SUMMARY
A simple epithelium forms a barrier between the outside and the inside of an organism, and is the first organized multicellular tissue found in evolution. We examine the relationship between the evolution of epithelia and specialized cell-cell adhesion proteins comprising the classical cadherin/β-catenin/α-catenin complex (CCC). A review of the divergent functional properties of the CCC in metazoans and non-metazoans, and an updated phylogenetic coverage of the CCC using recent genomic data reveal: 1) The core CCC likely originated before the last common ancestor of unikonts and their closest bikont sister taxa. 2) Formation of the CCC may have constrained sequence evolution of the classical cadherin cytoplasmic domain and β-catenin in metazoa. 3) The α-catenin binding domain in β-catenin appears to be the favored mutation site for disrupting β-catenin function in the CCC. 4) The ancestral function of the α/β-catenin heterodimer appears to be an actin-binding module. In some metazoan groups, more complex functions of α-catenin were gained by sequence divergence in the non-actin binding (N-, M-) domains. 5) Allosteric regulation of α-catenin, rather than loss of function mutations, may have evolved for more complex regulation of the actin cytoskeleton.
Keywords: Protein Evolution, Metazoan Evolution, Epithelia, Cell-cell Adhesion, Adherens Junction, α-catenin, β-catenin, Classical Cadherin, Actin Cytoskeleton, Vinculin
I. INTRODUCTION
A simple epithelium is a conserved feature of all metazoans and is essential for organized multicellularity. It is comprised of a closed monolayer, often a tube, of polarized cells that surround a luminal space (Figure 1A), thus separating the inside of the organism from its surrounding environment. The cytoskeleton, cytoplasmic organelles, and plasma membrane domains are organized asymmetrically, with the apical plasma membrane facing the luminal space and the basolateral membrane contacting opposing cells and an extracellular matrix (ECM) (Bryant and Mostov, 2008; Gumbiner, 2005; Nelson et al., 2013). Cell-cell adhesion complexes hold epithelial cells together, and an ECM surrounds epithelial tubes. Disruptions in epithelial polarity and cell-cell adhesion cause developmental defects and are found in diseases in adult tissues (Benjamin and Nelson, 2008; Bullions et al., 1997; Kane et al., 1996; Larue et al., 1996; Larue et al., 1994; Marchiando et al., 2010; Stepniak et al., 2009; Torres et al., 1997; Watabe et al., 1994).
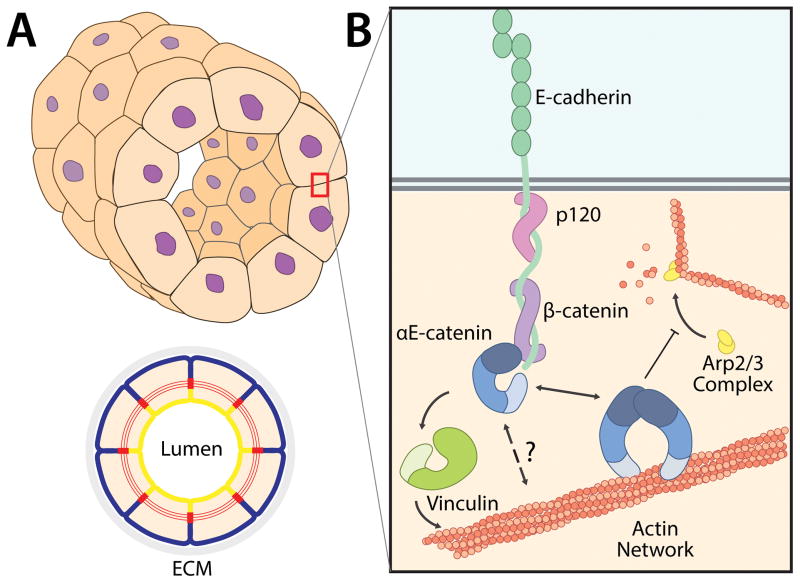
Figure 1. Cadherin/Catenin Complex at Mammalian Cell-Cell Contacts.
(A) A simple, tubular epithelium composed of a closed monolayer of polarized cells. The schematic, below demonstrates cell polarity: the apical membrane (yellow) faces the lumen and the basolateral membrane (blue) contacts the underlying ECM and serosa (grey). Cells have a cortical actin belt, which is connected between cells at adherens junctions (red).
(B) The cadherin-catenin complex mediates cell-cell adhesion. Classical cadherins contain extracellular repeat domains that mediate trans-interactions with the extracellular domain of cadherins on opposing cells, and a cytoplasmic domain that binds p120 and β-catenin. Monomeric α-catenin localizes to the cadherin-catenin complex by binding β-catenin. The mechanism by which α-catenin associates with the actin cytoskeleton is not well understood. Vinculin binds both α-catenin and actin, and may facilitate linkage between the cadherin-catenin complex and actin. Homodimeric α-catenin binds and bundles actin filaments and inhibits Arp2/3 complex-mediated nucleation of actin.
Simple epithelia predate the origin of metazoans and are the first organized tissues found in evolution. They are present in two distinct lineages of unikonts: the amoebozoans (Dictyostelium slime molds and their relatives), and the opisthokonts (eukaryotes) that include metazoans, choanoflagellates and fungi, which are thought to have developed multicellularity independently. First, the amoebozoan Dictyostelium discoideum requires a polarized tip epithelium to form a fruiting body from aggregated amoebae (Dickinson et al., 2011a). Second, simple epithelia constitute the core tissues of all metazoans: the feeding chambers of porifera (sponges) are lined with an epithelium (choanoderm) comprised of polarized choanocytes that directionally absorb nutrients from seawater (Leys and Hill, 2012), and the placozoan Trichoplax adhaerens consists of several thousand cells arranged in an epithelial bilayer of which the ventral layer is required for nutrient absorption (Schierwater et al., 2009; Srivastava et al., 2008). In morphologically complex animals, such as mammals, epithelia define tissue architecture and regulate functionally diverse organs such as the lung, gut, kidney, and epidermis. Thus, formation of a simple polarized epithelium is a principal requirement for the evolution of organized multicellularity and the functional diversification of tissues (Cereijido et al., 2004).
Classical cadherins are the primary molecules that mediate epithelial cell-cell adhesion in metazoans (Halbleib and Nelson, 2006; Harris and Tepass, 2010). Classical cadherins are defined by a cytoplasmic domain that binds adaptor proteins (catenins) that regulate downstream signaling and actin cytoskeleton dynamics. Classical cadherins establish cell-cell contacts, often in a discrete structure termed the Adherens Junction (AJ) located at the boundary between the apical and basolateral membrane domains (Figure 1A) (Nelson, 2003). AJs are linked to a circumferential actomyosin belt, which generates dynamic forces important for epithelial sheet remodeling and tissue morphogenesis, and epithelial tissue integrity (Costa et al., 1998; Nagafuchi et al., 1994; Wessells et al., 1971). During cell-cell contact formation, the actin and microtubule cytoskeletons are remodeled to mechanically strengthen contacts, facilitate polarized vesicle trafficking, and maintain cell shape (Adams and Nelson, 1998; Mellman and Nelson, 2008; Nejsum and Nelson, 2007).
The evolution of cadherin-mediated cell-cell adhesion coincided with the formation of different body plans derived from epithelial sheets. Members of the cadherin and catenin protein families [the cadherin-catenin complex (CCC)] are present in all metazoans and many pre-metazoan unikonts (Abedin and King, 2008; Hulpiau et al., 2013; Hulpiau and van Roy, 2009; Oda and Takeichi, 2011) (this study). Non-metazoan lineages do not possess a complete set of CCC protein orthologs, and the ancestral function of these cell-cell adhesion proteins in unicellular organisms is unclear. Furthermore, recent functional studies demonstrate divergent properties of the CCC within bilaterians (Desai et al., 2013; Dickinson et al., 2011b; Drees et al., 2005; Kwiatkowski et al., 2010; Miller et al., 2013).
The CCC mechanically couples neighboring cells by trans interactions between cadherins on opposing cells and linkage to the underlying actin cytoskeletons (Huveneers and de Rooij, 2013; Shapiro and Weis, 2009). In general, the cadherin protein superfamily consists of transmembrane proteins that contain extracellular cadherin repeat domains (CADs) (Boggon et al., 2002; Shapiro and Weis, 2009). Adhesive contacts between classical cadherins require extracellular Ca2+ for protein confirmation (Koch et al., 1997), and are mediated by a strand swap dimer formed between the opposed N-terminal EC1 domains (Harrison et al., 2011). Classical cadherins have a conserved cytoplasmic domain that binds catenins (Huber and Weis, 2001; Hulpiau and van Roy, 2009). The catenins are responsible for transducing force and molecular signals from the CCC to the actin cytoskeleton (Borghi et al., 2012; Weis and Nelson, 2006; Yonemura et al., 2010). The armadillo repeat family proteins p120-catenin and β-catenin bind directly to the cadherin cytoplasmic domain; p120-catenin interacts with Rho GTPases that control cytoskeletal dynamics, and regulates cadherin endocytosis (for recent reviews see (Davis et al., 2003; Pieters et al., 2012)) and will not be discussed further (for recent reviews see (Carnahan et al., 2010; Menke and Giehl, 2012; Pieters et al., 2012)). In turn, β-catenin binds to α-catenin (Herrenknecht et al., 1991) thereby forming the core cytoplasmic protein complex of the CCC (Figures 1B, 2A).
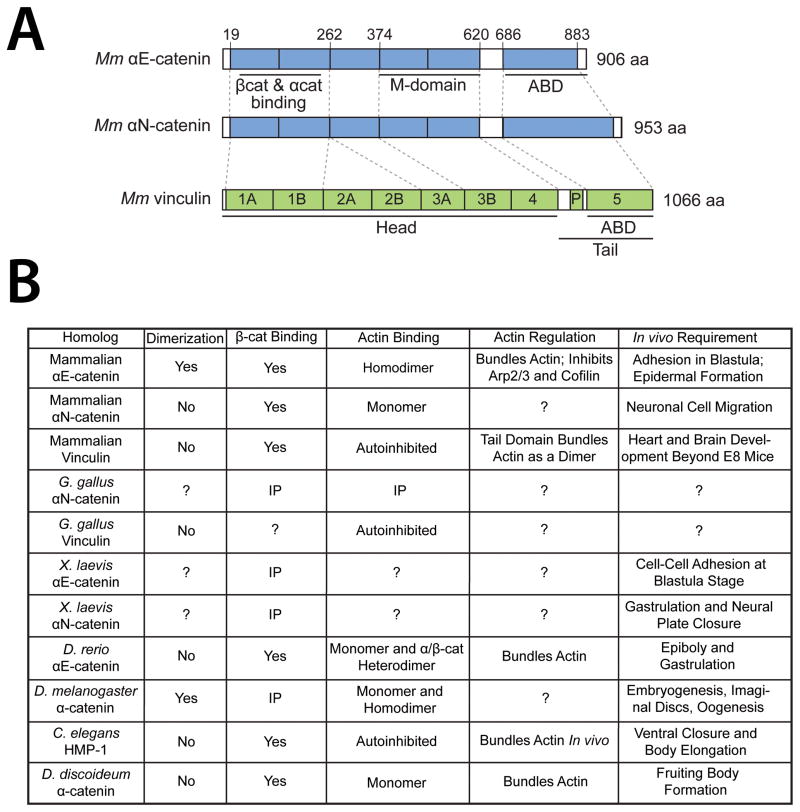
Figure 2. Functional Properties of α-Catenin/Vinculin Family Proteins.
(A) Domain organization of mammalian α-catenin/vinculin family proteins. Mammalian vinculin is composed of 7 four-helix bundles, a proline-rich hinge region, and a C-terminal five-helix bundle. α-Catenins share a similar structure but lack the D2 domain. Head, tail, and actin-binding domains of vinculin as well as β-catenin binding/dimerization, modulation, and F-actin binding domains in Mm αE- and αN-catenin are annotated. Regions of homology are indicated by dashed lines. Adapted from (Miller et al., 2013)
(B) Functional properties of characterized α-catenin/vinculin family proteins across unikonta. Homodimerization, β-catenin binding, and F-actin binding and regulation in vitro using purified proteins is indicated. Indirect evidence of binding by coimmunoprecipitation (IP) is also noted. Developmental and/or in vivo requirement for each homolog is summarized on the right. Question marks signify untested properties or inconclusive data.
α-Catenin is an F (filamentous)-actin-binding protein (Rimm et al., 1995), and is a key protein in the CCC that links cadherin-mediated cell-cell contacts to the underlying actin cytoskeleton. α-Catenin is a paralog of vinculin, which is an F-actin-binding protein at cell-ECM and cell-cell adhesions (Peng et al., 2011); we refer to α-catenin/vinculin proteins as VIN-family proteins. Mammalian αE-catenin is composed of a series of four-helix bundles connected to a C-terminal five-helix bundle, and the conformation and accessibility of these domains regulate αE-catenin function (Ishiyama et al., 2013; Pokutta et al., 2002; Pokutta and Weis, 2000; Rangarajan and Izard, 2013; Yang et al., 2001; Choi et al., 2012) (Figure 2A). Mammalian αE-catenin has binding sites for β-catenin and F-actin in the N-terminal and C-terminal domains, respectively (Figure 2A). Mammalian αE-catenin also binds several F-actin-binding proteins, including vinculin (Watabe-Uchida et al., 1998), α-actinin (Knudsen et al., 1995), ZO-1 (Itoh et al., 1997; Maiers et al., 2013), l-afadin (Pokutta et al., 2002), and EPLIN (Abe and Takeichi, 2008); whether non-mammalian α-catenin orthologs bind these proteins has not been studied. Mammalian αE-catenin bundles actin filaments (Rimm et al., 1995), and inhibits Arp2/3-mediated nucleation of actin filament assembly (Drees et al., 2005) (Figure 1B).
Here, we focus on evolution of the core components of the CCC - classical cadherins, β-catenin and α-catenin – with particular focus on α-catenin. The evolution of classical cadherins and β-catenin has been reviewed (Abedin and King, 2008; Hulpiau et al., 2013; Hulpiau and van Roy, 2009; Schneider et al., 2003). First, we synthesize recent structure-function studies of α-catenin/vinculin family proteins across unikonta (Section II. Functional Analysis of CCC Evolution). Second, we use bioinformatic analysis to identify putative orthologs of the core CCC components by sequence alignment and domain architecture (Section III. Genomic Analysis of CCC Evolution). Finally, we combine information about the divergence of α-catenin/vinculin function with bioinformatic observations to provide new insights into how the CCC may have evolved (Section IV. Conclusion and Synthesis).
II. FUNCTIONAL ANALYSIS OF CCC EVOLUTION
Functional characterization of the α-catenin/vinculin family across unikonta
Mammals
Mammals possess three isoforms of α-catenin, termed αE-, αN-, and αT-catenin, which originated from the same ancestral gene and share the same location on human chromosome 10 (Janssens et al., 2003). αE-, αN-, and αT-catenin are expressed predominantly, but not exclusively, in epithelia, neurons, and heart/testis, respectively (Herrenknecht et al., 1991; Shapiro and Weis, 2009; Uchida et al., 1994). Mammals also express vinculin, which localizes to both integrin-ECM adhesions and AJs (le Duc et al., 2010; Peng et al., 2011; Watabe-Uchida et al., 1998; Ziegler et al., 2006). The functional properties of mammalian αE-catenin have been characterized in detail (Figures 1B, 2B; see below). Less is known about αN-catenin, and we are not aware of any detailed biochemical analysis of αT-catenin.
αE-Catenin is essential for the formation of epithelia and morphogenetic cell movements during mammalian development. Mus musculus null mutants of αE-catenin (Mm αE-catenin) loose cell-cell adhesion in the trophoblast epithelium of the preimplantation embryo, and development is arrested at the blastula stage (Torres et al., 1997). Conditional knockout of Mm αE-catenin in keratinocytes disrupts AJ formation and is embryonic lethal due to epithelial hyperproliferation and tumor formation (Vasioukhin et al., 2001).
Mm αE-catenin contains 3 proteolytically-defined domains (Figure 2A) homologous to the domain structure of vinculin: an N-terminal domain (NTD) that mediates homodimerization and β-catenin binding, a conformationally flexible M-domain that interacts with several actin binding proteins (see above), and a C-terminal domain that binds F-actin (ABD) (Aberle et al., 1996; Herrenknecht et al., 1991; Pokutta and Weis, 2000; Rangarajan and Izard, 2013; Yang et al., 2001).
Mm αE-catenin exists in three oligomeric states; a monomer, an αE-catenin/β-catenin heterodimer, and a homodimer (Drees et al., 2005) (Figure 2B). All of these oligomeric forms are found in cell extracts from MDCK epithelial cells (Benjamin et al., 2010). The Kd for binding between β-catenin and Mm αE-catenin is 25 –100 nM (Koslov et al., 1997) (Figure 2B; Sabine Pokutta and W.I.W., unpublished data). The Kd for Mm αE-catenin homodimerization is weaker than for αE-catenin-β-catenin binding and is in the single micromolar range (Shapiro and Weis, 2009) (Sabine Pokutta and W.I.W, unpublished data).
Mammalian αE-catenin binds F-actin (Rimm et al., 1995), but this interaction is regulated by αE-catenin conformation (Figure 2B) (Drees et al., 2005). The Mm αE-catenin monomer binds F-actin weakly (Drees et al., 2005; Yamada et al., 2005), whereas homodimerization potentiates actin binding (Kd 0.3 μM) (Drees et al., 2005; Rangarajan and Izard, 2013; Rimm et al., 1995). Since the αE-catenin homodimerization and β-catenin binding sites overlap (Pokutta and Weis, 2000), αE-catenin can bind β-catenin as a monomer or F-actin as a homodimer, but the E-cadherin/β-catenin/αE-catenin complex binds F-actin weakly in bulk assays in vitro (Yamada et al., 2005) (Figure 2A).
The weak binding of Mm CCC to F-actin led to a model of αE-catenin function in which clustering of the CCC at cell-cell contacts would produce a high local concentration of actin-binding αE-catenin homodimers, thereby facilitating dynamic interactions between the CCC and actin (Yamada et al., 2005) (Figure 3C). Recent work, however, demonstrated that the CCC is under constitutive actomyosin-generated tension (Borghi et al., 2012), which could regulate αE-catenin conformation at cell-cell adhesions (Yonemura et al., 2010). Thus, an alternative model is that cytoplasmic forces relieve αE-catenin auto-inhibition such that the αE-catenin monomer can directly couple the CCC to the cortical actin cytoskeleton. Further work is needed to test this model and define how the CCC interacts with the actin cytoskeleton.
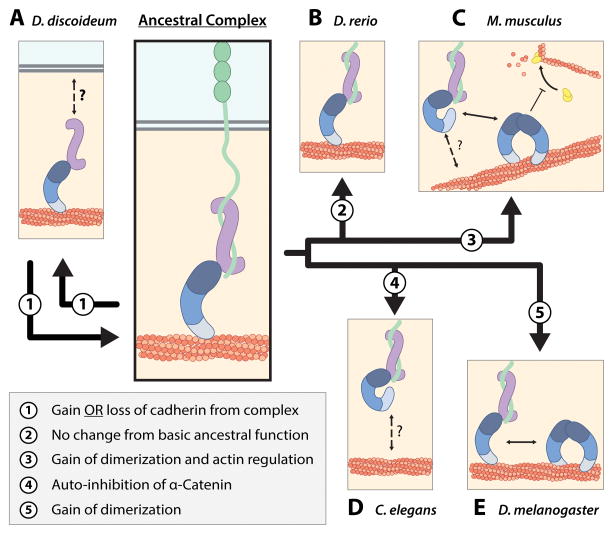
Figure 3. Functional Divergence of Cadherin-Catenin Complex.
(A) We propose that ancestral cadherin-catenin complex interactions consisted of monomeric actin-binding α-catenin that coupled the classical cadherin-β-catenin complex to the cortical actin network (center panel). Divergence of the complex in each species from the ancestral complex is depicted in the box on bottom left. D. discoideum α-catenin is an monomer that forms a heterodimer complex with Aardvark (β-catenin homolog), and localizes to cell-cell contacts and the cortical actin cytoskeleton. Dd α-catenin bundles actin, but the mechanism by which the Dd α-catenin-Aardvark complex associates with the cell membrane is not known. D. discoideum does not possess cadherin homologs and the adhesion proteins to which the heterodimer complex is attached at cell-cell contacts have not been identified.
(B) D. rerio αE-catenin is a monomer that is not auto-inhibited, and can simultaneously bind β-catenin and F-actin, but does not regulate actin dynamics via Arp2/3 inhibition.
(C) The mammalian cadherin-catenin complex possesses divergent regulatory properties from the ancestral complex. Monomeric αE-catenin associates with the E-cadherin-β-catenin complex, but is allosterically regulated and binds actin weakly. The mechanism by which the cadherin-catenin complex is linked to the cortical actin cytoskeleton is poorly understood. Homodimerization of αE-catenin potentiates bundling of actin and inhibition of Arp2/3 complex-mediated actin polymerization.
(D) C. elegans HMP-1 forms a ternary complex with HMR-1 (classical cadherin homolog) and HMP-2 (β-catenin homolog), but is autoinhibited and cannot bind F-actin in vitro. The mechanism by which actin binding is activated in vivo is not known.
(E) D. melanogaster α-catenin exists as monomer and homodimer species, both of which bind actin. D. melanogaster α-catenin forms a ternary complex with DE-cadherin and Armadillo.
Mm αE-catenin also regulates actin dynamics independently of its role in the CCC. Mm αE-catenin homodimers bundle actin filaments (Rimm et al., 1995), and inhibit Arp2/3 complex-mediated nucleation of F-actin in vitro (Drees et al., 2005) (Figures 2B, 3C). Regulation of actin dynamics by cytoplasmic αE-catenin in vivo is important: depletion of the cytosolic pool of αE-catenin homodimers increases actin-dependent membrane dynamics and cell migration rate of MDCK epithelial cells (Benjamin et al., 2010), and deletion of αE-catenin from keratinocytes in M. musculus embryos results in a hyper-migratory phenotype (Vasioukhin et al., 2001).
Less is known about the functional and biochemical properties of αN-catenin. Two splice variants of αN-catenin have been identified (Uchida et al., 1994), but their significance is unknown. M. musculus null mutants of αN-catenin have defects in proper cell layering in the cerebellum and hippocampus (Park et al., 2002). This phenotype may be due to defects in neuronal cell migration and cell-cell contacts, since depletion of Mm αN-catenin from dendritic spines increases membrane activity while αN-catenin over-expression reduces membrane dynamics (Abe et al., 2004). Although Mm αN-catenin appears to regulate cell-cell adhesion and cell migration like αE-catenin, initial studies indicate that it has different biochemical properties. Multi-angle light scattering and small-angle X-ray scattering show that Mm αN-catenin is monomeric with a less compact conformation than Mm αE-catenin (Ishiyama et al., 2013). However it remains unclear if Mm αN-catenin homodimerizes at higher concentrations or whether it is allosterically regulated like Mm αE-catenin. Mm αN-catenin binds β-catenin and F-actin (Ishiyama et al., 2013) (S. Pokutta and WIW, unpublished data), but it is not known whether it binds simultaneously to β-catenin and the actin cytoskeleton. Therefore, there are insufficient data to propose a model of Mm αN-catenin function in cadherin-mediated cell-cell adhesion.
The function of mammalian vinculin in cadherin-mediated adhesion is not well understood, compared to its well-known roles in integrin-mediated cell adhesion to the ECM (Bakolitsa et al., 2004; Plotnikov et al., 2012; Ziegler et al., 2006). Vinculin knockout mice exhibit gross defects in neural tube closure and heart development, and reduced cell-substrate adhesion to the ECM, but no specific defects in AJ formation or cell-cell contacts were described (Xu et al., 1998) (Figure 2B). Nevertheless, vinculin has biochemical properties similar to Mm αE-catenin. The N-terminal head domain of mammalian vinculin has binding sites for β-catenin and αE-catenin, and the C-terminal tail domain binds actin (Bakolitsa et al., 1999; Craig and Johnson, 1996; Janssen et al., 2006; Peng et al., 2010) (Figure 2). Vinculin can bundle actin filaments through homodimerization of the C-terminal tail domain (Janssen et al., 2006), but the full length protein is autoinhibited by a strong intramolecular head-tail interaction (Bakolitsa et al., 2004; Choi et al., 2012; Rangarajan and Izard, 2012; Ziegler et al., 2006) (Figure 2B). It has been proposed that vinculin mediates binding between αE-catenin at the CCC and the cortical actin cytoskeleton (Huveneers and de Rooij, 2013; Peng et al., 2011; Yonemura et al., 2010), and Mm αE-catenin can activate vinculin binding to actin in vitro (Choi et al., 2012). Further work is needed to define how αE-catenin and vinculin auto-inhibition is relieved, and how these paralogs bind the CCC to the actin cytoskeleton in vivo.
Gallus gallus
Like mammals and other amniotes, G. gallus (chicken) expresses αE-, αN-, and αT-catenin isoforms, and a vinculin. There are no published functional or in vivo studies of Gg αE- or αT-catenin. Gg αN-catenin and vinculin have been used in place of their mammalian orthologs in several studies and exhibit conserved functional properties. Gg αN-catenin co-immunoprecipitates with β-catenin and actin (Figure 2B) (Hirano and Takeichi, 1994), and can rescue cadherin-mediated cell-cell adhesion in human lung adenocarcinoma cells, which express E-cadherin and β-catenin but not α-catenin (Hirano and Takeichi, 1994). Functional properties of Gg vinculin are also conserved with mammals: Gg vinculin binds αE-catenin and actin in vitro, and is auto-inhibited by strong head–tail domain interactions (Figure 2B) (Chen et al., 2006; Choi et al., 2012).
Xenopus laevis
Amphibians and other anamniote vertebrates possess two isoforms of α-catenin, αE-catenin and αN-catenin, and a vinculin. The function of both α-catenin isoforms has been investigated in X. laevis following α-catenin depletion with morpholino antisense oligonucleotides, but direct biochemical studies of interactions between α-catenin and other CCC members have not been performed. Xl vinculin has not been studied.
Xl αE-catenin co-localizes with E-cadherin at cell-cell contacts in early embryos and is redistributed to the cytoplasm upon depletion of E-cadherin (Kurth et al., 1999). Xl αE-catenin morphants have defects in cell-cell adhesion at the blastula stage and are arrested in further development (Kofron et al., 1997) (Figure 2B). Xl αE-catenin and αN-catenin co-immunoprecipitate with β-catenin and E-cadherin in whole embryo lysates (Kurth et al., 1999; Nandadasa et al., 2012) (Figure 2B). Xl αN-catenin morphants and null mutants have defects in neural plate closure and gastrulation, respectively (Nandadasa et al., 2012; Sehgal et al., 1997). Whether Xl αE-catenin or αN-catenin binds actin or regulates cytoskeletal dynamics has not been studied.
Danio rerio
D. rerio (zebrafish) contains orthologs of αE- and αN-catenin, and vinculin. D. rerio αE-catenin (Dr αE-catenin) has been characterized in vivo by depletion with morpholinos (Schepis et al., 2012), and in vitro using purified recombinant proteins (Miller et al., 2013). Biochemical properties and cellular functions of D. rerio αN-catenin and vinculin have not been described.
Dr αE-catenin is expressed at cell-cell contacts between enveloping layer cells (EVL) and deep cells at the mid-blastula transition in wild-type embryos. αE-catenin morphants are delayed in epiboly due to defects in cell migration and adhesion in both the EVL and deep cells (Figure 2B) (Schepis et al., 2012). EVL cells become elongated at 80% epiboly in control embryos, whereas they retain a spherical morphology in αE-catenin morphants (Schepis et al., 2012). At 50% epiboly, deep cells in αE-catenin morphants display reverse radial intercalation at a rate approximately equal to normal radial intercalation (Schepis et al., 2012). Defects in radial intercalation may be related to increased plasma membrane blebbing and loss of cadherin-mediated adhesion in αE-catenin-depleted deep cells (Schepis and Nelson, 2012; Schepis et al., 2012). αE-catenin is required to maintain cortical tension and normal adhesive forces (Maitre et al., 2012). Taken together, these data indicate that αE-catenin regulates cell-cell adhesion and membrane dynamics by anchoring E-cadherin to the cortical actin cytoskeleton. Interestingly, Mm αE-catenin only partially rescues epiboly in Dr αE-catenin morphants (Schepis et al., 2012). This suggests that functional and/or regulatory properties of D. rerio and mammalian αE-catenin have diverged (see below).
Despite 90% amino acid sequence identity, D. rerio and Mm αE-catenin have surprisingly different biochemical properties (Miller et al., 2013). Dr αE-catenin is monomeric in solution (Figure 2B), and binds F-actin as a monomer or as a αE-catenin/β-catenin heterodimer and, therefore, is not autoinhibited (Figure 2B). However, Dr αE-catenin binds F-actin >20x more weakly when bound to β-catenin, similar to the Mm αE-catenin/β-catenin heterodimer (Miller et al., 2013). Dr αE-catenin bundles F-actin poorly compared to Mm αE-catenin and does not inhibit Arp2/3 complex nucleation of F-actin (Miller et al., 2013) (Figure 2B). Thus, Dr αE-catenin may directly link the CCC to the cortical actin cytoskeleton, and has a limited effect on actin dynamics (Figure 3B). This is consistent with in vivo data, in which depletion of Dr αE-catenin disrupts cortical tension (Maitre et al., 2012), and induces protracted membrane blebbing (Schepis and Nelson, 2012; Schepis et al., 2012) that does not involve the Arp2/3 complex (Charras and Paluch, 2008).
Drosophila melanogaster
D. melanogaster, like all other invertebrates so far examined, expresses one homolog of α-catenin and vinculin. Sequence analysis indicates that invertebrate α-catenin is most similar to vertebrate αN-catenin (Hulpiau and van Roy, 2009), although this has not been confirmed biochemically. D. melanogaster α-catenin (Dm α-catenin) has been studied extensively in vivo, but detailed biochemical experiments in vitro of the DE-cadherin/Armadillo (β-catenin)/α-catenin complex have not been performed. Deletion of vinculin, by inversion of the X-chromosome, results in viable and fertile adults indicating that it is not essential (Alatortsev et al., 1997).
Although the binding affinities between proteins in the Dm CCC have not been determined, it appears that Dm DE-cadherin, Armadillo, and α-catenin form a ternary complex at cell-cell junctions. Dm α-catenin localizes to cell-cell contacts throughout embryogenesis, and co-immunoprecipitates with Armadillo and DE-cadherin (Oda et al., 1993) (Figure 2B). Depletion of α-catenin disrupts embryogenesis, oogenesis and imaginal disc development (Sarpal et al., 2012), and causes the collapse of epithelia (Cavey et al., 2008). Dm α-catenin function in cell-cell adhesion requires binding to Armadillo (Desai et al., 2013), indicating that it may function as an anchor between DE-cadherin/Armadillo and the actin cytoskeleton (Sarpal et al., 2012). A DE-cadherin:α-catenin chimera can rescue cell-cell adhesion in an α-catenin weak allele background but the embryos die during larval stages (Sarpal et al., 2012), suggesting that DE-cadherin:α-catenin fusion proteins cannot replace all functions of endogenous α-catenin. Dm α-catenin mutants with deletions of either the N-terminal domain (NTD) or F-actin-binding domain (ABD) do not rescue embryos with a weak α-catenin allele (Desai et al., 2013). Interestingly, Desai et al. suggest that Mm αN-catenin can rescue a weak Dm α-catenin phenotype (Desai et al., 2013). Dm α-catenin also appears to regulate the dynamics of the actin cytoskeleton: RNAi-mediated depletion of α-catenin inhibits the accumulation of Rho GTPase near the apical membrane (Magie et al., 2002), and disruption of the Arp2/3 complex or its activator SCAR ameliorates the hypermigratory phenotype of a α-catenin weak allele (Sarpal et al., 2012).
Dm α-catenin is a monomer or a homodimer (Desai et al., 2013), but unlike Mm α-catenin, both monomeric and homodimeric forms of Dm α-catenin bind actin (Figure 2B, 3E) (Desai et al., 2013). While these results and the rescue experiments (above) indicate that Dm α-catenin directly links the CCC to the actin cytoskeleton (Figure 3E) this has not been confirmed biochemically. That Dm α-catenin homodimerizes indicates that it has a cytoplasmic function similar to Mm αE-catenin homodimer, since Mm αE-catenin homodimer cannot interact with β-catenin in the CCC.
Caenorhabditis elegans
C. elegans possesses α-catenin (HMP-1) and vinculin (DEB-1) orthologs which are expressed in different cell types (Hardin et al., 2013). HMP-1 has been investigated in loss of function mutants, by biochemical characterization in vitro, and through genetic dissection of actin binding. Little is known about the biochemical or cellular functions of DEB-1, although null mutations are lethal due to defects in body elongation and muscle development (Barstead and Waterston, 1989, 1991).
The C. elegans CCC comprises HMR-1 (E-cadherin), HMP-2 (β-catenin) and HMP-1, localizes to cell-cell contacts and the actin cytoskeleton, and is required for proper morphogenetic cell shape changes in early embryogenesis (Costa et al., 1998; Hardin et al., 2013). During epidermal morphogenesis, actomyosin-driven contractile forces are transmitted along circumferential actin filament bundles to cell-cell junctions and are required for cell shape changes and elongation of the embryo (Costa et al., 1998). In HMP-1 loss of function mutants, circumferential actin filament bundles are detached from the plasma membrane, which results in dorsal folding of the epidermis (the Humpback phenotype) (Costa et al., 1998) (Figure 2B). Strong loss of function HMP-1 mutants also display defects in closure of the ventral midline between ventral hypodermal cells and the posterior of the body (Costa et al., 1998).
Purified HMP-1 is a monomer even at high concentrations, but the full-length protein does not bind actin in vitro (Figure 2B), although the C-terminal ABD binds F-actin directly with an affinity similar to Mm αE-catenin homodimer (Kwiatkowski et al., 2010). This indicates that the ABD is inaccessible in the full-length protein, perhaps due to autoinhibitory interactions between the NTD and ABD. HMP-1 forms a ternary complex with HMP-2 and Casein kinase1-phosphorylated HMR-1, but this in vitro reconstituted CCC also does not bind actin (Kwiatkowski et al., 2010) (Figures 2B, 3D). In vivo, however, HMP-1 must bind both HMP-2 and actin directly as deletion of either the HMP-2 binding region or ABD recapitulates the Humpback phenotype (Kwiatkowski et al., 2010). HMP-1 mutants lacking residues 687–742, 802, or 826–927 in the C-terminal ABD have a phenotype similar to embryos lacking HMP-1 or HMP-2 (Maiden et al., 2013), indicating that these C-terminal amino acids are critical to HMP-1 interactions with the actin cytoskeleton. However, the regulatory mechanism involved in activating actin binding by HMP-1 in vivo remains unknown (Figure 2B, 3D).
Dictyostelium discoideum
The genome of the amoebozoan D. discoideum encodes two VIN proteins. One is a divergent sequence with a ~1000 amino acid insertion at the C-terminus that has not been characterized. The other is a putative homolog of α-catenin with equal sequence identity to both metazoan α-catenin and vinculin that has been characterized; this homolog is more like α-catenin than vinculin since it localizes to cell-cell contacts and not to cell-ECM contacts, and in vitro characterization of recombinant protein shows that it does not bind talin or form strong NTD-ABD interactions (Dickinson et al., 2011a).
D. discoideum lives as a unicellular amoeba, but upon starvation develops into a multicellular fruiting body through a process called culmination (Grimson et al., 2000; Schaap et al., 2006). The fruiting body comprises a spore head supported by a rigid vertical stalk and surrounded by a single layer of cells called the tip epithelium (Dickinson et al., 2011a). The tip epithelium has actin-associated intercellular junctions reminiscent of metazoan AJs (Grimson et al., 2000), and Dd α-catenin localizes to these cell-cell contacts (Dickinson et al., 2011a). D. discoideum possesses a protein with multiple armadillo repeats termed Aardvark (Grimson et al., 2000) that binds to Dd α-catenin through a sequence similar to Mm β-catenin (Dickinson et al., 2011a). Knockdown of either Aardvark or Dd α-catenin inhibits the organization of cells in culminants, resulting in the disruption of stalk and spore head formation (Dickinson et al., 2011a) (Figure 2B). Aardvark appears to mediate the association of Dd α-catenin and intercellular junctions, since Dd α-catenin does not localize to cell-cell contacts in the absence of Aardvark (Dickinson et al., 2011a). Thus in the context of binding Dd α-catenin, Aardvark has properties of a β-catenin homolog (Dickinson et al., 2011a). The presence of a polarized epithelium in D. discoideum that is organized by α-catenin and β-catenin homologs suggests that multicellularity may be a more ancient evolutionary development than previously thought (Dickinson et al., 2012).
D. discoideum is the only pre-metazoan organism in which a VIN protein has been characterized. Dd α-catenin is a monomer, and binds either Aardvark or Mm β-catenin in vitro indicating that this interaction is evolutionarily conserved (Dickinson et al., 2011a) (Figure 2B). Dd α-catenin is not autoinhibited, as the monomer binds and bundles F-actin, but it does not inhibit the Arp2/3 complex (Dickinson et al., 2011a) (Figure 2B). Currently, it is unclear how the Dd α-catenin/Aardvark complex couples the cortical actin cytoskeleton to the plasma membrane, since homologs of classical cadherins are absent in D. discoideum (Figure 3A). Nevertheless, like the monomeric Dr α-catenin homolog, Dd α-catenin appears to be an actin binding protein that links adhesive junctions and the actin cytoskeleton (Dickinson et al., 2011a).
Summary: an evolutionary perspective of how the CCC formed
The CCC has properties essential for organized multicellular development, tissue organization, and physiology of metazoans and, to date, at least one non-metazoan (an amoebozoan). The core CCC components - classical cadherins, β-catenin and α-catenin - are present in all metazoans, and it might be expected that there would be significant functional conservation between homologs. The biochemical properties of classical cadherins and β-catenin have not been studied in detail across a phylogenetically diverse range of organisms. Nevertheless, existing data indicate high conservation of homotypic interactions between cadherin extracellular cadherin repeat domains, binding between the cadherin cytoplasmic domain and the β-catenin armadillo repeat domain, and the formation of heterodimers between β-catenin and α-catenin (Abedin and King, 2008; Dickinson et al., 2011a; Grimson et al., 2000; Nichols et al., 2006; Nichols et al., 2012; Schneider et al., 2003). In contrast, α-catenin homologs have surprisingly divergent functional properties across metazoans and a non-metazoan, which raises 2 questions: 1) Did the CCC as a whole evolve different functions that were tailored to specific requirements in different organisms; or 2) Is α-catenin functionally more variable than other CCC components, and hence α-catenin variability evolved to regulate CCC interactions with the actin cytoskeleton?
Answers to these questions require analysis of CCC protein sequences during evolution. Therefore we screened for CCC proteins from phylogenetically diverse genomes to begin to reconstruct the ancient origins of the core components of the CCC, investigate whether domain organization was preserved between diverse homologs, and speculate whether molecular interactions between CCC members restricted or expanded their evolution.
III. GENOMIC ANALYSIS OF CCC EVOLUTION
Ancient origins of core cadherin-catenin complex components
Each core member of the CCC has characteristic domains: the extracellular cadherin repeat (CAD) of classical cadherins; the armadillo repeat domain of β-catenin comprised of multiple ARM repeats; and the vinculin/α-catenin (VIN) family proteins comprised of several helical bundle domains (Figure 2A). Protein sequences containing these domains are found in a diverse range of eukaryotes, bacteria and archaea, but outside the metazoa their presence seems to vary independently between clades (Figure 4A). Recent genomic evidence indicates that the core components of the CCC predate the origin of metazoa.
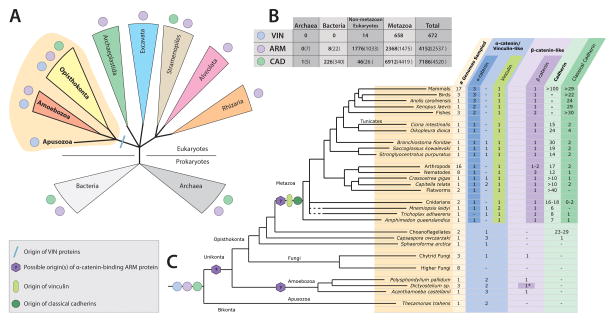
Figure 4. Distribution of Cadherin/Catenin Complex Proteins In Cellular Organisms.
(A) A cladogram indicating the presence of the VIN, ARM, and CAD proteins (blue, purple, and green circles, respectively) in bacteria, archaea, and major eukaryotic lineages. The blue line indicates the lineage-specific origin of VIN proteins.
(B) Numbers of CCC proteins found within each domain of life found in the pFam (in bold) and SMART (in parentheses) online databases (Letunic et al., 2009; Punta et al., 2012).
(C) Distribution of CCC proteins within metazoans and opisthokont and unikont outgroups, deliniating between true orthologs and similar sequences.
The presence or absence of these characteristic CCC protein domains within a proteome does not necessarily indicate they mediate protein-protein interactions and functions of the CCC (see Section II, above), but it does provide evidence for the potential evolutionary point of origin of each CCC component. To determine evolutionary relationships more definitively, bioinformatics analysis must be extended to the identification of putative orthologs, structure-based sequence alignment and domain architecture, and ultimately functional characterization. Here we report the presence of CCC protein domains across cellular organisms, with a number of putative orthologs found within opisthokonts and their near ancestors. We used non-exhaustive Hidden-Markov Model (HMM)–based searches for the pFam profiles for CAD, ARM, and VIN (PF00028, PF00514, and PF01044, respectively) (Punta et al., 2012).
The CAD and ARM repeat domains are found in a large number of proteins in highly variable contexts. CADs are adhesive domains responsible for hetero- and homotypic interactions in many trans-membrane and secreted proteins, and are dependent on calcium for their conformation and adhesive function (Harrison et al., 2011; Ivanov et al., 2001; Koch et al., 1997). Bacterial CAD proteins have calcium-dependent homotypic and heterotypic interactions in vitro, and bind to the cell surface when added exogenously to bacterial cultures (Fraiberg et al., 2010). Within metazoans, CADs are essential for cell-cell adhesion and specificity of cell-cell contacts (Hulpiau and van Roy, 2011; Oda and Takeichi, 2011; Wheelock and Johnson, 2003). An ARM domain comprises multiple copies of a 3-helix ARM repeat motif that associate to form a rigid protein-binding structure (Huber et al., 1997) involved in different cellular processes, including cytoskeleton organization, growth factor signaling, nuclear import, and gene transcription (Nelson and Nusse, 2004). ARM domains may contain a variable number of ARM repeats (Coates, 2003; Tewari et al., 2010). Multi-CAD and multi-ARM repeat proteins are found in small numbers in both archaea and bacteria, indicating that the domains themselves likely evolved prior to the divergence of the last common ancestor of eukaryotes and prokaryotes. Thus multi-CAD and multi-ARM repeat proteins were part of the ancestral proteome of cellular organisms (Figure 4A).
The vinculin-homology family (VIN) comprises a series of four-helix bundles connected to a C-terminal five-helix bundle. VIN proteins bind F-actin, and link actin filaments to proteins at the plasma membrane (see Section II, above). Multiple VIN-containing proteins are detected in the proteomes of all metazoans, and are found in some opisthokont relatives of metazoa (choanoflagellates, chytridomyctes), as well as in the nearest eukaryotic sister clades, apusozoa and amoebozoa (Abedin and King, 2008; Dickinson et al., 2011a; Grimson et al., 2000) (Figure 4C). A VIN has not been detected in organisms outside of this larger clade of opisthokonts and their near relatives, indicating a lineage-specific origin near the base of the eukaryotic branch leading to metazoans (Figure 4A).
The basic protein repertoire necessary for the assembly of the core CCC - a multi-CAD protein, a multi-ARM protein and a VIN protein - likely originated sometime before the last common ancestor of unikonts and their closest bikont sister taxa. Therefore, it can be inferred that in opisthokont clades, such as fungi and choanoflagellates, the absence of some or all of these proteins represents a loss of ancestral sequence diversity. It also indicates that the evolution of the CCC was not driven by the emergence of novel domains, but rather through domain rearrangement and the accumulation of functional interactions between CCC proteins within certain taxa.
Pre-metazoan assembly of a functional cadherin-catenin complex
Three changes in the CCC coincided with the advent of multicellularity: 1) Modification of the unstructured cytoplasmic domain of cadherin to contain a binding motif for β-catenin; 2) Gain of α-catenin and cadherin binding sites in an ARM protein (β-catenin); 3) Duplication of an ancestral VIN-containing protein resulting in separate α-catenin and vinculin protein families. When compared to the nearest outgroup to metazoa, the choanoflagellates, each of these changes appears to be an evolutionary gain of function, but their novelty becomes uncertain when examined in a broader context. First, a more complete search reveals that most non-metazoans have two or more VIN proteins, and that choanoflagellates are an exception (Figure 4C). Second, D. discoideum has a multi-ARM repeat protein, Aardvark, that binds to a α-catenin ortholog (Dickinson et al., 2011a). The presence of the β-catenin-binding motif in classical cadherins, however, appears to be a metazoan novelty. Still, the unexpected functional diversity of VIN-containing proteins (see Section II, above), and the observation of functional CCC protein interactions in an amoebozoan support the hypothesis that the core actin-binding functions of the CCC complex arose before the advent of metazoan multicellularity (Figure 4). Below, we discuss these trends in light of more complete genomic evidence.
Cadherins
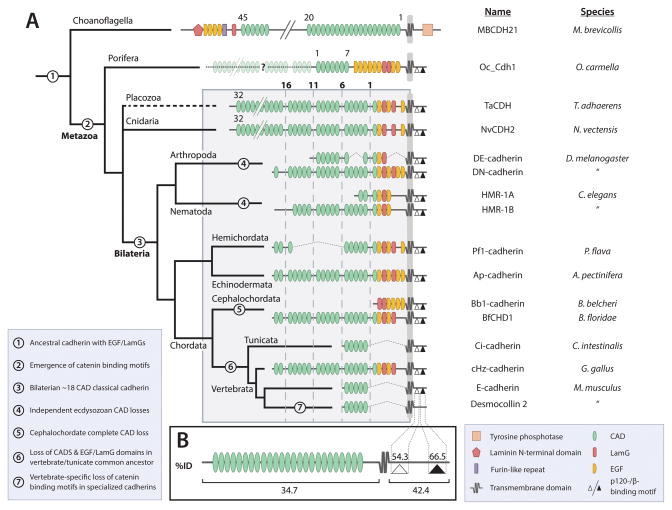
An interesting trend can be observed in the abundance of cadherins across the opisthokont-metazoan boundary (Figure 4, 5). A larger diversity of cadherins is found in choanoflagellates than in any invertebrate phylum outside deuterostomes and platyhelminthes, but none of the choanoflagellate cadherins contain the characteristic catenin-binding motif found in metazoan classical cadherins (Figure 5). As this motif is observed in the classical cadherins of sponges, which are currently thought to be the most basally branching metazoan group, a β-catenin-binding cadherin should be considered an ancestral character of the metazoa (Fahey and Degnan, 2010; Nichols et., 2012). Interestingly, the cadherins in two invertebrate phyla, ctenophora and platyhelminthes, do not have a discernable β-catenin-binding domain. It is possible that the lack of a β-catenin-binding site in these cadherins is an artifact due to low genomic coverage in the available sequence data for these groups. However, it is also possible that ancestors of these phyla secondarily lost a classical cadherin. If these phyla truly do not possess a classical cadherin, determination of the transmembrane component of the CCC (or the functional substitute for the entire complex) in flatworms and comb jellies could be informative. Nevertheless, if we exclude these two phyla, either on grounds of insufficient data or the principle of parsimony, we can conclude that members of the metazoan lineage inherited a classical cadherin with a conserved cytoplasmic domain that bound β-catenin.
Figure 5. Evolution of Classical Cadherin Domain Architecture from Porifera to Vertebrates.
(A) A phylogeny of representative classical cadherin proteins from multiple animal phyla, indicating the modification of the extracellular region by loss and/or rearrangement of conserved domains. Key transitions are noted by number in the legend in the lower left.
(B) Schematic representation of a classical cadherin, indicating the average percent amino acid sequence identity of the catenin binding motifs and the cytoplasmic and extracellular regions of the protein.
β-Catenin
Analysis of non-metazoan ARM proteins reveals a lack of clear orthologs of β-catenin, although β-catenin-like proteins are present (Figure 4). All three available chytridomycte genomes (Allomyces macrogynus, Batrachochytrium dendrobatidis, and Spizellomyces punctatus) each have a 12–13 ARM repeat protein that is ~20% identical to Mm β-catenin (19%, 20%, and 20% identity, respectively), but each is more similar to the mammalian protein Armadillo-repeat Protein 4 (48%, 22%, and 51% identical, respectively). The D. discoideum protein Aardvark has a single ortholog in each of the other amoebozoans analyzed. With the exception of D. discoideum Aardvark, none of these ARM proteins have been assayed for binding to their corresponding VIN protein (α-catenin), nor analyzed to determine their evolutionary relationships, so inferring anything about their role in a putative CCC is impossible. Without further functional evidence to guide us, these data produce two equally probable hypotheses: 1) An α-catenin-binding ARM protein evolved independently twice (i.e., D. discoideum Aardvark is convergent with metazoan β-catenin); or 2) A single gain of function event occurred in an ancestral opisthokont, followed by domain rearrangement or loss in some clades. Additional sequence data may be necessary to determine the evolutionary relationships between identified β-catenin-like ARM proteins, but the hypothesis that the known sequences interact with their corresponding VIN protein is readily testable.
α-Catenin
Previous studies of α-catenin evolution have examined only a sparse phylogeny of metazoans (Zhao et al., 2011). This resulted in the perspective that only a single α-catenin existed prior to evolution of the chordate lineage, and that there were two subsequent duplication events that gave rise to the three known isoforms of mammalian α-catenin (αE, αN, and αT; see above) and vinculin. Expanding the analysis to include additional invertebrate phyla confirms this initial finding in terms of direct α-catenin orthologs (Figure 4). However, deeper analysis shows that one or more additional α-catenin-like proteins are found in many metazoan phyla, including ctenophores, mollusks, annelids and basal deuterostome phyla (Figure 4). Interestingly, the basal metazoan Trichoplax does not have a direct α-catenin ortholog, but it does have a single vinculin protein and an additional α-catenin-like protein with ambiguous orthology (19% and 17% identity to α-catenin and vinculin, respectively). Another previously unexamined basal metazoan, the ctenophore Mnemiopsis leiydi, also has a α-catenin-like protein, and two vinculins. The discovery of additional α-catenin-like proteins raises questions about our current understanding of their evolutionary relationships – this will be addressed briefly in the following sections, but should also be considered an important question for further investigation.
Classical cadherins: variation and constraint mediated by catenin interactions
Of the three core components of the CCC, classical cadherins display the most diversity in terms of domain composition and organization (Oda and Takeichi, 2011). Much is known about cadherin evolution in terms of the relatedness of the different classes and of the architectural diversity of the extracellular region (Hulpiau et al., 2013; Hulpiau and van Roy, 2011; Oda and Takeichi, 2011). Here, we focus on functional variation between metazoan classical cadherins in relation to the CCC, and make a general comparison between variability in the extracellular and cytoplasmic domains.
A general trend in classical cadherin evolution is that organismal complexity is positively correlated with cadherin diversity and inversely correlated with cadherin complexity - nearly all bilaterians have a larger number of classical cadherins, each with fewer extracellular domains than non-bilaterians (Figure 5A). For example, the placozoan classical cadherin (T. adhaerens only has one discernable isoform) contains 32 CADs, while the classical cadherins of bilaterians contain between 2 and 17 CADs (Hulpiau and van Roy, 2009) (Figure 5). The length of classical cadherins in sponge is unclear, as all published sequences are annotated as fragments; however, sponge are thought to have only one classical cadherin with at least 10 or more CADs (Fahey and Degnan, 2010; Nichols et al., 2012).
Additionally, non-chordate classical cadherins contain multiple repeats of epidermal growth factor-like domains (EGFs), laminin A globular domains (LamGs), and a region homologous to the extracellular proteolytic cleavage site of Dd DE-cadherin in the C-terminus of the extracellular domain proximal to the plasma membrane (Oda and Takeichi, 2011; Oda and Tsukita, 1999). The function of these additional domains is poorly understood, but initial in vivo evidence from D. melanogaster indicates that they may play a signaling or regulatory role in development (Haruta et al., 2010). Thus non-chordate classical cadherins may have multiple functions beyond cell-cell adhesion.
From this analysis, we posit that the specialized cell/tissue-type classical cadherins of bilaterians evolved by duplication and sub-functionalization of a ubiquitously expressed classical cadherin with a multi-functional extracellular region. However, studies of expression patterns and functional interactions of large classical cadherins in basal metazoans are needed to test this hypothesis.
Within the highly variable cadherin superfamily, some elements of the protein sequence remained static in order to maintain necessary functions at cell-cell contacts. In order to participate in the CCC, a classical cadherin in a given species must maintain over evolutionary time a β-catenin binding motif within the cytoplasmic domain. There are two binding motifs in the cytoplasmic domain of classical cadherins for p120-catenin and β-catenin, respectively (Hulpiau and van Roy, 2011). Using a heuristic method suitable for general comparison, we can state that the average percent amino acid sequence identity within the β-catenin binding motif is nearly two-fold higher than across the entire protein (Figure 5B). The p120 binding motif is also more conserved than the whole protein, though to a lesser extent than the β-catenin binding motif. An important caveat is that the algorithms used to calculate pair-wise identity are designed to return an optimal alignment, and they ignore large gaps and terminal regions in order to optimize the alignment score. Therefore, the numbers reported here should not be considered indicative of conservation within a region, but rather a means of holistic comparison between regions of a protein. Nevertheless, it is clear that the β-catenin binding motif is more conserved than the rest of cadherin sequence, indicating that in spite of large rearrangements of the extracellular domain of classical cadherins, the sequence in cadherins required for participation in the CCC (i.e., binding to β-catenin) is evolutionarily constrained to maintain this critical function.
β-catenin evolution – domain homology, evidence for evolutionary constraint by α-catenin interaction, and a consensus α-catenin binding motif
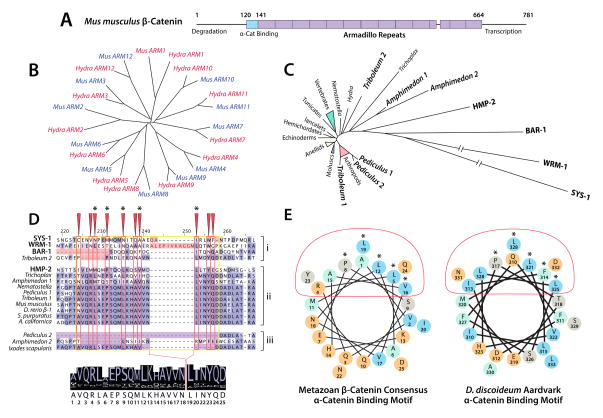
β-Catenin sequence and function appear to be highly conserved within metazoans (Figure 6). β-Catenin has a characteristic structure consisting of unstructured N- and C-terminal regions involved in the regulation of its degradation and transcriptional activity, respectively, a central region consisting of twelve ARM repeats, and a α-catenin binding domain located partially N-terminal to the first ARM (Huber et al., 1997; Shapiro and Weis, 2009) (Figure 6A). The order and identity of the ARM repeats within β-catenin are highly conserved. When sequences of metazoan β-catenin orthologs are subdivided based upon their individual ARM repeats and clustered algorithmically according to sequence similarity, it is clear that each ARM repeat groups exclusively, even between distantly related taxa (Schneider et al., 2003) (Figure 6B).
Figure 6. Evolutionary Constraint of β-Catenin by the α/β-Catenin Interaction.
(A) A schematic representation of Mm β-catenin, indicating the position of the 〈-catenin binding region and Armadillo repeats.
(B) A phylogeny of Hydra and Mus Armadillo repeats 1–12 (after (Schneider et al., 2003)) indicating homology of each repeat.
(C) A neighbor-joining phylogeny of metazoan β-catenins, indicating divergent paralogs in bold. Branch length correlates to number of amino acid substitutions between proteins.
(D) An alignment generated using MUSCLE (Edgar, 2004), visualized in JalView (Waterhouse et al., 2009), highlighting the α-catenin binding motif (yellow box), conserved binding surface (red boxes/arrows), and structurally important residues (asterisks). Paralogs that fail to bind α-catenin (i) and untested paralogs and orthologs with abberant binding motifs (iii) are segregated from α-catenin-binding paralogs and single orthologs (ii) for clarity. A consensus sequence generated from the proteins in group ii is displayed below.
(E) Helical wheel representations of the consensus α-catenin binding helix from D, and that of D. discoideum (after (Dickinson et al., 2011a)). Charged residues are colored orange, and hydrophobic residues are colored cyan. Red ovals and asterisks indicate the α-catenin binding surface and structurally important residues as in D.
In spite of this strong sequence conservation across the metazoa, there are a small number of β-catenin orthologs that are quite divergent. On a neighbor-joining phylogeny in which branch length corresponds to the number of amino acid substitutions, several proteins stand apart as clear outliers. Interestingly, most of these occur in species in which there has been a lineage-specific duplication of β-catenin: A. queenslandica, T. casteneum, and most notably in the three additional C. elegans proteins BAR-1, SYS-1, and WRM-1 (Liu et al., 2008; Zhao et al., 2011) (Figure 6C). In A. queenslandica, the two β-catenin paralogs have different mRNA expression patterns, but neither their subcellular localization nor function have been examined (Adamska et al., 2010). The C. elegans β-catenin paralogs HMP-2, BAR-1, SYS-1 and WRM-1 also exhibit sub-functionalization. Of the four, only HMP-2 binds to mammalian α-catenin and HMP-1 (Kwiatkowski et al., 2010; Natarajan et al., 2001), while BAR-1, SYS-1 and WRM-1 are involved in Wnt signaling and gene transcription (Liu et al., 2008; Natarajan et al., 2001). In T. castaneum, β-catenin paralogs exhibit partial sub-functionalization in cell-cell adhesion (β-1), and centrosomal regulation (β-2) (Bao et al., 2012) that is also a function of mammalian β-catenin (Mbom et al., 2013). Additionally, T. castaneum β-2 has a 6-amino-acid deletion within the conserved α-catenin binding motif, but maintains a high level of sequence conservation in residues important for E-cadherin binding (Bao et al., 2012; Pai et al., 1996). These data indicate that β-catenin participation in the CCC may be a form of evolutionary constraint that is partially responsible for the high level of sequence conservation within β-catenin. Release from this selective pressure might have been achieved through disruption of α-catenin binding in β-catenin paralogs.
A multiple alignment of single and duplicated β-catenin paralogs across metazoa reveals that all highly divergent paralogs possess significant insertions or deletions within the α-catenin binding domain (Figure 6D). The paralogs for which there is evidence supporting a loss of α-catenin binding function (Figure 6D, top) all possess insertions or deletions in this region. In contrast, α-catenin-binding paralogs align closely with those organisms that have only one single β-catenin ortholog (Figure 6D, center). A. queenslandica and P. humanus possess ®-catenin paralogs with significant alterations in the α-catenin binding domain (Figure 6D, bottom), indicating that these proteins may have lost the capacity to bind α-catenin. Interestingly, I. scapularis has a single β-catenin ortholog with a deletion within the α-catenin binding domain. One hypothesis is that the α-catenin binding site in β-catenin is the favored mutation site for disrupting β-catenin function in the CCC, as this would produce only a single adhesive phenotype (Brembeck et al., 2004; Hoffmans and Basler, 2007; Huber and Weis, 2001; Roura et al., 1999; Taurin et al., 2006); in contrast, a mutation of the central ARM repeat region would have multiple affects on adhesion and Wnt signaling due to the significant overlap of binding sites for E-cadherin (Huber and Weis, 2001), TCF (Graham et al., 2001; Graham et al., 2000), Axin (Spink et al., 2000; Xing et al., 2003) and Adenomatous Polyposis Coli (Ha et al., 2004; Spink et al., 2000; Xing et al., 2004).
From this sequence alignment, we are also able to derive a consensus α-catenin binding motif in β-catenin (depicted on a helical wheel in Figure 6E). Excluding C. elegans sequences and other divergent paralogs, the β-catenin residues shown to form structurally important contacts with α-catenin (consensus residues 5, 8, 12, 16, and 19) are entirely conserved within bilaterians, and mostly conserved across metazoans (Aberle et al., 1996; Pokutta and Weis, 2000). These key residues are also conserved in the α-catenin binding motif of D. discoideum Aardvark, demonstrating their importance for α/β heterodimer interaction (Dickinson et al., 2011a) (Figure 6E). A more detailed comparison of these two motifs may yield a consensus search sequence usable for the detection of putative α-catenin-binding ARM proteins in divergent non-metazoan eukaryotes. This α-catenin binding region should also be considered a candidate region for studies of intermolecular co-evolution between α- and β-catenin.
Evolutionary history of the α-catenin/vinculin family
In light of recent studies that have identified unexpected diversity in functional properties of α-catenins (Figures 1–3) and the sequences reported above that were not described previously, a more thorough review of VIN protein phylogeny is merited. Phylogenetic analysis of distantly related sequences is often inconclusive, but when combined with functional evidence and domain architecture we can draw conclusions about the ancestry of α-catenin and related proteins. Here we describe the presence of a previously unknown group of metazoan α-catenin-like proteins that may be representative of an ancestral metazoan α-catenin, and discuss the origin of the vinculin family as a duplication event from a α-catenin-like protein in an ancestral metazoan.
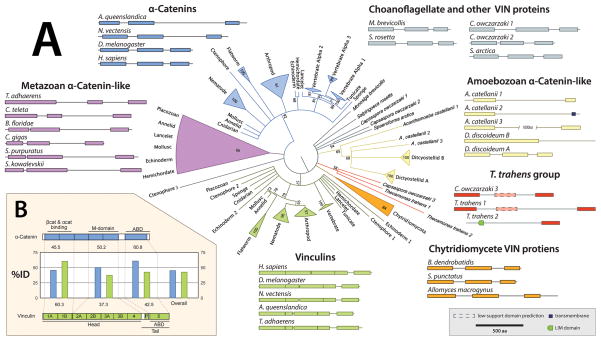
To determine the relatedness of metazoan α-catenins and vinculins and non-metazoan VIN proteins, a maximum-likelihood phylogeny based on the sequence of 96 VIN proteins was made from diverse metazoan and non-metazoan phyla (Figure 7A). Known α-catenins and vinculins group as distinct clades, but non-metazoan sequences do not group in either of these clades. The chytridomycete and amoebozoan VIN proteins also group as independent clades. These clades arise from a polytomy of indeterminate relatedness that also contains sequences from choanoflagellates and their near relative, the apusozoan Thecamonas trahens, and all the metazoan sequences that were not described previously. These metazoan sequences group together as a clade (0.66 ML posterior probability), but their relatedness to the described α-catenins can not be accurately determined as this clade groups unreliably at the base of the α-catenin branch (0.28 ML posterior probability, depicted in Figure 7A as collapsed to the polytomy). This clade has both protostome and deuterostome VIN members, and also contains the only α-catenin-like sequence found in Trichoplax, indicating that it is both widely distributed and ancestral in the metazoa. As this clade was until now not described, the function of these α-catenin-like proteins is not known; a comparison between organisms that possess both α-catenin and a member of this clade with organisms possessing only one or the other may provide important data for understanding the evolution of the α-catenin protein family. This phylogeny demonstrates that the α-catenin and vinculin families are distinct metazoan lineages that diverged at the base of the animal tree. However, the relatedness of divergent ancestral groups to these two clades cannot be readily determined from the phylogeny alone.
Figure 7. Evolutionary History of VIN Families.
(A) A maximum likelihood phylogeny of 96 VIN-containing proteins generated using RAxML (Stamatakis, 2006) with 1000 rapid bootstrap iterations and best-fit model parameters as determined by ProtTest3 (PROTGAMMALGF; (Darriba et al., 2011)) on a trimmed alignment (trimAl; Capella-Gutierrez et al. 2009). α-catenins are colored blue, vinculins are colored green, amoebozoan sequences are colored yellow, and ungrouped sequences are colored grey. Corresponding schematics of domain architecture are grouped adjacent to appropriate clades.
(B) A general schematic of the domain architecture of α-catenin and vinculin, with a bar graph indicating the average percent amino acid sequence identity of the highlighted regions within each protein family.
As indicated by previous studies of the α-catenin-related protein in D. discoideum, general domain architecture may be more indicative of functional relatedness than strict sequence comparison – Dd α-catenin is equally related to H. sapiens αN-catenin and vinculin by sequence (17% identity), but the arrangement of its functional domains and biochemical properties corresponds more closely to α-catenin than to vinculin (Dickinson et al., 2011a). Similar comparison of domain architecture between the groups represented here reveals both a surprising diversity within divergent VIN sequences, as well as the presence of a conserved architectural pattern. Within opisthokonts, all VIN proteins contain only helical bundle domains characteristic of the VIN family and no other recognizable functional domains.
Outside of opisthokonts, other protein motifs are included such as a LIM domain in T. trahens VIN 2 and the inclusion of a transmembrane domain in A. castellani VIN 2. The LIM domain is a tandem zinc-finger structure that functions as a protein-binding interface, and is associated with cytoskeletal organization and signal transduction from the plasma membrane to the nucleus (Kadrmas and Beckerle, 2004). Many known LIM proteins have been shown to bind F-actin or actin-binding proteins (Khurana et al., 2002), but none have been described that contain a VIN domain. T. trahens VIN 2 appears to be a novel VIN-LIM-containing protein that represents a class of eukaryotic LIM proteins that were not described previously (Koch et al., 2012). The inclusion of a transmembrane domain in A. castellani VIN 2 is also novel – no other described VIN protein has been observed to have a transmembrane domain.
There is significant variation in domain architecture within previously described VIN proteins. Even amongst VIN proteins there are clear outliers in which we can observe domain loss or the addition of large insertions (Figure 7A). The loss of one or more VIN domains is observed in several amoebozoans, including Dictyostelium sp. and A. castellani, and putatively within the opisthokont C. owczarzaki and the apusozoan T. trahens, where a third VIN domain is identified with unreliable significance (Figure 7A, dashed outline). Insertion sequences are observed in α-catenins and α-catenin-like sequences of many basal metazoans. For instance, both N. vectensis and T. adhaerens have large insertions (~400 and 600 amino acids, respectively) between the M- and actin-binding domains. There are multiple phosphorylation sites in this region in Mm α-catenin (Huttlin et al., 2010) that are conserved within the insertions of basal metazoans, but the functional significance of these sites is not understood. Nothing is known of how these inserted sequences affect the conformation or functional properties of α-catenin such as actin binding, homodimerization capacity, or auto-inhibition. However, the location of these insertions provide a natural study system in which to test hypotheses of the roles of internal residues in regulating α-catenin structure and function.
As observed with classical cadherins and β-catenin, we would expect to find signs of constraint within α-catenin sequences resulting from its necessary function as a link between the CCC and the actin cytoskeleton. However, as α-catenin plays multiple roles at the plasma membrane in the mammalian CCC (see Section II), it is unclear which regions would be expected to have high sequence conservation. In a comparison of α-catenin sequences, we observe that the C-terminal ABD is more conserved relative to the rest of the protein, whereas the N-terminal (homo- and hetero-) dimerization domain is more variable (Figure 7B). This observation indicates that the actin-binding domain may be under selection to maintain the capacity to bind actin, whereas the N-terminal dimerization and M domains have varied more significantly over evolutionary time.
Several related hypotheses can be posited to explain why the interaction between the β-catenin/cadherin complex and α-catenin varies significantly between metazoan groups. 1) The interaction with the actin cytoskeleton is the more ancestral function of an α/β-catenin heterodimer. 2) Greater sequence divergence occurred in the N- and M-domains as more functions at the plasma membrane are gained in some metazoan groups. 3) Regulation of actin binding may have evolved by auto-inhibition/allostery rather than loss of function mutations due to overlapping functional regions/binding surfaces of VIN domains. These hypotheses are not mutually exclusive nor are they the only interpretation, but they stand out in light of recent functional evidence in diverse organisms.
IV. CONCLUSION AND SYNTHESIS
Functional divergence within a highly conserved protein complex
It is generally accepted that homologous genes and proteins have conserved functions between different organisms, and molecular evolution occurs through changes in gene regulatory elements (Carroll, 2008). A common assumption made in sequence comparison is that shifts in protein function will correspond to large differences in sequence, but at the molecular level the importance of a single residue cannot be underestimated. Few studies have tested whether protein homology, as determined by sequence comparison, is indicative of conservation of biochemical, cellular or developmental properties, and those that have tried had a false positive rate of over 50% when calling orthologs with equivalent functions (Ponting, 2001; Watson et al., 2005; Yu et al., 2012). Indeed similarities in sequence are not predictive of conserved protein function throughout the cadherin superfamily, hedgehog signaling pathway components, and vertebrate glucocorticoid receptor homologs (Dickinson et al., 2011b; Ortlund et al., 2007). As demonstrated here, in vitro biochemical experiments make it possible to directly test hypotheses of conservation of protein function and, by careful selection of a set of homologs for analysis, can reveal how proteins acquired novel functions in evolution.
The CCC is highly conserved throughout metazoans, where evolutionary assembly of the complex likely occurred in unicellular ancestors. Molecular interactions between the cytoplasmic domain of classical cadherins and ARM repeats of β-catenin have not been investigated over as diverse a set of organisms as α-catenin, but cadherin/β-catenin complex formation appears to be highly conserved in animals and positively regulated by phosphorylation of the cadherin cytoplasmic domain (Kwiatkowski et al., 2010; Nichols et al., 2006; Nichols et al., 2012; Schneider et al., 2003) (this study). Direct binding between α-catenin and β-catenin is conserved throughout metazoans (Aberle et al., 1996; Hirano and Takeichi, 1994; Koslov et al., 1997; Kwiatkowski et al., 2010; Miller et al., 2013; Schneider et al., 2003).
While metazoan α-catenin homologs share >20% sequence identity and conserved domain architecture, regulatory mechanisms of actin binding are divergent between unikonts, and novel functional properties have developed in some homologs. Auto-inhibition reduces the actin binding affinity of mammalian αE-catenin, C. elegans HMP-1 and vertebrate vinculin. However, auto-inhibition is absent from Dd α-catenin, Dm α-catenin, and Dr αE-catenin (see Section II, above). Dm α-catenin and Mm αE-catenin dimerize in solution. β-Catenin binding also negatively regulates Mm α-catenin binding to F-actin in vitro (Miller et al., 2013; Yamada et al., 2005), but it is not known whether this occurs in other species. Future in vivo experiments should determine how these distinct biochemical properties cater to specific developmental or physiological requirements. Between unikonts, α-catenin appears to have sequence divergence as well as variable function. Functional implications of changes in domain structure of non-bilaterian and premetazoan VIN homologs in organisms that do not contain all core components of the CCC will require biochemical characterization of purified proteins.
Sequence vs. function of α-catenin/vinculin family proteins
With available bioinformatics tools, it is not possible to determine the specific biochemical and functional properties of α-catenin/vinculin family proteins from sequence alone. For instance, it is not possible to use a comparison of sequence identity to classify pre-metazoan VIN proteins as members of either the α-catenin or vinculin protein families. Dd α-catenin, which has an equal level of sequence identity to metazoan α-catenin and vinculin, possesses biochemical and functional properties of α-catenin and not vinculin (Dickinson et al., 2011a; Dickinson et al., 2011b). Furthermore, as described above, a large polytomy of α-catenin-like proteins results from phylogenetic analysis of VIN family proteins (Figure 7A). While domain organization may make stronger predictions of protein function and relatedness than strict sequence identity, functional predictions based on domain organization can be confounded by deletion, convergent evolution, or duplication of domains. As a result, in the absence of functional characterization one cannot predict specific functions of these proteins in cell-cell or cell-substrate adhesion.
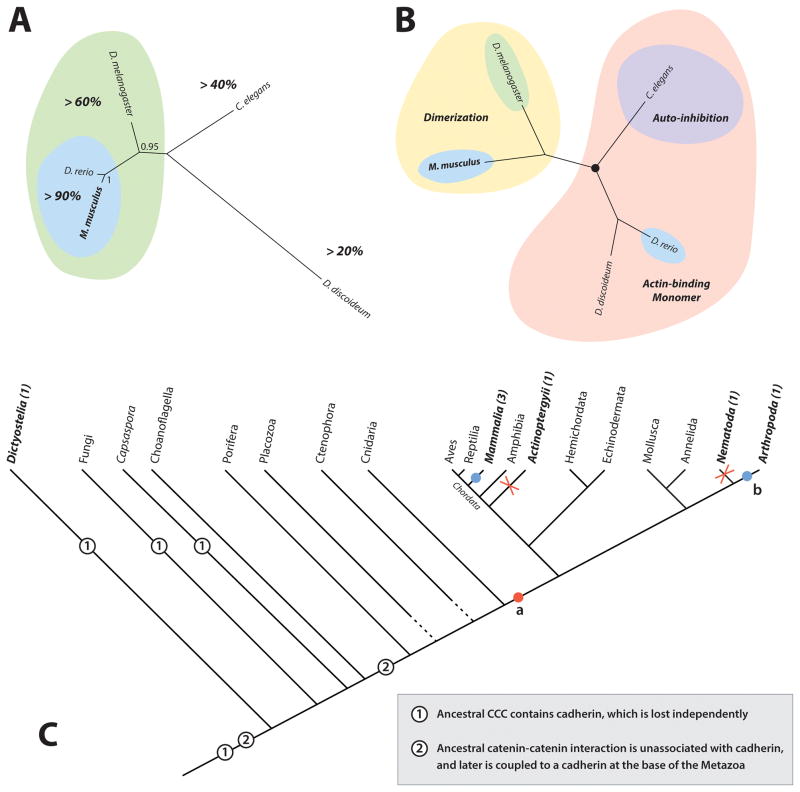
In a sequence-based phylogeny of biochemically characterized α-catenin orthologs, mammalian and D. rerio αE-catenin cluster together with 90% sequence identity (Figure 8A). Dm α-catenin clusters with mammalian αE-catenin with ~60% sequence identity, while C. elegans HMP-1 and Dd α-catenin are outgroups with <40% and <20% identity, respectively, to the mammalian orthologs (Figure 8A). To directly juxtapose sequence-based methods with functional comparisons of α-catenin family proteins, we generated an alternative cladogram in which orthologs are clustered together based on common functional properties (Figure 8B). Based upon functional properties, we would group Mm αE-catenin and Dm α-catenin due to their shared character of dimerization, and place Dd α-catenin and Dr αE-catenin together in a separate group because they are monomers that bind and bundle actin constitutively, with C. elegans HMP-1 forming a divergent outgroup to these due to its auto-inhibition of actin-binding. Another possible functional grouping might place mammalian and C. elegans α-catenins together because they both appear to be inhibited in their capacity to bind actin as monomers, but auto-inhibition of actin binding in mammalian α-catenin is less well supported. Regardless of how the tree is drawn, one distinction is clear – when grouped functionally, the two proteins most similar by sequence, Mm and Dr α-catenin, cannot be grouped together, and must be grouped with proteins to which they have much lower sequence identity (Figure 8B). Thus, sequence similarity is a poor indicator of functional similarity between studied α-catenin orthologs. These examples highlight the necessity of biochemical characterization as a means of assessing functional equivalence between orthologs. Current bioinformatic methods can identify putative orthologs with remarkable speed and accuracy, but our capacity to predict tertiary structure and biochemical function based on protein sequence is quite limited. Only through in vitro characterization and in vivo observation can we describe in detail the subtleties of protein activity. As demonstrated here, such observations can vastly increase the power of bioinformatic analysis: knowledge of binding sites, structurally important residues, and other functional properties, when synthesized with sequenced-based methods, can yield deeper inferences about protein evolution. By using known biochemical properties and domain boundaries to inform sequence analysis it may be possible to understand the evolution of α-catenin at a deeper level.
Figure 8. Sequence Identity versus Functional Equivalence, and CCC Evolutionary Hypotheses.
(A) A neighbor-joining tree of the 5 characterized metazoan α-catenin orthologs. Percent identity to Mm 〈E-catenin is indicated in bold. Orthologs sharing > 90% identity are grouped in blue, > 60% in green, and < 60% left uncolored.
(B) A cladogram of the same proteins, as grouped by function. The red oval groups actin-binding monomers, the yellow groups proteins capable of homodimerization, and the purple indicates auto-inhibition of actin binding.
(C) A phylogeny of unikonts, indicating two evolutionary hypotheses of CCC evolution (numbered circles), and two hypotheses for the evolution of dimerization in metazoan 〈-catenins. See text for detailed explanations of hypotheses.
Evolutionary context of the cadherin-catenin complex
Classical cadherins are clearly unique to the metazoa, but as the last common ancestor of all eukaryotes necessarily had some form of cadherin, the role of cadherins during the formation of the ancestral CCC is unclear. The loss of cadherins from amoebozoans, apusozoans, and fungi can be seen either as a loss of cadherin from an ancestral complex, or as an indication that cadherins did not constitute the original membrane component of the CCC. Evidence from D. discoideum indicates that the α-catenin/Aardvark heterodimer links actin to the plasma membrane at cell contacts, but with only a single data point we are unable to say if this is an amoebazoan-specfic (or species-specific) loss of the cadherin component of the CCC due to derivation of other novel junction complexes, or if cadherin was absent from the ancestral CCC altogether. Further investigation of the CCC components of choanoflagellates and C. owczarzaki, the only non-metazoan opthistokonts known to have proteins of all three CCC protein families, is necessary to elucidate the evolution of cadherin-catenin interactions.
The evolution of β-catenin appears to be due to gain of function. Based on the homology of α-catenin binding motifs between metazoan β-catenins and D. discoideum Aardvark (Figure 6E), the capacity to bind α-catenin, and thereby potentially link the actin cytoskeleton to the plasma membrane, was the ancestral function of the common ancestor of these proteins. The role of β-catenin in Wnt signaling is a metazoan novelty acquired through cooption following the evolution of the Wnt pathway. Based on this evidence, we can infer that a α-catenin-binding ARM protein was part of the shared inheritance of opthistokonts, which has either been lost in multiple groups (capsaspora, ichthyosporeans, choanflagellates, fungi, etc.), or remains to be discovered. As previous search attempts have focused on finding full β-catenin orthologs, and have not utilized a strategy based on identification of α-catenin binding motifs, the latter is likely true, but only further analysis of the ARM proteins of unicellular organisms can determine how ancient this interaction truly is.
We propose a model of α-catenin evolution in which the ancestral homolog was a constitutively active F-actin binding monomer that gained β-catenin binding in a unicellular ancestor to amoebozoa and opisthokonta (Figure 8C). In unicellular organisms, the α/β-catenin heterodimer may have supported cell-cell contact formation during mating and feeding behavior. Duplication of a α-catenin-like protein in an ancestral unikont resulted in multiple VIN-family proteins, which developed distinct functions in cell-cell and cell-ECM adhesion in metazoans. Innovation of vinculin-like proteins likely facilitated cell-substrate adhesion during crawling in unicellular organisms, and perhaps strengthened cell-cell contacts in early multicellular organisms.
α-Catenin has not been characterized in a basal, non-bilaterian metazoan and we can only speculate about its regulatory properties. Nevertheless, we posit that it was likely a monomeric, β-catenin and F-actin binding protein that gained additional functions for complex development and tissue construction through association with classical cadherins. Given the limited phylogenetic coverage of currently characterized orthologs (groups in bold, Figure 8C), there are two equally probable evolutionary histories of bilaterian α-catenin: 1) α-Catenin developed the ability to homodimerize in an ancestor to the bilaterian clade, and this function was subsequently lost in C. elegans and D. rerio (Figure 8C: red dot and X, respectively). 2) The innovation of homodimerization is convergent between D. melanogaster and mammals (Figure 8C; blue dots). The fact that homodimerization in mammals, but not in Drosophila, is necessary to potentiate F-actin binding by relieving auto-inhibition, supports a hypothesis of convergent evolution. Similarly auto-inhibition may have originated in a common ancestor to bilateria and was subsequently disrupted in D. melanogaster and D. rerio, or may have been independent innovations of C. elegans and mammals. Future characterization of orthologs from other bilaterians will allow a fuller explanation of the evolutionary origin of these regulatory properties. In addition, while αE-, αN-, and αT-catenin isoforms originated from a single gene in higher vertebrates, the significance of three isoforms is not well understood, and future biochemical and cell biological studies will reveal the properties underlying their distinct developmental functions.
In light of evidence reviewed here, the assembly of the CCC can be viewed under two alternative hypotheses: 1) The ancestral CCC included a cadherin, which was lost in some non-metazoan lineages (1 on Figure 8C); or 2) An ancestral interaction between an ARM protein and a VIN protein linked the actin cytoskeleton to the plasma membrane by way of a currently unknown protein, and a cadherin was co-opted to replace the membrane portion of this complex at the base of the metazoa (2 on Figure 8C). Both hypotheses assert that the core α-catenin/β-catenin interaction of the CCC is ancestral and homologous within unikonts, but differs in how they regulate cell-cell adhesion. The first hypothesis stipulates that the capacity to form cell-cell contacts mediated by the CCC was ancestral to unikonts, and therefore that extant non-colonial unicellular lineages may be secondarily simplified. The second hypothesis requires fewer evolutionary transitions.
Interpreting the validity of these hypotheses based on only parsimony belies the evolutionary complexity of cell-cell junction formation. An ancestral capacity to form cell-cell contacts is in line with the prevalence and diversity of colonial, aggregate, and multicellular organisms within the Unikonta, but without additional data points from fungi, choanoflagellates, and other non-metazoan opthistokonts we can only speculate about the importance of the ancestral CCC in the evolution of these organisms. Given the presence of CCC complex in all metazoans, it is likely that conserved residues and domains at binding interfaces mediated refinement of the ancestral CCC into the complex seen in bilaterians, and thus constrained the independent evolution of its constituent proteins. Studies of co-evolution of classical cadherins, β-catenin, and α-catenin homologs will provide further understanding of CCC function within metazoans, and investigation of distant homologs in non-metazoans will provide insight into the evolution of the CCC and the origins of multicellularity.
Acknowledgments
This work was supported by NIH Ruth L. Kirschstein National Research Service Award GM007276 (PWM), NSF Pre-doctoral Fellowship (DNC), NIH GM035527 (WJN), NIH GM56169 (WIW), and NIH U01 GM094663 (WJN, WIW).
References
- Abe K, Chisaka O, Van Roy F, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat Neurosci. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- Abe K, Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci U S A. 2008;105:13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- Aberle H, Schwartz H, Hoschuetzky H, Kemler R. Single amino acid substitutions in proteins of the armadillo gene family abolish their binding to alpha-catenin. J Biol Chem. 1996;271:1520–1526. doi: 10.1074/jbc.271.3.1520. [DOI] [PubMed] [Google Scholar]
- Adams CL, Nelson WJ. Cytomechanics of cadherin-mediated cell-cell adhesion. Curr Opin Cell Biol. 1998;10:572–577. doi: 10.1016/s0955-0674(98)80031-8. [DOI] [PubMed] [Google Scholar]
- Adamska M, Larroux C, Adamski M, Green K, Lovas E, Koop D, Richards GS, Zwafink C, Degnan BM. Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica. Evol Dev. 2010;12:494–518. doi: 10.1111/j.1525-142X.2010.00435.x. [DOI] [PubMed] [Google Scholar]
- Alatortsev VE, Kramerova IA, Frolov MV, Lavrov SA, Westphal ED. Vinculin gene is non-essential in Drosophila melanogaster. FEBS Lett. 1997;413:197–201. doi: 10.1016/s0014-5793(97)00901-0. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- Bao R, Fischer T, Bolognesi R, Brown SJ, Friedrich M. Parallel duplication and partial subfunctionalization of beta-catenin/armadillo during insect evolution. Mol Biol Evol. 2012;29:647–662. doi: 10.1093/molbev/msr219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. Vinculin is essential for muscle function in the nematode. J Cell Biol. 1991;114:715–724. doi: 10.1083/jcb.114.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JM, Kwiatkowski AV, Yang C, Korobova F, Pokutta S, Svitkina T, Weis WI, Nelson WJ. AlphaE-catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. J Cell Biol. 2010;189:339–352. doi: 10.1083/jcb.200910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JM, Nelson WJ. Bench to bedside and back again: molecular mechanisms of alpha-catenin function and roles in tumorigenesis. Semin Cancer Biol. 2008;18:53–64. doi: 10.1016/j.semcancer.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Essential role of BCL9-2 in the switch between beta-catenin’s adhesive and transcriptional functions. Genes Dev. 2004;18:2225–2230. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Mostov KE. From cells to organs: building polarized tissue. Nat Rev Mol Cell Biol. 2008;9:887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullions LC, Notterman DA, Chung LS, Levine AJ. Expression of wild-type alpha-catenin protein in cells with a mutant alpha-catenin gene restores both growth regulation and tumor suppressor activities. Mol Cell Biol. 1997;17:4501–4508. doi: 10.1128/mcb.17.8.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan RH, Rokas A, Gaucher EA, Reynolds AB. The molecular evolution of the p120-catenin subfamily and its functional associations. PLoS One. 2010;5:e15747. doi: 10.1371/journal.pone.0015747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Cavey M, Rauzi M, Lenne PF, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453:751–756. doi: 10.1038/nature06953. [DOI] [PubMed] [Google Scholar]
- Cereijido M, Contreras RG, Shoshani L. Cell adhesion, polarity, and epithelia in the dawn of metazoans. Physiol Rev. 2004;84:1229–1262. doi: 10.1152/physrev.00001.2004. [DOI] [PubMed] [Google Scholar]
- Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J Biol Chem. 2006;281:40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Pokutta S, Cadwell GW, Bobkov AA, Bankston LA, Liddington RC, Weis WI. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci U S A. 2012;109:8576–8581. doi: 10.1073/pnas.1203906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates JC. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 2003;13:463–471. doi: 10.1016/s0962-8924(03)00167-3. [DOI] [PubMed] [Google Scholar]
- Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig SW, Johnson RP. Assembly of focal adhesions: progress, paradigms, and portents. Curr Opin Cell Biol. 1996;8:74–85. doi: 10.1016/s0955-0674(96)80051-2. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R, Sarpal R, Ishiyama N, Pellikka M, Ikura M, Tepass U. Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat Cell Biol. 2013;15:261–273. doi: 10.1038/ncb2685. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Nelson WJ, Weis WI. A polarized epithelium organized by beta- and alpha-catenin predates cadherin and metazoan origins. Science. 2011a;331:1336–1339. doi: 10.1126/science.1199633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Nelson WJ, Weis WI. An epithelial tissue in Dictyostelium challenges the traditional origin of metazoan multicellularity. Bioessays. 2012;34:833–840. doi: 10.1002/bies.201100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Weis WI, Nelson WJ. Protein evolution in cell and tissue development: going beyond sequence and transcriptional analysis. Dev Cell. 2011b;21:32–34. doi: 10.1016/j.devcel.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey B, Degnan BM. Origin of animal epithelia: insights from the sponge genome. Evol Dev. 2010;12:601–617. doi: 10.1111/j.1525-142X.2010.00445.x. [DOI] [PubMed] [Google Scholar]
- Fraiberg M, Borovok I, Weiner RM, Lamed R. Discovery and characterization of cadherin domains in Saccharophagus degradans 2–40. J Bacteriol. 2010;192:1066–1074. doi: 10.1128/JB.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- Grimson MJ, Coates JC, Reynolds JP, Shipman M, Blanton RL, Harwood AJ. Adherens junctions and beta-catenin-mediated cell signalling in a non-metazoan organism. Nature. 2000;408:727–731. doi: 10.1038/35047099. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Hardin J, Lynch A, Loveless T, Pettitt J. Cadherins and their partners in the nematode worm Caenorhabditis elegans. Prog Mol Biol Transl Sci. 2013;116:239–262. doi: 10.1016/B978-0-12-394311-8.00011-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJC, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta T, Warrior R, Yonemura S, Oda H. The proximal half of the Drosophila E-cadherin extracellular region is dispensable for many cadherin-dependent events but required for ventral furrow formation. Genes Cells. 2010 doi: 10.1111/j.1365-2443.2010.01389.x. [DOI] [PubMed] [Google Scholar]
- Herrenknecht K, Ozawa M, Eckerskorn C, Lottspeich F, Lenter M, Kemler R. The uvomorulin-anchorage protein alpha catenin is a vinculin homologue. Proc Natl Acad Sci U S A. 1991;88:9156–9160. doi: 10.1073/pnas.88.20.9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Takeichi M. Differential expression of alpha N-catenin and N-cadherin during early development of chicken embryos. Int J Dev Biol. 1994;38:379–384. [PubMed] [Google Scholar]
- Hoffmans R, Basler K. BCL9-2 binds Arm/beta-catenin in a Tyr142-independent manner and requires Pygopus for its function in Wg/Wnt signaling. Mech Dev. 2007;124:59–67. doi: 10.1016/j.mod.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, Gul IS, van Roy F. New insights into the evolution of metazoan cadherins and catenins. Prog Mol Biol Transl Sci. 2013;116:71–94. doi: 10.1016/B978-0-12-394311-8.00004-2. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F. New insights into the evolution of metazoan cadherins. Mol Biol Evol. 2011;28:647–657. doi: 10.1093/molbev/msq233. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, de Rooij J. Mechanosensitive systems at the cadherin-F-actin interface. J Cell Sci. 2013;126:403–413. doi: 10.1242/jcs.109447. [DOI] [PubMed] [Google Scholar]
- Ishiyama N, Tanaka N, Abe K, Yang YJ, Abbas YM, Umitsu M, Nagar B, Bueler SA, Rubinstein JL, Takeichi M, et al. An Autoinhibited Structure of α-Catenin and Its Implications for Vinculin Recruitment to Adherens Junctions. J Biol Chem. 2013;288:15913–15925. doi: 10.1074/jbc.M113.453928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Moroi S, Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. J Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov DB, Philippova MP, Tkachuk VA. Structure and functions of classical cadherins. Biochemistry (Mosc) 2001;66:1174–1186. doi: 10.1023/a:1012445316415. [DOI] [PubMed] [Google Scholar]
- Janssen MEW, Kim E, Liu H, Fujimoto LM, Bobkov A, Volkmann N, Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Mol Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Janssens B, Mohapatra B, Vatta M, Goossens S, Vanpoucke G, Kools P, Montoye T, van Hengel J, Bowles NE, van Roy F, et al. Assessment of the CTNNA3 gene encoding human alpha T-catenin regarding its involvement in dilated cardiomyopathy. Hum Genet. 2003;112:227–236. doi: 10.1007/s00439-002-0857-5. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kane DA, Maischein HM, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, et al. The zebrafish early arrest mutants. Development. 1996;123:57–66. doi: 10.1242/dev.123.1.57. [DOI] [PubMed] [Google Scholar]
- Khurana B, Khurana T, Khaire N, Noegel AA. Functions of LIM proteins in cell polarity and chemotactic motility. EMBO J. 2002;21:5331–5342. doi: 10.1093/emboj/cdf550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of alpha-actinin with the cadherin/catenin cell-cell adhesion complex via alpha-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AW, Pokutta S, Lustig A, Engel J. Calcium binding and homoassociation of E-cadherin domains. Biochemistry. 1997;36:7697–7705. doi: 10.1021/bi9705624. [DOI] [PubMed] [Google Scholar]
- Koch BJ, Ryan JF, Baxevanis AD. The diversification of the LIM superclass at the base of the metazoa increased subcellular complexity and promoted multicellular specialization. PLoS One. 2012;7:e33261. doi: 10.1371/journal.pone.0033261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M, Spagnuolo A, Klymkowsky M, Wylie C, Heasman J. The roles of maternal alpha-catenin and plakoglobin in the early Xenopus embryo. Development. 1997;124:1553–1560. doi: 10.1242/dev.124.8.1553. [DOI] [PubMed] [Google Scholar]
- Koslov ER, Maupin P, Pradhan D, Morrow JS, Rimm DL. Alpha-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with beta-catenin. J Biol Chem. 1997;272:27301–27306. doi: 10.1074/jbc.272.43.27301. [DOI] [PubMed] [Google Scholar]
- Kurth T, Fesenko IV, Schneider S, Munchberg FE, Joos TO, Spieker TP, Hausen P. Immunocytochemical studies of the interactions of cadherins and catenins in the early Xenopus embryo. Dev Dyn. 1999;215:155–169. doi: 10.1002/(SICI)1097-0177(199906)215:2<155::AID-DVDY8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Maiden SL, Pokutta S, Choi HJ, Benjamin JM, Lynch AM, Nelson WJ, Weis WI, Hardin J. In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:14591–14596. doi: 10.1073/pnas.1007349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Antos C, Butz S, Huber O, Delmas V, Dominis M, Kemler R. A role for cadherins in tissue formation. Development. 1996;122:3185–3194. doi: 10.1242/dev.122.10.3185. [DOI] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci U S A. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys SP, Hill A. The physiology and molecular biology of sponge tissues. Adv Mar Biol. 2012;62:1–56. doi: 10.1016/B978-0-12-394283-8.00001-1. [DOI] [PubMed] [Google Scholar]
- Liu J, Phillips BT, Amaya MF, Kimble J, Xu W. The C. elegans SYS-1 protein is a bona fide beta-catenin. Dev Cell. 2008;14:751–761. doi: 10.1016/j.devcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden SL, Harrison N, Keegan J, Cain B, Lynch AM, Pettitt J, Hardin J. Specific conserved C-terminal amino acids of Caenorhabditis elegans HMP-1/α-catenin modulate F-actin binding independently of vinculin. J Biol Chem. 2013;288:5694–5706. doi: 10.1074/jbc.M112.438093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiers JL, Peng X, Fanning AS, Demali KA. ZO-1 recruitment to alpha-catenin: a novel mechanism for coupling the assembly of tight junctions to adherens junctions. J Cell Sci. 2013 doi: 10.1242/jcs.126565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre JL, Berthoumieux H, Krens SFG, Salbreux G, Julicher F, Paluch E, Heisenberg CP. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science. 2012;338:253–256. doi: 10.1126/science.1225399. [DOI] [PubMed] [Google Scholar]
- Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Annu Rev Pathol. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- Mbom BC, Nelson WJ, Barth A. beta-catenin at the centrosome: Discrete pools of beta-catenin communicate during mitosis and may co-ordinate centrosome functions and cell cycle progression. Bioessays. 2013 doi: 10.1002/bies.201300045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9:833–845. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke A, Giehl K. Regulation of adherens junctions by Rho GTPases and p120-catenin. Arch Biochem Biophys. 2012;524:48–55. doi: 10.1016/j.abb.2012.04.019. [DOI] [PubMed] [Google Scholar]
- Miller PW, Pokutta S, Ghosh A, Almo SC, Weis WI, Nelson WJ, Kwiatkowski AV. Danio rerio αE-catenin is a Monomeric F-actin Binding Protein With Distinct Properties from Mus musculus αE-catenin. J Biol Chem. 2013 doi: 10.1074/jbc.M113.458406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A, Ishihara S, Tsukita S. The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin-alpha catenin fusion molecules. J Cell Biol. 1994;127:235–245. doi: 10.1083/jcb.127.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandadasa S, Tao Q, Shoemaker A, Cha S-W, Wylie C. Regulation of classical cadherin membrane expression and F-actin assembly by alpha-catenins, during Xenopus embryogenesis. PLoS One. 2012:7. doi: 10.1371/journal.pone.0038756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan L, Witwer NE, Eisenmann DM. The divergent Caenorhabditis elegans beta-catenin proteins BAR-1, WRM-1 and HMP-2 make distinct protein interactions but retain functional redundancy in vivo. Genetics. 2001;159:159–172. doi: 10.1093/genetics/159.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejsum LN, Nelson WJ. A molecular mechanism directly linking E-cadherin adhesion to initiation of epithelial cell surface polarity. J Cell Biol. 2007;178:323–335. doi: 10.1083/jcb.200705094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ, Dickinson DJ, Weis WI. Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog Mol Biol Transl Sci. 2013;116:3–23. doi: 10.1016/B978-0-12-394311-8.00001-7. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci U S A. 2006;103:12451–12456. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc Natl Acad Sci U S A. 2012;109:13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Takeichi M. Evolution: structural and functional diversity of cadherin at the adherens junction. J Cell Biol. 2011;193:1137–1146. doi: 10.1083/jcb.201008173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Tsukita S. Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins. Dev Biol. 1999;216:406–422. doi: 10.1006/dbio.1999.9494. [DOI] [PubMed] [Google Scholar]
- Oda H, Uemura T, Shiomi K, Nagafuchi A, Tsukita S, Takeichi M. Identification of a Drosophila homologue of alpha-catenin and its association with the armadillo protein. J Cell Biol. 1993;121:1133–1140. doi: 10.1083/jcb.121.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai LM, Kirkpatrick C, Blanton J, Oda H, Takeichi M, Peifer M. Drosophila alpha-catenin and E-cadherin bind to distinct regions of Drosophila Armadillo. J Biol Chem. 1996;271:32411–32420. doi: 10.1074/jbc.271.50.32411. [DOI] [PubMed] [Google Scholar]
- Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat Genet. 2002;31:279–284. doi: 10.1038/ng908. [DOI] [PubMed] [Google Scholar]
- Peng X, Cuff LE, Lawton CD, DeMali KA. Vinculin regulates cell-surface E-cadherin expression by binding to beta-catenin. J Cell Sci. 2010;123:567–577. doi: 10.1242/jcs.056432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters T, van Roy F, van Hengel J. Functions of p120ctn isoforms in cell-cell adhesion and intracellular signaling. Front Biosci. 2012;17:1669–1694. doi: 10.2741/4012. [DOI] [PubMed] [Google Scholar]
- Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J Biol Chem. 2002;277:18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Weis WI. Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol Cell. 2000;5:533–543. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Issues in predicting protein function from sequence. Brief Bioinform. 2001;2:19–29. doi: 10.1093/bib/2.1.19. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Izard T. The cytoskeletal protein alpha-catenin unfurls upon binding to vinculin. J Biol Chem. 2012;287:18492–18499. doi: 10.1074/jbc.M112.351023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Izard T. Dimer asymmetry defines α-catenin interactions. Nat Struct Mol Biol. 2013;20:188–193. doi: 10.1038/nsmb.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci U S A. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura S, Miravet S, Piedra J, Garcia de Herreros A, Dunach M. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J Biol Chem. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- Sarpal R, Pellikka M, Patel RR, Hui FYW, Godt D, Tepass U. Mutational analysis supports a core role for Drosophila α-catenin in adherens junction function. J Cell Sci. 2012;125:233–245. doi: 10.1242/jcs.096644. [DOI] [PubMed] [Google Scholar]
- Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, Cavender J, Milano-Curto A, Rozen DE, Dingermann T, et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepis A, Nelson WJ. Adherens junction function and regulation during zebrafish gastrulation. Cell Adh Migr. 2012;6:173–178. doi: 10.4161/cam.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepis A, Sepich D, Nelson WJ. αE-catenin regulates cell-cell adhesion and membrane blebbing during zebrafish epiboly. Development. 2012;139:537–546. doi: 10.1242/dev.073932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B, de Jong D, Desalle R. Placozoa and the evolution of Metazoa and intrasomatic cell differentiation. Int J Biochem Cell Biol. 2009;41:370–379. doi: 10.1016/j.biocel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Schneider SQ, Finnerty JR, Martindale MQ. Protein evolution: structure-function relationships of the oncogene beta-catenin in the evolution of multicellular animals. J Exp Zool B Mol Dev Evol. 2003;295:25–44. doi: 10.1002/jez.b.6. [DOI] [PubMed] [Google Scholar]
- Sehgal RN, Gumbiner BM, Reichardt LF. Antagonism of cell adhesion by an alpha-catenin mutant, and of the Wnt-signaling pathway by alpha-catenin in Xenopus embryos. J Cell Biol. 1997;139:1033–1046. doi: 10.1083/jcb.139.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009:1. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink KE, Polakis P, Weis WI. Structural basis of the Axin-adenomatous polyposis coli interaction. EMBO J. 2000;19:2270–2279. doi: 10.1093/emboj/19.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol. 2009:1. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem. 2006;281:9971–9976. doi: 10.1074/jbc.M508778200. [DOI] [PubMed] [Google Scholar]
- Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Torres M, Stoykova A, Huber O, Chowdhury K, Bonaldo P, Mansouri A, Butz S, Kemler R, Gruss P. An alpha-E-catenin gene trap mutation defines its function in preimplantation development. Proc Natl Acad Sci U S A. 1997;94:901–906. doi: 10.1073/pnas.94.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Shimamura K, Miyatani S, Copeland NG, Gilbert DJ, Jenkins NA, Takeichi M. Mouse alpha N-catenin: two isoforms, specific expression in the nervous system, and chromosomal localization of the gene. Dev Biol. 1994;163:75–85. doi: 10.1006/dbio.1994.1124. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- Watabe M, Nagafuchi A, Tsukita S, Takeichi M. Induction of polarized cell-cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J Cell Biol. 1994;127:247–256. doi: 10.1083/jcb.127.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Uchida N, Imamura Y, Nagafuchi A, Fujimoto K, Uemura T, Vermeulen S, van Roy F, Adamson ED, Takeichi M. alpha-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J Cell Biol. 1998;142:847–857. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr Opin Struct Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Weis WI, Nelson WJ. Re-solving the cadherin-catenin-actin conundrum. J Biol Chem. 2006;281:35593–35597. doi: 10.1074/jbc.R600027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells NK, Spooner BS, Ash JF, Bradley MO, Luduena MA, Taylor EL, Wrenn JT, Yamaa K. Microfilaments in cellular and developmental processes. Science. 1971;171:135–143. doi: 10.1126/science.171.3967.135. [DOI] [PubMed] [Google Scholar]
- Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003;19:207–235. doi: 10.1146/annurev.cellbio.19.011102.111135. [DOI] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 2003;17:2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Le Trong I, Hinds TR, Stenkamp R, Kimelman D, Xu W. Crystal structure of a beta-catenin/APC complex reveals a critical role for APC phosphorylation in APC function. Mol Cell. 2004;15:523–533. doi: 10.1016/j.molcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of alpha-catenin: implications for modulation of cell adhesion. EMBO J. 2001;20:3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Yu DS, Lee DH, Kim SK, Lee CH, Song JY, Kong EB, Kim JF. Algorithm for predicting functionally equivalent proteins from BLAST and HMMER searches. J Microbiol Biotechnol. 2012;22:1054–1058. doi: 10.4014/jmb.1203.03050. [DOI] [PubMed] [Google Scholar]
- Zhao ZM, Reynolds AB, Gaucher EA. The evolutionary history of the catenin gene family during metazoan evolution. BMC Evol Biol. 2011;11:198. doi: 10.1186/1471-2148-11-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]