b anti-C. jejuni plasma antibodies. Kruskal Wallis ANOVA on ranks indicated the presence of statistically significant differences between groups (P ≤ 0.001 for all three experiments). In experiment 1, post hoc pairwise comparisons were significantly different between mice infected with all strains and control mice. In experiment 2, significant differences were observed between mice infected with strains 11168, HB93-13, and RM1221 and control mice and between mice infected with strain 269.97 and mice infected with strains 11168, HB93-13, and RM1221. In experiment 3, significant differences were observed between mice infected with strain CG8486 and control mice and mice infected with strains 11168, CG8421, and 84-25; between mice infected with strain 81–176 and control mice; and between mice infected with strain CF93-6 and control mice.
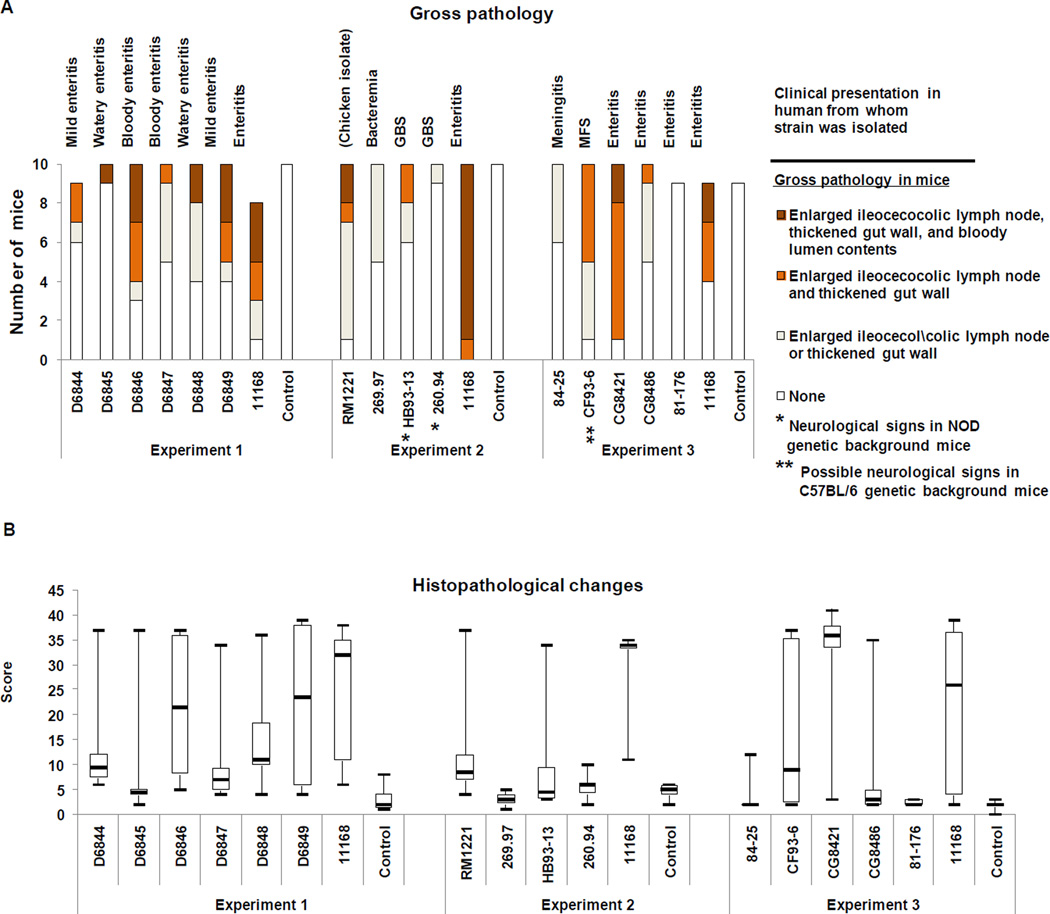
3.2.4. Gross pathology
Gross pathology was observed at the time of necropsy in at least one infected mouse for all C. jejuni strains except 81–176 (Table 1; Figure 2A). (Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology). Gross pathology was minimal (enlarged ileocecocolic lymph node only) in mice infected with strains 260.94, 269.97, and 84-25. Enteritis (thickened cecum or colon wall) was observed in at least one mouse infected with all other strains. Hemorrhagic enteritis (bloody lumen contents in the GI tract) was observed in at least one mouse infected with strains 11168, D6845, D6846, D6848, D6849, RM1221, and CG8421. We note that some mice recorded as producing bloody feces in clinical evaluations were not recorded as having bloody contents in the lumen of the GI tract at the time of necropsy because the cecum and colon were empty or filled with watery fluid by that time. In all cases, histopathology assessments were used to confirm the presence or absence of blood. No broth-inoculated mice exhibited gross pathology.
Figure 2. Gross pathology findings and level of histopathological changes at the ileocecocolic junction in C57BL/6 IL-10−/− mice.
A. Gross pathology. Findings are as indicated in the legend. B. Histopathological changes. Histopathology scores were determined as previously described [15]. Boxes enclose values falling between the first and third quartiles of the distribution; whiskers indicate maximum and minimum scores, and the heavy bar within each box indicates the median score. In experiment 1, the following pairwise comparisons were significant: (1) mice infected with C. jejuni 11168 vs control mice (Pcorrected = 0.0001) and mice infected with strain D6845 (Pcorrected = 0.0033); (2) mice infected with strain D6848 vs control mice (Pcorrected = 0.0080) and mice infected with strains D6845, (Pcorrected = 0.0014) and D6847 (Pcorrected = 0.0372); and mice infected with strain D6846 vs control mcie (Pcorrected = 0.0154). In experiment 2, the following pairwise comparisons were significant: mice infected with strain 11168 vs control mice (Pcorrected = 0.00001), and mice infected with strains RM1221 (Pcorrected = 0.0076), 269.97 (Pcorrected = 0.00001), HB93-13 (Pcorrected = 0.0036), and 260.94 (Pcorrected = 0.0001). In experiment 3, the following pairwise comparisons were significant: (1) mice infected with strain 11168 vs control mice (Pcorrected = 0.032) and mice infected with strain 81–176 (Pcorrected = 0.032) and (2) mice infected with strain CG8421 vs control mice (Pcorrected = 0.0001, and mice infected with strains 84-25, (Pcorrected = 0.0001), CG8486, (Pcorrected = 0.0056), and 81–176 (Pcorrected = 0.0001).
3.2.5. Histopathological changes
Histopathological scoring indicated that at least one mouse infected with all strains except 269.97. 260.94, 84-25, and 81–176 developed severe histologic lesions at the ileocecocolic junction (histopathology score ≥ 20; Grade 2 as previously defined [15] (Figure 2B). Strains 11168, D6846, D6849, and CG8421 caused the greatest degree of typhlocolitis in the largest number of mice, as shown by high median scores. No control mice exhibited histologic lesions at the ileocecocolic junction or in spleen. Kruskal Wallis ANOVA on ranks indicated the presence of statistically significant differences between groups (P ≤ 0.001 for all three experiments).
In experiment 1, pairwise post hoc comparisons showed significant differences between mice infected with C. jejuni 11168 and control mice and mice infected with strain D6845; between mice infected with strain D6848 and control mice and mice infected with strains D6845 and D6847; and between mice infected with strain D6846 and control mice. Significant pairwise differences in experiment 2 were observed for mice infected with strain 11168 vs control mice and mice infected with strains RM1221, 269.97, HB93-13, and 260.94. Significant pairwise differences were observed in experiment 3 for mice infected with strain11168 vs control mice and mice infected with strain 81–176 as well as for mice infected with strain CG8421 vs control mice and mice infected with strains 84-25, CG8486, and 81–176.
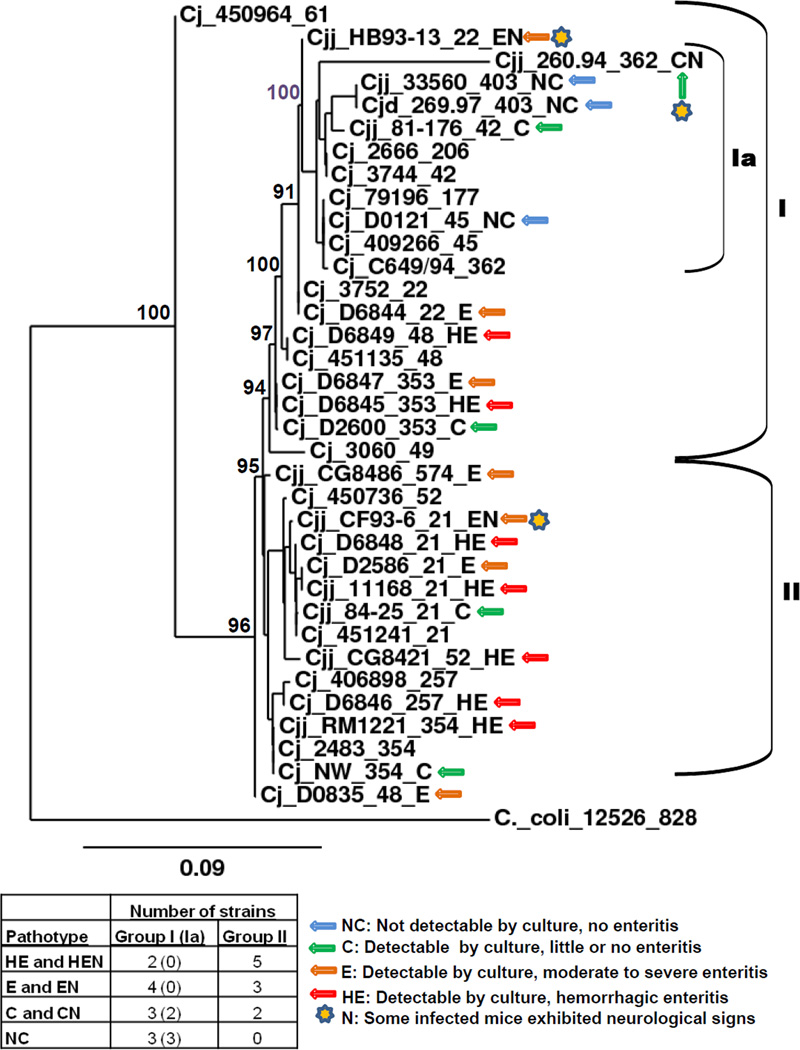
3.3. Phylogenetic relationships among strains (MLST)
Sequencing of seven housekeeping loci was performed for the six minimally passaged CDC strains and the resulting sequences compared to those in the Campylobacter jejuni and Campylobacter coli MLST database at the University of Oxford [21] and are shown in Table 1. All matches were 100% to the reference alleles except for the pgm alleles in strains D6845 and D6846, which differed from their respective reference alleles by a single nucleotide. MLST sequence types of other strains are as specified by Sails et al. [47] or were determined using sequences from GenBank and BLAST utilities at the Campylobacter jejuni and Campylobacter coli MLST database. Figure 3 shows a maximum likelihood phylogenetic tree based on the seven locus MLST scheme of Dingle et al. [48]. The thirteen C. jejuni reference isolates of Wareing et al. [24] were included, and the tree was rooted with C. coli 12526. Most strains belonging to the same clonal complex clustered together; however, strain 11168 fell into cluster I, while the four other clonal complex 21 strains tested in mice fell into cluster II. Similarly, strain D0835 fell into cluster II, while other clonal complex 48 strains fell into cluster I. The strains tested in mice fell into two closely related clusters; there was no differentiation between the two clusters in the array of disease phenotypes represented in them, except that all three non-colonizing strains fell into group I. Cluster I contained three non-colonizing strains, three colonizing strains not causing enteritis, three strains colonizing and causing enteritis, and three strains colonizing and causing hemorrhagic enteritis. Cluster II contained two colonizing strains not causing enteritis, four strains colonizing and causing enteritis, and four strains colonizing and causing hemorrhagic enteritis. The different pathotypes were distributed throughout both clusters, although Cluster I did contain a subcluster (Ia) containing the three non-colonizing strains (33560T, 269.97, D0121) and two of five strains that colonized but did not cause enteritis (81–176, 260.94). Five MLST clonal complexes (21, 353, 354, and 403) were represented by more than one strain; four clonal complex 21 and three 353 strains fell into three pathotypes; two clonal complex 354 and two clonal complex 48 strains, into two pathotypes, and two clonal complex 403 strains into one pathotype.
Figure 3. Phylogenetic relationships of C. jejuni strains (MLST).
Strain designations are coded as follows: species/subspecies name_strain name_MLST clonal complex_pathotype. Abbreviations: Cjj, Camplyobacter jejuni subsp. jejuni; Cjd, Camplyobacter jejuni subsp. doylei; Cj, Camplyobacter jejuni, no subspecies indicated; NC, no colonization; C, colonization with no enteritis; E, colonization with enteritis; HE, colonization with hemorrhagic enteritis; CN, colonization with neurological signs but no enteritis; EN, colonization with enteritis and neurological signs; HEN, colonization with hemorrhagic enteritis and neurological signs. Pathotypes of C. jejuni strains 33560T, D0121, D0835, D2586, D2600, and NW were derived from passage 1 data in a previous serial passage study reported by Bell et al. [17]. Maximum likelihood tree generated using utilities at (http://www.phylogeny.fr/; [23]. Bootstrap support values (% of 1000 runs) are shown for major nodes; tree rooted with C. coli strain 12526.
3.4. Gene content and pathotype
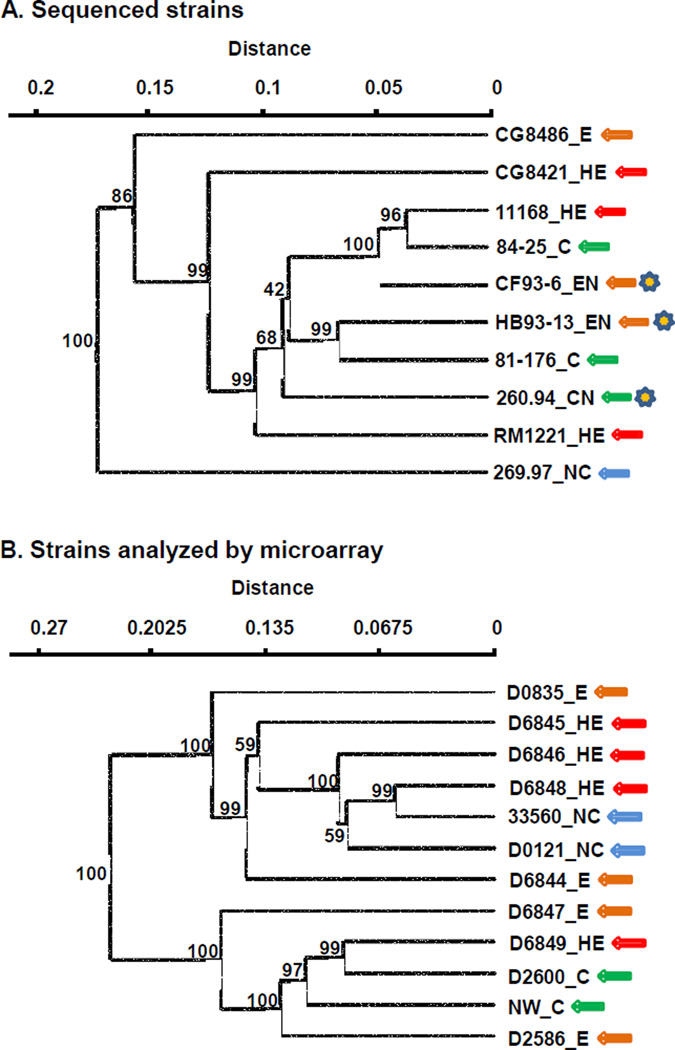
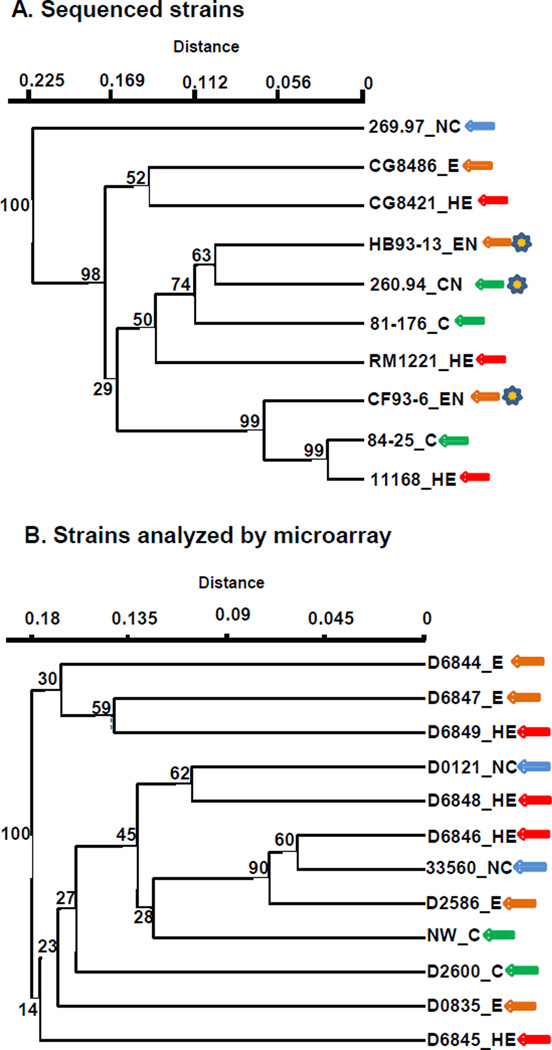
The two sets of strains used in this study (sequenced strains and strains analyzed by microarray) were examined for possible correlations between both total gene content and virulence gene content and virulence phenotype in C57BL/6 IL-10−/− mice. Results for the sequenced strains and the strains analyzed by microarray are presented separately because different kinds of analyses were used for the two sets of strains. Results of total and virulence gene content analyses are shown in Figures 4 and 5.
Figure 4. Pathotypes in C57BL/6 IL-10−/− mice and genomic content.
Virulence phenotypes, MLST clonal complex (CC), and heat-stable serotype (HS) are shown in parentheses. Abbreviations: NC, no colonization; C, colonization with no enteritis; E, colonization with enteritis; HE, colonization with hemorrhagic enteritis; CN, colonization with neurological signs but no enteritis; EN, colonization with enteritis; HEN, colonization with hemorrhagic enteritis and neurological signs. Hamming distance with UPGMA clustering; bootstrap support values (% of 1000 runs) are shown for all nodes Panel A, Genome-sequenced strains (COGS). Data for genome-sequenced strains was from DOE Joint Genome Institute, Integrated Microbial Genomes (http://img.jgi.doe.gov/cgi-bin/w/main.cgi) [28]. Panel B, strains analyzed by microarray.
Figure 5. Pathotypes in C57BL/6 IL-10−/− mice and virulence gene content.
Virulence phenotypes are indicated after the strain designation. Abbreviations: NC, no colonization; C, colonization with no enteritis; E, colonization with enteritis; HE, colonization with hemorrhagic enteritis; CN, colonization with neurological signs but no enteritis; EN, colonization with enteritis; HEN, colonization with hemorrhagic enteritis and neurological signs. Hamming distance with UPGMA clustering; bootstrap support values (% of 1000 runs) are shown for all nodes. Panel A, Genome-sequenced strains. Panel B, strains analyzed by microarray.
3.4.1 Total genome content
Figure 4A shows a cluster analysis based on abundance profiles of clusters of orthologous groups of proteins (COGS) of the ten genome-sequenced C. jejuni strains tested in C57BL/6 IL-10−/− mice. C. jejuni subsp. doylei strain 269.97 was distant from the nine C. jejuni subsp. jejuni strains in this analysis; the three clonal complex 21, heatstable serotype 2 strains (11168, CF-93-6, and 84-25) clustered together. A similar analysis using pfam domains also showed a distant relationship between strain 269.97 and the nine subsp. jejuni strains with 100% bootstrap support as well as clustering of the three clonal complex 21, heat-stable serotype 2 strains with bootstrap support of 73%, but with poor bootstrap support for other interior nodes (results not shown). The two clusters each contain strains of three different pathotypes. The Mantel test did not give a significant result for correlation of the COGS content of the ten sequenced strains with their colonization and enteritis phenotypes.
A whole-ORF C. jejuni 11168 microarray was expanded to include 683 ORFs detected by bioinformatic analysis in at least one of the nine sequenced strains examined in mice but not in strain 11168 (Supplementary Table 1). Complete results for microarray genomic comparisons are given in Supplementary Table 2. Genetic variability was high among the twelve strains analyzed by microarray: of the 2391 chromosomal ORFs represented in the array, only about half (1209) were found unambiguously in all strains; 805 were unambiguously absent in at least one strain. Only four ORFs were not found in at least one strain. Three of twelve isolates analyzed by microarray contained sequences hybridizing to at least one-third of the ORFs in C. jejuni plasmid pVir [49–50]; nearly full or partial copies of the four C. jejuni chromosomal insertion elements (CJIEs) described by Fouts et al. [51] were present in a number of strains (CJIE1 in seven strains; CJIE2 in five strains; CJIE3 in six strains; CJIE4 in 5 strains). Figure 4B shows a cluster analysis based on presence or absence of all ORFs contained in the expanded C. jejuni microarray, excluding those ORFs belonging to pVir and the CJIEs. The two clusters each contain strains of three different pathotypes. The Mantel test did not give a significant result for correlation of the complete genomes of the twelve strains analyzed by microarray with their colonization and enteritis phenotypes.
3.4.2. Virulence-associated gene content
A literature search was conducted to identify genes, complex loci, and functions experimentally implicated in virulence in C. jejuni using criteria described in Materials and Methods. Three hundred fourteen genes were identified (Supplementary Table 3) that are found in C. jejuni 11168 or in other strains and that have functions in virulence mechanisms such as colonization, motility (including flagellar modification), adherence, invasion, iron acquisition, lipooligosaccharide (LOS) biosynthesis, and capsule biosynthesis. Complete virulence gene profiles for both genome-sequenced strains and strains analyzed by microarray are shown in Supplementary Table 3, sections A and B. Cluster analyses of virulence gene content of sequenced strains and strains analyzed by microarray are shown in Figure 5, panels A and B, respectively. The Mantel test did not give a significant result for correlation of the virulence gene profiles of either the ten sequenced strains or the twelve strains analyzed by microarray with their colonization and enteritis phenotypes. The Mantel test was also used to test for correlation between colonization and disease phenotypes and the set of loci found by Cornelius et al. [56] to be related to disease risk; no significant correlations were found.
Of the 172 general virulence loci (that is, loci not involved in N- and O-linked glycosylation, LOS biosynthesis, and capsule biosynthesis), 82 were present in all strains with strong matches to the corresponding C. jejuni 11168 allele and 90 were variable. Of the 90 variable loci, 64 exhibited sequence divergence only, 10 exhibited presence/absence variation only, and 16 exhibited both sequence divergence and presence/absence variation. The four complex loci associated with production of surface molecules (N- and O-linked glycosylation, LOS biosynthesis, and capsule biosynthesis) account for 107 of the 284 virulence ORFs and, as expected, their gene content was variable. Only 19 of the 107 ORFs involved in N- and O-linked glycosylation, LOS biosynthesis, and capsule biosynthesis were present in all strains with strong matches to the corresponding C. jejuni 11168 allele. Of the 88 variable loci involved in N- and O-linked glycosylation, LOS biosynthesis, and capsule biosynthesis, 31 exhibited sequence divergence only, 12 exhibited presence/absence variation only, and 45 exhibited both sequence divergence and presence/absence variation.
Table 3 shows the presence, absence or divergence in colonizing and non-colonizing strains of virulence-associated ORFs present in all enteritis-causing strains. Detailed comparisons of presence/absence variation and sequence divergence in virulence-related ORFs and contingency genes are given in the next four sections, along with a detailed comparison of the three clonal complex 21, HS serotype 2 strains.
Table 3. Presence, absence or divergence in colonizing and non-colonizing strains of virulence-associated ORFs present in all enteritis-causing strains.
| 11168 ORF | Gene name |
84-25 | 260.94 | 81–176 | 269.97 | D0121 | D2600 | NW | 33560 | Function or affected phenotype |
|---|---|---|---|---|---|---|---|---|---|---|
| Cj0019c | docB | P | ? | ? | ? | P | P | P | P | Invasion, colonization, acid resistance |
| Cj0020c | docA | P | P | P | ? | P | P | P | P | Detoxification, peroxidase, colonization |
| Cj0042 | flgD | P | P | P | ? | P | P | P | P | Motility |
| Cj0061c | fliA | P | P | P | P | P | ? | P | P | Motility, σ28 |
| Cj0078c | cdtB | P | P | P | A | P | ? | P | P | Toxin |
| Cj0079c | cdtA | P | P | P | A | P | P | P | P | Toxin |
| Cj0091 | P | P | P | A | P | P | P | P | Adherence, colonization | |
| Cj0178 | ctuA | P | P | A | A | P | P | P | P | Iron acquisition |
| Cj0358 | P | P | P | ? | P | P | P | P | Possible peroxidase | |
| Cj0415 | P | P | P | ? | P | P | P | P | Colonization | |
| Cj0428 | P | P | P | ? | P | P | P | P | Motility | |
| Cj0537 | oorB | P | P | P | P | P | P | ? | P | Colonization, respiration |
| Cj0561c | P | P | P | A | P | P | P | P | Fitness in chickens | |
| Cj0571 | P | P | ? | A | P | P | P | P | Transcriptional regulator | |
| Cj0588 | tlyA | P | P | P | P | P | A | P | P | Adherence, hemolysin? |
| Cj0596 | PEB4 | P | P | P | A | P | P | P | P | Adherence |
| Cj0670 | rpoN | P | P | P | P | P | ? | P | P | σ54, colonization, invasion |
| Cj0780 | napA | P | P | P | ? | P | P | P | P | Colonization, electron transport |
| Cj0887c | flaD | P | P | P | P | P | ? | P | P | Motility, invasion |
| Cj0917 | cstA | P | P | P | P | P | ? | P | P | Carbon starvation |
| Cj0921c | peb1A | P | P | P | P | P | ? | P | P | Transport, binding |
| Cj1169c | OMP50 | P | P | P | ? | P | P | P | P | Major outer membrane protein, adherence |
| Cj1222c | dccS | P | P | P | ? | P | P | P | P | Signalling, colonization |
| Cj1229 | cbpA | P | P | P | P | P | ? | P | P | Stress response |
| Cj1242 | ciaC | P | P | P | ? | P | ? | P | P | Secreted into host cells |
| Cj1261 | racR | P | P | P | P | P | A | P | P | Regulator, colonization |
| Cj1262 | racS | P | P | P | P | P | A | P | P | Colonization |
| Cj1272c | spoT | P | P | P | P | P | A | P | P | Global regulator |
| Cj1279c | flpA | P | P | P | P | P | A | P | P | Adherence, colonization |
| Cj1293 | pseB | P | P | P | P | P | ? | P | P | O-linked glycosylation, motility |
| Cj1298 | P | P | P | P | ? | P | A | P | O-linked glycosylation | |
| Cj1299 | acpP2 | P | P | P | P | ? | P | A | P | O-linked glycosylation |
| Cj1303 | fabH2 | P | P | P | P | P | P | A | P | O-linked glycosylation |
| Cj1309c | P | A | A | P | P | A | A | P | O-linked glycosylation | |
| Cj1310c | P | P | P | P | P | ? | P | P | O-linked glycosylation | |
| Cj1354 | ceuD | P | P | P | A | P | P | P | P | Siderophore transport |
| Cj1355 | ceuE | P | P | P | A | P | P | P | P | Siderophore transport |
| Cj1397 | feoA | P | P | P | ? | P | P | P | P | Iron acquisition |
Abbreviations: P, present; A, absent; ?, divergent by TBLASTN or ambiguous in microarray. Gene names from [25] and [40]; citations for functions and/or affected phenotypes for individual ORFs are given in Supplementary Table 3.
3.4.1. Presence/absence variation
Strains 33560T (non-colonizing) and 84-25 (colonizing) possessed all 314 virulence-associated C. jejuni 11168 ORFs (Supplementary Table 3); however, strain 33560T carries a specific mutation that may account for its lack of virulence [59]. Eight strains only lacked one or more ORFs in one or more of the four variable complex loci and possessed copies of all other virulence-associated ORFs. These strains exhibited non-colonizing (D0121), colonizing but non-enteritis-producing (84-25, 260.94, NW (strain NW produced enteritis in 1 of 25 mice over four serial passages [17]), and watery enteritis-producing (CF93-6, CG8486, D0835, and D6847) pathotypes. Other strains lacked from one to nine virulence-associated ORFs found in all enteritis-causing strains; the array of missing ORFs was different for each strain. As expected, many C. jejuni 11168 ORFs belonging to the four complex loci encoding proteins involved in N- and O-linked protein glycosylation, LOS biosynthesis, and capsule biosynthesis were missing in other strains.
In addition to ORFs comprising the four complex loci, virulence-associated ORFs not found in one or more strains causing enteritis included Cj0170/1 (hypothetical protein), Cj0276 (mreB, adherence), Cj0379c (colonization), Cj0486 (putative transport/binding protein), Cj0526c (fliE; motility), Cj0628/9 (capA; possible membrane lipoprotein; adherence), Cj0698 (flgG; motility), Cj0755 (cfrA, transport/binding protein), Cj1055c (mucin degradation), Cj1510c (fdhA, respiration) Cj1555c (invasion), Cj1626c (colonization), Cj1647 (iamA, invasion), and Cj1677/8 (capB; adherence). All sequenced strains possessed either fspA1 or fspA2 (Cj0859c; apoptosis induction [52]); there was no correlation of pathotype in C57BL/6 IL-10−/− mice with fspA allele among the sequenced strains. Further details and citations regarding these loci can be found in Supplementary Table 3, Sections A and B.
General virulence ORFs missing in some strains showing reduced virulence included Cj0178 (iron acquisition; missing in strain 81–176), Cj0588 (hemolysin, D2600), Cj1261and Cj1262 (racR, racS; D2600), Cj1272c (spoT, regulator; D2600), and Cj1279c (flpA, colonization, adherence, D2600), in addition to ORFs in the four variable complex loci. On the other hand, strain NW, which also colonized without producing enteritis, did not lack any of the general virulence loci, and yet was unable to evolve to greater virulence during three serial passages in C57BL/6 IL-10−/− mice [17].
As previously shown [41], C. jejuni subsp. doylei strain 269.97, which colonized one of ten mice and caused mild enteritis in that mouse had the greatest number of missing virulence ORFs. ORFs missing in strain 269.97 included cytolethal distending toxin genes cdtA, cdtB, and cdtC; the phopholipase A gene pldA; siderophore transport genes ceuB, ceuC, ceuD, and ceuE; iron acquisition genes Cj0177 and ctuA (Cj0178), and the putative transcriptional regulators Cj0571 and Cj1555c. These results are consistent with those obtained in a microarray analysis conducted by Parker et al. [41], who found that cytolethal distending toxin genes and iron transport genes were missing in many C. jejuni subsp. doylei strains.
3.4.2. Genes exhibiting sequence divergence
Fifty-one general virulence-associated ORFs were present in all strains producing either hemorrhagic or watery enteritis but divergent in TBLASTN or ambiguous in microarray analysis in one or more strains that colonized without causing enteritis. Strains 84-25, 260.94, and 81–176 had one, five, and seven ORFs, respectively, classified as divergent in TBLASTN analysis; strains NW, D2586, D2600 had five, eleven, and sixteen ORFs, respectively, classified as ambiguous in the microarray analysis. Thirty-two general virulence-associated ORFs were present in all colonizing strains (including strains producing either hemorrhagic or watery enteritis) but divergent in TBLASTN or ambiguous in microarray analysis in one or more strains that did not colonize. Strain 33560T had five ORFs and strain D0121 twelve ORFs classified as ambiguous in microarray analysis; strain 269.97 had 22 ORFs classified as divergent in TBLASTN analysis.
3.4.3 Contingency genes
Contingency genes, which contain potentially phase-variable homopolynucleotide tracts and some of which are known or putative virulence factors, were also variable among the strains; Sections A and C of Supplementary Table 3 show the presence, absence, or divergence of homologues to thirty-one C. jejuni 11168 contingency genes among the other strains. As expected, the pseudogenes Cj0046 and Cj0565c were divergent in most strains, and homologues to contingency genes in the LOS and capsule biosynthesis loci were more likely to be absent in some strains. Three C. jejuni 11168 contingency ORFs in the O-linked glycoslyation locus: Cj1305c, Cj1310c, and Cj1342c; were detected in all twenty-one other strains. These ORFs are likely paralogues; therefore we note that microarray and BLAST analyses might not distinguish whether a genome has a single or multiple highly similar copies. ORFs Cj0031/0032 (putative restriction/modification enzyme), Cj0045c (iron acquisition), Cj0275 (clpX protease), Cj0617 and Cj0618 (function unknown), Cj0676 (kdpA; cation transport/binding), Cj1295 and Cj1306c (O-linked glycosylation), and Cj1367c (function unknown) were detected in nineteen or twenty of the twenty-one strains. Homologues of contingency loci Cj0170/0171 (unknown function), and Cj1335/1336 (O-linked glycosylation) were detected in most strains; while loci Cj0628/0629 (capA; adherence) and Cj1667/1668 (capB; adherence) were absent or divergent in many strains. We do not know at this time whether the detected homologues to contingency genes detected in the strains analyzed by microarray actually contain homopolynucleotide tracts capable of phase variation.
3.4.4. Genes in three clonal complex 21, HS serotype 2 strains with different disease phenotypes
Finally, genome contents of virulence loci differing among the three sequenced MLST clonal complex 21, HS serotype 2 C. jejuni subsp. jejuni strains (11168, CF93-6, and 84-25). These strains are the most closely related phylogenetically and most similar in total and virulence-associated gene content in the entire set of strains examined. Strain 11168 caused hemorrhagic enteritis, strain CF93-6 caused watery enteritis, and strain 84-25 colonized but did not cause enteritis. The genomes of strains CF93-6 and 84-25 are reported by NCBI as not yet complete; detailed comparisons based on the available sequences are thus somewhat provisional.
Again, as expected, the three strains differed in ORFs involved in O-linked glycosylation, LOS biosynthesis, and capsule biosynthesis. Among other virulence loci, the three strains differ at the ORF homologous to Cj0262c (docC, putative chemotaxis protein [53]). Strain 84-25 apparently does not possess a full-length copy of this C. jejuni 11168 ORF; strain CF93-6 does. The predicted protein sequence of the strain 84-25 ORF homologous to Cj0486 (putative fucose transport protein implicated in invasion [54]) contains two non-conservative amino acid substitutions compared to the identical predicted sequences for the strain 11168 and CF93-6 ORFs; however, this ORF is absent from other strains producing enteritis. Both C. jejuni 84-25 and CF93-6 possessed fspA2, rather than C. jejuni 11168 ORF Cj0859 (fspA1, apoptosis induction). ORF1564 (tlp3, invasion) was divergent in strain CF-93-6. All of these ORFs are absent or divergent in one or more strains causing enteritis.
Among general virulence loci that are also contingency loci, the N-terminal portion of the amino acid sequence of ORF Cj00628/0629 (capA, probable lipoprotein implicated in colonization and adherence [55]) in strain 84-25 is divergent from the corresponding sequences of both strains CF93-6 and 11168, most notably by a twentyone amino acid deletion. However, ORF Cj00628/0629 is apparently absent from strain 8421, which produced hemorrhagic enteritis. Strain CF93-6 homologues of all other strain 11168 contingency loci known to be involved in general virulence, LOS biosynthesis, and capsule biosynthesis do have homopolynucleotide tracts. All but one of the C. jejuni 11168 O-linked glycosylation contingency loci in strain CF93-6 do have homopolynucleotide tracts; the homologue of the Cj1310c locus in strain CF93-6 lacks a poly-C tract. Strain 84-25 homologues of all other strain 11168 contingency loci known to be involved in general virulence, LOS biosynthesis, O-linked glycosylation, and capsule biosynthesis have homopolynucleotide tracts.
3.4.5. Summary of genome and “virulome” comparisons
The preceding results indicate that each strain examined has a unique complement of virulence-associated loci and that there are no broad patterns in sets of loci that either promote or reduce the ability of an individual strain to cause disease.
3.5. Genes up- or down-regulated in the germ-free mouse cecum
In order to identify additional ORFs potentially involved in the ability of C. jejuni to colonize and cause enteritis in C57BL/6 IL-10−/− mice, C. jejuni 11168 RNA isolated from the cecal contents of germ-free C57BL/6 IL-10−/− mice was compared to RNA isolated from C. jejuni cells grown in vitro by hybridization of derived cDNA to the whole-ORF microarray described below. Two hundred eighty-one C. jejuni 11168 ORFs exhibited statistically significant changes (P ≤ 0.05) in mRNA levels (Supplementary Table 4). Ninety of these 285 ORFs exhibited both P values ≤ 0.05 and fold changes ≥ 2.0; fifty-five of these ninety ORFs were up-regulated and thirty-five down-regulated (Supplementary Table 5). Of the fifty-five up-regulated ORFs, ten have functions associated with virulence, particularly motility and chemotaxis. Two regulatory proteins known to be involved in colonization and virulence, cmeR and cheY, were up-regulated, as were two stress response proteins, cbpA and groEL (groEL was up-regulated three-fold with a P value of 0.052). CheY affects motility [57], while CmeR is a pleiotrophic regulatory protein [58]. One ORF, Cj0437 (sdhA; subunit of succinate dehydrogenase [40]) that was reported by Guo et al. [58] as being up-regulated by CmeR in in vitro studies was also up-regulated more than two-fold in the mouse cecum. Ten ORFs associated with energy metabolism were up-regulated; as were eight ORFs encoding bacterial cell surface molecules such as lipoproteins and porins and five ORFs encoding transport and binding proteins, including one putative iron-binding protein (Cj1224). ORFs Cj1293 (O-linked glycosylation) and Cj1450 (intracellular survival) were also upregulated. Finally, two contingency loci encoding hypothetical proteins thought to be involved in virulence exhibited greater than two-fold changes in transcript levels. ORF Cj0618 was up-regulated in the mouse cecum, while ORF Cj0170 was down-regulated compared to levels in C. jejuni 11168 cells grown in broth. Forty-nine of the up- or down-regulated ORFs belong to sixteen described C. jejuni regulons; many are subject to control by more than one regulatory protein (Table 3).
Among the fifty-five ORFs up-regulated in mice, five were missing in one or more strains causing hemorrhagic enteritis: Cj0698 (flgG), Cj1511c (fdhA; putative formate dehydrogenase), Cj1721c (putative outer membrane protein), and Cj1723c (putative periplasmic protein) (protein functions from [40]). Six up-regulated ORFs were missing in one or more strains able to cause watery enteritis: Cj0122 (function unknown), Cj0385c (putative membrane protein), Cj0427 (function unknown), Cj0485 (putative oxidoreductase), and Cj1668c (putative periplasmic protein) (protein functions from [40]). Among the thirty-five down-regulated ORFs, two were missing in strains causing hemorrhagic enteritis: Cj0181 (ribosome processing) and Cj1677/8 (hypothetical protein) (protein functions from [40]). Two down-regulated ORFs were missing in one or more strains able to cause watery enteritis: Cj0170/1 (hypothetical protein) and Cj0303 (transport protein modA) (protein functions from [40]).
Genome content of the ninety significantly up- and down-regulated ORFs in the twenty-two strains is given in Supplementary Table 3. Twenty-eight ORFs are variable in the set of sequenced strains, while twenty-five ORFs are variable in the set of strains analyzed by microarray. Ten ORFs were variable in both sets of strains (Cj0122, unknown function; Cj0171/1, unknown function; Cj0181, transport/binding protein; Cj0265, energy metabolism; Cj0428, unknown function; Cj1193, periplasmic protein; Cj1322, unknown function; Cj1626c, periplasmic protein; Cj1721, membrane protein; Cj1723, periplasmic protein) (protein functions from [40]).
The Mantel test was used to examine possible correlation between colonization and disease phenotypes and the ninety ORFs up- or down-regulated in the mouse cecum [45]. For sequenced strains, R was 0.7264 (P = 0.025). For the strains analyzed by microarray, R was 0.1797 (P = 0.1984). However, Hyytiainen and Hanninen [59] have recently found that two independent isolates of strain 33560T contain a protein-truncating, inactivating frameshift mutation in the gene encoding the pleiotropic regulatory protein CmeR [58]. Expression of the locus encoding this protein was up-regulated in germ-free mice, and inactivation of the protein seems a likely cause of the inability of the inability of strain 33560T to colonize C57BL/6 IL-10−/− mice. No differences in known virulence gene content between strains 11168 and 33560T were detectable by microarray analysis (Supplementary Table 3). When the Mantel test was repeated for the strains analyzed by microarray with strain 33560T omitted from the data set, R was 0.5237 (P = 0.0182). To examine this correlation further, up- and down-regulated loci were tested separately in both sets of C. jejuni strains. For the sequenced strains the set of down-regulated loci in sequenced strains gave a result significant at the 0.05 level (R = 0.7591; P = 0.005); up-regulated loci in sequenced strains gave an R value of 0.6069 (P = 0.0658). In strains analyzed by microarray (strain 33560T omitted), up-regulated loci gave a result significant at the 0.05 level (R = 0.4219; P = 0.0454), while down-regulated loci in strains analyzed by microarray (strain 33560T omitted) gave R = 0.3429 (P = 0.1202).
4. DISCUSSION
4.1. C. jejuni pathotypes in C57BL/6 IL-10−/− mice
These studies constituted a test of whether disease in murine models induced by C. jejuni strains from a large panel of human cases adequately mimics the course of disease, symptoms/clinical signs, and histopathologic lesions observed in patients. We did observe an array of virulence phenotypes (pathotypes) in C57BL/6 IL-10−/− mice infected with the 16 patient strains, including our known positive control C. jejuni 11168 that causes severe disease in IL-10 deficient mice. Most infected mice had diarrhea that was either watery or bloody. It is possible that all cases of watery diarrhea would have progressed to bloody diarrhea if the duration of infection had been extended, but this information was not obtainable because of the need to euthanize the mice at a standard humane endpoint. However, our previous work examining adaptation of particular C. jejuni strains to the IL-10 deficient murine host showed that disease may shift from watery to bloody diarrhea upon serial passage in IL-10 deficient mice for some but not all strains, suggesting that the ability to cause bloody diarrhea is a bacterial attribute [17].
In this study, clinical signs in mice did not correspond exactly to the clinical presentation in the human from whom the strain was isolated; in general, C. jejuni infected IL-10 deficient mice tended to have more severe disease than reported in the human case from which the strain originated. This result is not surprising because IL-10 is known to down-modulate intestinal inflammation [16]. Despite this deficiency, IL-10−/− mice showed a spectrum of phenotypes that was C. jejuni strain dependent. We also note that it is possible and even probable that many of the genome-sequenced strains have lost virulence during laboratory passage. Despite these uncertainties, all six of the newly studied minimally passaged strains from the CDC were able to produce hemorrhagic enteritis, based on the observation of red blood cells in lumen contents in sections of the ileocecocolic junction in at least one infected mouse. However, previously studied minimally passaged isolates (D0121, D0835, D2586, D2600, and NW) did display different degrees of pathogenicity [17].
It is also important to note that the available information regarding the consistency of clinical presentations associated with C. jejuni infection in humans indicates that symptoms caused by a single strain can vary between individuals. For example, studies of experimental infection of humans with strains 81–176 and A3249 have shown that infection with a single strain can result in a spectrum of symptoms [1, 60–61]. Hence, the variability of clinical signs within groups of C57BL/6 IL-10−/− mice infected with the same C. jejuni strain is not surprising. We also recognized at the start of these experiments that no histologic information is available for most human campylobacteriosis cases; this lack of information could lead to false assumptions about the severity of lesions in humans. Finally, we also know that the fat and/or linoleic acid content of the diet affects the severity of C. jejuni enterits in C57BL/6 IL-10−/− mice, possibly through modulation of the immune response by linoleic acid [17]. Thus, human and murine disease manifestations might differ due to subtle differences between the species in immune responses and the modulation of those responses by diet. The fact that C. jejuni strain 260.94, which does not cause enteritis in C57BL/6 IL10−/− mice, does so in NOD IL-10−/− mice (unpublished observation) supports a potential role for host immune system genetics in the outcome of C. jejuni disease.
The further question of pathogen interactions with and immune modulation by the microbiota is also just beginning to be explored in both humans and mice. Given the many and variable potential influences on the outcome of an infection, it seems likely that the outcome of infections of individual hosts in nature is the result of microevolutionary processes within each individual host. Selective pressures that vary with host tissue, host immune responses, the resident microbiota, environmental factors such as diet, and interactions among these host components would act simultaneously on genetically distinct founding pathogen populations. A recent detailed study of withinhost microevolution of Burkholderia pseudomallei in naturally-occurring infections illustrates the complexities involved [62]. Even though it does not exactly replicate human disease, the experimentally tractable murine model offers the best opportunity to tease out some of the factors affecting the outcome of C. jejuni infection, particularly since the immune responses of the mouse can be studied in great detail.
4.2. Genome content of strains belonging to different pathotypes
These studies were also designed to determine whether C. jejuni phylogeny or strain-specific gene content was associated with a particular pathotype in the mice. The result of MLST analysis suggested a potential association of pathotype with phylogeny, but this result should be viewed with caution for several reasons. Although several colonization deficient and non-enteritis-causing C. jejuni subsp. jejuni strains did cluster with the single C. jejuni subsp. doylei strain in the MLST analysis (cluster 1a), we cannot exclude the possibility that some or all strains in cluster Ia are potentially pathogenic in C57BL/6 IL-10−/− mice. First, except for the bovine isolate 33560T, all of the non-colonizing and colonizing but non-disease-producing strains were isolated from humans exhibiting clinical disease. (Some data in Table 1 are reproduced from [17] under open license agreement with BioMed Central and BMC Microbiology). Second, strains may have lost virulence during laboratory passage; this outcome is especially likely for the sequenced strains and the type strain 33560, which have been studied extensively in the laboratory. As noted in section 3.4.5, the inability of strain 33560T to colonize C57BL/6 IL-10−/− mice is likely due to mutation in the gene for the regulatory protein CmeR [59], which is up-regulated in the germ-free mouse cecum. Third, the two strains most closely related to the colonization and enteritis deficient strains of cluster 1a, strains D6844 and HB93-13, were able to cause enteritis in C57BL/6 IL-10−/− mice. Finally, several strains, including 11168, did increase in their ability to cause enteritis after serial passage in mice [17]; therefore, other strains also may be capable of causing disease following adaptation to the mouse gastrointestinal tract. Therefore, the potential association of pathotype with phylogeny in group Ia must be viewed as intriguing but in need of verification by further work with minimally passaged isolates.
Neither comparisons of gene content of the ten genome-sequenced strains nor microarray comparisons of eleven additional strains using a whole open reading frame C. jejuni pan-genome microarray revealed genetic differences that corresponded to the defined pathotypes. The extensive genetic variability for which C. jejuni is known was apparent even in this limited set of 22 strains. However, in cluster analyses based on either on COGS content of the entire genomes of the ten C. jejuni subsp. jejuni genome-sequenced strains or on the entire genome content of strains analyzed by microarray, there was no apparent relationship between clustering pattern and disease phenotype. Similarly, when the presence or absence of virulence-associated genes was examined, there was no consistent lack of particular known or putative virulence-associated ORFs that would explain the different disease phenotypes in C57BL/6 IL-10−/− mice; each strain had a unique combination of missing virulence-associated C. jejuni 11168 ORFs.
The genetic bases of the demonstrated ability of several strains and the contrasting inability of strain NW to evolve to greater pathogenicity in previously reported serial passage experiments are largely unknown [17], although work in our laboratory has implicated both phase variation of contingency genes and changes in gene expression of several additional contingency gene loci in the increased pathogenicity of C. jejuni 11168 after host adaptation by serial passage in C57BL/6 IL-10−/− mice [20]. Since the acquisition of appropriate genes from resident microbiota is unlikely, the occurrence or non-occurrence of particular point mutations, phase variation in contingency genes, or changes in gene expression patterns in these C. jejuni strains are likely responsible for the differing observed evolutionary outcomes [17].
Contingency genes contain homopolymeric tracts subject to high-frequency slip-strand mutagenesis; furthermore, growth in the mouse GI tract has been shown to exert selective effects on genetic diversity at C. jejuni loci wlaN and flgR, which contain homopolymeric tracts [84]. It is therefore noteworthy that most strains examined had detectable homologues to four contingency ORFs belonging to the O-linked glycosylation locus, this complex locus plays a role in motility and therefore virulence due to the activity of its genes in modification of flagellin [85]. Phase variation in flagellin glycosylation patterns may also contribute to immune system evasion. Similarly, the C. jejuni 11168 contingency ORF Cj1342c, which was detected in all strains, encodes the motility factor maf7 [32, 86]. Finally, the contingency ORF Cj0045c, detected in nineteen strains, resembles iron acquisition genes and is co-regulated with flagellar genes [32]. All of these observations strongly suggest a role for contingency genes affecting motility in the adaptation of C. jejuni to the host.
4.3. C. jejuni 11168 gene expression in germ-free mice
Unlike total genome content and content of known virulence-associated ORFs, the genome content of ninety C. jejuni 11168 ORFs significantly up- and down-regulated in germ-free C57BL/6 IL-10−/− mice was significantly correlated by the Mantel test with the percentages of conventional C57BL/6 IL-10−/− mice colonized and experiencing enteritis after inoculation with each of the twenty-two C. jejuni strains. Both up- and down-regulated ORFs contributed to the significance of the result; no particular ORFs were associated with the ability or inability to colonize or cause enteritis.
Gene expression in C. jejuni 11168 has been studied in other in vivo models, including ceca of newly hatched chicks [87], empty piglet stomachs [88], rabbit ileal loops [75], and subcutaneously implanted rabbit tissue chambers [89]. Comparison of results in mice to results from other in vivo studies is somewhat problematic because of differences between the studies in host species, anatomical location, duration of infection (15 minutes to 96 hours), and presence or absence of digesta and gut microbiota. Nevertheless, the pattern of gene expression in the germ-free mouse cecum more closely resembled that in newly hatched chick ceca and empty piglet stomachs than that in rabbit ileal loops or tissue chambers. Expression levels of most ORFs showing differential regulation in both mice and chick ceca (9 of 11 ORFs) or piglet stomachs (21 of 26 ORFs) changed in the same direction (i.e., either up or down). Expression levels of considerably fewer ORFs showing differential regulation in both mice and rabbit ileal loops (1 of 11 ORFs) or subcutaneous tissue culture chambers in rabbits (12 of 34 ORFs) changed in the same direction. This comparison thus indicates that both host species and resident microbiota contribute substantially to gene expression patterns in C. jejuni.
This set of ninety ORFs surely does not comprise the minimal gene complement needed for C. jejuni to colonize and/or cause enteritis in C57BL/6 IL-10−/− mice, nor do all the up- or down-regulated ORFs necessarily play a direct role in C. jejuni pathogenesis. Nevertheless, the fact that the significance of the observed correlation of colonization and enteritis phenotype with the presence or absence of differentially expressed ORFs depends on both up- and down-regulated ORFs reinforces the conclusion that the regulatory circuits that control C. jejuni gene expression patterns play a crucial role in these processes. This conclusion is reinforced by the prior observation that regulatory patterns evolve during adaptation of C. jejuni to the mouse intestinal tract [20]. Taken together, our results on genome content and gene expression in this C. jejuni mouse model suggest that virulence is a combinatorial property of gene content in C. jejuni, as it is in Pseudomonas aeruginosa [90]. Pathotype expression thus likely depends differences in gene expression patterns as well as on strain-specific presence, absence or divergence of particular ORFs, phase variation, and point mutations. Genetic regulatory circuits may thus present opportunities for development of novel control measures.
4.4. The C. jejuni virulome
Supplementary Table 6 contains 201 ORFs that comprise a first approximation of the C. jejuni “virulome,” or set of genes necessary to cause disease, as expressed in C57BL/6 IL-10−/− mice, based on the known colonization- and virulence-associated ORFs listed in Supplementary Table 2 and the ninety ORFs shown in this study to be differentially regulated in the mouse cecum (Supplementary Table 5). It should be noted that proof of the essentiality of all ORFs listed as part of the C. jejuni virulome would require testing by another method, such as screening in mice of strains carrying targeted mutations. The proposed virulome includes three groups of ORFs present in all strains that colonized and caused either bloody or watery enteritis in C57BL/6 IL-10−/− mice: 1) 130 virulence-associated ORFs, 2) 27 ORFs belonging to the highly variable four complex loci (N-linked and O-linked glycosylation, LOS, and capsule present in all enteritis-causing strains. and 3) 43 additional ORFs differentially regulated more than two-fold in the mouse cecum.
A number of caveats apply to this initial delineation of the C. jejuni virulome, which should not be regarded as either exhaustive or as containing only virulence-related ORFs. It is possible that some ORFs in each category of Supplementary Table 6 may not be required for virulence, because no strain lacking any of these ORFs was tested in this work. In addition, there are known to be many potential combinations of ORFs in the four complex loci found in virulent strains. Some of the additional ORFs that were differentially regulated less than two-fold in the mouse cecum may be important for virulence. Finally, there are likely to be as yet undescribed genes necessary for virulence among the numerous hypothetical ORFs in the C. jejuni pan-genome. However, this approximate virulome should serve a useful purpose as a starting point for studies focussed on particular loci and functions.
5. CONCLUSIONS
We conclude that for the C. jejuni subsp. jejuni strains studied here, genetic differences between strains such as individual genetic complements of virulence loci, point mutations, phase variation in contingency genes, differences in gene expression patterns are likely responsible for the observed differences in the virulence phenotypes displayed by these strains. Furthermore, the facts that (1) strains producing hemorrhagic enteritis can lack documented virulence-associated genes and (2) strains which lack documented virulence-associated genes can increase in pathogenicity over a few passages indicate that in spite of a small genome size, C. jejuni possesses multiple, redundant genes or operons encoding particular virulence properties, such as the well-known multiple iron acquisition systems [74], that contribute to its ability to harm its human host. This genetic versatility surely accounts for the evolutionary success of this common multi-host pathogen and will make the development of effective control measures–both in medical contexts and in the food chain–extremely challenging. Nevertheless, the vital role of gene expression patterns in host colonization and disease induction indicates that regulatory circuits provide a promising point of attack for development of control measures for C. jejuni.
Supplementary Material
Highlights.
22 Campylobacter jejuni strains produced 5 disease phenotypes in IL-10−/− mice.
Disease phenotype (pathotype) was not correlated with total gene content.
Pathotype was not correlated with genome content of known virulence genes.
90 open reading frames (ORFS) were up or down regulated in ceca of germ-free mice.
Pathotype was significantly correlated with genome content of these 90 ORFS.
ACKNOWLEDGEMENTS
FUNDING
We thank Drs. Martin Blaser, Linda Demma, Patricia Fields, Collette Fitzgerald, Patricia Guerry, Nicole Iovine, and William Miller for C. jejuni strains and Drs. Shahida Baqar and Titus Brown for comments on the manuscript. This project was funded in whole with federal funds from NIAID, NIH, Department of Health and Human Services, under Contract No. N01-AI-30058 and Co-operative Agreement 1U19-AI-09087.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS
The authors declare that they have no competing interests.
Contributor Information
J. A. Bell, Email: bellj@msu.edu.
J.P. Jerome, Email: jeromejo@msu.edu.
A. E. Plovanich-Jones, Email: hrvatska@msu.edu.
E. J. Smith, Email: ejsmith@msu.edu.
J. R. Gettings, Email: jrgettin@ncsu.edu.
H. Y. Kim, Email: kimhahyu@msu.edu.
J. R. Landgraf, Email: landgra1@msu.edu.
T. Lefébure, Email: tristan.lefebure@univ-lyon1.fr.
J. J. Kopper, Email: kopperja@msu.edu.
V. A. Rathinam, Email: Vijay.Rathinam@umassmed.edu.
J. L. St. Charles, Email: stcharl5@msu.edu.
B. A. Buffa, Email: buffabia@msu.edu.
A. P. Brooks, Email: brook183@msu.edu.
S. A. Poe, Email: psara@umich.edu.
K. A. Eaton, Email: kateaton@umich.edu.
M. J. Stanhope, Email: mjs297@cornell.edu.
L. S. Mansfield, Email: mansfie4@cvm.msu.edu.
REFERENCES
- 1.Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- 2.Perkins D, Newstead G. Campylobacter jejuni enterocolitis causing peritonitis, ileitis and intestinal obstruction. Aust N Z J Surg. 1994;64:55–58. doi: 10.1111/j.1445-2197.1994.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 3.Farmer R. Infectious causes of diarrhea in the differential diagnosis of inflammatory bowel disease. Med Clin North Am. 1990;74:29–38. doi: 10.1016/s0025-7125(16)30584-3. [DOI] [PubMed] [Google Scholar]
- 4.Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 5.Butzler JP. Campylobacter, from obscurity to celebrity. Clin Microbiol Infect. 2004;10:868–876. doi: 10.1111/j.1469-0691.2004.00983.x. [DOI] [PubMed] [Google Scholar]
- 6.Snijders F, Kuijper J, de Wever B, van der Hoek L, Danner S, Dankert J. Prevalence of Campylobacter-associated diarrhea among patients infected with human immunodeficiency virus. Clin Infect Dis. 1997;24:1107–1113. doi: 10.1086/513643. [DOI] [PubMed] [Google Scholar]
- 7.Tee W, Mijch A. Campylobacter jejuni bacteremia in human immunodeficiency virus (HIV)-infected and non-HIV-infected patients: comparison of clinical features and review. Clin Infect Dis. 1998;26:91–96. doi: 10.1086/516263. [DOI] [PubMed] [Google Scholar]
- 8.Skirrow MB, Jones D, Sutcliffe E, Benjamin J. Campylobacter bacteraemia in England and Wales, 1981–91. Epidemiol Infect. 1993;110:567–573. doi: 10.1017/s0950268800050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engberg J, Nachamkin I, Fussing V, McKhann GM, Griffin JW, Piffaretti JC, et al. Absence of clonality of Campylobacter jejuni in serotypes other than HS: 19 associated with Guillain-Barre syndrome and gastroenteritis. J Infect Dis. 2001;184:215–220. doi: 10.1086/322010. [DOI] [PubMed] [Google Scholar]
- 10.Nachamkin I, Engberg J, Gutacker M, Meinersman RJ, Li CY, Arzate P, et al. Molecular population genetic analysis of Campylobacter jejuni HS: 19 associated with Guillain-Barre syndrome and gastroenteritis. J Infect Dis. 2001;184:221–226. doi: 10.1086/322008. [DOI] [PubMed] [Google Scholar]
- 11.Godschalk PC, Kuijf ML, Li J, St Michael F, Ang CW, Jacobs BC, et al. Structural characterization of Campylobacter jejuni lipooligosaccharide outer cores associated with Guillain-Barre and Miller Fisher syndromes. Infect Immun. 2007;75:1245–1254. doi: 10.1128/IAI.00872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlyshev AV, Champion OL, Churcher C, Brisson JR, Jarrell HC, Gilbert M, et al. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol Microbiol. 2005;55:90–103. doi: 10.1111/j.1365-2958.2004.04374.x. [DOI] [PubMed] [Google Scholar]
- 13.Fearnhead P, Smith NG, Barrigas M, Fox A, French N. Analysis of recombination in Campylobacter jejuni from MLST population data. J Mol Evol. 2005;61:333–340. doi: 10.1007/s00239-004-0316-0. [DOI] [PubMed] [Google Scholar]
- 14.Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, et al. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol Biol Evol. 2009;26:385–397. doi: 10.1093/molbev/msn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansfield LS, Bell JA, Wilson DL, Murphy AJ, Elsheikha HM, Rathinam VA, et al. C57BL/6 and congenic interleukin-10-deficient mice can serve as models of Campylobacter jejuni colonization and enteritis. Infect Immun. 2007;75:1099–1115. doi: 10.1128/IAI.00833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansfield LS, Patterson JS, Fierro BR, Murphy AJ, Rathinam VA, Kopper JJ, et al. Genetic background of IL-10(−/−) mice alters host-pathogen interactions with Campylobacter jejuni and influences disease phenotype. Microb Pathog. 2008;45:241–257. doi: 10.1016/j.micpath.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell JA, St Charles JL, Murphy AJ, Rathinam VA, Plovanich-Jones AE, Stanley EL, et al. Multiple factors interact to produce responses resembling spectrum of human disease in Campylobacter jejuni infected C57BL/6 IL-10−/− mice. BMC Microbiol. 2009;9:57. doi: 10.1186/1471-2180-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M, et al. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schell MA, Lipscomb L, DeShazer D. Comparative genomics and an insect model rapidly identify novel virulence genes of Burkholderia mallei. Journal of bacteriology. 2008;190:2306–2313. doi: 10.1128/JB.01735-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jerome JP, Bell JA, Plovanich-Jones AE, Barrick JE, Brown CT, Mansfield LS. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS One. 2011;6:e16399. doi: 10.1371/journal.pone.0016399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolley KA, Chan MS, Maiden MC. mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolley K, Maiden M. BIGSdb: Scalable analysis of bacterial genome variation at the population level. [Accessed 6/15/2012];BMC Bioinformatics. 2010 11:595. doi: 10.1186/1471-2105-11-595. http://pubmlst.org/campylobacter/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dereeper A, G V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. [Accessed 6/15/2012];Nucleic Acids Research. 2008 36:W465–W469. doi: 10.1093/nar/gkn180. http://www.phylogeny.fr/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wareing DR, Ure R, Colles FM, Bolton FJ, Fox AJ, Maiden MC, et al. Reference isolates for the clonal complexes of Campylobacter jejuni. Lett Appl Microbiol. 2003;36:106–110. doi: 10.1046/j.1472-765x.2003.01270.x. [DOI] [PubMed] [Google Scholar]
- 25.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 26.Lefebure T, Stanhope MJ. Pervasive, genome-wide positive selection leading to functional divergence in the bacterial genus Campylobacter. Genome research. 2009;19:1224–1232. doi: 10.1101/gr.089250.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: K S, K S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. [Accessed 6/15/2012]. pp. 365–386. http://frodo.wi.mit.edu/ [DOI] [PubMed] [Google Scholar]
- 28.DOE_Joint_Genome_Institute. [Accessed 6/15/2012];1997–2010 http://img.jgi.doe.gov/cgi-bin/w/main.cgi.
- 29.Hammer O, Harper DAT, Ryan PD. PAST: Paleontological Statistics Software Package for Education and Data Analysis. [Accessed 6/15/2012];Palaeontologia Electronica. 2001 4 http://folk.uio.no/ohammer/past/ [Google Scholar]
- 30.Kim CC, Joyce EA, Chan K, Falkow S. Improved analytical methods for microarray-based genome-composition analysis. [Accessed 6/12/2012.];Genome biology. 2002 3 doi: 10.1186/gb-2002-3-11-research0065. RESEARCH0065. http://falkow.stanford.edu/whatwedo/software/software.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpaij N, Fluit AC, Lindsay JA, Bonten MJ, Willems RJ. New methods to analyse microarray data that partially lack a reference signal. BMC Genomics. 2009;10:522. doi: 10.1186/1471-2164-10-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrillo CD, Taboada E, Nash JH, Lanthier P, Kelly J, Lau PC, et al. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem. 2004;279:20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
- 33.Gaynor EC, Cawthraw S, Manning G, MacKichan JK, Falkow S, Newell DG. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J Bacteriol. 2004;186:503–517. doi: 10.1128/JB.186.2.503-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guccione E, Leon-Kempis Mdel R, Pearson BM, Hitchin E, Mulholland F, van Diemen PM, et al. Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol Microbiol. 2008;69:77–93. doi: 10.1111/j.1365-2958.2008.06263.x. [DOI] [PubMed] [Google Scholar]
- 35.Novik V, Hofreuter D, Galan JE. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect Immun. 2010 doi: 10.1128/IAI.00109-10. Epub: 2010/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weerakoon DR, Borden NJ, Goodson CM, Grimes J, Olson JW. The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb Pathog. 2009;47:8–15. doi: 10.1016/j.micpath.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Weingarten RA, Taveirne ME, Olson JW. The dual-functioning fumarate reductase is the sole succinate:quinone reductase in Campylobacter jejuni and is required for full host colonization. J Bacteriol. 2009;191:5293–5300. doi: 10.1128/JB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hermans D, Van Deun K, Martel A, Van Immerseel F, Messens W, Heyndrickx M, et al. Colonization factors of Campylobacter jejuni in the chicken gut. Veterinary research. 2011;42:82. doi: 10.1186/1297-9716-42-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S, Madden TL, Schäffer A, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. [Accessed 6/15/2012];Nucleic Acids Res. 1997 25:3389–3402. doi: 10.1093/nar/25.17.3389. http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gundogdu O, Bentley SD, Holden MT, Parkhill J, Dorrell N, Wren BW. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics. 2007;8:162. doi: 10.1186/1471-2164-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker CT, Miller WG, Horn ST, Lastovica AJ. Common genomic features of Campylobacter jejuni subsp. doylei strains distinguish them from C. jejuni subsp. jejuni. BMC Microbiol. 2007;7:50. doi: 10.1186/1471-2180-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel R, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. [Accessed 6/15/2012];Nucleic Acids Res. 2003 31:3784–3788. doi: 10.1093/nar/gkg563. http://web.expasy.org/translate/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowrey R. [Accessed 6/15/2012];1998–2010 http://vassarstats.net/ [Google Scholar]
- 44.Ludbrook J. Multiple comparison procedures updated. Clinical and Experimental Pharmacology and Physiology. 1998;25:1032–1037. doi: 10.1111/j.1440-1681.1998.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 45.Shannon WD, Watson MA, Perry A, Rich K. Mantel statistics to correlate gene expression levels from microarrays with clinical covariates. Genet Epidemiol. 2002;23:87–96. doi: 10.1002/gepi.1115. [DOI] [PubMed] [Google Scholar]
- 46.St. Charles J, Bell J, Mansfield L. Development of murine model to study Guillain Barré Syndrome following Campylobacter jejuni infection [abstract] Abstracts of the 110th General Meeting, American Society for Microbiology. 2010 Abstract D-1211. [Google Scholar]
- 47.Sails AD, Swaminathan B, Fields PI. Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing correlate with strain associations identified by multilocus enzyme electrophoresis. J Clin Microbiol. 2003;41:4058–4067. doi: 10.1128/JCM.41.9.4058-4067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, et al. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infect Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bacon DJ, Alm RA, Hu L, Hickey TE, Ewing CP, Batchelor RA, et al. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81–176. Infection and Immunity. 2002;70:6242–6250. doi: 10.1128/IAI.70.11.6242-6250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biology. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poly F, Ewing C, Goon S, Hickey TE, Rockabrand D, Majam G, et al. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect Immun. 2007;75:3859–3867. doi: 10.1128/IAI.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vegge CS, Brondsted L, Li YP, Bang DD, Ingmer H. Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl Environ Microbiol. 2009;75:5308–5314. doi: 10.1128/AEM.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Javed MA, Grant AJ, Bagnall MC, Maskell DJ, Newell DG, Manning G. Transposon mutagenesis in a hyper-invasive clinical isolate of Campylobacter jejuni reveals a number of genes with potential roles in invasion. Microbiology (Reading, England) 2010;156:1134–1143. doi: 10.1099/mic.0.033399-0. [DOI] [PubMed] [Google Scholar]
- 55.Ashgar SS, Oldfield NJ, Wooldridge KG, Jones MA, Irving GJ, Turner DP, et al. CapA, an autotransporter protein of Campylobacter jejuni, mediates association with human epithelial cells and colonization of the chicken gut. J Bacteriol. 2007;189:1856–1865. doi: 10.1128/JB.01427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornelius AJ, Gilpin B, Carter P, Nicol C, On SLW. Comparison of PCR binary typing (P-BIT), a new approach to epidemiological subtyping of Campylobacter jejuni, with serotyping, pulsed-field gel electrophoresis, and multilocus sequence typing methods. Applied and Environmental Microbiology. 2010;76:1533–1544. doi: 10.1128/AEM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 58.Guo B, Wang Y, Shi F, Barton YW, Plummer P, Reynolds DL, et al. CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J Bacteriol. 2008;190:1879–1890. doi: 10.1128/JB.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyytiainen H, Hanninen ML. Quality control strain Campylobacter jejuni ATCC 33560 contains a frameshift mutation in the CmeR regulator. Antimicrobial agents and chemotherapy. 2012;56:1148. doi: 10.1128/AAC.06228-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prendergast MM, Tribble DR, Baqar S, Scott DA, Ferris JA, Walker RI, et al. In vivo phase variation and serologic response to lipooligosaccharide of Campylobacter jejuni in experimental human infection. Infect Immun. 2004;72:916–922. doi: 10.1128/IAI.72.2.916-922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tribble DR, Baqar S, Scott DA, Oplinger ML, Trespalacios F, Rollins D, et al. Assessment of the duration of protection in Campylobacter jejuni experimental infection in humans. Infect Immun. 2010;78:1750–1759. doi: 10.1128/IAI.01021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price EP, Hornstra HM, Limmathurotsakul D, Max TL, Sarovich DS, Vogler AJ, et al. Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS pathogens. 2010;6:e1000725. doi: 10.1371/journal.ppat.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bang DD, Borck B, Nielsen EM, Scheutz F, Pedersen K, Madsen M. Detection of seven virulence and toxin genes of Campylobacter jejuni isolates from Danish turkeys by PCR and cytolethal distending toxin production of the isolates. J Food Prot. 2004;67:2171–2177. doi: 10.4315/0362-028x-67.10.2171. [DOI] [PubMed] [Google Scholar]
- 64.Bang DD, Nielsen EM, Scheutz F, Pedersen K, Handberg K, Madsen M. PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J Appl Microbiol. 2003;94:1003–1014. doi: 10.1046/j.1365-2672.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- 65.Bang DD, Scheutz F, Ahrens P, Pedersen K, Blom J, Madsen M. Prevalence of cytolethal distending toxin (cdt) genes and CDT production in Campylobacter spp. isolated from Danish broilers. J Med Microbiol. 2001;50:1087–1094. doi: 10.1099/0022-1317-50-12-1087. [DOI] [PubMed] [Google Scholar]
- 66.AbuOun M, Manning G, Cawthraw S, Ridley A, Ahmed I, Wassenaar T, et al. Cytolethal distending toxin (CDT)-negative Campylobacter jejuni strains and anti-CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infect Immun. 2005;73:3053–3062. doi: 10.1128/IAI.73.5.3053-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeon B, Itoh K, Ryu S. Promoter analysis of cytolethal distending toxin genes (cdtA, B, and C) and effect of a luxS mutation on CDT production in Campylobacter jejuni. Microbiol Immunol. 2005;49:599–603. doi: 10.1111/j.1348-0421.2005.tb03651.x. [DOI] [PubMed] [Google Scholar]
- 68.Lara-Tejero M, Galan JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290:354–357. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 69.Lara-Tejero M, Galan JE. CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect Immun. 2001;69:4358–4365. doi: 10.1128/IAI.69.7.4358-4365.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Purdy D, Buswell CM, Hodgson AE, McAlpine K, Henderson I, Leach SA. Characterisation of cytolethal distending toxin (CDT) mutants of Campylobacter jejuni. J Med Microbiol. 2000;49:473–479. doi: 10.1099/0022-1317-49-5-473. [DOI] [PubMed] [Google Scholar]
- 71.Whitehouse CA, Balbo PB, Pesci EC, Cottle DL, Mirabito PM, Pickett CL. Campylobacter jejuni cytolethal distending toxin causes a G2-phase cell cycle block. Infect Immun. 1998;66:1934–1940. doi: 10.1128/iai.66.5.1934-1940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziprin RL, Young CR, Byrd JA, Stanker LH, Hume ME, Gray SA, et al. Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis. 2001;45:549–557. [PubMed] [Google Scholar]
- 73.Holmes K, Mulholland F, Pearson BM, Pin C, McNicholl-Kennedy J, Ketley JM, et al. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology. 2005;151:243–257. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- 74.Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stintzi A, Marlow D, Palyada K, Naikare H, Panciera R, Whitworth L, et al. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect Immun. 2005;73:1797–1810. doi: 10.1128/IAI.73.3.1797-1810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller CE, Rock JD, Ridley KA, Williams PH, Ketley JM. Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J Bacteriol. 2008;190:1900–1911. doi: 10.1128/JB.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fox JG, Rogers AB, Whary MT, Ge Z, Taylor NS, Xu S, et al. Gastroenteritis in NF-kappaB-deficient mice is produced with wild-type Campylobacter jejuni but not with C. jejuni lacking cytolethal distending toxin despite persistent colonization with both strains. Infect Immun. 2004;72:1116–1125. doi: 10.1128/IAI.72.2.1116-1125.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tu QV, McGuckin MA, Mendz GL. Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J Med Microbiol. 2008;57:795–802. doi: 10.1099/jmm.0.47752-0. [DOI] [PubMed] [Google Scholar]
- 79.Javed MA, Grant AJ, Bagnall MC, Maskell DJ, Newell DG, Manning G. Transposon mutagenesis in a hyper-invasive clinical isolate of Campylobacter jejuni reveals a number of genes with potential roles in invasion. Microbiology. 2010;156:1134–1143. doi: 10.1099/mic.0.033399-0. [DOI] [PubMed] [Google Scholar]
- 80.Salamaszynska-Guz A, Klimuszko D. Functional analysis of the Campylobacter jejuni cj0183 and cj0588 genes. Curr Microbiol. 2008;56:592–596. doi: 10.1007/s00284-008-9130-z. [DOI] [PubMed] [Google Scholar]
- 81.Bras AM, Chatterjee S, Wren BW, Newell DG, Ketley JM. A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J Bacteriol. 1999;181:3298–3302. doi: 10.1128/jb.181.10.3298-3302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol. 2003;52:345–348. doi: 10.1099/jmm.0.05056-0. [DOI] [PubMed] [Google Scholar]
- 83.Flanagan RC, Neal-McKinney JM, Dhillon AS, Miller WG, Konkel ME. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect Immun. 2009;77:2399–2407. doi: 10.1128/IAI.01266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilson DL, Rathinam VAK, Qi W, Wick LM, Landgraf J, Bell JA, et al. Genetic diversity in Campylobacter jejuni is associated with differential colonization of broiler chickens and C57BL/6J IL10-deficient mice. Microbiology. 2010;156:2046–2057. doi: 10.1099/mic.0.035717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, et al. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 2006;60:299–311. doi: 10.1111/j.1365-2958.2006.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karlyshev AV, Linton D, Gregson NA, Wren BW. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology. 2002;148:473–480. doi: 10.1099/00221287-148-2-473. [DOI] [PubMed] [Google Scholar]
- 87.Woodall CA, Jones MA, Barrow PA, Hinds J, Marsden GL, Kelly DJ, et al. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infection and immunity. 2005;73:5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Applied and Environmental Microbiology. 2008;74:1583–1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flint A, Butcher J, Clarke C, Marlow D, Stintzi A. Use of a rabbit soft tissue chamber model to investigate Campylobacter jejuni-host interactions. Frontiers in microbiology. 2010;1:126. doi: 10.3389/fmicb.2010.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome biology. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gundogdu O, Mills DC, Elmi A, Martin MJ, Wren BW, Dorrell N. The Campylobacter jejuni transcriptional regulator Cj1556 plays a role in the oxidative and aerobic stress response and is important for bacterial survival in vivo. Journal of bacteriology. 2011;193:4238–4249. doi: 10.1128/JB.05189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hwang S, Kim M, Ryu S, Jeon B. Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PloS one. 2011;6:e22300. doi: 10.1371/journal.pone.0022300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Svensson SL, Davis LM, MacKichan JK, Allan BJ, Pajaniappan M, Thompson SA, et al. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Molecular microbiology. 2009;7 doi: 10.1111/j.1365-2958.2008.06534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.MacKichan JK, Gaynor EC, Chang C, Cawthraw S, Newell DG, Miller JF, et al. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Molecular microbiology. 2004;54:1269–1286. doi: 10.1111/j.1365-2958.2004.04371.x. [DOI] [PubMed] [Google Scholar]
- 95.Wösten MM, van Dijk L, Veenendaal AK, de Zoete MR, Bleumink-Pluijm NM, van Putten JP. Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Molecular microbiology. 2010;75:1577–1591. doi: 10.1111/j.1365-2958.2010.07079.x. [DOI] [PubMed] [Google Scholar]
- 96.Wösten MM, Wagenaar JA, van Putten JP. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. Journal of biological chemistry. 2004;279:16214–16222. doi: 10.1074/jbc.M400357200. [DOI] [PubMed] [Google Scholar]
- 97.Holmes CW, Penn CW, Lund PA. The hrcA and hspR regulons of Campylobacter jejuni. Microbiology. 2010;156:158–166. doi: 10.1099/mic.0.031708-0. [DOI] [PubMed] [Google Scholar]
- 98.Andersen MT, Brøndsted L, Pearson BM, Mulholland F, Parker M, Pin C, et al. Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology. 2005;151:905–915. doi: 10.1099/mic.0.27513-0. [DOI] [PubMed] [Google Scholar]
- 99.Jeon B, Itoh K, Misawa N, Ryu S. Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiology and immunology. 2003;47:833–839. doi: 10.1111/j.1348-0421.2003.tb03449.x. [DOI] [PubMed] [Google Scholar]
- 100.He Y, Frye JG, Strobaugh TP, Chen CY. Analysis of AI-2/LuxS-dependent transcription in Campylobacter jejuni strain 81–176. Foodborne pathogens and disease. 2008;5:399–415. doi: 10.1089/fpd.2008.0106. [DOI] [PubMed] [Google Scholar]
- 101.Palyada K, Sun YQ, Flint A, Butcher J, Naikare H, Stintzi A. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC genomics. 2009;10:481. doi: 10.1186/1471-2164-10-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wösten MM, Parker CT, van Mourik A, Guilhabert MR, van Dijk L, van Putten JP. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Molecular microbiology. 2006;62:278–291. doi: 10.1111/j.1365-2958.2006.05372.x. [DOI] [PubMed] [Google Scholar]
- 103.Gaynor EC, Wells DH, MacKichan JK, Falkow S. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Molecular microbiology. 2005;56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.