Abstract
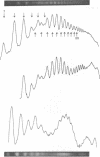
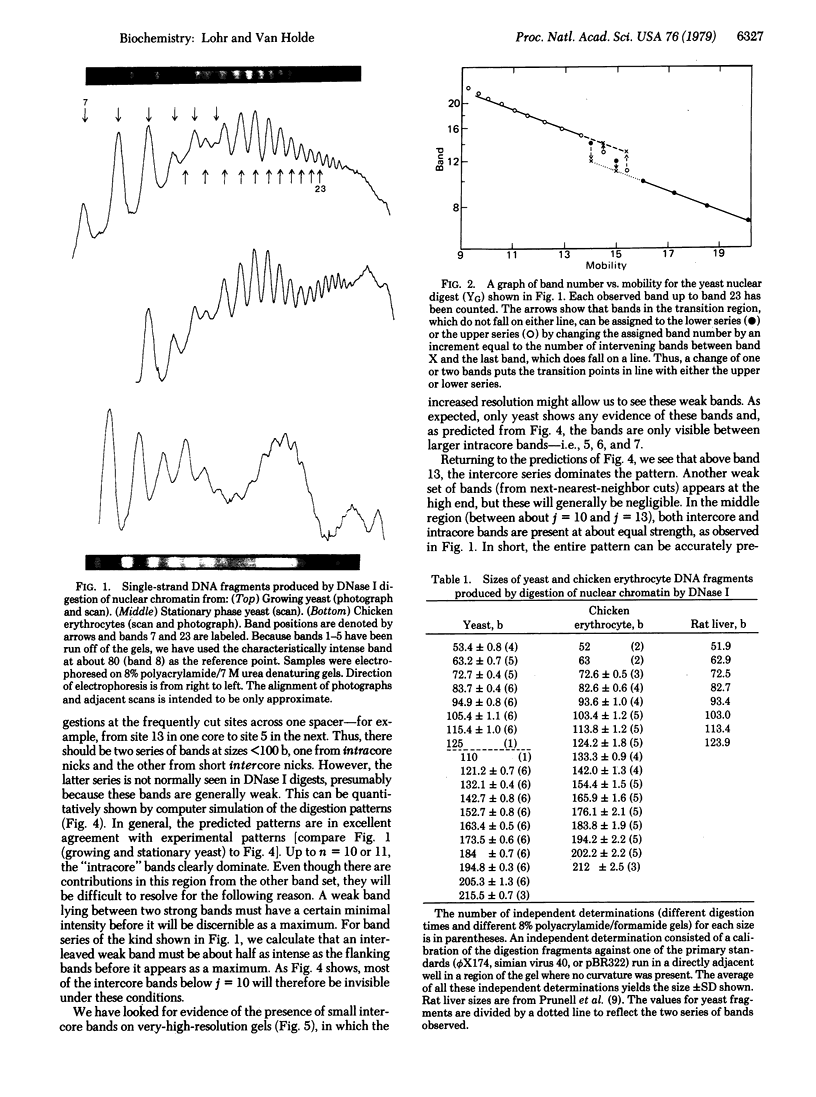
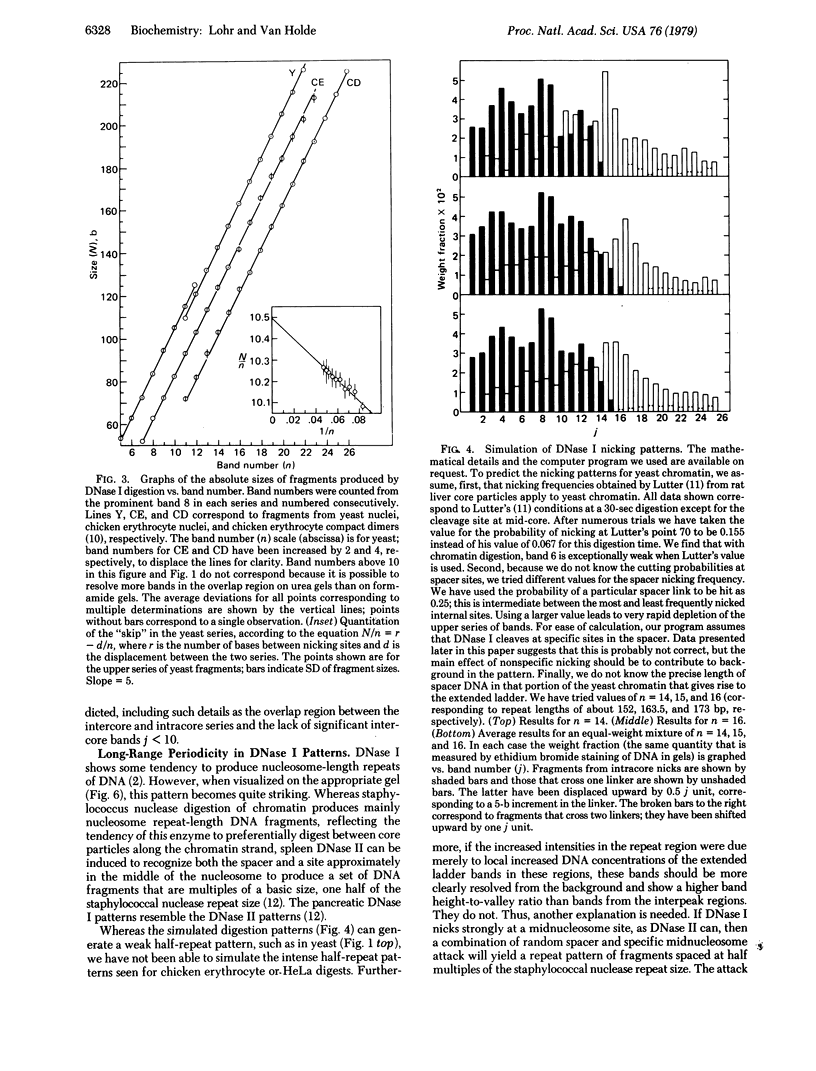
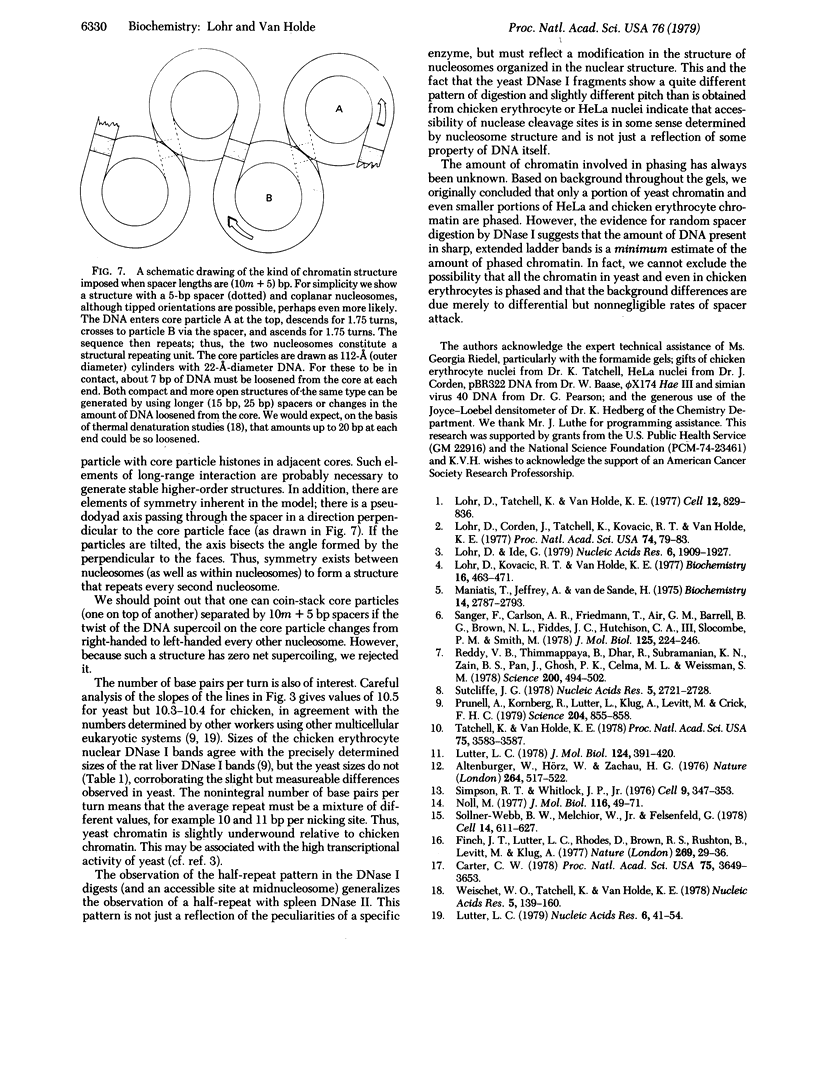
Detailed analysis of the DNA fragment patterns produced by DNase I digestion of yeast, HeLa, and chicken erythrocyte nuclei reveals surprising features of nucleosome phasing. First, the spacer regions in phased yeast chromatin must be of lengths (10m + 5) base pairs, where m = 0, 1, 2,....This feature is not seen in parallel studies of chicken erythrocyte chromatin. The 5-base pair increment in the yeast spacer imposes interesting restraints on the higher order structure of yeast chromatin. Second, we have been able to simulate the DNase I cutting patterns and get good agreement with the observed yeast patterns. Third, three different chromatins show a long range periodicity in the DNase I digest pattern, with a period half that of the staphylococcal nuclease repeat. These results suggest that the amount of chromatin observed in discrete extended-ladder bands is a minimum estimate of phasing and in fact phasing may be a more general feature.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenburger W., Hörz W., Zachau H. G. Nuclease cleavage of chromatin at 100-nucleotide pair intervals. Nature. 1976 Dec 9;264(5586):517–522. doi: 10.1038/264517a0. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr Histone packing in the nucleosome core particle of chromatin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3649–3653. doi: 10.1073/pnas.75.8.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Ide G. Comparison on the structure and transcriptional capability of growing phase and stationary yeast chromatin: a model for reversible gene activation. Nucleic Acids Res. 1979;6(5):1909–1927. doi: 10.1093/nar/6.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lohr D., Tatchell K., Van Holde K. E. On the occurrence of nucleosome phasing in chromatin. Cell. 1977 Nov;12(3):829–836. doi: 10.1016/0092-8674(77)90281-1. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Precise location of DNase I cutting sites in the nucleosome core determined by high resolution gel electrophoresis. Nucleic Acids Res. 1979 Jan;6(1):41–56. doi: 10.1093/nar/6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. DNA folding in the nucleosome. J Mol Biol. 1977 Oct 15;116(1):49–71. doi: 10.1016/0022-2836(77)90118-8. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Melchior W., Jr, Felsenfeld G. DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell. 1978 Jul;14(3):611–627. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Compact oligomers and nucleosome phasing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3583–3587. doi: 10.1073/pnas.75.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]