Abstract
The envelope of human cytomegalovirus (HCMV) consists of a large number of glycoproteins. The most abundant glycoprotein in the HCMV envelope is the glycoprotein M (UL100) which together with glycoprotein N (UL73) form the gM/gN protein complex. Using yeast two hybrid screening, we found that the gM carboxy-terminal cytoplasmic tail (gM-CT) interacts with FIP4, a Rab11-GTPase effector protein. Depletion of FIP4 expression in HCMV infected cells resulted in a decrease of infectious virus production that was also associated with an alteration of the HCMV assembly compartment (AC) phenotype. A similar phenotype was also observed in HCMV infected cells that expressed dominant negative Rab11(S25N). Recently, it has been shown that FIP4 interactions with Rab11 and additionally with Arf6/Arf5 are important for the vesicular transport of proteins in the endosomal recycling compartment (ERC) and during cytokinesis. Surprisingly, FIP4 interaction with gM-CT limited binding of FIP4 with Arf5/Arf6, however, FIP4 interaction with gM-CT did not prevent recruitment of Rab11 into the ternary complex. These data argued for a contribution of the ERC during cytoplasmic envelopment of HCMV and revealed a novel FIP4 function independent of Arf5 or Arf6 activity.
Keywords: HCMV, assembly compartment (AC), gM/gN, endocytic recycling compartment (ERC), FIP4, Rab11
HCMV is a ubiquitous pathogen which can result in considerable morbidity and mortality in immunocompromised individuals (transplant recipients, HIV infected individuals) (1). HCMV is a major cause of the congenital infection of infants, an infection associated with severe brain damage and progressive hearing lost (2).
HCMV is the largest member of the herpesvirus family, and its structure resembles other herpesviruses. HCMV contains a dsDNA genome within an icosahedral capsid, which in turn is surrounded by a complex layer of tegument proteins. The tegumented capsid is enclosed in a glycoprotein-rich lipid envelope (3–6). The morphogenesis of the HCMV particle is a multi-step process which begins in the nucleus of the infected cell and results in the formation of the nucleocapsid (7). During the nuclear stage of the infection, when nucleocapsids are produced, tegument and envelope proteins are expressed and accumulate in the cytoplasm of infected cells. It has been shown that the expression of HCMV proteins in the cytoplasm is accompanied by the localization of the HCMV-expressed structural proteins within a cellular compartment, termed the assembly compartment (AC) (8–11). The structure of the AC is thought to be formed from the cellular secretory system including Golgi, Trans-Golgi Network (TGN), and possibly endosomes, which undergo morphological and structural changes upon HCMV infection (12–19). Tegumented nucleocapsids exported from the nucleus possibly bud into the lipid membranes of the AC enriched with viral proteins to acquire tegument and envelope that leads to the final step of maturation of the infectious particles. To date, there have been limited studies focusing on the formation of the AC, the trafficking of virus proteins within the AC, and finally, the cellular components contributing to cytoplasmic stages of the HCMV maturation.
Sequence analysis of the AD169 HCMV strain has revealed that approximately 50 open reading frames (ORFs) can potentially encode glycoproteins (20). The HCMV envelope has been shown to contain highly conserved glycoproteins that are also present in the envelope of other herpesviruses, including gB, gH/gL/gO and gM/gN as well as a large number of less well defined virus-encoded proteins and some proteins of cellular origin (3). The gM/gN complex is the most abundant component of the HCMV envelope (21). The homologues of the gM and gN protein are encoded by all herpesviruses, suggesting an important role in the virus life cycle. Although deletion of the ORF encoding either gM or gN from the genome of PRV, EHV-1, or VZV results only in the decrease of infectious virus production when compared to the wild-type virus, HCMV gM and gN are essential for the production of the infectious virus (22–25).
HCMV gM is a product of the UL100 ORF, and is composed of 372 amino acids (aa) with an apparent molecular weight of 42 kDa. It is a type III glycoprotein with seven membrane-spanning domains and a C-terminal cytoplasmic tail. gM forms a protein complex with the product of the UL73 ORF, gN, that is stabilized by a disulfide bond as well as hydrophobic interactions of the transmembrane domains. In addition, gM/gN complex formation is required for the export of gM and gN from the endoplasmic reticulum (ER) during intracellular trafficking of the complex to the TGN and endosomal compartment (26, 27).
Recently, we demonstrated that replication of HCMV depends on the presence of the gM C-terminal cytoplasmic tail sequence (gM-CT) and that deletion of this sequence resulted in the loss of infectious virus production. In addition, we observed that the acidic cluster of amino acids (aa) present in the gM-CT domain influenced the rate of the gM/gN trafficking within the AC of the HCMV infected cells (28). Combined, this data suggested that gM-CT could interact with cellular components that regulate and direct gM/gN complex intracellular transport. To identify cellular components that interact with gM-CT, we performed a yeast-two hybrid screening and found that the gM-CT interacted with the Rab11 effector protein FIP4 (family of interacting protein 4).
Rab11 is a small GTPase which cycles between GTP-bound active and GDP-bound inactive states and has been suggested to regulate protein trafficking/sorting in the endosomal recycling compartment (ERC) (29). Furthermore, Rab11 has been shown to be crucial for several other membrane transport pathways, including phagocytosis, apical targeting in epithelial cells, and retrograde protein transport from endosomes to TGN (30–32). It has also been observed that Rab11 has a major role in membrane sorting during cytokinesis (33–35). The diversity of Rab11 functions is thought to depend on the fact that Rab11 is recruited to the different cellular compartments by a variety of specific effector proteins. The family of the Rab11 family interacting proteins (Rab11-FIPs) is represented by six members. All Rab11-FIPs share a highly conserved 20 amino acid long motif at the C-terminus of the protein, known as the Rab11 binding domain (RBD) (36). All Rab11 effectors can be further classified according to the protein structure and homology. Class II FIPs (eferin/arfophilin-1/FIP3 and arfophilin-2/FIP4) are characterized by the presence of EF hands at the N-terminus and by homology with Drosophila nuclear fallout protein (Nuf) which is required for the cellularization of Drosophila embryos (37–39). Consistent with this finding is the observation that FIP3 and possibly FIP4 are important for recruitment of Rab11 during cytokinesis (34, 40). FIP4 has been shown to concentrate in the ERC of HeLa cells; however expression of FIP4 deletion mutants had no effect on the protein recycling from the cell surface (37, 41). Finally, FIP3 and FIP4 have been shown to bind simultaneously to Rab11 and Arf6 or Arf5, although both FIP3 and FIP4 proteins exhibit lower affinity for Arf5 than Arf6 (40, 42, 43). Arfs also belong to the family of small GTPases, which are required for the recruitment and assembly of numerous coat proteins during vesicular transport (44). The ability of FIP3 and FIP4 to bind both types of GTPases is a rarely described function which links multiple trafficking pathways by interactions with two different GTPases families.
In this report, we described interactions between HCMV gM and FIP4. We showed that FIP4 bound to gM is capable of recruiting Rab11, but failed to bind either Arf5 or Arf6. The intracellular ternary complex in HCMV infected cells possibly consists of gM/gN-FIP4-Rab11. In addition, shRNA depletion of FIP4 expression or expression of dominant negative Rab11(S25N) in virus infected cells led to a significant decrease in the infectious virus yields. We also observed that the mature AC formation in the HCMV infected cells was associated with accumulation of the Transferrin-TRITC marker of ERC and exclusion of EGF-Alexa488 to the cell periphery, presumably in late endosomes/lysosomes. Together this data suggested that gM/gN localization in the ERC dependent on the Rab11 GTPase function and that the ERC contributes to the AC formation and plays a significant role during final steps of HCMV envelopment.
Results
gM cytoplasmic tail binds to FIP4
gM (UL100) is a type III integral membrane protein consisting of 7 predicted trans-membrane domains and a 50 amino acids (aa) C-terminal cytoplasmic tail (gM-CT) (26). In our previous study, we showed that the gM-CT was essential for the replication of HCMV, suggesting that the cytoplasmic tail of gM has key role in the functions of gM, potentially through the protein-protein interactions (28). In order to identify potential protein interactions, we performed yeast two hybrid screening. We utilized a human liver cDNA library and as a bait a gM (UL100) fragment that encoded full-length C-terminal tail of gM plus the last predicted transmembrane domain (region 300-372 aa) (Fig. 1A). It is important to note that initially we used the gM construct containing only the predicted gM cytoplasmic tail sequence as bait (region 323-373 aa). This screening failed to identify any interacting proteins, possibly due to the misfolding of this protein domain (data not shown). In the second yeast two hybrid screen using as bait a 300-372 aa region of gM, we isolated a 300 bp long cDNA clone encoding fragment of the Rab11 effector protein-FIP4 corresponding to aa 454-543 of FIP4 (Fig. 1B).
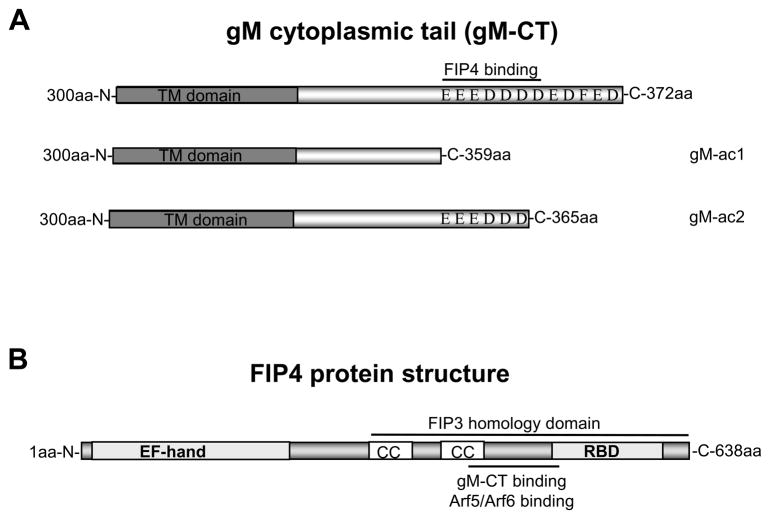
Figure 1. The gM cytoplasmic tail (gM-CT) interacts with FIP4.
(A) Diagram representing gM protein fragment (300–372aa) which was cloned and used as a bait during yeast two hybrid screening and N-terminal GST tagged constructs of gM-CT: gM-ac1 and gM-ac2 which were used for the pull down assays of FIP4-myc.
(B) Diagram of the FIP4 protein with depicted domains: N-terminal EF-hand, CC coil-coiled domain responsible for the FIP4 homo- and heterodimerization with FIP3, Rab Binding Domain (RBD) which interacts with Rab11, 454-543 aa region which is essential for the interaction with Arf5/Arf6 and was recovered as a cDNA clone of gM-CT interactions during yeast two hybrid screening.
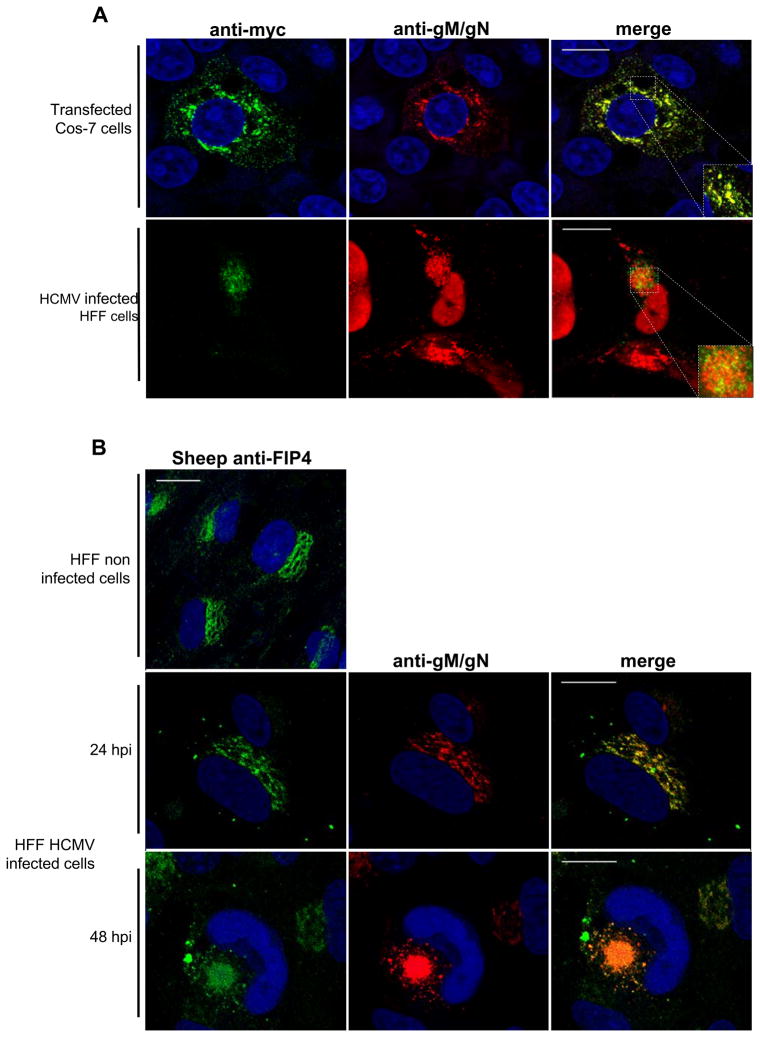
To further explore gM and FIP4 interactions, we obtained the full-length cDNA clone of FIP4, amplified the FIP4 gene by PCR, and cloned the PCR product into an expression plasmid encoding a myc tag. As a first step to confirm the gM and FIP4 interaction, we used immunofluorescence assay to co-localize the gM/gN complex and FIP4 in transfected and HCMV-infected cells (Fig. 2). The FIP4-myc construct was used for the cotransfection of Cos7 cells with gM and gN constructs as well as in the transfection/HCMV-infection experiment performed in HFF cells. In the transfected Cos7 cells the fluorescent signal generated by FIP4-myc strongly co-localized in vesicles stained with the anti-gM/gN mab 14-16A (Fig. 2A, top panel). Similarly, in HCMV infected HFF cells, the signal from gM/gN co-localized with transfected FIP4-myc (Fig. 2A, bottom panel). Additionally, we used a sheep anti-FIP4 antibody to co-localize endogenous FIP4 with gM/gN complex in the infected HFF cells (anti-FIP4 antibody kindly provided by Dr. G.W. Gould, Glasgow, UK) (Fig. 2B). In uninfected HFF cells, FIP4 was localized in a ‘ribbon like’ structure of the TGN (Fig. 2B top panel). Previously, we have reported that gM/gN also co-localized in the TGN of transfected cells [28]. Interestingly, in HFF infected cells at early time points post infection gM/gN and FIP4 co-localized in the similar structure, but at later time points post infection immunofluorescent signal of both antigens co-localized in the mature assembly compartment. The antibody that recognized endogenous as well as the myc-tagged form of FIP4 co-localized with signal of the gM/gN complex in the HCMV assembly compartment and confirmed that tagging of FIP4 with myc did not alter its intracellular localization and interaction with gM (Fig. 2).
Figure 2. Immunofluorescence co-localization of FIP4 and gM/gN complex in transfected and HCMV infected cells.
(A) Cos7 cells were co-transfected with constructs expressing FIP4-myc, gM and gN, 48 hours post transfection cells were fixed and reacted with anti-myc and anti-gM/gN mab followed with FITC and TRTC labeled secondary antibody as described in Material and Methods (top panel). HFF cells were electroporated with FIP4-myc construct and then infected with HCMV, 6 dpi cells were fixed and reacted with anti-myc, anti-gM/gN and anti-IE1 mab (detecting immediate early antigen-1 expressed in the nucleus of the infected cells) antibody and appropriate secondary antibody (bottom panel).
(B) To detect co-localization of endogenous FIP4 in HCMV infected cells with gM/gN complex. HFF cells were infected with HCMV, 24 and 48 hpi cells were fixed and reacted with a polyclonal anti-FIP4 sheep antibodies and anti-gM/gN mab and appropriate secondary antibody. Note that before staining cells were reacted with anti-FC receptor antibody to block non-specific FC receptor staining (data not shown). In addition, non-immune goat antibodies did not stain infected cells (data not shown). Bar in all merged images indicates 20 μm.
To provide biochemical evidence of a gM and FIP4 interaction, we performed a GST pull down assay. We generated a GST fusion protein with the gM C-terminal tail corresponding to the same fragment that was used as the bait in the yeast two hybrid screening. Additionally, we generated GST-tagged gM-CT deletion mutants including: a deletion of the C-terminal 7 aa (gM-ac2), and a deletion of the 13 C-terminal aa (gM-ac1) (Fig. 1A). These GST-tagged gM constructs were used to verify and map interactions with FIP4 using cell lysates prepared from human kidney cells (HK293) cells transfected with a plasmid expressing myc-tagged FIP4 protein. The results of these assays indicated that gM-CT pulled down FIP4-myc (Fig. 3A, lane 3), confirming the interaction between gM-CT and FIP4. Furthermore, the gM-CT deletion construct gM-ac2 which lacked only the 7 C-terminal acidic aa of gM also pulled down FIP4-myc (Fig. 3A, lane 5). However, recombinant GST-tagged construct of gM-ac1 that lacked the 13 C-terminal aa of gM disrupted the interaction between gM-CT and FIP4 (Fig. 3A lane 4). These findings indicated that the region of gM-CT protein that was required for the FIP4 binding was between aa position 359-365 of the gM-CT. An irrelevant myc-epitope tagged protein failed to interact with GST-tagged gM-CT and, FIP4-myc did not interact with the GST-tagged pp28, an acidic cluster containing protein encoded by HCMV (data not shown).
Figure 3. FIP4 interact with the gM-CT.
(A) Pull down assay in which glutathione sepharose beads containing gM-CT constructs (GST-gM-CT, GST-gM-ac1, GST-gM-ac2) or purified GST alone were incubated with the HK293 cell lysate expressing FIP4-myc. After extensive washing protein samples were boiled in sample buffer, resolved by SDS-PAGE and assayed by western analysis. Western blots were probed with anti-myc (9E10) mab and detected with the HRP (top panel). The lower panel shows the Coomassie blue stained gels of the GST purified input of proteins used in pull down assay.
(B) Pull down assay of FIP3 or FIP4 by cytoplasmic tail of gM. Glutathione sepharose beads containing gM-CT constructs or GST alone were incubated with HK293 cell lysates expressing FIP3-myc or FIP4-myc. After extensive washing, beads were boiled and eluted proteins resolved by SDS-PAGE followed by western blot analysis. Western blots were probed with anti-myc mab and detected with the HRP.
(C) Fluorescence Resonance Energy Transfer (FRET) indicating strong FIP4 and gM interaction in HCMV infected cells. For FRET assays, HFF cells were electroporated with FIP4 or FIP3 myc-tagged constructs and plated on the coverslips. 24 hours later, the cells were infected with HCMV. The cells were fixed in 4% PFA and stained with IMP anti-gM monoclonal antibodies (specific for the C-terminal tail of gM) or anti-myc antibody followed by labeling with secondary antibody conjugated with FITC or TxRed. A non-bleaching region was selected as an internal control of FRET analysis detail description of the assay is provided in Materials and Methods.
To investigate if gM-CT interaction was specific to FIP4 we also performed GST-gM-CT pull down assay with FIP3-myc, the isoform of the FIP family that is most closely related to FIP4 by sequence alignment (Fig. 3B). As in the previous experiment, FIP3-myc was expressed in HK293 cells and the cell lysate was incubated with GST-gM-CT or with GST alone (Fig. 3B lower panel). The results of this experiment provided no evidence of FIP3-myc binding to gM-CT or GST alone, indicating that gM-CT interaction is specific to FIP4. Additional experiments utilizing E. coli expressed gM-CT, FIP4 and FIP3 confirmed that gM-CT specifically interacted with FIP4 and not FIP3 (data not shown). In order to validate FIP4 and gM interaction in an in vivo experiment we utilized available FIP3 and FIP4 myc-tagged constructs in static Fluorescence Resonance Energy Transfer (FRET) to investigate FIP3/4 gM interactions in infected HFF cells. The FRET efficiency in these experiments demonstrated protein-protein interaction and indicated an interaction between gM and FIP4 in the assembly compartment of infected cells (average FRET efficiency 27%), while only minimal FRET efficiency was generated with FIP3 and gM indicating the lack of significant protein-protein interaction (average FRET efficiency less than 2%) (Fig. 3C).
shRNA depletion of the FIP4 reduces infectious virus yield
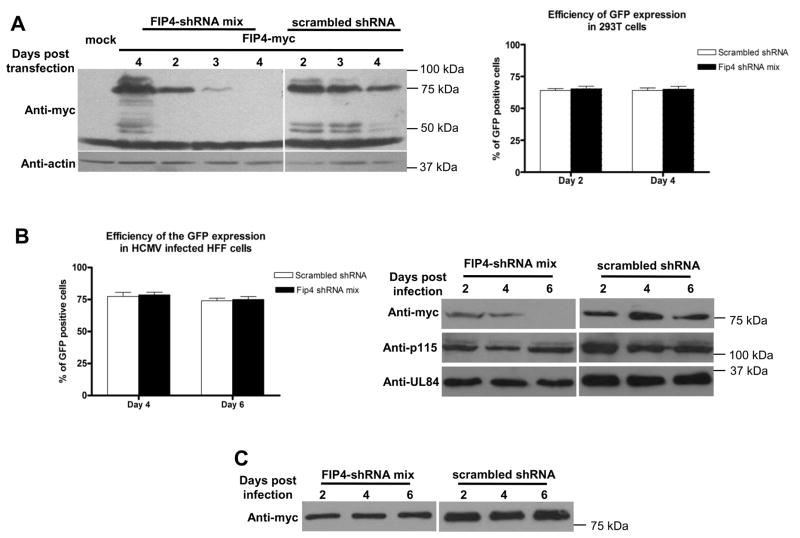
In order to further explore the relationship between the gM-CT, FIP4 and virus assembly, we used shRNA depletion to knockdown expression of FIP4. A panel of four shRNA sequences to reduce expression of FIP4 and one shRNA scrambled sequence control that were cloned into an EGFP expression vector were used. We chose this system because it allowed us to monitor cells expressing shRNA by GFP expression. Additionally, we have chosen to use the mixture of shRNA vectors to deplete FIP4 because this approach proved to be the most efficient for the long term inhibition of FIP4 expression (data not shown). Efficiency of shRNA depletion was monitored over time using western blot of HK293 cells expressing FIP4-myc and shRNA containing vectors. By day 4 after transfection, FIP4 expression was reduced by 80–100% (Fig. 4A).
Figure 4. shRNA depletion of the FIP4 expression.
(A) HK293 cells were co-transfected with FIP4-myc together with either shRNA to deplete FIP4 expression or with scrambled control shRNA. The protein samples were collected 2, 3, and 4 days post transfection and frozen in −80°C. The control cell sample transfected only with FIP4-myc was harvested on the day 4 post transfection. All cell pellets were lysed in equal amounts of sample buffer and then resolved by SDS-PAGE, followed by analysis by western blotting. Western blots were probed with anti-myc to detect expression of the FIP4-myc (top panel). Additionally, the membranes were probed with anti-actin antibody as a loading control (bottom panel). The histogram represents efficiency of shRNA transfection in HK293 cells. The percentage of transfected cells was calculated as number of GFP expressing cells, 2 and 4 days post transfection per 100 cells counted stained with DAPI as described in Materials and Methods.
(B) HFF cells were electroporated using Amaxa with shRNA to deplete FIP4 expression or with scrambled control shRNA and the cells were plated on 13 mm coverslips. On day 4 and 6 post electroporation cells were fixed and stained with DAPI to visualized total number of cells on the cover slips. The efficiency of shRNA expression was estimated by calculating number of GFP expressing cells per 100 cells stained nuclei as described in the Materials and Methods.
HFF cells were electroporated with FIP4-myc and either with shRNA mix to deplete FIP4 or a control scrambled shRNA. One day post electroporation, cells were infected with AD169 HCMV. On 2, 4, and 6 days post infection HFF cells were harvested in lysis buffer, subjected to SDS-PAGE and analyzed by western blotting. FIP-4 expression was detected with anti-myc mab (top panel). The membrane was stripped and than probed with anti-p115 (middle panel) and anti-UL85 mab (bottom panel) as an off target control for the shRNA.
(C) HFF cells were electroporated with FIP3-myc and the FIP4 shRNA mix or control scrambled shRNA. One day post electroporation cells were infected with AD169 HCMV. On 2, 4, and 6 days post infection HFF cells were harvested in lysis buffer, resolved by SDS-PAGE gel, and analyzed by western blotting. FIP3 was detected with anti-myc mab.
Because most of our experiments were performed in HFF cells which are known to be difficult to transfect, we also assessed the efficiency of the shRNA expression in HFF cells. After electroporation of HFF cells with a total 4ug DNA of the vectors containing shRNA targeting FIP4 expression or with the construct containing scrambled shRNA sequence, we allowed cells to recover for 4 or 6 days, at which time the percent of the cells expressing detectable levels of GFP was calculated vs. the total number of cells. In this experiment we observed that the number of cells expressing GFP was maintained at 70–80% of total HFF cells (Fig. 4B). This data was also confirmed by flow cytometery (data not shown).
In addition to test if the HCMV infection of HFF cells altered shRNA depletion of FIP4 we tested FIP4-myc expression in cells co-electroporated with either with FIP4-shRNA mix or shRNA scrambled construct (Fig. 4B). Following electroporation, the HFF cells were infected with HCMV strain AD169, cells lysates collected in lysis buffer at 2, 4 and 6 days post infection and analyzed by western blot. As shown in Figure 4B shRNA FIP4 depletion was very efficient (Fig. 4B top panel). In addition, we did not observe a decrease in the expression of an endogenous protein of the early secretory pathway, p115 (Fig 4B middle panel) and no change in the expression of the HCMV encoded minor capsid protein, UL 85 (Fig 4B bottom panel) (45, 46). Since there is high genomic sequence similarity between FIP4 and FIP3 we also tested an influence of FIP4-shRNA expression on the FIP3-myc expression. Similarly as in above experiment we tested FIP3-myc expression in virus infected cells co-electroporated with FIP4-shRNA mix or with shRNA scrambled construct (Fig. 4C). Western blot analysis of FIP3 expression revealed that FIP4-shRNA depletion had no effect on the FIP3-myc expression levels in HFF infected cells.
Additionally, in order to measure relative levels of FIP2 and FIP3 expression in comparison to FIP4 in FIP4-shRNA depleted cells we performed quantitative RT-PCR (Fig. S1). This experiment was performed 24 hours post infection in HFF cells and demonstrated that FIP4 depletion did not alter FIP2 or FIP3 RNA expression but resulted in substantially lower FIP4 RNA expression (Fig S1). Together these experiments indicated that knockdown of FIP4 expression was efficient and little if any cytotoxicity was observed for at least 6 days post transfection.
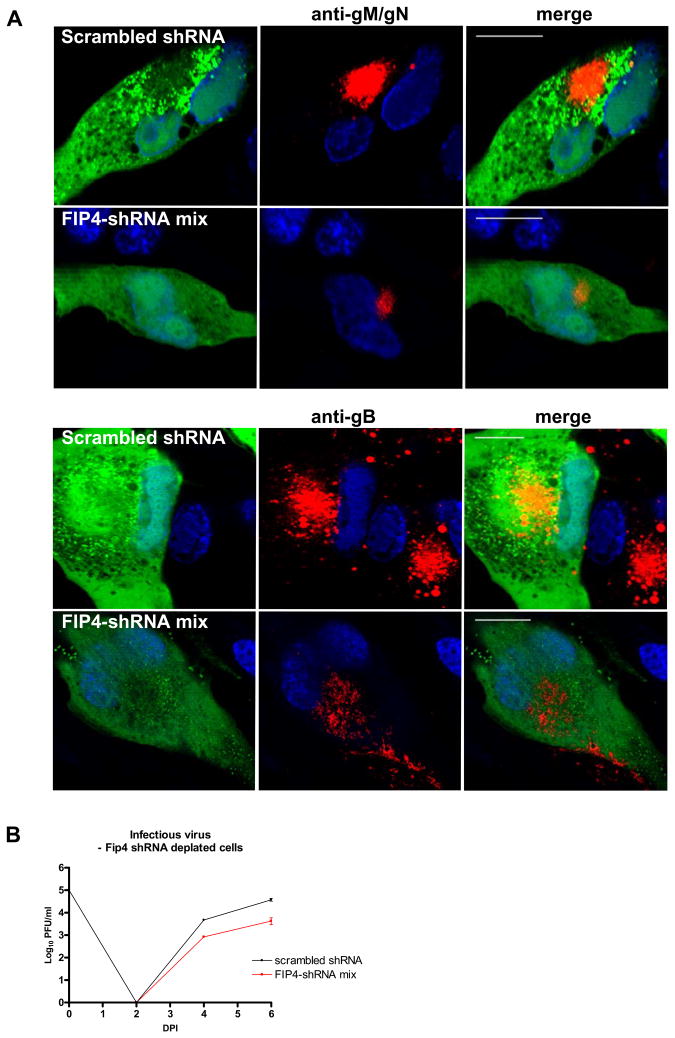
To assess the effect of the FIP4 depletion on the HCMV assembly, HFF cells expressing shRNA targeted to deplete FIP4 or scrambled shRNA sequence were infected with HCMV. In these cells, we examined the AC phenotype as well as the yield of infectious virus production over 6 days of infection. In cells in which FIP4 expression was reduced by shRNA, the AC appeared smaller and less compacted at 4 days post infection as compared with cells transfected with the scrambled sequence or with GFP alone (Fig. 5A). The AC phenotype defect was observed when HCMV infected cells were labeled with anti-gM/gN mab 14-16A (Fig. 5A top panels) as well as with anti-gB mab (Fig. 5A bottom panels). It is important to note that imaging data was collected using the same laser intensity and gain in FIP4 depleted and non-depleted cells. Thus, these imaging findings indicated that concentration of either gM/gN or gB in the AC was decreased following expression of shRNA which targeted FIP4 expression.
Figure 5. Depletion of the FIP4 expression alters the assembly compartment formation and leads to decrease in HCMV virus production.
(A-B) HFF cells were electroporated with shRNA to deplete FIP4 or with control scrambled shRNA sequence and plated on 13 mm glass coverslips. On the following day, the cells were infected with HCMV at the multiplicity of infection (moi) of 1.0 and 4 days later cells were fixed and stained with anti-gM/gN mab (A) or anti-gB mab (B) as described in Materials and Methods. Note that all images were collected with the identical laser intensity and gain settings, thus allowing for comparison of signal intensity. The assembly compartment is not well formed in cells depleted FIP4 expression by shRNA. Bar in all merged images indicates 20 μm.
(C) HFF expressing shRNA vectors to deplete FIP4 or control scrambled shRNA were infected with AD169 at an moi of 0.1 and total cultures (supernatant and cells) in 1.5 ml media were harvested at the indicated time points post infection. Cell supernatant and disrupted cells were combined and assayed for the amount of the infectious particles using a fluorescence based infectivity assay as described in Materials and Methods. Results are expressed as a log10 infectious units/ml of sample.
Our previous studies also demonstrated a defect in the maturation of the AC which appeared in cells infected with the mutant virus that lacked the acidic cluster of gM (28). Interestingly, this region of gM was also required for the interaction with FIP4. Additionally, this alternation in the phenotype of AC was accompanied by a reduced yield of infectious virus. Following electroporation of shRNA to deplete FIP4, cells were infected with HCMV and cell cultures harvested and quantified the yield of infectious virus. As shown in Figure 5B, cells depleted of FIP4 expression yielded 1 log less infectious virions when compared to the cells expressing shRNA scrambled sequence over 6 days of infection. These results were consistent with the imaging data, suggested that depletion of FIP4 affected the localization of the gM/gN to the AC, the maturation of the AC, and consequently infectious virus production.
FIP4 interacting with gM recruits Rab11 but fails to bind Arf5 or Arf6
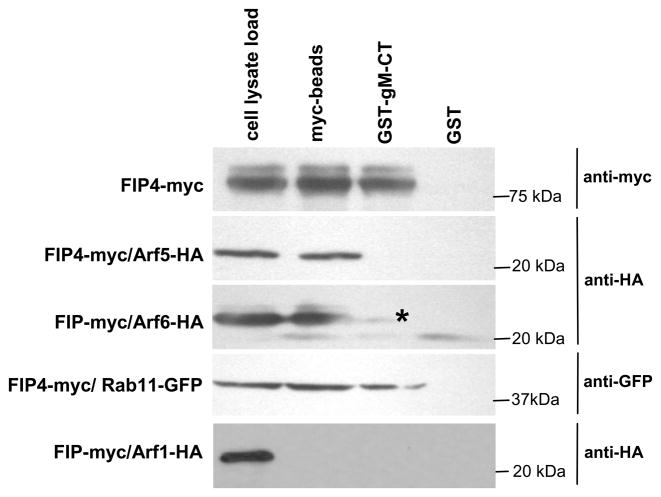
In previous reports, it has been shown that FIP4 was an effector of Rab11 and simultaneously recruited not only Rab11 but also Arf GTPases (Arf5 or Arf6) (40, 42, 43). This cooperative binding of more than one GTPase was required for the function of Rab11-FIP4 in vesicular transport and during cytokinesis. The FIP4 domain which binds Arf5/6 overlaps with the domain of the interaction with gM-CT which we identified in the yeast two hybrid screening (Fig. 1B). To determine if FIP4 was capable of simultaneously binding to gM-CT and Arf5/6 or Rab11, we performed GST pull down assays. The GST-tagged gM-CT was purified on glutathione sepharose beads and used in the pull down experiments with HK293 cell lysates overexpressing FIP4-myc alone, or FIP4-myc co-transfected with HA-tagged Arf5, HA-tagged Arf6, GFP-tagged Rab11 or HA-tagged Arf1 as controls. Initially, after lysis of transfected HK293 cells, clarified supernatants were divided into three equal portions. One of these portions was immunoprecipitated with anti-myc-magnetic beads to insure that FIP4-myc was binding Arf5/6-HA as well as Rab11-GFP. As is shown in Figure 6 (lane 2), all of these GTPases except Arf1 were efficiently bound by FIP4-myc expressed in mammalian cells. The remaining two portions of each cell lysate were mixed with glutathione agarose beads with purified GST-gM-CT or GST alone as a control. As in previous experiments, GST-tagged gM-CT but not GST alone bound FIP4-myc from the cell lysate (lane 3). Following GST pull down the western blot was probed with anti-HA antibody to detect Arf5 or Arf6. Although both proteins efficiently bound to FIP4 (indicated by the myc-pull down, lane 2) neither produced a strong signal in the presence of GST-gM-CT or GST alone (Fig. 6, lanes 3 and 4). In the same experiment we demonstrated pull down of Rab11-GFP with gM-CT in the presence of FIP4 (Fig. 6 lane 3). These results suggested that the gM-CT binds to FIP4 and FIP4 in turn recruits Rab11 to form ternary complex in vitro. Interestingly, after long exposure of the film during development of the immunoblots of Arf6 pull downs with gM-CT we detected very weak binding of Arf6 to FIP4 in the presence of the gM-CT as indicated by a faint band detected with anti-HA antibody (Fig. 6, lane 3). The results of these experiments suggested that gM-CT could inhibit the binding of the Arf5 and Arf6 GTPases to FIP4. In a second study, we performed FIP4 pull down assay as described above in the presence of constant amounts of transiently expressed Arf5 or Arf6 and then incubated these cell lysates with increasing amounts of the GST-gM. Results from this experiment demonstrated that binding of Arf5 and Arf6 to FIP4 both were inhibited by increasing amounts of GST-gM (Figure S2).
Figure 6.
FIP4 bound to the gM-CT recruits Rab11 but fails to bind Arf5 or Arf6. HK293 cells were transfected with FIP4-myc alone or co-transfected with FIP4-myc and either with HA-Arf5, HA-Arf6, GFP-Rab11, and Arf1-HA as a control. Two days post transfection cells were lysed and analyzed by immunoprecipitation with myc antibody-tagged magnetic beads as described in the Materials and Methods or incubated with sepharose beads containing purified GST-gM-CT or GST alone. After immunoprecipitation and extensive washing, precipitated protein were resolved by SDS-PAGE, transferred to membranes and probed with appropriate antibody. The asterisk indicates HA-Arf6 band which was visualized only after prolonged exposure of the membrane that had been probed with anti-HA and developed with HRP.
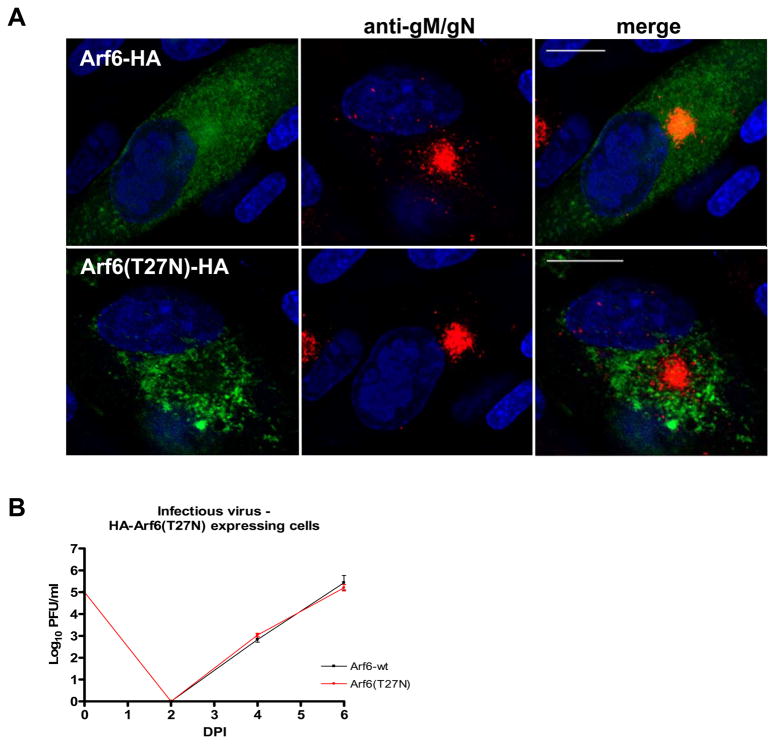
Expression of the dominant negative Arf6(T27N) has no effect on the HCMV assembly and yield
Our biochemical data indicated that FIP4 failed to recruit Arf5 and possibly only a limited amount of Arf6 to FIP4 in the presence of the GST tagged gM-CT. This result did not exclude the possibility of the in vivo functional involvement of Arf6 in HCMV assembly. To test this possibility, we examined the phenotype of the AC formation and the yield of infectious HCMV from cells expressing a dominant negative Arf6(T27N). HFF cells were electroporated with HA-Arf6 or HA-Arf6(T27N), and infected with HCMV. After 4 days of infection, cells were examined by confocal microscopy for the phenotype of the AC and for the yield of infectious virus. As shown, a small pool of the HA-Arf6 that accumulated in the AC co-localized with gM/gN (Fig. 7A top panel). In cells expressing dominant negative form of Arf6(T27N), the HA-Arf6(T27N) stained with anti-HA antibody was virtually excluded from the AC and failed to exhibit co-localization with the gM/gN complex (Fig. 7A bottom panel). Moreover, the expression of HA-Arf6(T27N) had no effect on the appearance or size of the AC and in these cells, as the AC appeared to be well formed (Fig. 7A bottom panel). HCMV virus titers obtained from the cells expressing HA-Arf6(T27N) were similar to those obtained from the cells expressing the wild type HA-Arf6 (Fig. 7B). These results confirmed our biochemical data which suggested that gM binding to FIP4 excluded Arf6 and also argued that the assembly of the HCMV was independent of Arf6 function.
Figure 7. Expression of the dominant negative HA-Arf6(T27N) does not limit HCMV virus assembly and production.
(A) Analysis of gM/gN localization in the HCMV infected cells expressing HA-Arf6 or dominant negative HA-Arf6 (T27N). HFF cells were electroporated with plasmids encoding HA-Arf6 or HA-Arf6 (T27N) and plated on 13 mm glass coverslips. Twenty-four hours later cells were infected with HCMV at an moi of 1.0 then 4 days post infection (dpi) cells were fixed and stained with anti-gM/gN and with anti-HA antibody, as described in the Methods. Bar in all merged images indicates 20 μm.
(B) HCMV growth kinetics in HFF cells expressing HA-Arf6 or dominant negative HA-Arf6 (T27N). HFF expressing HA-Arf6 or HA-Arf6 (T27N) were infected with HCMV at a moi of 0.1 and total cultures (supernatant and cells) in 1.5 ml media were harvested at the indicated time points post infection. Cell supernatant and disrupted cells were combined and assayed for the amount of the infectious particles using a fluorescence based infectivity assay described in the Materials and Methods. Results were expressed as a log10 infectious units/ml of sample.
The expression of dominant negative Rab11 Rab11(S25N) alters HCMV assembly and yield
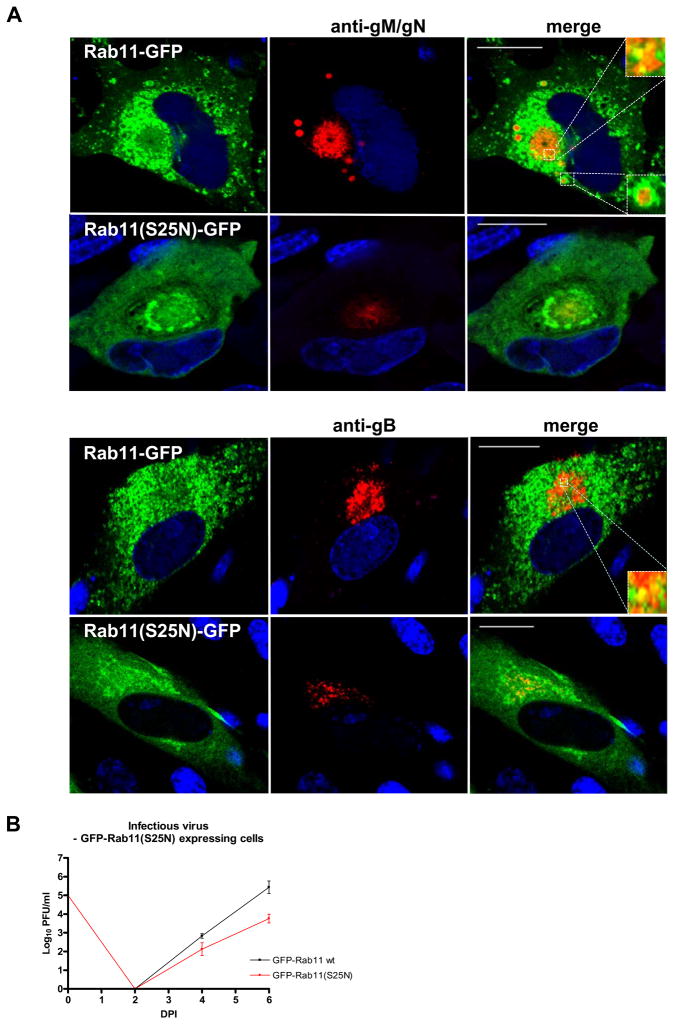
FIP4 is an effector protein of the small GTPase Rab11 which is a GTPase regulating morphogenesis of the endocytic recycling compartment (ERC). A major role of the Rab11 in the ERC is thought to be associated with vesicular export from the ERC (29, 44, 47). Diversity of the intracellular function of the Rab11 depends on its ability to be recruited to different intracellular compartments by the effector proteins such as FIPs. Our experiments this far demonstrated that gM-CT binds FIP4 which recruits Rab11, thus we anticipated that these interactions could influence HCMV envelopment and assembly. To directly investigate this possibility we expressed a dominant negative of GFP-Rab11(S25N) in virus infected HFF cells and assayed these cells for the phenotype of the AC by imaging and infectious virus production. The wild-type GFP-Rab11 signal in the HCMV infected cells was strongly concentrated around the periphery of the AC and in vesicles that were present around the AC that were labeled the anti-gM/gN mab or anti-gB mab but less heavily concentrated in the AC (Fig 8A). In contrast, in cells expressing the dominant negative GFP-Rab11(S25N), the fluorescent signal of the GFP was more diffusely expressed throughout the cytoplasm and only a minimal fraction accumulated in the AC. Importantly, we observed a significant decrease in the localization of the gM/gN complex in the AC in cells expressing GFP-Rab11(S25N) (Fig. 8A). This result indicated that Rab11 contributed to the formation and localization of the gM/gN complex in the AC, arguing for a potentially important role of this GTPase and the ERC in the process of HCMV envelopment.
Figure 8. Expression of the dominant negative GFP-Rab11(S25N) alters formation of the assembly compartment and leads to a decrease in HCMV virus yield.
(A–B) HFF cells were electroporated with GFP-Rab11 or dominant negative GFP-Rab11(S25N) and plated on 13 mm glass coverslips. On the following day, cells were infected with HCMV at an moi of 1.0 and 4 dpi cells were fixed and stained with anti-gM/gN (14-16A) (A) or anti-gB mab (B), as described in the Material and Methods. Note that all images were collected with the identical laser intensity and gain. In cells expressing dominant negative GFP-Rab11(S25N) visualized by the GFP expression there was a decrease in the signal from the gM/gN complex as well as gB within the AC as compared to the cells that did not express the dominant negative Rab11(S25N). Bar in all merged images indicates 20 μm.
(C) HCMV growth kinetics in HFF cells expressing Rab11 or GFP-Rab11(S25N). HFF expressing GFP constructs of Rab11 and Rab11(S25N) were infected with HCMV at an moi of 0.1 and total cultures (supernatant and cells) in 1.5 ml media were harvested at the indicated time points post infection. Cell supernatant and disrupted cells were combined and assayed for the amount of the infectious particles using a fluorescence based infectivity assay described in the Materials and Methods. Results were expressed as a log10 infectious units/ml of sample.
Based on the observed loss of localization of gM/gN in the AC in cells expressing GFP-Rab11(S25N), we quantified the yield of infectious particles from these cultures. Cells expressing dominant negative Rab11(S25N) yielded 2 logs less infectious particles, as compared to the wild-type GFP-Rab11 or GFP alone, a finding that was consistent with an important role of Rab11 during HCMV assembly (Fig. 8B).
Formation of the assembly compartment in the HCMV infected cells is associated with ERC but not late endosomes
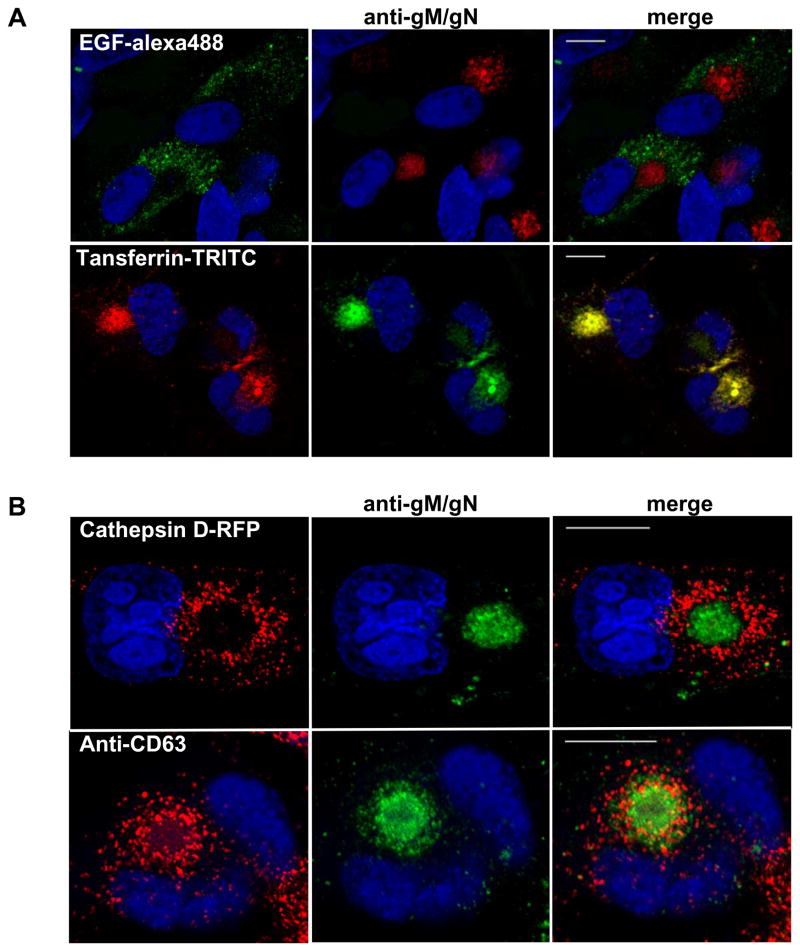
To date there have been several studies attempting to elucidate the role of the cellular secretory pathway in the formation of the HCMV induced AC (8, 12, 13, 16, 22). Most studies have suggested that the TGN plays a major role in the AC formation (14, 48). There have been no reports which have investigated the role of the ERC in the formation of the AC. Our results suggested that the gM/gN complex engages Rab11 GTPase during AC morphogenesis through gM-CT interactions with FIP4. In order to examine the role of different endosomal compartments in the biogenesis of the AC, we used immunofluorescence assay to study the relationship of different endosomal compartments in HCMV infected cells. To distinguish between late endosomes/lysosomes and ERC in the HCMV infected cells we utilized epidermal growth factor (EGF) and transferrin (Tf) to label specific compartments (49, 50). EGF and Tf are the ligands which bind to cell surface receptors and are subsequently internalized by endocytosis upon interaction with their respective ligands. Following internalization, EGF traffics through early/late endosomes to lysosomes. In contrast, upon internalization, Tf is directed from the early endosomes to the ERC. Based on this information we used fluorescent conjugates of EGF-Alexa488 and Tf-TRITC to label late/lysosomes and ERC vesicles respectively. After 6 days of infection, HFF cells were extensively washed and than incubated for 2 hours in serum free medium containing EGF-Alexa488, or Tf-TRITC or both. As shown in Figure 9A, EGF stained late endososmes/lysosomes were excluded from the AC whereas Tf-TRITC labeled ERC was concentrated in the AC and co-localized with the gM/gN complex.
Figure 9. Assembly compartment formation in HFF cells is associated with ERC markers and does not contain markers of the late endocytic pathway.
(A) HFF cells were infected with HCMV and on day 6 post infection, cells were washed extensively in serum free media and incubated for two hours with the EGF-Alexa488 or transferrin (Tf)-TRITC in serum free media. After washing cells were fixed and stained with anti-gM/gN mab followed with anti-IgM FITC or TRITC respectively. Cells were imaged by confocal microscopy. Bar in all merged images indicates 20 μm.
(B) HFF were electroporated with RFP-tagged cathepsin-D, infected with HCMV, and on day 6 post infection, cells were stained with anti-gM/gN mab (top panel). Alternatively, on day 6 following infection with HCMV, HFF cells were stained with anti-CD63 and anti-gM/gN and developed with TRTC goat-anti-mouse IgG1 or FITC goat anti-mouse IgM respectively (bottom panel). All images were collected at 100× magnification.
Previous reports demonstrated colocalization of late endosomal and multivesicular bodies (MVB) markers within the AC of infected cells (13, 17). To further examine relationship between the AC and late endosomes/lysosomes in the HFF infected cells we electroporated HFF cells with RFP tagged cathepsin-D, an endosomal/lysosomal marker, followed by infection of the cells with HCMV (51, 52). Figure 9B (top panel) indicated that cathepsin-D containing vesicles were virtually excluded from the assembly compartment labeled with anti-gM/gN mab 14-16A. In addition HCMV infected cells were co-stained with antibodies reactive with the MVB marker, CD63, and anti-gM/gN mab 14-16A (13, 17, 18). Imaging of these cells revealed only partial co-localization of MVB marker CD63 within the AC, whereas majority of the CD63 containing vesicles were less heavily concentrated within the center of the AC and more highly represented on the periphery of the AC (Fig. 9B bottom panel).
Together this data suggested that the core of the AC is likely formed from vesicles of the ERC origin, but does not exclude the possible contribution of the MVBs during the maturation of the AC.
Discussion
Studies of the intracellular trafficking of the HCMV envelope glycoproteins have been instrumental in understanding of final stages of HCMV cytoplasmic assembly and envelopment. In our pervious study of the HCMV gM/gN glycoprotein complex, we specifically disrupted expression of the two trafficking motifs in the gM C-terminal cytoplasmic tail (gM-CT). We generated HCMV mutants: gMΔAC (deletion of the 13 aa C-terminal acidic cluster) and gMΔYQAL (alanine substitutions in the YXXØ tyrosine-based motif) both of which exhibited a decrease in infectious virus production secondary to defects in HCMV envelope assembly (28). In these studies, we also demonstrated that gM cytoplasmic tail was essential for the assembly of infectious virus presumably secondary to its role in the transport of the gM/gN complex to the AC of infected cells (28).
Trafficking of the transmembrane cargo proteins along a secretory pathway is facilitated by vesicular transport which involves a number of the cellular proteins recruited by trafficking/sorting motifs such as a cluster of acidic aa, tyrosine-based motifs, and others including di-leucine signals present in the cytoplasmic domains of these proteins (53, 54). Protein-protein interactions between cargo proteins and secretory pathway machinery regulate vesicles formation, transport and fusion in different cellular compartment (53, 54). Several HCMV expressed glycoproteins contain acidic cluster and/or tyrosine based motifs and the deletion of these motifs has been associated with defects in virus assembly and replication. As an example, the acidic cluster in the cytoplasmic tail of HCMV expressed glycoprotein gB has been shown to interact with phosphofurin acidic cluster sorting protein-1 (PACS-1) and deletion of the acidic cluster in gB disrupts accumulation of the gB in the TGN resulting in about a 2 fold decrease of virus production (48). Deletion of the acidic cluster in HCMV membrane associated tegument protein pp28 has also been shown to have an even more dramatic phenotype that not only prevented trafficking of this tegument protein to the AC but also recovery of infectious virus (11, 55).
Based on findings from our previous studies which revealed that trafficking motifs in the gM-CT were functional, we performed yeast two hybrid screening to identify cellular proteins interacting with gM-CT. We identified the cellular protein - FIP4 as a protein that interacted with the cytoplasmic tail of gM. Additionally, using bacterially expressed and purified, GST-tagged gM-CT in pull down assay, we demonstrated that acidic cluster aa 359-365 (EEEDDD) in the C-terminal cytoplasmic tail of gM was required for the gM and FIP4 interaction. These findings together with yeast two hybrid screening data indicated that gM did not require other virus expressed proteins to interact with FIP4. Interestingly, pp28 tegument protein which contains a functional acidic cluster did not interact with FIP4 indicating that length and aa composition of the gM acidic cluster apparently provided selectivity for the interaction with FIP4. However, it is also possible that the conformation of the gM-CT dictated by the presence of the acidic cluster and its negative charge provide the specificity for the interactions of the gM-CT with FIP4.
To confirm the functional importance of the FIP4 and gM interaction we used shRNA to decrease FIP4 expression in the HCMV infected cells. Depletion of the FIP4 expression led to an approximate 1 log decrease in the infectious virus production that was also accompanied by an observable defect in the AC formation. In cells expressing shRNA targeting FIP4 expression, the AC was not completely developed and appeared less morphologically mature when compared to the AC in control, scrambled shRNA expressing cells. Interestingly, a similar defect in the AC formation and virus yield was previously observed in the cells infected with gMΔAC virus mutant that lacked the acidic cluster (28). These data argued that a defect in the formation of the AC associated with the disruption of the gM and FIP4 interaction could be explained by the delay in the gM/gN trafficking and accumulation of this abundant envelope glycoprotein complex in the AC. In addition, in cells depleted in FIP4 expression, a defect in the AC phenotype was observed when cells were labeled with anti-gM/gN as well as with anti-gB mab. These results may also indicate that lack of FIP4 expression and consequently delayed accumulation of glycoproteins in the AC disrupts AC formation which leads to the changes in its appearance. Taken together, our findings raised the possibility that gM/gN expression and accumulation in the AC plays a key role in formation and maturation of the AC during HCMV infection.
FIP4 is a member of the family of the Rab11 interacting proteins which is composed of six members (56). All members of this family contain 20 aa long C-terminal Rab binding domain (RBD), which is specific for the interactions with Rab11 (36).
Fip4 (arphofilin-2) is closely related to FIP3 (eferine/arphofilin-1) because unlike other members of this family of proteins, both FIP3 and FIP4 contain an EF-hand, coil-coiled domain responsible for homo and hetero dimerization (37, 40). FIP3 and FIP4 also have a Drosophila homolog, nuclear fallout protein (Nuf) (36, 37).
Intriguingly, in previous analysis of the tissue mRNA expression profile it was shown that high levels of FIP4 expression were present in neuronal tissue including brain, testis and only minimal FIP4 expression was detected in other tissues (37, 57–59). Our studies presented in Figure S1B. indicate that HCMV infection of HFF cells leads to significant up regulation of FIP4 expression and at the same time HCMV infection had no influence on FIP2 or FIP3 expression levels. This result not only validate the importance of FIP4 expression in virus infected cells but also provides a new perspective of the importance of cell expressed components during HCMV infection.
FIP3 and FIP4 have been shown to bind Arf5/Arf6 and Rab11 simultaneously, and are the only known human proteins that link the distinct functional activities of Arf and Rab GTPases families (40, 42, 43). We utilized the cytoplasmic tail of gM expressed in bacteria to determine if FIP4 bound to the gM-CT tail also recruited Arf5/6 and Rab11 GTPases. In the presence of the gM-CT, FIP4 binding to Arf5 or Arf6 was inhibited, but FIP4 still efficiently bound Rab11. This result was consistent with yeast two hybrid data because the cDNA fragment of FIP4 that interacted with gM-CT overlapped with the sequence encoding the domain of FIP4 that recruited Arf5 and Arf6. Because, neither Arf5 nor Arf6 contain an acidic cluster motif in the domains of interaction with FIP4, it is unlikely that gM-CT and Arf5/6 target similar sequences during their interactions with FIP4. However, the inhibition of Arf5/6 binding to FIP4 by the gM-CT suggests several possible mechanisms such as induction of a conformational change in FIP4 following gM-CT binding that could limit Arf5/6 binding to FIP4 (Fig. S2). Importantly, the gM-CT interaction did not alter recruitment of Rab11 by FIP4.
To confirm the relevance of these biochemical data in vivo, we decided to specifically target the function of FIP4 interacting GTPase Arf5, Arf6, and Rab11 in HCMV infected cells. The intracellular localization of Arf5 is sensitive to the effect of brefeldin A (BFA). In HeLa cells, the FIP4 has been shown to be BFA sensitive as a consequence of FIP4 interaction with Arf5 (37, 60). However, the localization of FIP4 and gM/gN in the AC was not sensitive to BFA treatment, which indicated that the gM/gN and FIP4 interaction and localization to the AC of virus infected cells are independent of Arf5 function (data not shown) (37, 60). Arf6 is a GTPase that controls the internalization of vesicles containing proteins retrieved from the plasma membrane during endocytosis (61–63). In cells expressing dominant negative Arf6(T27N) we did not observe any perturbation in the AC formation nor differences in the production of infectious virus when compared with cells expressing wild-type Arf6. Our data demonstrated that the dominant negative Arf6 did not alter infectious virus production. This finding was of interest because it not only confirmed that FIP4 did not interact with Arf6 in the presence of the gM but also suggested that endocytosis was not a critical pathway during HCMV assembly in fibroblasts. This was consistent with previous studies by Jarvis et. al. who noted that following inhibition of endocytosis by the expression of the dominant negative dynamin-I in virus infected cells, infectious virus production was not significantly decreased (60). These results suggested that retrieval of HCMV glycoproteins from cells surface by the clathrin dependent endocytic machinery was not required for efficient HCMV assembly. It is important also to note that in HCMV infected cells expressing Arf6, a substantial amount of this GTPase co-localized within the AC indicating that Arf6 coated vesicles enter the AC. However, inhibition of the Arf6 function had no influence on the localization of the gM/gN complex within the AC because the phenotype of the gM/gN complex was not altered in cells expressing dominant negative Arf6(T27N). This finding further suggested that gM/gN does not require internalization from the plasma membrane in order to be localized in the assembly compartment. This possibility is consistent with experiment in which we observed that expression of Arf6(T27N) in the HCMV infected cells did prevent internalization and accumulation of fluorescently labeled Tf in the ERC of cells feed with this ligand (data not shown).
In contrast to these findings, expression of the dominant negative Rab11(S25N) in the HCMV infected cells, resulted in a defect in the phenotype of the AC formation and a 2 log decrease in infectious virus production. In addition, in cells expressing dominant negative Rab11(S25N), the accumulation of the gM/gN complex within the AC appeared delayed and was associated with a defect in maturation of the AC. The effect of the dominant negative Rab11(S25N) on infectious virus production was even more dramatic than that observed in cells with decreased FIP4 expression, suggesting a central role of Rab11 GTPase activity in the HCMV assembly. This result is consistent with a primary function of the FIP4 acting as a scaffold protein to facilitate formation of a FIP4-Rab11 ternary complex which then engages gM/gN containing vesicles through interactions with gM-CT.
There has been limited amount of information describing role of the FIP4 in intracellular trafficking of proteins. It has been shown that FIP3 and FIP4 are abundantly localized in the ERC and possibly also in the TGN of HeLa cells (37). In addition, it has been shown that expression of an N-terminal deletion mutant of FIP4, resulted in the accumulation of FIP4 in a Rab11 positive endosomal recycling compartment but that the accumulation of FIP4 did not alter transferrin uptake in these cells, a result suggesting that FIP4 was responsible for the influx of vesicles into the ERC (37, 64). Nuf, the Drosophila counterpart of FIP4 had a similar effect when expressed in mammalian cells (37). The presence of Nuf in the cytoplasm of daughter cells after asymmetric cell division of Drosophila embryos has been shown to be essential in the morphogenesis of the ERC (39, 65). Following this study it was also demonstrated that FIP3 was critical for the structural integrity of the ERC in mammalian cells (64).
To investigate the role of the ERC in HCMV assembly we utilized transferrin (Tf) labeling of the ERC compartment in the HCMV infected cells. In our experiments, Tf labeled ERC strongly co-localized with the AC labeled with anti-gM/gN or anti-gB antibody (data not shown for gB staning). In contrast, late endososmal/lysosomes vesicles stained with fluorescently labeled epidermal growth factor (EGF) or cathepsin-D were excluded from the AC of the infected cells. This data indicated that accumulation the gM/gN protein complex within the cytoplasmic AC coincided with the site of the Tf accumulation, which has been shown to be the ERC. This result suggested that gM/gN containing vesicles are transported from the TGN and accumulated in the ERC, since we observed that internalization of the glycoproteins from the cell surface had little to no influence on the AC formation and consequently on HCMV assembly. Moreover, this result indicated that influx of the vesicles to the ERC was not affected during HCMV assembly. Our results also indicated that Arf6 labeled vesicles localized in the AC during HCMV infection. In addition when virus infected HFF cells were stained with anti-CD63 mab, MVB marker. CD63 positive vesicles overlapped with the periphery of the AC. This suggests that AC may also contain some MVB vesicles. Interestingly in the recent publication it was shown that Rab11 function is essential for the formation and fusion of MVBs in certain cell lines (19). Lastly, in these studies we have not been able to explore the precise function of the gM-FIP4-Rab11 complex in the assembly of HCMV; however, it is possible that exclusion Arf5 and Arf6 by gM bound to FIP4 favored influx of the gM/gN containing vesicles from the TGN to the ERC (Fig. 10).
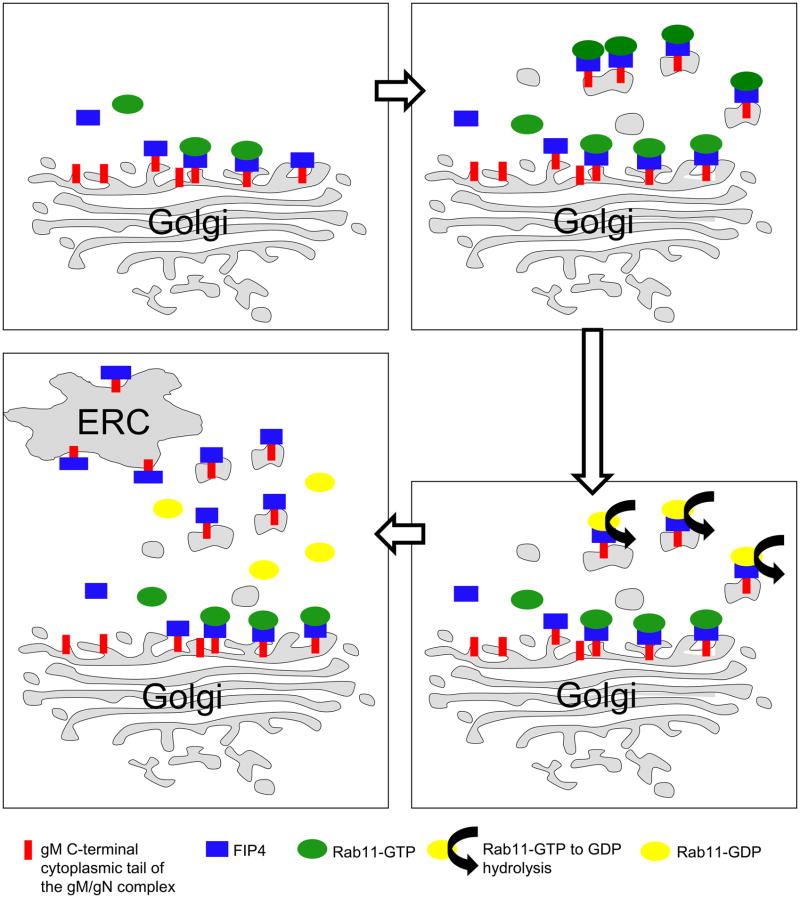
Figure 10. Model of FIP4 function and gM/gN trafficking to the AC of HCMV infected HFF cells.
(A) gM/gN complex transits through secretory pathway after reaching TGN, the gM cytoplasmic tail interacts with FIP4.
(B) Fip4 interacting with gM recruits Rab11-GTP and facilitates export from TGN to ERC.
(C) Vesicles exported from TGN induces Rab11 GTP-GDP hydrolysis on gM/gN-FIP4 coated vesicles and Rab11-GDP dissociation from the vesicles.
(D) gM-FIP4 containing vesicles reach ERC. Progressive accumulation of the gM/gN in the ERC and possibly other glycoproteins leads to the formation of the AC a cellular compartment containing host cell proteins of the ERC.
From the results provided in these studies, we propose a model in which the gM cytoplasmic tail interacts with FIP4 at the TGN, this interaction allows for the recruitment of Rab11 and leads to the formation of a ternary complex (Fig. 10). This gM-FIP4-Rab11 interaction allows efficient vesicles formation and transport from TGN and accumulation of the gM/gN containing vesicles in the ERC. Accumulation of the glycoproteins in the ERC is a vectorial event, because our previous FRAP data indicated that gM/gN which enters the center of the AC did not traffic back to the periphery of the AC, a pathway that has been demonstrated for the tegument protein pp28 in HCMV infected cells (66). This accumulation of the gM/gN glycoproteins and presumably other HCMV glycoproteins in the ERC vesicles provides a platform for the budding of tegumented capsids into glycoprotein enriched structures to yield enveloped HCMV particles (Fig. 10).
Materials and methods
Virus and virus titration, cell lines
HCMV strain AD169 was propagated in primary human foreskin fibroblasts (HFF) grown in Dulbecco’s modified Eagles medium (DMEM) supplemented with 5% fetal calf serum (FCS) and penicillin/streptomycin. Infectious HCMV virus stocks were prepared from the supernatant of infected HFF cells which exhibited 100% cytopathic effect and were stored at −80°C until use. To assay the amount of infectious particles in the samples, HFF cells monolayer grown in 96 well microtiter plates were infected with the serial dilutions of the virus sample. After adsorption of virus in room temperature for 1 hour, cells were washed and fresh medium was added. Virus titers were determined after 24hours of infection by an indirect immunfluorescence assay using a monoclonal antibody (mab) reactive with the immediate early protein 1 (IE1) of HCMV as described previously (67).
Human embryonic kidney HK293 cells were cultured in DMEM media supplemented with 10% FCS, 0.1% non-essential amino acids and 50 mg/l G418. Cos-7 cells were passage in DMEM supplemented with 10% FCS, and penicillin/streptomycin.
Plasmids
pEGFP-Rab11A, and its dominant negative form pEGFP-Rab11(S25N), were kindly donated to Addgene (Cambridge, MA) by Dr. R. Pagano (Mayo Clinic, Rochester, MN). We obtained these constructs from Addgene. Similarly, pcDNA3-HA-Arf6 and dominant negative pcDNA3-HA-Arf6(T27N) were purchased from Addgene (Cambridge, MA). The original clones were generated by Dr. T. Roberts (Florida State University, Tallahassee, FL). pcDNA3-HA-Arf5, pc-DNA3-HA-Arf1 construct was a kind gift of Dr. E. Sztul (University of Alabama at Birmingham, Birmingham, AL). The RFP-cathepsin-D construct was kindly provided by Dr. M. Ballestas (University of Alabama at Birmingham, Birmingham, AL).
FIP4 cDNA clone (NIH_MGC_287) was purchased from OpenBiosystems (Huntsville, AL), and used as a template to amplify by PCR full length FIP4 gene with BamHI and EcoRI overhangs what allowed for direct cloning of FIP4 into pEF1A-myc/his expression vector (Invitrogen, Carlsbad, CA). pcDNA-gM (UL100) and pcDNA-gN (UL73) have been described previously (26).
Yeast two hybrid system
To identify proteins interacting with gM (UL100) C-terminal cytoplasmic tail (gM-CT aa 300-372), we used the ProQuest yeast two-hybrid system with Gateway technology (Invitrogen, Carlsbad, CA). The gM C-terminal 216 bp fragment was amplified by PCR with primers containing attB1 and attB2 recombination sites that allowed cloning of a PCR product into “bait” vector pDEST32. To search for potential interactions, we used the ProQuest cDNA human liver library, cloned into the “prey” vector pDEST22 (Invitrogen, Carlsbad, CA). Finally, we transformed the pDEST22-cDNA library along with pDEST32-gM-CT into MaV203 yeast strain. Positive clones containing gM-CT interactors were identified by growth of transformants on histidine-and uracil-deficient medium and by assay with X-gal for the induction of the lacZ gene. Transformants that responded positively to the selective conditions were amplified by colony PCR using primers complementary to the pDEST22 vector outside of the multiple cloning sites (MCS). PCR products were sequenced and then analyzed using the NCBI BLAST software.
shRNA depletion of FIP4
To deplete FIP4 expression we used SureSilencing shRNA plasmids (SuperArray, Frederick, MD). This system consists of 5 shRNA sequences (4 shRNA sequences to deplete FIP4 and shRNA scrambled sequence) cloned into hMGFP vector that allows identification of shRNA expressing cells by a immunofluorescence assay. In experiments with HFF cells, 4 ug of vector DNA were electroporated using Amaxa and basic nucleofector kit according to the manufacture guidelines (Amaxa Bioscience, Gaithersburg, MD). The efficiency of electroporation was evaluated by an immunofluorescence assay. In brief, after electroporation HFF cells were plated on 13 mm coverslips. Two and four days post electroporation cells were washed in Dulbecco’s Phosphate-Buffered Saline (DPBS) and fixed in 4% para-formaldehyde (PFA). Cells were additionally stained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA) to visualize total number of the cells on the coverslips. We counted the number of cells expressing GFP per 100 cells stained with DAPI to determine the efficiency of electroporation.
FIP4 depletion efficiency was assessed by western blot analysis of cells lysates from transfected HK293 cells. HK293 cells were co-transfected using Trans-LT1 (Invitrogen, Carlsbad, CA), with pEF-FIP4-myc and shRNA depleting FIP4 expression or control shRNA scrambled sequence. As a control we used HK293 cells transfected only with pEF-FIP4-myc. 2, 4, and 6 days post transfection, cell pellets were harvested and stored in −80°C until use. Cell pellets were lysed in the SDS-PAGE loading buffer (20% SDS, 500mM Tris, pH 7.6, 1% Bromophenol Blue, 50% Glycerol, 1M DTT), boiled and resolved on the SDS-PAGE. After transfer to the nitrocellulose membrane (Pierce Rockford, IL), immunoblots were blocked in 5% BSA and probed with anti-myc (9E10) (Iowa Developmental Biology Core, Iowa City, IA). Western blots were then probed with anti-mouse horseradish conjugated antibody, followed by incubation with ECL reagents (Pierce, Rockford, IL). Additionally, membranes were striped with Restore PlusStripping buffer (Pierce, Rockford, IL) and reprobed with anti-actin antibody (Chemicon International, Temecula, CA). To asses FIP4 depletion in the infected cells, HFF cells were transfected with pEF-myc-FIP4 or pEF-myc-FIP3 as well as with shRNA to deplete FIP4 or shRNA scrambled sequence control. After 24 hours of recovery cells were infected with AD169 strain of HCMV and harvested 2, 4 and 6 days post infection in the SDS-page loading buffer. Cell lysates were subjected to SDS-PAGE, western blot transfer and followed with the membrane antibody labeling anti-myc (9E10), anti-p115 kindly provided by Dr. E. Sztul (University of Alabama at Birmingham, Birmingham, AL), anti-UL85. Western blots were developed using HRP conjugated secondary antibody and ECL reagents (Pierce, Rockford, IL).
GST pull down assay
A series of N-terminal glutathione S-transferase tagged gM-CT (GST-gM) constructs were generated using PCR and cloning into pGEX-4T vector (GE Healthcare, Piscataway, NJ). All gM GST tagged constructs were transformed into E. coli BL-21 bacterial strain and expression of the GST-recombinant protein was induced with 1mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 hours in 30 ml cultures at 30°C. After centrifugation, the bacterial pellet was lysed in STE buffer (10mM tris, 1mM EDTA, 150mM NaCL) containing lysozyme, DTT, 0.7% sarcosyl and 2% TritonX-100. After sonication and centrifugation supernatant was incubated for 30 minutes with glutathione-sepharose beads (GE Healthcare, Piscataway, NJ). Sepharose beads with bound recombinant protein were washed extensively with DPBS. Quality and quantity of the recombinant protein was evaluated following SDS-PAGE analysis of beads and Coomassie blue staining. For pull down assays, HK293 cells were transfected in 10 cm cell culture dishes with pEF1A-FIP4-myc tagged construct. After 48 hours, cells were lysed in 1ml of lysis buffer (150mM NaCl, 1% Triton X-100, 50 mM Tris-HCl, pH8.0) for 1 hour and the lysate cleared by centrifugation. This supernatant was mixed with equivalent amounts of the purified GST fusion protein bound to the sepharose beads, and incubated for 1 hour in 4°C. The beads were pelleted and washed with lysis buffer, followed by 2 washes with DPBS. Finally, the beads were resuspended in the SDS-PAGE loading buffer (20% SDS, 500mM Tris, pH 7.6, 1% Bromophenol Blue, 50% Glycerol, 1M DTT), boiled for 5 minutes. Samples were then subjected to SDS-PAGE. The gels were than either stained with Coomassie blue or visualized by western blotting with anti-myc (9E10) followed by HRP conjugated goat anti-mouse antibody and ECL (Pierce, Rockford, IL).
FIP4 binding assay
For the FIP4 binding assay HK293 cells were co-transfected with pEF-FIP4-myc and pcDNA3-HA-Arf-5, pcDNA3-HA-Arf6, pcDNA3-HA-Arf1 or pEGFP-Rab11 using Trans-LT1 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer directions. 48 hours post transfection cells were lysed in lysis buffer (150mM NaCl, 1% Triton X-100, 50 mM Tris-HCl, pH8.0), centrifuged and the supernatant of each sample was divided into three equal samples. One of the samples was subjected to immunoprecipitation with myc-tagged magnetic beads (Miltenyi Biotec, Auburn, CA) according the manufactures protocol. The remaining samples were mixed with GST alone or GST purified gM-CT recombinant protein, and GST pull down was completed as described above. All protein samples were resolved on SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% BSA and probed with anti-myc (9E10) (Iowa Developmental Biology Core, University of Iowa), or anti-HA (Covance, Richmond, CA) or anti-GFP (Invitrogen, Carlsbad, CA). The membranes were then probed with anti-mouse or anti-rabbit horseradish conjugated antibody, followed by incubation with ECL reagents (Pierce, Rockford, IL).
Immunofluorescence
Cos-7 cells were grown on 13 mm glass cover slips in 24 well plates and transfected at 60% of confluence with 0.5μg of each plasmid DNA using Trans-LT1 reagent (Invitrogen, Carlsbad, CA) according to the manufactures directions. Cells were fixed and immunostained 24 hours post transfection. The HFF cells were electoporated with 4ug of plasmid DNA using Amaxa and basic nucleofector kit (Amaxa Bioscience, Gaithersburg, MD) and seeded on 13 mm glass cover slips in 24 well plates. After recovery 24 hours post electroporation HFF cells were infected with HCMV AD169 strain and immunostaining was carried on day 4 or 6 post infection. To assay protein expression by static immunofluorescence, coverslips were washed with PBS and fixed in 4% PFA. The cells were permeabilized with 0.1% Triton X-100 and blocked with 10–50% goat serum (Invitrogen, Carlsbad, CA). The coverslips were then incubated with primary antibodies, including a anti-myc (9E10) (Iowa Developmental Biology Core, University of Iowa), anti-gM/gN (14-16A), anti-IE1 (p63-27), anti-gB (7-17), anti-FIP4 (provided by Dr. G. Gould), anti-HA (Covance, Richmond, CA). Following incubation with primary antibody and washing with PBS buffer containing 0.1% Tween20, antibody binding was detected with the appropriate secondary antibody conjugated with either FITC, or TRITC (Southern Biotech, Birmingham, AL). For the labeling of HCMV infected cells with TRITC-transferrin or FITC-EGF (Invitrogen, Carlsbad, CA), cells were washed three times in DPBS and incubated for an hour in serum free media. Then cells were labeled for two hours with media containing transferring-TRITC (100ug/ml) or EGF-FITC (10ug/ml). After incubation cells were washed and fixed in 4% PFA, and followed with the immunostaining as described above. All images were collected using an Olympus Fluoview confocal microscope.
Fluorescence recovery after photobleaching (FRET) assay
For FRET assays, HFF cells were electroporated with FIP4 or FIP3 myc-tagged construct and plated on the coverslips and 24 hours later infected with AD169 HCMV strain. The cells were then fixed in 4% PFA and stained with IMP anti-gM (anti -gM mab which recognizes C-terminal tail of gM) or anti-myc antibody followed by labeling with secondary antibody conjugated with FITC (donor) or TxRed (acceptor). FRET was obtained using a donor dequenching and acceptor photobleaching protocol on a confocal laser scanning microscope (Leica SP2; Leica, Bannockburn, IL). An image of FITC fluorescence and TxRed fluorescence before and after photobleaching was obtained by using the respective filter sets. Such data were collected from at least 10 different cells. Three to five regions of interest (ROI) in the photobleached area were selected per cell and the mean FITC fluorescence before and after photobleaching was obtained by using Leica software. FRET efficiency was calculated by using the following relationship: FRET efficiency (%) = [(Dpost−Dpre)/Dpost]·100. Here, Dpost is the fluorescence intensity of the FITC (donor [D]) after photobleaching, and Dpre is the fluorescence intensity of the FITC before photobleaching. A non-bleaching region was selected as an internal control on FRET analysis.
Supplementary Material
HFF cells were electroporated with sh-RNA to deplete FIP4 or control scrambled shRNA and infected with AD169 HCMV strain at moi 1.0. Twenty-four hours post infection, mRNA was harvested according Cell-to-CT (Ambion, Applied Biosystems, Austin, TX) protocol. cDNA was generated and the amount of the FIP2, FIP3, and FIP4 transcripts in FIP4-shRNA depleted versus non-depleted samples was quantified using TaqMan technology and Applied Biosystem designed primers. Results are presented as a graph comparing relative number of cDNA transcripts in FIP4 shRNA depleted versus scrambled shRNA expressing cells.
(B) As described above mRNA was extracted from non-infected and virus infected cells after 4 days of infection according to the Cell-to-CT (Ambion, Applied Biosystems, Austin, TX) protocol. Following cDNA generation using FIP2, FIP3 and FIP4 specific primers, transcripts were quantified in the infected versus non-infected cells.
GST-gM inhibits Arf5 and Arf6 binding to FIP4 in high concentrations. HK293 cells were co-transfected with FIP4-myc and either with HA-Arf5 or HA-Arf6. Two days post transfection cells were lysed divided for equal fractions and incubated with sepharose beads containing purified GST-gM-CT. After precipitation and extensive washing, proteins remaining on the glutathione sepharose beads were resolved by SDS-PAGE, transferred to membranes and probed with anti-HA, anti-myc antibody and developed with HRP.
Acknowledgments
This work was supported by grants from the HHS, NIH (AI49537, AI50189 to WJB). We would like to thank Dr. M. Ballestas and UAB Neuroscience NINDS Protein Core P30 NS47466 for their assistance.
References
- 1.Yoshikawa T. Significance of human herpesviruses to transplant recipients. Curr Opin Infect Dis. 2003 Dec;16(6):601–6. doi: 10.1097/00001432-200312000-00014. [DOI] [PubMed] [Google Scholar]
- 2.Halwachs-Baumann G. Recent developments in human cytomegalovirus diagnosis. Expert Rev Anti Infect Ther. 2007 Jun;5(3):427–39. doi: 10.1586/14787210.5.3.427. [DOI] [PubMed] [Google Scholar]
- 3.Mocarski ES, Courcelle CT. Cytomegaloviruses and their replication. 4. Philadelphia, Pa: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 4.Trus BL, Gibson W, Cheng N, Steven AC. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999 Mar;73(3):2181–92. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X, Shah S, Atanasov I, Lo P, Liu F, Britt WJ, et al. Three-dimensional localization of the smallest capsid protein in the human cytomegalovirus capsid. J Virol. 2005 Jan;79(2):1327–32. doi: 10.1128/JVI.79.2.1327-1332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science. 2000 May 5;288(5467):877–80. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 7.Mettenleiter TC. Herpesvirus assembly and egress. J Virol. 2002 Feb;76(4):1537–47. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez V, Greis KD, Sztul E, Britt WJ. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000 Jan;74(2):975–86. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez V, Sztul E, Britt WJ. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J Virol. 2000 Apr;74(8):3842–51. doi: 10.1128/jvi.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Pellett PE. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology. 2007 May 10;361(2):263–73. doi: 10.1016/j.virol.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Seo JY, Britt WJ. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J Virol. 2006 Jun;80(11):5611–26. doi: 10.1128/JVI.02630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S, Vasanji A, Pellett PE. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol. 2007 Nov;81(21):11861–9. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraile-Ramos A, Kledal TN, Pelchen-Matthews A, Bowers K, Schwartz TW, Marsh M. The human cytomegalovirus US28 protein is located in endocytic vesicles and undergoes constitutive endocytosis and recycling. Mol Biol Cell. 2001 Jun;12(6):1737–49. doi: 10.1091/mbc.12.6.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homman-Loudiyi M, Hultenby K, Britt W, Soderberg-Naucler C. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-golgi network 46, and mannosidase II. J Virol. 2003 Mar;77(5):3191–203. doi: 10.1128/JVI.77.5.3191-3203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Hubinger A, et al. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch Virol. 1996;141(3–4):557–72. doi: 10.1007/BF01718317. [DOI] [PubMed] [Google Scholar]
- 16.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993 Feb;60(1):163–78. [PubMed] [Google Scholar]
- 17.Fraile-Ramos A, Pelchen-Matthews A, Kledal TN, Browne H, Schwartz TW, Marsh M. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic. 2002 Mar;3(3):218–32. doi: 10.1034/j.1600-0854.2002.030307.x. [DOI] [PubMed] [Google Scholar]
- 18.Fraile-Ramos A, Pelchen-Matthews A, Risco C, Rejas MT, Emery VC, Hassan-Walker AF, et al. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol. 2007 Dec;9(12):2955–67. doi: 10.1111/j.1462-5822.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 19.Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005 Feb;6(2):131–43. doi: 10.1111/j.1600-0854.2004.00257.x. [DOI] [PubMed] [Google Scholar]
- 20.Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–69. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 21.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004 Oct;78(20):10960–6. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol. 2000 Sep;74(17):7720–9. doi: 10.1128/jvi.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brack AR, Dijkstra JM, Granzow H, Klupp BG, Mettenleiter TC. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999 Jul;73(7):5364–72. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lake CM, Hutt-Fletcher LM. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J Virol. 2000 Dec;74(23):11162–72. doi: 10.1128/jvi.74.23.11162-11172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudolph J, Seyboldt C, Granzow H, Osterrieder N. The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M. J Virol. 2002 Mar;76(6):2952–63. doi: 10.1128/JVI.76.6.2952-2963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mach M, Kropff B, Dal Monte P, Britt W. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73) J Virol. 2000 Dec;74(24):11881–92. doi: 10.1128/jvi.74.24.11881-11892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mach M, Osinski K, Kropff B, Schloetzer-Schrehardt U, Krzyzaniak M, Britt W. The carboxy-terminal domain of glycoprotein N of human cytomegalovirus is required for virion morphogenesis. J Virol. 2007 May;81(10):5212–24. doi: 10.1128/JVI.01463-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krzyzaniak M, Mach M, Britt WJ. The cytoplasmic tail of glycoprotein M (gpUL100) expresses trafficking signals required for human cytomegalovirus assembly and replication. J Virol. 2007 Oct;81(19):10316–28. doi: 10.1128/JVI.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996 Nov;135(4):913–24. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox D, Lee DJ, Dale BM, Calafat J, Greenberg S. A Rab11-containing rapidly recycling compartment in macrophages that promotes phagocytosis. Proc Natl Acad Sci U S A. 2000 Jan 18;97(2):680–5. doi: 10.1073/pnas.97.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Kumar R, Navarre J, Casanova JE, Goldenring JR. Regulation of vesicle trafficking in madin-darby canine kidney cells by Rab11a and Rab25. J Biol Chem. 2000 Sep 15;275(37):29138–46. doi: 10.1074/jbc.M004410200. [DOI] [PubMed] [Google Scholar]
- 32.Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network. J Cell Biol. 2000 Dec 11;151(6):1207–20. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelissier A, Chauvin JP, Lecuit T. Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Curr Biol. 2003 Oct 28;13(21):1848–57. doi: 10.1016/j.cub.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 34.Riggs B, Fasulo B, Royou A, Mische S, Cao J, Hays TS, et al. The concentration of Nuf, a Rab11 effector, at the microtubule-organizing center is cell cycle regulated, dynein-dependent, and coincides with furrow formation. Mol Biol Cell. 2007 Sep;18(9):3313–22. doi: 10.1091/mbc.E07-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, et al. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005 Feb;16(2):849–60. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prekeris R, Davies JM, Scheller RH. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J Biol Chem. 2001 Oct 19;276(42):38966–70. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- 37.Hickson GR, Matheson J, Riggs B, Maier VH, Fielding AB, Prekeris R, et al. Arfophilins are dual Arf/Rab 11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Mol Biol Cell. 2003 Jul;14(7):2908–20. doi: 10.1091/mbc.E03-03-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin OH, Ross AH, Mihai I, Exton JH. Identification of arfophilin, a target protein for GTP-bound class II ADP-ribosylation factors. J Biol Chem. 1999 Dec 17;274(51):36609–15. doi: 10.1074/jbc.274.51.36609. [DOI] [PubMed] [Google Scholar]
- 39.Riggs B, Rothwell W, Mische S, Hickson GR, Matheson J, Hays TS, et al. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J Cell Biol. 2003 Oct 13;163(1):143–54. doi: 10.1083/jcb.200305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, et al. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 2005 Oct 5;24(19):3389–99. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace DM, Lindsay AJ, Hendrick AG, McCaffrey MW. Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem Biophys Res Commun. 2002 Dec 20;299(5):770–9. doi: 10.1016/s0006-291x(02)02720-1. [DOI] [PubMed] [Google Scholar]
- 42.Schonteich E, Pilli M, Simon GC, Matern HT, Junutula JR, Sentz D, et al. Molecular characterization of Rab11-FIP3 binding to ARF GTPases. Eur J Cell Biol. 2007 Aug;86(8):417–31. doi: 10.1016/j.ejcb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Shin OH, Couvillon AD, Exton JH. Arfophilin is a common target of both class II and class III ADP-ribosylation factors. Biochemistry. 2001 Sep 11;40(36):10846–52. doi: 10.1021/bi0107391. [DOI] [PubMed] [Google Scholar]
- 44.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006 May;7(5):347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez C, Fujita H, Hubbard A, Sztul E. ER to Golgi transport: Requirement for p115 at a pre-Golgi VTC stage. J Cell Biol. 1999 Dec 13;147(6):1205–22. doi: 10.1083/jcb.147.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson W, Baxter MK, Clopper KS. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J Virol. 1996 Nov;70(11):7454–61. doi: 10.1128/jvi.70.11.7454-7461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saraste J, Goud B. Functional symmetry of endomembranes. Mol Biol Cell. 2007 Apr;18(4):1430–6. doi: 10.1091/mbc.E06-10-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crump CM, Hung CH, Thomas L, Wan L, Thomas G. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J Virol. 2003 Oct;77(20):11105–13. doi: 10.1128/JVI.77.20.11105-11113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benveniste M, Schlessinger J, Kam Z. Characterization of internalization and endosome formation of epidermal growth factor in transfected NIH-3T3 cells by computerized image-intensified three-dimensional fluorescence microscopy. J Cell Biol. 1989 Nov;109(5):2105–15. doi: 10.1083/jcb.109.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hopkins CR, Gibson A, Shipman M, Miller K. Movement of internalized ligand-receptor complexes along a continuous endosomal reticulum. Nature. 1990 Jul 26;346(6282):335–9. doi: 10.1038/346335a0. [DOI] [PubMed] [Google Scholar]
- 51.Pohlmann R, Boeker MW, von Figura K. The two mannose 6-phosphate receptors transport distinct complements of lysosomal proteins. J Biol Chem. 1995 Nov 10;270(45):27311–8. doi: 10.1074/jbc.270.45.27311. [DOI] [PubMed] [Google Scholar]
- 52.Press B, Feng Y, Hoflack B, Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol. 1998 Mar 9;140(5):1075–89. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prag G, Lee S, Mattera R, Arighi CN, Beach BM, Bonifacino JS, et al. Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding proteins. Proc Natl Acad Sci U S A. 2005 Feb 15;102(7):2334–9. doi: 10.1073/pnas.0500118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tortorella LL, Schapiro FB, Maxfield FR. Role of an acidic cluster/dileucine motif in cation-independent mannose 6-phosphate receptor traffic. Traffic. 2007 Apr;8(4):402–13. doi: 10.1111/j.1600-0854.2007.00541.x. [DOI] [PubMed] [Google Scholar]
- 55.Jones TR, Lee SW. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J Virol. 2004 Feb;78(3):1488–502. doi: 10.1128/JVI.78.3.1488-1502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem. 2004 Aug 6;279(32):33430–7. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- 57.Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O. Prediction of the coding sequences of unidentified human genes. XX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2001 Apr 27;8(2):85–95. doi: 10.1093/dnares/8.2.85. [DOI] [PubMed] [Google Scholar]
- 58.Muto A, Aoki Y, Watanabe S. Mouse Rab11-FIP4 regulates proliferation and differentiation of retinal progenitors in a Rab11-independent manner. Dev Dyn. 2007 Jan;236(1):214–25. doi: 10.1002/dvdy.21009. [DOI] [PubMed] [Google Scholar]
- 59.Muto A, Arai K, Watanabe S. Rab11-FIP4 is predominantly expressed in neural tissues and involved in proliferation as well as in differentiation during zebrafish retinal development. Dev Biol. 2006 Apr 1;292(1):90–102. doi: 10.1016/j.ydbio.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 60.Jarvis MA, Fish KN, Soderberg-Naucler C, Streblow DN, Meyers HL, Thomas G, et al. Retrieval of human cytomegalovirus glycoprotein B from cell surface is not required for virus envelopment in astrocytoma cells. J Virol. 2002 May;76(10):5147–55. doi: 10.1128/JVI.76.10.5147-5155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chavrier P, Goud B. The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol. 1999 Aug;11(4):466–75. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 62.Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003 Oct 24;278(43):41573–6. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- 63.Donaldson JG, Honda A. Localization and function of Arf family GTPases. Biochem Soc Trans. 2005 Aug;33(Pt 4):639–42. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- 64.Horgan CP, Oleksy A, Zhdanov AV, Lall PY, White IJ, Khan AR, et al. Rab11-FIP3 is critical for the structural integrity of the endosomal recycling compartment. Traffic. 2007 Apr;8(4):414–30. doi: 10.1111/j.1600-0854.2007.00543.x. [DOI] [PubMed] [Google Scholar]
- 65.Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, et al. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005 Sep 9;122(5):763–73. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 66.Seo JY, Britt WJ. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J Virol. 2007 Jun;81(12):6536–47. doi: 10.1128/JVI.02852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andreoni M, Faircloth M, Vugler L, Britt WJ. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods. 1989 Feb;23(2):157–67. doi: 10.1016/0166-0934(89)90129-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HFF cells were electroporated with sh-RNA to deplete FIP4 or control scrambled shRNA and infected with AD169 HCMV strain at moi 1.0. Twenty-four hours post infection, mRNA was harvested according Cell-to-CT (Ambion, Applied Biosystems, Austin, TX) protocol. cDNA was generated and the amount of the FIP2, FIP3, and FIP4 transcripts in FIP4-shRNA depleted versus non-depleted samples was quantified using TaqMan technology and Applied Biosystem designed primers. Results are presented as a graph comparing relative number of cDNA transcripts in FIP4 shRNA depleted versus scrambled shRNA expressing cells.
(B) As described above mRNA was extracted from non-infected and virus infected cells after 4 days of infection according to the Cell-to-CT (Ambion, Applied Biosystems, Austin, TX) protocol. Following cDNA generation using FIP2, FIP3 and FIP4 specific primers, transcripts were quantified in the infected versus non-infected cells.
GST-gM inhibits Arf5 and Arf6 binding to FIP4 in high concentrations. HK293 cells were co-transfected with FIP4-myc and either with HA-Arf5 or HA-Arf6. Two days post transfection cells were lysed divided for equal fractions and incubated with sepharose beads containing purified GST-gM-CT. After precipitation and extensive washing, proteins remaining on the glutathione sepharose beads were resolved by SDS-PAGE, transferred to membranes and probed with anti-HA, anti-myc antibody and developed with HRP.