Abstract
Once programmed to acquire a specific identity and function, cells rarely change in vivo1. Neurons of the mammalian central nervous system (CNS) in particular are a classic example of a stable, terminally differentiated cell type. With the exception of the adult neurogenic niches, where a limited set of neuronal subtypes continue to be generated throughout life2,3, CNS neurons are only born during embryonic and early postnatal development. Once generated, neurons become permanently postmitotic and do not change their identity for the life span of the organism. Here, we have investigated whether excitatory neurons of the neocortex can be instructed to directly reprogram their identity postmitotically from one subtype into another, in vivo. We show that embryonic and early postnatal callosal projection neurons (CPN) of layer II/III can be postmitotically lineage reprogrammed into layer V/VI corticofugal projection neurons (CFuPN) upon expression of the transcription factor Fezf2. Reprogrammed callosal neurons acquire molecular properties of corticofugal projection neurons and change their axonal connectivity from interhemispheric, intracortical projections to corticofugal projections directed below the cortex. The data indicate that during a window of postmitotic development neurons can change their identity, acquiring critical features of alternate neuronal lineages.
Seminal work has challenged the idea that differentiated cells cannot change their identity and has underscored the powerful role played by transcription factors in instructing lineage reprogramming 4–6. However, whether all cells possess the same ability to change identity is still unclear. In the central nervous system, neurons represent a quintessential example of a permanently postmitotic and differentiated cell type. Despite the tight control over maintenance of neuronal identity, it is not known whether neurons can be lineage reprogrammed to a new cell fate under appropriate stimuli.
Prior work has demonstrated that an entire mouse can be cloned via the transfer of nuclei of at least some neuron types into enucleated oocytes7,8. This work supports the argument that in the CNS neurons resist phenotypic changes without irreversible loss of genetic material and that epigenetic mechanisms that may be in place to maintain cell identity, can be reset under appropriate conditions. In support of this notion is the demonstration that spinal motor neurons can undergo a class switch if manipulated immediately after exit from the cell cycle9–11.
To determine whether excitatory neurons of the cerebral cortex, one of the most complex regions of the CNS, can undergo direct lineage reprogramming after they have become postmitotic and whether this can occur in vivo, we have investigated if a lineage conversion between layer II/III callosal projection neurons and layer V/VI corticofugal projection neurons12, can be instructed within the brain. Both of these neuronal subtypes are generated during development and do not regenerate in the adult13.
The molecular programs controlling CFuPN development are emerging12,14. In particular, the transcription factor Fezf2 is a powerful master gene necessary for the development of subcerebral projection neurons, a class of CFuPN15–17, and whose overexpression is sufficient to confer corticofugal neuronal identity on neural progenitors fated to generate different neuronal populations17,18.
Here, we developed two different approaches to ectopically express Fezf2 in postmitotic layer II/III CPN. First, to determine whether at the earliest stages of postmitotic development CPN retain the ability to lineage reprogram, we have taken advantage of the Cdk5r gene promoter19 to drive Fezf2 expression in “young” postmitotic CPN. In the developing cerebral cortex, Cdk5r is restricted to migratory, postmitotic neurons20,21. To confirm the specificity of its minimal promoter, we co-electroporated Cdk5r-DsRed and Nestin-eGFP expression vectors in E13.5 cortical progenitors and determined their expression patterns. As expected, we found that eGFP was restricted to progenitors while DsRed was restricted to postmitotic CPN (Supplementary Fig. 1).
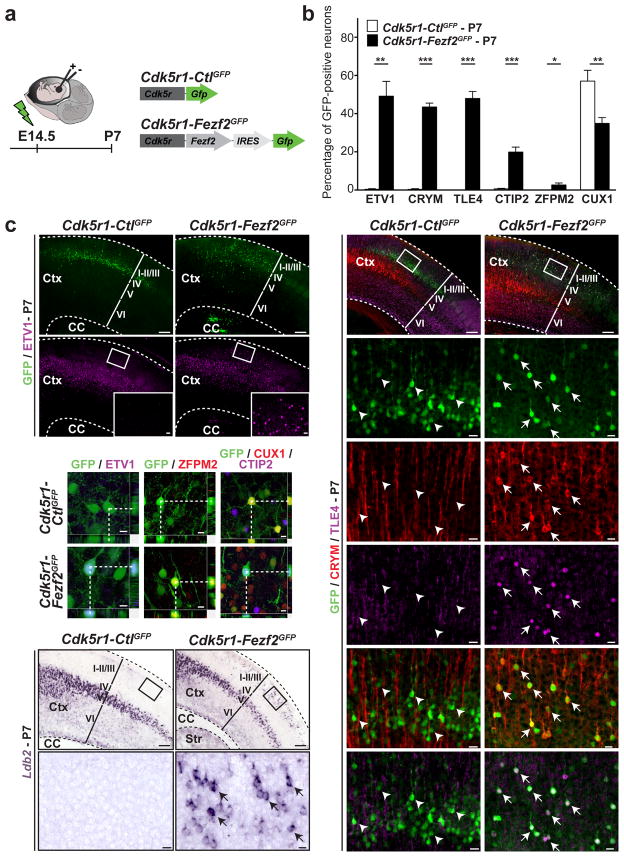
Cdk5r-Fezf2eGFP or control Cdk5r-CtleGFP expression vectors were then electroporated in utero into cortical progenitors at E14.5 and mice were sacrificed at E18.5, P7, P14 and P28. In both control and Fezf2-electroporated embryos, GFP-positive neurons were visible in the upper layers; however, neurons that expressed Fezf2 showed a reproducible change in migration, with a subset of neurons located ectopically in the deep layers (Figure 1c). We thus concentrated our analysis only on GFP-positive neurons located within layer II/III. We first analyzed the molecular identity of the neurons generated. We found that ER81, CRYM, TLE4, CTIP2, ZFPM2 and Clim1 – all markers of CFuPN excluded from layer II/III CPN12,14,16,22 – were present in layer II/III of the Cdk5r-Fezf2eGFP cortex (n=4) but not in control Cdk5r-CtleGFP-electroporated cortex (n=4) at all ages examined (Fig. 1, 2, 3). Next we quantified at the single neuron level the percentage of GFP-positive neurons in layers II/III that expressed CFuPN and CPN proteins. We found that, at P7, CPN that had received Fezf2 expressed ER81, a protein normally restricted to layer V22 (49.18 +/− 7.7% versus 0.41 +/− 0.31% in Cdk5r-CtleGFP); CRYM, a protein selectively expressed in CFuPN14 (43.51 +/− 2.02% versus 0.36 +/− 0.36% in Cdk5r-CtleGFP); TLE4, a protein restricted to corticothalamic neurons12 (47.98 +/− 3.62% versus 0% in Cdk5r-CtleGFP); CTIP2, a protein specific for subcerebral projection neurons and corticothalamic neurons14 (19.90 +/− 2.61% versus 0.61 +/− 0.25% in Cdk5r-CtleGFP); and ZFPM2, a protein only expressed in CFuPN16 (2.70 +/− 1.01% versus 0% in Cdk5r-CtleGFP). Notably, CUX1, a protein selectively expressed in upper layer CPN12,23 was expressed in significantly fewer CPN that had received Cdk5r-Fezf2eGFP compared to controls (34.92 +/− 3.02% versus 57.03 +/− 5.7% in Cdk5r-CtleGFP) (Fig. 1b). These results show that young postmitotic CPN are able to acquire molecular features of CFuPN.
Figure 1. Expression of Fezf2 in migratory, postmitotic CPN induces them to acquire molecular features of CFuPN.
a, Schematic representation of the experimental approach. Fezf2 or control vectors were electroporated at E14.5 and expression was restricted to CPN at the earliest stages of postmitotic development using the promoter of the Cdk5r gene. b, Quantification of the percentage of electroporated CPN expressing CFuPN-specific markers (ER81, CRYM, TLE4, CTIP2 and ZFPM2) and downregulating CPN-specific markers (CUX1). Results are expressed as the mean ± s.e.m. The paired, two-tailed t test was used for statistical analysis. * p<0.05; ** p<0.01; *** p<0.001. c, Immunocytochemistry and in situ hybridization for a panel of CFuPN and layer II/III CPN markers. Ctx, cortex; CC, corpus callosum; Str, striatum. Scale bars, 200 μm, 20 μm in high magnification panels and 10 μm in confocal images.
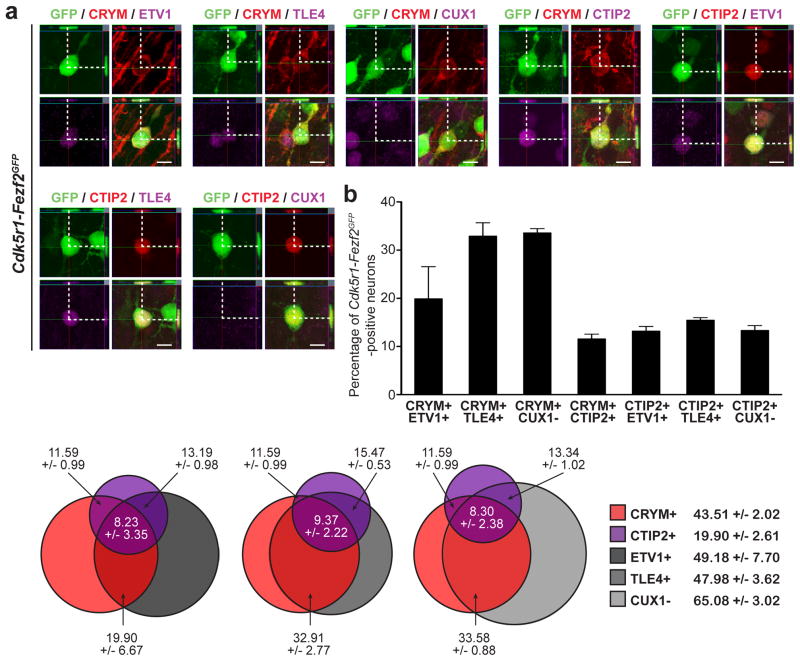
Figure 2. Fezf2 expression in postmitotic CPN induces a program of CFuPN-specific genes.
a, Confocal analysis showing that CFuPN-specific genes are co-expressed in Cdk5r-Fezf2eGFP- electroporated CPN. Scale bars, 10 μm. b, Quantification of the percentage of Cdk5r-Fezf2eGFP-electroporated CPN expressing a combination of two CFuPN-specific markers. Results are expressed as the mean ± s.e.m. c, Proportional Venn diagrams representing an estimate of the percentage of Cdk5r-Fezf2eGFP-electroporated CPN co-expressing combinations of three CFuPN-specific markers.
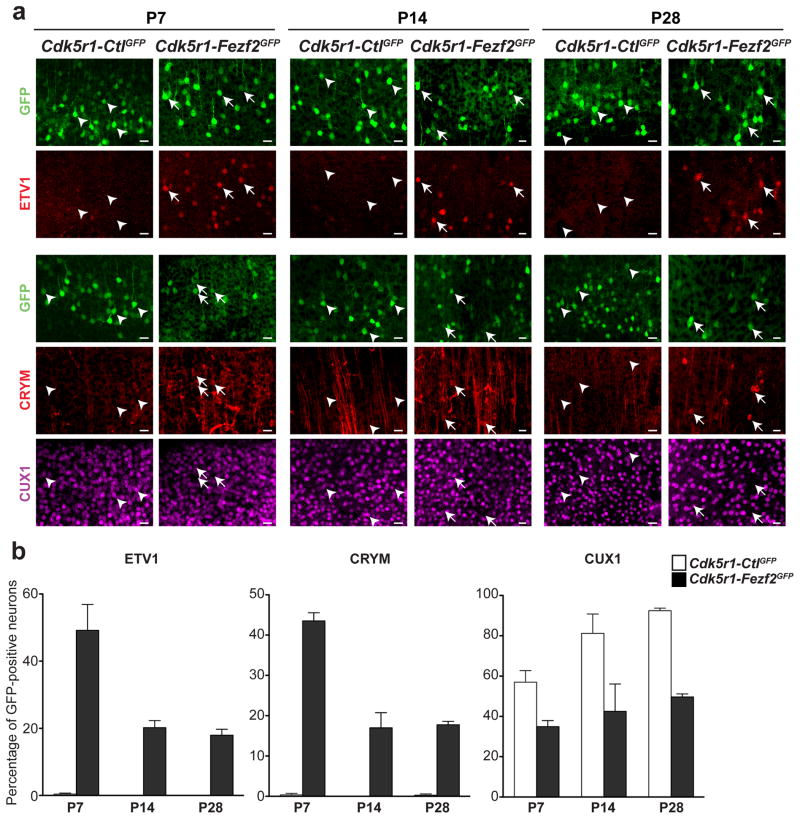
Figure 3. Reprogrammed CPN maintain stable expression of CFuPN markers.
a, Immunocytochemistry for ER81, CRYM and CUX1 in Cdk5r-Fezf2eGFP-electroporated CPN versus control (Cdk5r-CtleGFP) at P7, P14 and P28. Scale bars 20 μm. b, Quantification of the percentage of Cdk5r-Fezf2eGFP- and Cdk5r-CtleGFP-electroporated CPN maintaining ER81, CRYM and CUX1 expression at P7, P14 and P28. Results are expressed as the mean ± s.e.m.
In order to further assess the extent of these molecular changes, we investigated the combinatorial profile of expression of these subtype-specific proteins. We found that up to 75.6% of Fezf2-expressing CPN that expressed one CFuPN protein also co-expressed a second CFuPN marker. Specifically, Fezf2-expressing CPN co-expressed CRYM and ER81 (19.90 +/− 6.67%; n=4), CRYM and TLE4 (32.91 +/− 2.77%; n=4), CRYM and CTIP2 (11.59 +/− 0.99%; n=3), CTIP2 and ER81 (13.19 +/− 0.98%; n=4), and CTIP2 and TLE4 (15.47 +/− 0.53%; n=4) (Fig. 2a,b). Notably, a distinct fraction of Fezf2-expressing CPN that was negative for CUX1 (65.08 +/− 3.02%) was also positive for CRYM (33.58 +/− 0.88%; n=3) and CTIP2 (13.34 +/− 1.02%; n=4) (Fig. 2b), indicating that they had made a clear lineage choice. Fezf2-expressing CPN were also calculated to co-express CRYM, CTIP2 and ER81 (8.23 +/− 3.35%), CRYM, CTIP2 and TLE4 (9.37 +/− 2.22%) and to be positive for CRYM and CTIP2 but negative for CUX1 (8.30 +/− 3.28%) (Fig. 2c). These results demonstrate that, in an early period of postmitotic life, CPN of layer II/III are able to reprogram their molecular identity, acquiring combinatorial expression of CFuPN proteins and repressing CPN proteins.
Interestingly, reprogrammed CPN also expressed the transcription factor Clim 1, (Fig. 1c), which is not a direct target gene of Fezf2 (Lodato et al., unpublished data), providing evidence that the reprogramming process goes beyond activation of a first line of Fezf2 direct target genes to induce a broader program of CFuPN gene expression.
It is possible that reprogrammed CPN may not be stable and, over time, revert to their original identity or die. We therefore assessed the expression of ER81, CRYM and CUX1 in Fezf2-expressing CPN over time. ER81 remained expressed in Fezf2-expressing CPN at P14 (20.21 +/− 2.10%, n=2 versus 0% in Cdk5r-CtleGFP, n=3) and P28 (17.96 +/− 1.78%, n= 3 versus 0%, in Cdk5r-CtleGFP n=2) (Fig. 3a,b). Similarly, CRYM was maintained at P14 (16.99 +/− 3.74%, n=2 versus 0% in Cdk5r-CtleGFP, n=4) and P28 (17.74 +/− 0.83%, n=3 versus 0.29 +/− 0.29%, in Cdk5r-CtleGFP n=2) (Fig. 3a,b). Finally, we found that the percentage of CUX1-positive neurons remained low in the Cdk5r-Fezf2eGFP-targeted CPN population over time (Fig. 3a,b). Together, these data show that Cdk5r-Fezf2eGFP-electroporated, reprogrammed CPN retained CFuPN molecular identity more than four weeks after the initial expression of Fezf2. Importantly, there is a reduction in the fraction of Fezf2-expressing CPN that express ER81 and CRYM between P7 and P14, however these values stabilize after P14.
To investigate whether this decrease was due to cell death, we quantified the number of caspase-3 positive cells among GFP-expressing neurons, in both Cdk5r-Fezf2eGFP and Cdk5r-CtleGFP cortices at P7, P14 and P28. We found minimal cell death and no significant differences between groups (Supplementary Fig. 2a,b). We conclude that the reduction in the percentages of ER81- and CRYM-positive Cdk5r-Fezf2eGFP-expressing CPN observed by P14 is not due to cell death but rather to the inability to maintain expression of these markers. In agreement, the percentage of CUX1-negative Cdk5r-Fezf2eGFP-expressing CPN remained unchanged over time.
Together, the data indicate that upper layer II/III CPN can undergo an early postmitotic change of identity in response to cell autonomous signals instructive of a different neuronal lineage. Furthermore, a significant proportion of reprogrammed neurons maintain their new molecular identity in postnatal cortex.
We next investigated whether CPN can reprogram their molecular identity at later postmitotic ages, when they have already positioned in the upper layers and have acquired connectivity features of the CPN lineage. The reprogramming abilities of neurons at these later postmitotic stages of development have not been investigated before for any CNS neuron subtype.
We constructed a conditional expression vector where the Fezf2 expression cassette was cloned in antisense orientation between two pairs of inverted LoxP sites (DF-Fezf2eGFP). We electroporated cortical progenitors at E14.5 with DF-Fezf2eGFP or control DF-CtleGFP together with a 4-hydroxytamoxifen (TAM)-inducible form of Cre recombinase25 (CAG-ERT2-Cre-ERT2) (Fig. 4a). Recombination was induced by injections of TAM at either E17.5, P3 or P21 (Fig. 4 and Supplementary Fig. 3 and 4). TAM administration at P3 and P21, resulted in robust recombination and Fezf2 expression (95.24 +/− 0.43% and 93.32 +/− 1.99%, respectively) (Supplementary Fig. 4a,b). In parallel experiments, to increase nuclear plasticity, expression of Fezf2 was paired with either administration of Valproic Acid (VPA) a histone deacetylase inhibitor26, or with surgical transection of the corpus callosum (callosotomy) as a CPN-specific type of injury (Fig. 4a). Pups were sacrificed at P14, P28 and P42.
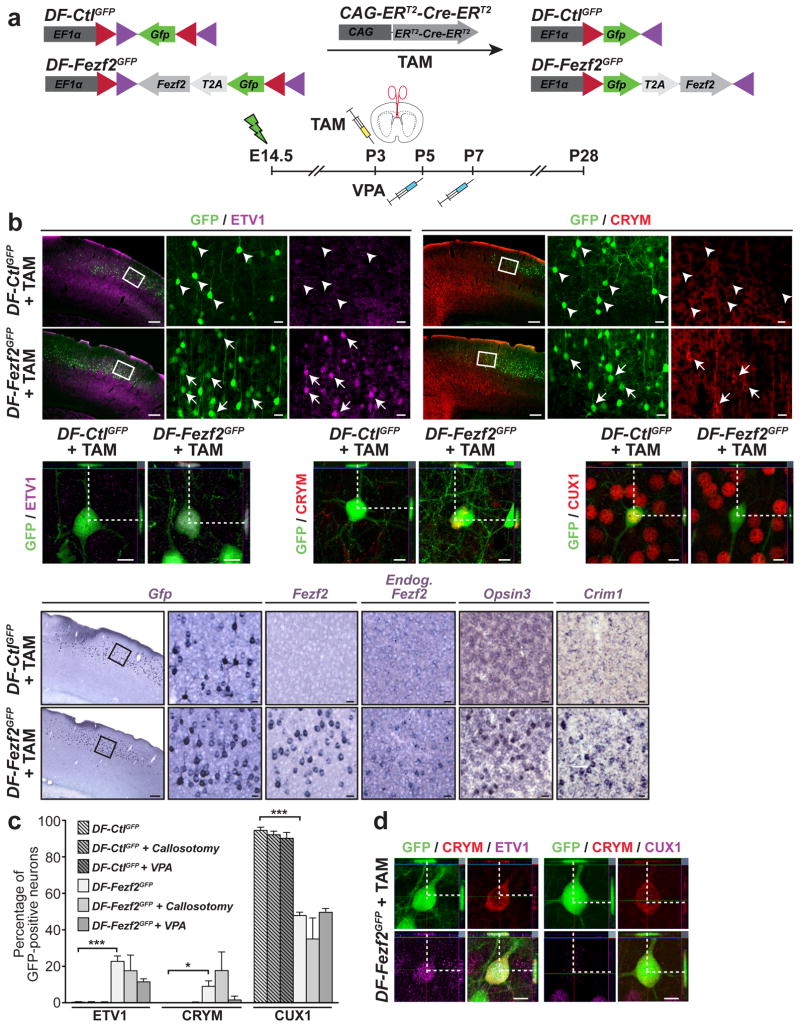
Figure 4. Callosal projection neurons can undergo molecular reprogramming of subtype-specific genes in response to Fezf2 expression at postnatal day P3.
a, Schematic of the constructs used to conditionally express Fezf2 at P3 and of experimental approach. CPN received Fezf2 (or control DNA) alone or in combination with either VPA treatment or callosotomy. b, Immunocytochemistry and in situ hybridization on coronal sections of cortex. c, Quantification of the percentage of electroporated CPN expressing CFuPN and CPN proteins. Results are expressed as the mean ± s.e.m. The paired, two-tailed t test was used for statistical analysis. * p<0.05; ** p<0.01; *** p<0.001. d, Confocal analysis showing that a subset of DF-Fezf2eGFP-electroporated CPN co-expressed CRYM and ER81 and are CRYM-positive and CU7-negative. Scale bars, 200 μm, 20 μm in high magnification panels and 10 μm in confocal images.
We found that CPN that began to express Fezf2 at P3 were still able to reprogram their molecular identity and expressed several CFuPN genes. Similar results were obtained when Fezf2 was induced at E17.5 (Supplementary Fig. 3 and data not shown). At P28, twenty five days after initial Fezf2 expression, these neurons expressed ER81 (22.72 +/− 3%, n=3 versus 0.29% +/− 0.29% in DF-CtleGFP, n=3); CRYM (8.94 +/− 3.46%, n=3 versus 0% in DF-CtleGFP, n=5); Opsin3 and Crim1, two late markers of subcerebral neurons14; and were notably able to induce transcription of the endogenous Fezf2 gene (Fig. 4b,c). In addition, CPN expressing DF-Fezf2eGFP downregulated expression of CUX1 (47.78 +/− 1.85%, n=3 versus 94.47 +/− 1.81% in DF-CtleGFP, n=3) (Fig. 4b,c). Double marker analysis further showed that 33.56% of the CRYM-positive neurons co-expressed ER81 and 76.51% downregulated CUX1 (Fig. 4d and data not shown). By P28 we could not detect expression of the CFuPN markers CTIP2, TLE4 and ZFPM2 proteins, which at P7 are present in CPN reprogrammed early postmitotically (Fig. 1). These markers might only be expressed by reprogrammed neurons soon after initial expression of Fezf2, or P3 neurons might differ from early postmitotic neurons at P28.
The efficiency of reprogramming remained unaltered by VPA treatment (Fig. 4c). However, callosotomy increased the percentage of Fezf2-expressing CPN that was positive for CRYM and negative for CUX1 (Fig. 4c).
These findings indicate that postnatal CPN, despite having been postmitotic for several days still retain the ability to reprogram key aspects of their molecular identity in vivo. Although a significant proportion of CPN targeted at P3 could reprogram, the efficiency was lower compared to early postmitotic neurons, suggesting that a progressive restriction of fate occurs with time during the postmitotic life of the neuron. To test this possibility, we induced Fezf2 expression in upper layer CPN at P21. By P42, we found that virtually all targeted CPN expressed exogenous Fezf2 (82.52 +/− 14.77%). However neither upregulation of CRYM and ER81 nor downregulation of CUX1 was detected (Supplementary Fig. 4a,b,c,d and data not shown). The data indicate that CPN gradually lose their ability to reprogram their molecular identity and suggest the existence of a critical period of nuclear plasticity for postmitotic neurons.
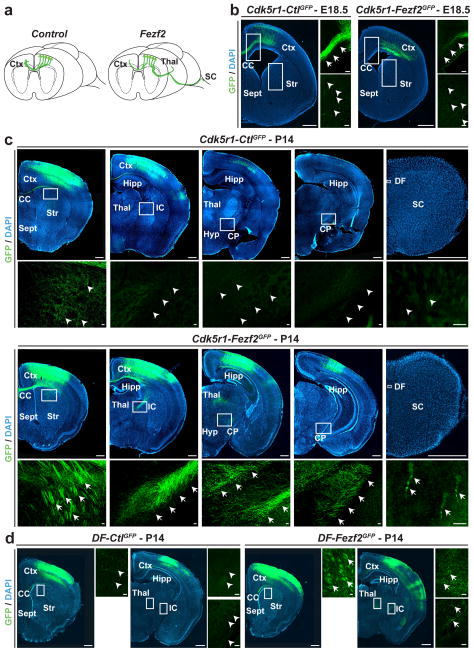
Neuronal identity is defined as the combination of molecular, morphological and connectivity features. Layer II/III CPN and CFuPN have distinct patterns of axonal connectivity. CPN axons extend across the corpus callosum, with subpopulations maintaining collaterals to the ipsi- and contralateral striatum and to the frontal cortex12,27,28. CFuPN, to the contrary, project axons below the cortex, through the internal capsule to targets in the thalamus, pons and spinal cord12. To determine whether CPN can reprogram their axonal projections from callosal to corticofugal targets, we first examined the axonal connectivity of pups that had received either Cdk5r-Fezf2eGFP or control DNA via electroporation at E14.5 and were sacrificed at E18.5, P7, P14, or P28 (Fig. 5). As expected, in Cdk5r-CtleGFP control animals, GFP-positive axon bundles were present crossing the corpus callosum and no axons entered the internal capsule (Fig. 5b, left and upper panels). In contrast, in Cdk5r-Fezf2eGFP mice, a reduction in GFP callosal axons was observed at E18.5 (Fig. 5b, right panels), and at P7, P14 and P28 distinct axon bundles had entered the internal capsule (Fig. 5c, lower panels and data not shown). In addition, axons of reprogrammed CPN, but not control CPN, were able to extend caudally to targets in the thalamus and the cerebral peduncle, and some axons reached the spinal cord (Fig. 5c, lower panels). Similar results were obtained when Fezf2 was expressed in CPN using the Doublecortin promoter, which also restricts expression to early postmitotic neurons19,29–31 (Supplementary Fig. 5). In all experiments axons that reached corticofugal targets were maintained at P28 (data not shown).
Figure 5. CPN can reprogram their axonal projections from callosal to corticofugal targets in response to Fezf2.
a, Graphic representation of changes in axonal connectivity by layer II/III CPN in response to Fezf2 versus control. b, GFP-positive CPN axons crossing the corpus callosum at E18.5 in Cdk5r-Fezf2eGFP and Cdk5r-CtleGFP-electroporated CPN. Callosal projections are greatly reduced in Cdk5r-Fezf2eGFP- versus Cdk5r-CtleGFP-electroporated CPN. c, At P14, GFP-positive axons are restricted to the corpus callosum in Cdk5r-CtleGFP-electroporated CPN. By contrast, axons of Cdk5r-Fezf2eGFP-electroporated CPN are also present in the internal capsule and extend caudally to the thalamus and the cerebral peduncle. A small number of GFP-positive axons are present in the dorsal funiculus of the spinal cord in Cdk5r-Fezf2eGFP- but not in control-electroporated animals. d, GFP-positive axons at P14 of DF-Fezf2eGFP- or DF-CtleGFP-electroporated CPN that received TAM postmitotically at E17.5. Callosal axons are restricted to the corpus callosum in DF-CtleGFP CPN while axons are also present within the internal capsule in DF-Fezf2eGFP CPN. CC, corpus callosum; CP, cerebral peduncle; Ctx, cortex; DF, dorsal funiculus; Hipp, hippocampus; Hyp, hypothalamus; IC, internal capsule; Str, striatum; Sept, septum; Thal, thalamus. Scale bars, 200 μm and 20 μm in high magnification panels.
To determine whether axonal projections could be reprogrammed at a later stage, we followed GFP-positive axons from postmitotic CPN that had received DF-Fezf2eGFP or control DF-CtleGFP together with CAG-ERT2-Cre-ERT2, and were induced by TAM at E17.5. This induction scheme corresponds to Fezf2 expression at a time when the bulk of CPN are still completing axonal extension across the corpus callosum and represents one of the latest time points available to investigate a change in axon pathfinding. By P14, GFP-positive axon bundles were present extending through the internal capsule and reaching the thalamus and cerebral peduncle of mice that received DF-Fezf2eGFP but not in DF-CtleGFP controls (Fig. 5d and data not shown). Together, the data indicate that CPN retain the ability to reprogram their axonal projections in response to a change of identity imposed postmitotically. The complexity of axon guidance decisions likely required to achieve such new long-distance connectivity provides further support of the molecular data indicating that a series of neuron subtype-specific genes, rather than individual molecules, is regulated as a result of reprogramming.
Recent work has demonstrated that differentiated cell types can be lineage reprogrammed to acquire a new cellular identity upon expression of transcription factors of alternative cell fates4–6,9–11,32. In the CNS, neurogenic transcription factors can reprogram cultured postnatal astrocytes into neural progenitors and neurons33–35. Pericytes isolated from the human cerebral cortex have also been recently shown to reprogram into neurons36. However, to what extent neurons have the potential to reprogram their identity is largely unknown. Here, we provide the first demonstration that in the neocortex a conversion between two subtypes of neurons can occur during a temporal window of embryonic and early postnatal development.
The data indicate that critical features of neuronal subtype-specific identity, including the acquisition of a new molecular identity and the ability to extend axons to a new target, can still be changed postmitotically, even several days after initial fate-specification. These changes occurred despite the ectopic location of the CFuPN-like neurons and through the use of a single transcription factor, suggesting that higher reprogramming efficiency might be obtained in more instructive environments or by combining Fezf2 with additional fate modifying signals. Importantly, the number of CPN able to reprogram is highest during the earliest stages of postmitotic development and, while this capacity persists at P3, it is eliminated by P21. This suggests that mechanisms are in place postmitotically to progressively restrict neuronal fate potential as neurons age. The work indicates that postmitotic neurons can undergo lineage reprogramming in vivo and paves the way for mechanistic studies aimed at enhancing the process of neuronal conversion by extending the period of neuronal reprogramming to the mature brain.
Methods
In utero electroporation
The Cdk5r, Dcx and Nestin minimal promoters used here have been previously described19. The Cdk5r-CtleGFP, Dcx-CtleGFP, Cdk5r-Fezf2eGFP and Dcx-Fezf2eGFP constructs were generated by subcloning an IRES-GFP or Fezf2-IRES-GFP cassette downstream of the Cdk5r and Dcx promoters. The DF-CtleGFP and DF-Fezf2eGFP constructs were generated by subcloning a GFP or GFP-T2A-Fezf2 cassette between two pairs of inverted LoxP sites within the AAV-DIO-ChR2-mCherry construct described previously37. CAG-ERT2-Cre-ERT2 has also been previously described25. In vivo electroporations were performed according to a previously published protocol17,38. Briefly, 800 μl of purified DNA (2μg/μl) mixed with 0.005% Fast Green in sterile PBS was injected in utero into the lateral ventricle of CD1 embryos (Charles River Laboratory) under ultrasound guidance (Vevo 770, VisualSonics). Five 35 volt pulses of 50 ms duration at 1 second intervals were delivered in appropriate orientation across the embryonic head using 1 cm-diameter platinum electrodes placed outside the uterus and a CUY21EDIT square wave electroporator (Nepa Gene). Embryonic TAM injections were performed as previously described25. P3 pups received a single dose of 0.5 mg of TAM (100 μl at 5 mg/ml) intra peritoneum (IP) and P21 mice received a single dose of 2 mg of TAM (200 μl at 10 mg/ml) IP. VPA-injected animals received two doses of 0.5 mg each (100 μl at 5 mg/ml) IP at P5 and P7. All animal studies were approved by the Massachusetts General Hospital and Harvard University Institutional Animal Care and Use Committee and performed in accordance with institutional and federal guidelines.
Immunocytochemistry and in situ hybridization
Brains for immunocytochemistry were processed as previously described14. Primary antibodies and dilutions were as follows: rabbit anti-ZFPM2 antibody16, 1:500 (Santa Cruz, sc-10755); rat anti-CTIP214 antibody, 1:1000 (Abcam, ab18465); rabbit anti-GFP antibody, 1:500 (Invitrogen, A-11122); rat anti-GFP antibody, 1:500 (Nacalai, 04404–84); rabbit anti-ER81 antibody22, 1:2500 (gift of Silvia Arber); rabbit anti-CRYM antibody14, 1:100 (Abcam, ab54669); rabbit anti-TLE4 antibody12, 1:100 (Santa Cruz, sc-9525); rabbit anti-cleaved Caspase-3, 1:400 (Cell Signaling Technology, 9661S). Appropriate secondary antibodies were from the Molecular Probes Alexa series and the Vectastain ACB system (Vector Labs). Tissue sections were imaged using a Nikon 90i fluorescence microscope equipped with a Retiga Exi camera (Q-IMAGING) and analyzed with Volocity image analysis software v6.0.1 (Improvision). Confocal images were obtained using a Nikon A1 confocal (Eclipse Ti) and a Zeiss 700 Confocal. Images were collected and analyzed with the NIS-Elements imaging (Nikon) and Zen 2010 programs, respectively. Nonradioactive in situ hybridizations were performed on 40 μm vibratome sections mounted on superfrost slides (Fisher) using reported methods39. Riboprobes were generated as previously described14. cDNA clones for Clim1, Opsin3, Crim1 and Gfp have been previously described14,18. cDNA clones to detect endogenous Fezf2 transcript were generated by RT-PCR using the following primer pairs: 5′-TCTTCCTGCCCTGTACCAAC-3′ and 5′-CCGGTGTCCCAAGAAAGTTA-3′.
Callosotomy
Callosotomy was performed at P3. Pups were anesthetized on wet ice and their head immobilized. The non-electroporated hemisphere was exposed and a 30G needle was inserted below the cortex starting at lambda and immediately lateral to the midline. The corpus callosum was then sectioned between bregma and lambda. Pups were resuscitated by bringing their body temperature to physiological levels.
Cell quantification and connectivity
For quantification of CFuPN and CPN marker gene expression, anatomically matched sections from Cdk5r-CtleGFP and Cdk5r-Fezf2eGFP were selected and processed by immunohistochemistry for subtype-specific markers (P7: n=5 mice, 4,053 total neurons for Cdk5r-CtleGFP and n=6 mice, 4,649 total neurons for Cdk5r-Fezf2eGFP; P14: n=3 mice, 2,608 total neurons for Cdk5r-CtleGFP and n=2 mice, 1,514 total neurons for Cdk5r-Fezf2eGFP; P28: n=2 mice, 1,182 total neurons for Cdk5r-CtleGFP and n=3 mice, 1,557 neurons for Cdk5r-Fezf2eGFP), DF-CtleGFP and DF-Fezf2eGFP with TAM injection at P3 (n=6 mice for DF-CtleGFP and for DF-Fezf2eGFP, n=2 mice for DF-Fezf2eGFP with callosotomy and n=3 mice for DF-CtleGFP with callosotomy, DF-CtleGFP with VPA and DF-Fezf2eGFP with VPA; 1,761 total neurons for DF-CtleGFP; 3,580 total neurons for DF-Fezf2eGFP; 2,271 total neurons for DF-CtleGFP with callosotomy; 2,199 total neurons for DF-CtleGFP with VPA; 1,243 total neurons for DF-Fezf2eGFP with callosotomy and 2,175 total neurons for DF-Fezf2eGFP with VPA) and DF-CtleGFP and DF-Fezf2eGFP with TAM injection at P21 (n=3 mice for DF-CtleGFP harvested at P28 and P42 and for DF-Fezf2eGFP harvested at P42 ; n=2 mice for DF-Fezf2eGFP harvested at P28; 1,633 and 1,123 total neurons for DF-CtleGFP harvested at P28 and P42, respectively and 3,021 and 2,035 for DF-Fezf2eGFP harvested at P28 and P42, respectively). Only GFP-positive neurons located in layer II/III and within matched areas were quantified. All the results are expressed as the mean ± s.e.m. The paired, two-tailed t test was used for statistical analysis. For analysis of long-distance connectivity, electroporated embryos were sacrificed at E18.5, P7, P14 and P28 for processing. eGFP-positive axons where visualized by immunocytochemistry for GFP and analyzed in coronal sections of the brain and spinal cord at different rostrocaudal locations.
Supplementary Material
Acknowledgments
We thank Doug Melton, Andy McMahon, Jeffrey Macklis, Bradley Molyneaux and Edward Stronge for their advice and comments on the manuscript; Connie Cepko, Silvia Arber, Feng Zhang and Qiang Lu for sharing of antibodies and expression vectors; Mohammed Mostajo Radji for help with data analysis; Amanda Merlino and Zachary Trayes-Gibson for technical support; and Claudio Mare for schematic drawings. This work was supported by the US National Institute of Health (NS062849) and the Harvard Stem Cell Institute to P.A.; C.R. was partially supported by a Milton-Safenowitz postdoctoral fellowship from the ALS Association.
References
- 1.Waddington CH. The strategy of the genes; a discussion of some aspects of theoretical biology. Allen & Unwin; 1957. [Google Scholar]
- 2.Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- 3.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Selvaraj V, Plane JM, Williams AJ, Deng W. Switching cell fate: the remarkable rise of induced pluripotent stem cells and lineage reprogramming technologies. Trends Biotechnol. 2010;28:214–223. doi: 10.1016/j.tibtech.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Eggan K, et al. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Ishii T, Feinstein P, Mombaerts P. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature. 2004;428:393–399. doi: 10.1038/nature02433. [DOI] [PubMed] [Google Scholar]
- 9.De Marco Garcia NV, Jessell TM. Early motor neuron pool identity and muscle nerve trajectory defined by postmitotic restrictions in Nkx6.1 activity. Neuron. 2008;57:217–231. doi: 10.1016/j.neuron.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- 11.Thaler JP, et al. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- 12.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 13.Bayer SA, Altman J. Neocortical Development. Raven Press; 1991. [Google Scholar]
- 14.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 18.Rouaux C, Arlotta P. Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nat Neurosci. 2010;13:1345–1347. doi: 10.1038/nn.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Qiu R, Tsark W, Lu Q. Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res. 2007;85:3567–3573. doi: 10.1002/jnr.21440. [DOI] [PubMed] [Google Scholar]
- 20.Delalle I, Bhide PG, Caviness VS, Jr, Tsai LH. Temporal and spatial patterns of expression of p35, a regulatory subunit of cyclin-dependent kinase 5, in the nervous system of the mouse. J Neurocytol. 1997;26:283–296. doi: 10.1023/a:1018500617374. [DOI] [PubMed] [Google Scholar]
- 21.Tsai LH, Delalle I, Caviness VS, Jr, Chae T, Harlow E. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature. 1994;371:419–423. doi: 10.1038/371419a0. [DOI] [PubMed] [Google Scholar]
- 22.Yoneshima H, et al. Er81 is expressed in a subpopulation of layer 5 neurons in rodent and primate neocortices. Neuroscience. 2006;137:401–412. doi: 10.1016/j.neuroscience.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 23.Molyneaux BJ, et al. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fishell G, Hanashima C. Pyramidal neurons grow up and change their mind. Neuron. 2008;57:333–338. doi: 10.1016/j.neuron.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huangfu D, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 27.Fame RM, MacDonald JL, Macklis JD. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozaki HS, Wahlsten D. Timing and origin of the first cortical axons to project through the corpus callosum and the subsequent emergence of callosal projection cells in mouse. J Comp Neurol. 1998;400:197–206. doi: 10.1002/(sici)1096-9861(19981019)400:2<197::aid-cne3>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Francis F, et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 30.Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 31.Matsuo N, Kawamoto S, Matsubara K, Okubo K. Cloning and developmental expression of the murine homolog of doublecortin. Biochem Biophys Res Commun. 1998;252:571– 576. doi: 10.1006/bbrc.1998.9698. [DOI] [PubMed] [Google Scholar]
- 32.Rouaux C, Bhai S, Arlotta P. Programming and reprogramming neuronal subtypes in the central nervous system. Dev Neurobiol. 2012 doi: 10.1002/dneu.22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berninger B, et al. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinrich C, et al. Directing Astroglia from the Cerebral Cortex into Subtype Specific Functional Neurons. Plos Biol. 2010;8 doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heins N, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 36.Karow M, et al. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell. 2012;11:471–476. doi: 10.1016/j.stem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Cardin JA, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 39.Lodato S, et al. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69:763–779. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.