Abstract
The methylation of B-cell CLL/lymphoma 6 member B (BCL6B) DNA promoter was detected in several malignancies. Here, we quantitatively detect the methylated status of CpG sites of BCL6B DNA promoter of 459 patients with gastric cancer (GC) by using bisulfite gene sequencing. We show that patients with three or more methylated CpG sites in the BCL6B promoter were significantly associated with poor survival. Furthermore, by using the Akaike information criterion value calculation, we show that the methylated count of BCL6B promoter was identified to be the optimal prognostic predictor of GC patients.
Keywords: stomach, neoplasm, B-cell CLL/lymphoma 6 member B, survival, methylation
2. Introduction
Gastric cancer is the third most common cancer in China, with an incidence rate 2–3 times higher than the global average. This rate, combined with the large population, means that gastric cancer in China accounts for more than 40% of new gastric cancer cases world wide [1]. Owing to the lack of highly specific biomarkers of carcinogenesis and the precisely prognostic predictors of GC, the overall survival (OS) of patients has not significantly improved [2]. B-cell CLL/lymphoma 6 member B (BCL6B), also known as BAZF, is a member of the proto-oncogene Bcl6 family genes that encodes a sequence-specific transcriptional repressor containing the BTB/POZ domain in NH2-terminal region and zinc finger motifs in COOH-terminal region [3]. The expression of BCL6B mRNA was ubiquitously detected in many kinds of human tissues. The expression patterns of BAZF mRNA suggest that BCL6B may regulate differentiation in stages or lineages [4]. The 17-amino-acid region in the middle portion of BCL6 is a functional domain of transcriptional repressor activity and is responsible for inducibility of apoptosis in NIH3T3 cells [5]. BCL6b is also identified to be required for the enhanced magnitude of the secondary response of memory CD8+ T cells [6]. Recently, researchers reported that BCL6B played a pivotal role as a potential tumour suppressor in GC, and the detection of methylation of the BCL6B DNA promoter might be deemed an independent biomarker for the prognosis of GC [7]. Hypermethylation of the CpG islands in BCL6B promoter was demonstrated to be detected in 42.5% plasma DNA samples from GC patients, while no methylation of BCL6B promoter was found in the plasma DNA of healthy controls [8].
In view of the small number of patients and the qualitative method of the promoter methylation in those studies, we intend to detect the quantitative methylated levels of BCL6B DNA promoter in a large-scale patient study for elaborate elucidation of the prognostic predicted value of BCL6B promoter methylation in GC.
3. Results
3.1. Patient demographics
All 459 GC patient clinicopathological characteristics are listed in table 1. The median OS of all patients was 21 months. Of 459 patients, 61 (13.26%) were alive when the follow-up was over.
Table 1.
Patient information in this study.
| gender | |
| male | 314 (68.41%) |
| female | 145 (31.59%) |
| age at surgery | |
| ≤60 | 270 (58.82%) |
| >60 | 189 (41.18%) |
| tumour size | |
| <4.0 | 66 (14.38%) |
| ≥4.0 | 393 (85.62%) |
| tumour location | |
| upper third | 113 (24.62%) |
| middle third | 117 (25.49%) |
| lower third | 201 (43.79%) |
| more than 2/3 stomach | 28 (6.10%) |
| depth of tumour invasion (T stage) | |
| T1 | 5 (1.09%) |
| T2 | 46 (10.02%) |
| T3 | 284 (61.87%) |
| T4 | 124 (27.02%) |
| number of metastatic lymph nodes (N stage) | |
| N0 | 110 (23.97%) |
| N1 | 164 (35.72%) |
| N2 | 107 (23.31%) |
| N3 | 78 (17.00%) |
| location of lymph node metastasis | |
| no | 110 (23.97%) |
| perigastric | 159 (34.64%) |
| extragastric | 190 (41.39%) |
| Lauren classification | |
| intestinal | 122 (26.57%) |
| diffuse | 319 (69.50%) |
| mixed | 18 (3.93%) |
| methylated CpG site count | |
| 2 or less | 220 (47.93%) |
| 3 or more | 239 (52.07%) |
| methylated status of CpG +79 | |
| unmethylated | 229 (49.89%) |
| methylated | 230 (50.11%) |
3.2. Protein and mRNA expression of B-cell CLL/lymphoma 6 member B in gastric cancer tissues and normal gastric mucosal tissues
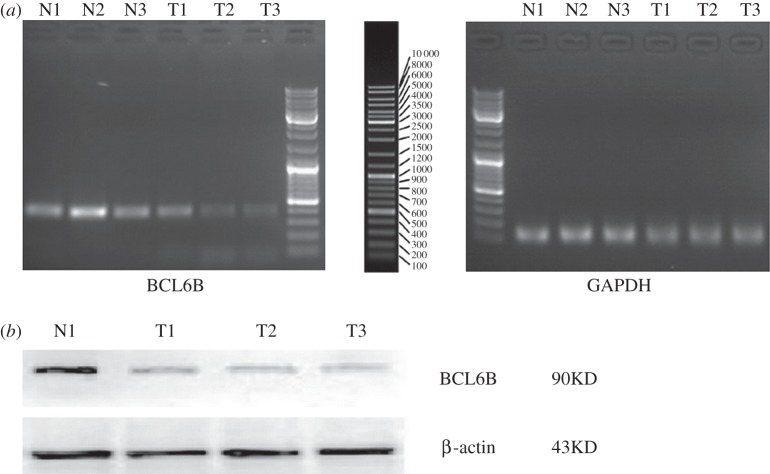
BCL6B mRNA expression was detected in 25 of 459 GC tissues and 25 normal gastric mucosal tissues by reverse transcription polymerase chain reaction (RT-PCR; figure 1a). We also found that there were significant differences of BCL6B mRNA expression among 25 GC tissues. The mean value of relative mRNA expression of BCL6B in 25 GC tissues was 0.410 ± 0.118, whereas the mean value of relative mRNA expression of BCL6B in 25 normal gastric mucosal tissues was 1.561 ± 0.406. The mean value of relative mRNA expression of BCL6B in 25 GC tissues was lower than that in 25 normal gastric mucosal tissues (p = 0.016).
Figure 1.
(a) BCL6B mRNA expression (RT-PCR) in GC tissues and in normal gastric mucosal tissues. (b) Western Blot analysis for BCL6B protein expression in GC tissues and in normal gastric mucosal tissues. T, GC tissues; N, normal gastric mucosal tissues.
Similarly, BCL6B protein expression was also detected in 25 of 459 GC tissues and 25 normal gastric mucosal tissues by Western blot, simultaneously (figure 1b). We found there were significant differences of BCL6B protein expression among 25 GC tissues. The mean value of relative protein expression of BCL6B in 25 GC tissues was 0.753 ± 0.144, whereas the mean value of relative protein expression of BCL6B in 25 normal gastric mucosal tissues was 1.662 ± 0.547. The mean value of relative protein expression of BCL6B in 25 GC tissues was much lower than that in 25 normal gastric mucosal tissues (p = 0.038).
3.3. Methylation detection of B-cell CLL/lymphoma 6 member B promoter
We detected the different levels of BCL6B promoter methylation (including methylation, non-methylation and partial methylation) in 25 of 459 GC tissues with the MSP analysis, whereas no BCL6B promoter methylation was found in 25 normal gastric mucosal tissues (figure 2). Of 25 GC tissues, five (20%) presented with the methylation of BCL6B promoter, 12 (48%) presented with the partial methylation of BCL6B promoter and eight (32%) presented with the non-methylation of BCL6B promoter.
Figure 2.
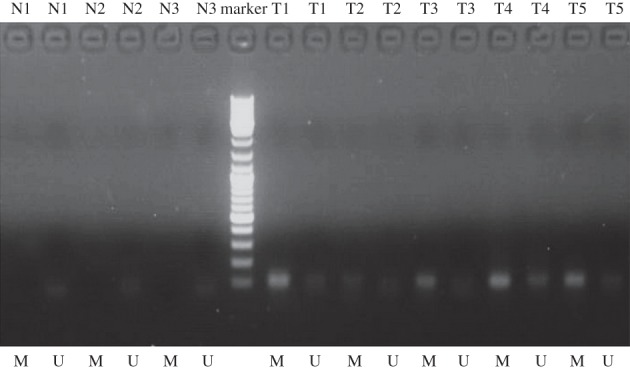
MSP detection of BCL6B promoter methylation in different GC tissues and normal gastric mucosal tissues. T, GC tissues; N, normal gastric mucosal tissues; M, methylated; U, unmethylated.
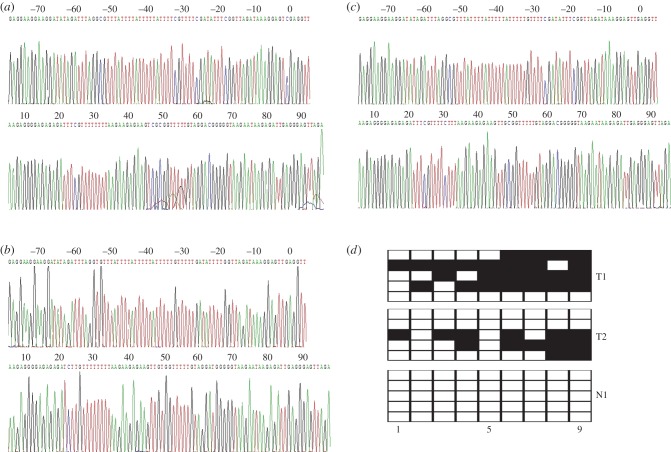
Subsequently, we adopted the bisulfite gene sequencing (BGS)-analysed methylated status of all CpG sites of BCL6B promoter to obtain the precise quantitative methylated degree of BCL6B promoter in all 459 GC patients. Methylated CpG site count of 459 GC patients ranged between 0 and 9. Of the 459 patients included in this study, 381 patients (83.01%) presented one or more methylated CpG sites and 78 patients (16.99%) presented no methylated CpG site. Patients without methylated CpG site had slightly longer mean OS than those with one or more methylated CpG sites (29.40 versus 28.08 months); there is no significant difference between the two groups of patients (p = 0.464). According to the result of cut-point analysis for the methylated CpG site count, 239 patients (52.07%) presented three or more methylated CpG sites and 220 patients (47.93%) presented two or fewer methylated CpG sites. No methylated CpG site was found in the normal gastric mucosal epithelial tissues. The methylation sequencing pictures and CpG site charts are shown in figure 3.
Figure 3.
(a) Bisulfite sequencing figure of BCL6B in GC tissue 1, (b) bisulfite sequencing figure of BCL6B in GC tissue 2, (c) bisulfite sequencing figure of BCL6B in normal gastric mucosal tissue and (d) bisulfite sequencing results in GC tissues and in normal gastric mucosal tissue. T, GC tissues; N, normal gastric mucosal tissues.
3.4. Survival analysis
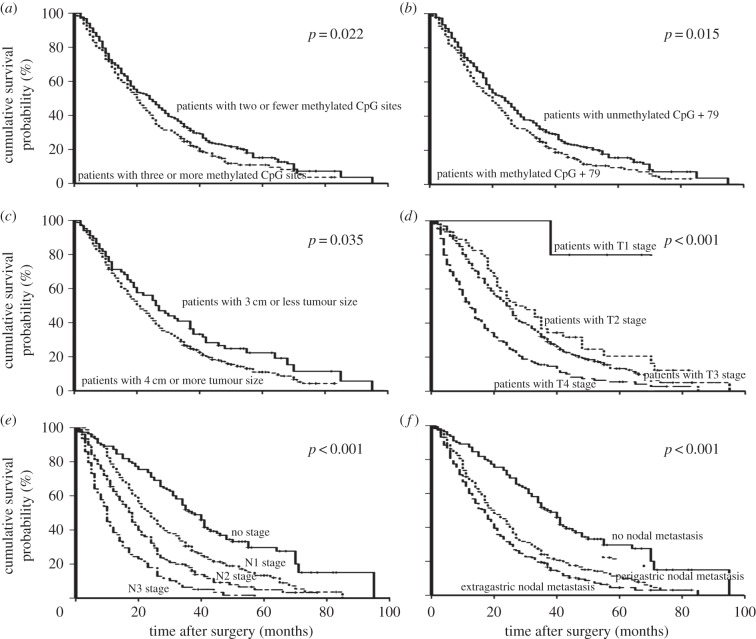
With the univariate survival analysis, four clinicopathological characteristics were found to have statistically significant associations with OS of 459 GC patients. They were as follows: T stage (p < 0.001), N stage (p < 0.001), tumour size (p = 0.035) and location of lymph node metastasis (p < 0.001) (table 2). We also demonstrated that the methylated status of CpG +79 of BCL6B promoter had significant association with the survival of 459 GC patients (p = 0.015). In addition, we found that patients with two or fewer methylated CpG sites had much longer median OS than those presenting with three or more methylated CpG sites of BCL6B promoter (p = 0.022) (table 2 and figure 4).
Table 2.
Survival analysis of 459 GC patients in this study.
| variables | median OS (mo) | χ2-value | univariate p-value | HR value | multivariate p-value | AIC value |
|---|---|---|---|---|---|---|
| gender | ||||||
| male | 22 | 0.633 | 0.426 | |||
| female | 20 | |||||
| age at surgery (years) | ||||||
| ≤60 | 20 | 0.007 | 0.934 | |||
| >60 | 23 | |||||
| tumour location | ||||||
| upper third | 21 | 7.685 | 0.053 | |||
| middle third | 18 | |||||
| lower third | 24 | |||||
| ≥2/3 stomach | 16 | |||||
| tumour size (cm) | ||||||
| <4.0 | 26 | 4.429 | 0.035 | |||
| ≥4.0 | 21 | |||||
| Lauren classification | ||||||
| intestinal | 26 | 5.576 | 0.062 | |||
| diffuse | 20 | |||||
| mixed | 17 | |||||
| depth of tumour invasion (T stage) | ||||||
| T1 | 70 | 41.108 | <0.001 | 1.510 (1.275–1.788) | <0.001 | 81.839 |
| T2 | 27 | |||||
| T3 | 24 | |||||
| T4 | 12 | |||||
| number of metastatic lymph nodes (N stage) | ||||||
| N0 | 37 | 101.047 | <0.001 | 1.548 (1.396–1.716) | <0.001 | 101.178 |
| N1 | 23 | |||||
| N2 | 18 | |||||
| N3 | 10 | |||||
| location of lymph node metastasis | ||||||
| no | 37 | 49.837 | <0.001 | |||
| perigastric | 20 | |||||
| extragastric | 17 | |||||
| methylated CpG site count | ||||||
| 2 or less | 24 | 5.218 | 0.022 | 1.263 (1.033–1.544) | 0.023 | 78.396 |
| 3 or more | 20 | |||||
| methylated status of CpG +79 | ||||||
| unmethylated | 24 | 5.880 | 0.015 | |||
| methylated | 20 | |||||
Figure 4.
Kaplan–Meier survival curves comparing months of survival in GC patients are shown for (a) methylated CpG site count of BCL6B promoter, (b) methylated status of CpG +79 of BCL6B promoter, (c) tumour size, (d) depth of tumour invasion (T stage), (e) number of metastatic lymph nodes and (f) location of lymph node metastasis.
All of the above six factors were included in a multivariate Cox proportional hazards model with bootstrapping performance to adjust for the effects of covariates. With the multivariate analysis, the independent predictors with the OS of all 459 GC patients were identified to be the methylated CpG site count (HR = 1.263, p = 0.023), N stage (HR = 1.548, p < 0.001) and T stage (HR = 1.510, p < 0.001) (table 2).
Lastly, we demonstrated that the methylated CpG site count of BCL6B promoter had a smaller AIC value than any of the other independent predictors calculated within the Cox proportional hazards regression model, representing the optimal prognostic predictor of GC (table 2).
4. Discussion
The human proto-oncogene BCL6 has been identified from chromosomal breakpoints in B-cell lymphomas. It functions as a sequence-specific transcriptional repressor owing to the NH2-terminal half of the BCL6 protein containing a repressor domain [9–12]. The BCL6 gene encodes a 92 to 98 kDa nuclear phosphoprotein [13,14] that contains the BTB/POZ domain in the NH2-terminal region and six Krüppel-type zinc finger motifs in the COOH-terminal region [9–11,15]. BCL6B, a homologue of BCL6, was identified to possess the 94% identical zinc finger motifs of BCL6 [16]. Several authors have reported relevant information for evaluation of the role of BCL6B in malignant disease. BCL6B was identified to be capable of repressing some STAT-induced transcription by binding to DNA sequences recognized by STAT factors [17]. It is believed that the JAK/STAT pathway plays a crucial role in carcinogenesis of GC [18], and repression of activation of STAT factors is a promising method for inhibition of the progress of GC [19]. Angiogenesis, based on the expression of angiogenic factors, is another key component of stomach canceration, and was demonstrated to significantly affect the progress, metastasis and prognosis of GC [20]. Recently, investigators reported that BCL6B, induced by vascular endothelial growth factor (VEGF)-A, supported angiogenic sprouting via BCL6B-CUL3-based poly-ubiquitination-dependent degradation of CBF1 to downregulate Notch signalling [21]. Subsequently, VEGF-A was reported to mediate BCL6B mRNA stabilization by activating heat shock protein 90β [22].
In 2011, BCL6B was demonstrated to be silenced or downregulated in all nine GC cell lines and readily expressed in normal gastric tissues [7]. Loss of BCL6B expression was regulated by promoter hypermethylation. Researchers demonstrated that re-expression of BCL6B in GC cell lines could inhibit colony formation, suppress cell viability, induce apoptosis and restrain tumorigenecity in nude mice [7]. They found that BCL6B hypermethylation was detected, respectively, in 49.0% (102/208) and 66.3% (67/101) of two independent cohorts of patients with GC, and demonstrated that the methylation of BCL6B promoter in GC tissues was an independent factor for the survival of patients by multivariate analysis [7]. Meanwhile, a similar conclusion was also drawn from plasmas of GC patients [8]. Therefore, the importance of BCL6B as a potential tumour suppressor in GC should be highlighted.
In this study, we initially found that 68% (17/25) GC tissues presented BCL6B promoter methylation by using MSP detection. Of those patients with BCL6B promoter methylation, we found that 12 presenting with the partial methylation of BCL6B promoter were detected by the MSP. However, we also found that the methylated degree of 12 patients presenting with the partial methylation of BCL6B promoter were inconsistent (figure 2). Therefore, we thought the detailed information of BCL6B promoter methylation should be quantitatively detected. Actually, many researchers proposed that methylated CpG site detection was appropriate for precise quantitative evaluation of the correlation between methylated levels of gene promoter and various abnormally biological events [23–26]. We meticulously analysed the methylated CpG sites of BCL6B promoter in 459 GC patients by using the bisulfite genomic sequencing (BGS) method with no fewer than five clones for each GC sample. After the methylated CpG site detection of BCL6B DNA promoter, 381 patients (83.01%) presented with one or more methylated CpG sites, and 78 patients (16.99%) presented without a methylated CpG site. However, the optimal cut-off value of the methylated CpG site count of BCL6B promoter was two, calculated by the cut-point survival analysis in 459 GC patients. Of 459 GC patients, 239 (52.07%) presenting with three or more methylated CpG sites had significantly shorter median OS than 220 (47.93%) with two or fewer methylated CpG sites of BCL6B promoter. Although we found that the methylated status of CpG +79 of BCL6B promoter, the methylated CpG site count of BCL6B promoter and other four clinicopathological variables were significantly associated with survival of GC patients, the methylated CpG site count of BCL6B promoter, N and T stages were identified to be the independent predictors of the OS of 459 GC patients by Cox regression with forward step procedures. Owing to its smaller AIC value, the methylated CpG site count of BCL6B promoter was ultimately demonstrated to be the optimal predictor of GC patients' prognosis by using the AIC value calculation within the Cox regression. The findings in this study indicate that quantitative BGS detection of BCL6B promoter methylation is appropriate to elucidate the methylated levels of BCL6B that can be applicable for accurate survival analysis.
5. Patients and methods
5.1. Data source
After the institutional review board of Tianjin Medical University Cancer Hospital (China) approved our study, data from the cancer registry of the Tianjin Cancer Institute were obtained. Data obtained from the registry were listed as follows: age, gender, tumour location, tumour size, depth of tumour invasion (T stage, according to the UICC TNM Classification for GC, 6th edn), number of metastatic lymph nodes (N stage, according to the UICC TNM Classification for GC, 6th edn), extent of lymph node metastasis, Lauren classification and follow-up vital status. Oral and written informed consents were also obtained from patients who were included in this study.
5.2. Patients and study samples
To analyse BCL6B promoter methylation, we collected 459 fresh GC tissues from patients with GC who underwent curative gastrectomy between April 2003 and December 2007 at the Department of Gastroenterology, Tianjin Medical University Cancer Hospital. A cohort of 25 normal gastric mucosal epithelial tissues from normal people was also obtained between 2004 and 2007 at the Department of Endoscopic Examination and Treatment, Tianjin Medical University Cancer Hospital. The tumour and normal gastric mucosal epithelial tissue samples were histologically verified. The patients were not subjected to radiation, chemical or biological treatment before potentially curative gastrectomy was performed. Adjuvant chemotherapy or radiotherapy was not routinely administered to the patients. The clinicopathological characteristics of these 459 GC patients are summarized in table 1. Consent regarding the use of tissue samples and records was obtained from each patient.
5.3. Surgical treatment
Curative resection was defined as the complete absence of grossly visible tumour tissues and metastatic lymph nodes remaining after resection was performed with pathologically negative resection margins. Primary tumours were resected en bloc with limited or extended lymphadenectomy (D1 or D2–3 according to the Japanese Gastric Cancer Association). Surgical specimens were evaluated according to the UICC TNM Classification for GC (6th edn).
5.4. DNA and RNA extractions
Genomic DNA was extracted from the 459 GC tissues and the 25 normal gastric mucosal tissues by using a QIAamp DNA mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Genomic DNA was modified using sodium bisulfite in EZ DNA Methylation-Gold kit (Zymo Research, Hornby, Canada). RNA was extracted from 25 of the 459 GC tissues and 25 normal gastric mucosal tissues by using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
5.5. Western blotting analysis
A total of 25 of the 459 GC tissues and 25 normal gastric mucosal tissues were each added to 1 ml of 100 mmol l−1 Tris/HCl (pH 7.5), 100 mmol l−1 NaCl, 0.5% sodium deoxycholate, 1 mmol l−1 ethylenediaminetetraacetic acid, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate and protease inhibitor. After blocking was performed, 50 µg of the sample was incubated for 60 min with mouse anti-BCL6B (Santa, sc56625, 1 : 1000 dilution) at room temperature. A gel imager system (Asia Xingtai Mechanical and Electrical Equipment Company, Beijing, China) was used to analyse images and determine grey values.
5.6. Semi-quantitative RT-PCR analysis
The mRNA expression of BCL6B was detected by subjecting 25 of the 459 GC tissues and 25 normal gastric mucosal tissues to RT-PCR. Total RNA was reverse-transcribed to cDNA in a 20 µl solution by using a reverse transcription kit (Invitrogen). The primers designed and used for BCL6B were listed as follows: forward sequence, 5′-TTGCTGTAGTTTGGTTGGGATT-3′; reverse sequence, 5′-ATGGGGAGAAAGAGGGAAGAG-3′. The GAPDH gene was used as an endogenous control of semi-quantitative DNA-PCR. Primers designed and used for GAPDH were listed as follows: forward sequence, 5′-GAAGGTGAAGGTCGGAGTC-3′; reverse sequence, 5′-GAAGATG GTGATGGGATTTC-3′. The following PCR cycling conditions were applied: 35 cycles of denaturation at 95°C for 3 min, annealing at 94°C for 30 s and extension at 56°C for 30 s, and a final extension at 72°C for 8 min. PCR products were electrophoresed on 2% agarose gel with ethidium bromide and visualized using a gel imager system (Asia Xingtai Mechanical and Electrical Equipment Company, Beijing, China).
5.7. Sodium bisulfite treatment
The genomic DNA was modified using sodium bisulfite in an EZ DNA Methylation-Gold kit.
5.8. Methylation-specific PCR
Twenty-five of 459 GC tissues and 25 normal gastric mucosal tissues were subjected to qualitative methylation analysis of the BCL6B promoter by methylation-specific PCR (MSP). The following BCL6B primers were used to detect the methylated (M) or unmethylated (U) alleles of the BCL6B promoter: for methylated alleles, BCL6B-MF, 5′-TTTTTATTTTCGTTTTCGATATTTC-3′ and BCL6B-MR, 5′-CGTCCTACAAAAACCGCG-3′; for unmethylated alleles, BCL6B-UF, 5′-TTATTTTTATTTTTGTTTTTGATATTTT-3′ and BCL6B-UR, 5′-CCCATCCTACAAAAACCACA-3′. A total of 25 cycles of MSP were performed using Ampli Taq-Gold (methylation-specific primers, annealing temperature 600°C; unmethylation-specific primers, annealing temperature 580°C). MSP primers were initially evaluated to verify whether or not any unbisulfited DNA is amplified, and the specificity of MSP was further confirmed by directly sequencing some PCR products. PCR was resolved using 2% agarose gel.
5.9. Bisulfite genomic sequencing
The methylation of the BCL6B promoter in 459 GC tissues and 25 normal gastric mucosal tissues was qualitatively analysed by the BGS. Hot start PCR with bisulfite-treated DNA was performed using a 173 bp PCR product spanning the promoter region from –78 bp to +95 bp relative to the transcription start site of BCL6B. The promoter region of BCL6B contains 11 CpG sites. The sequences of the PCR primers were listed as follows: forward sequence, 5′-GAGGAAGGAAGGATATAGATTTAGG-3′; reverse sequence, 5′-TCTAACTCCCTCAATCTCTTATTCTTAC-3′. The purified PCR products were cloned into the pUC18-T vector (Biodee, Beijing, China), and at least five clones from each sample were randomly selected and sequenced by Shanghai Sangon Co. (Shanghai, China).
5.10. Follow-up
After curative surgery, all of the patients were followed up every three months or six months for 2 years at the outpatient department; these patients were also followed up every year from the third year to the fifth year. Thereafter, these patients were followed up annually until their death. The median follow-up of the entire cohort was 44 months, ranging from 2 to 104 months. The follow-up of the patients included in this study was completed in December 2012. Ultrasonography, CT scans, chest X-ray and endoscopy were performed at every visit.
5.11. Statistical analysis
Median OS was determined using Kaplan–Meier method, and log-rank test was performed to determine significance. Potentially important factors in univariate analyses (p < 0.05) were included in multivariate analyses. OS was subjected to multivariate analysis by using the Cox proportional hazard model with forward step procedures. Hazard ratio (HR) and 95% confidence interval were calculated. Akaike information criterion (AIC) in a Cox proportional hazard regression model was calculated in terms of different categories to quantify their discriminatory ability. A small AIC value corresponds to an efficient model to predict outcomes [27]. With cut-point survival analysis [28], the optimal cut-off of the CpG site count was 2. Significance was set at p < 0.05. Statistical analyses were performed using SPSS 18.0.
Acknowledgements
The authors declare no competing financial interests.
The Institutional Research Ethics Committee of Tianjin Medical University Cancer Hospital approved the study protocol and provided permission to use patient clinical data.
Funding statement
This research was supported in part by grants from the National Basic Research Program of China (973 Program) 2010CB529301, the Anticancer Major Projects of Tianjin Municipal Science and Technology Commission 12ZCDZSY16400 and the Science Found Program of Tianjin Medical University 2012KYM01.
References
- 1.Rahman R, Asombang AW, Ibdah JA. 2014. Characteristics of gastric cancer in Asia. World J. Gastroenterol. 20, 4483–4490. (doi:10.3748/wjg.v20.i16.4483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer HJ, Wilke H. 2011. Treatment strategies in gastric cancer. Dtsch. Arztebl. Int. 108, 698–705. (doi:10.3238/arztebl.2011.0698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Laz EV, Waxman DJ. 2012. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell Biol. 32, 880–896. (doi:10.1128/MCB.06312-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakashita C, Fukuda T, Okabe S, Kobayashi H, Hirosawa S, Tokuhisa T, Miyasaka N, Miura O, Miki T. 2002. Cloning and characterization of the human BAZF gene, a homologue of the BCL6 oncogene. Biochem. Biophys. Res. Commun. 291, 567–573. (doi:10.1006/bbrc.2002.6481) [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Okada S, Hatano M, Okabe S, Tokuhisa T. 2001. A new functional domain of Bcl6 family that recruits histone deacetylases. Biochim. Biophys. Acta. 1540, 188–200. (doi:10.1016/S0167-4889(01)00128-8) [DOI] [PubMed] [Google Scholar]
- 6.Manders PM, et al. 2005. BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes. Proc. Natl Acad. Sci. USA 102, 7418–7425. (doi:10.1073/pnas.0501585102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu L, et al. 2012. Epigenetic inactivation of BCL6B, a novel functional tumour suppressor for gastric cancer, is associated with poor survival. Gut 61, 977–985. (doi:10.1136/gutjnl-2011-300411) [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Gao J, Xu L, Zeng Z, Sung JJ, Yu J. 2013. Promoter hypermethylation of BCL6B gene is a potential plasma DNA biomarker for gastric cancer. Biomarkers 18, 721–725. (doi:10.3109/1354750X.2013.853839) [DOI] [PubMed] [Google Scholar]
- 9.Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. 1993. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat. Genet. 5, 66–70. (doi:10.1038/ng0993-66) [DOI] [PubMed] [Google Scholar]
- 10.Akay OM, Aras BD, Isiksoy S, Toprak C, Mutlu FS, Artan S, Oner U, Gulbas Z. 2014. BCL2, BCL6, IGH, TP53, and MYC protein expression and gene rearrangements as prognostic markers in diffuse large B-cell lymphoma: a study of 44 Turkish patients. Cancer Genet. 207, 87–93. (doi:10.1016/j.cancergen.2014.02.001.) [DOI] [PubMed] [Google Scholar]
- 11.Chapuy B, et al. 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24, 777–790. (doi:10.1016/j.ccr.2013.11.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jardin F, Sahota SS. 2005. Targeted somatic mutation of the BCL6 proto-oncogene and its impact on lymphomagenesis. Hematology 10, 115–129. (doi:10.1080/10245330400026105) [DOI] [PubMed] [Google Scholar]
- 13.Phan RT, Dalla-Favera R. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432, 635–639. (doi:10.1038/nature03147) [DOI] [PubMed] [Google Scholar]
- 14.Chung KM, Chang ST, Huang WT, Lu CL, Wu HC, Hwang WS, Chang KY, Chuang SS. 2013. Bcl-6 expression and lactate dehydrogenase level predict prognosis of primary gastric diffuse large B-cell lymphoma. J. Formos. Med. Assoc. 112, 382–389. (doi:10.1016/j.jfma.2012.07.031) [DOI] [PubMed] [Google Scholar]
- 15.Hartmann S, Eray M, Döring C, Lehtinen T, Brunnberg U, Kujala P, Vornanen M, Hansmann ML. 2014. Diffuse large B cell lymphoma derived from nodular lymphocyte predominant Hodgkin lymphoma presents with variable histopathology. BMC Cancer 14, 332 (doi:10.1186/1471-2407-14-332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broxmeyer HE, Sehra S, Cooper S, Toney LM, Kusam S, Aloor JJ, Marchal CC, Dinauer MC, Dent AL. 2007. Aberrant regulation of hematopoiesis by T cells in BAZF-deficient mice. Mol. Cell. Biol. 27, 5275–5285. (doi:10.1128/MCB.01967-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartatik T, Okada S, Okabe S, Arima M, Hatano M, Tokuhisa T. 2001. Binding of BAZF and Bc16 to STAT6-binding DNA sequences. Biochem. Biophys. Res. Commun. 284, 26–32. (doi:10.1006/bbrc.2001.4931) [DOI] [PubMed] [Google Scholar]
- 18.Lee IO, Kim JH, Choi YJ, Pillinger MH, Kim SY, Blaser MJ, Lee YC. 2010. Helicobacter pylori CagA phosphorylation status determines the gp130-activated SHP2/ERK and JAK/STAT signal transduction pathways in gastric epithelial cells. J. Biol. Chem. 285, 16 042–16 050. (doi:10.1074/jbc.M110.111054.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.To KF, et al. 2004. Constitutional activation of IL-6-mediated JAK/STAT pathway through hypermethylation of SOCS-1 in human gastric cancer cell line. Br. J. Cancer 91, 1335–1341. (doi:10.1038/sj.bjc.6602133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth EC, Cunningham D. 2012. Targeted therapy for gastric cancer. Curr. Treat. Options. Oncol. 13, 377–389. (doi:10.1007/s11864-012-0192-6) [DOI] [PubMed] [Google Scholar]
- 21.Ohnuki H, et al. 2012. BAZF, a novel component of cullin3-based E3 ligase complex, mediates VEGFR and Notch cross-signaling in angiogenesis. Blood 119, 2688–2698. (doi:10.1182/blood-2011-03-345306) [DOI] [PubMed] [Google Scholar]
- 22.Miwa D, Sakaue T, Inoue H, Takemori N, Kurokawa M, Fukuda S, Omi K, Goishi K, Higashiyama S. 2013. Protein kinase D2 and heat shock protein 90 beta are required for BCL6-associated zinc finger protein mRNA stabilization induced by vascular endothelial growth factor-A. Angiogenesis 16, 675–688. (doi:10.1007/s10456-013-9345-x) [DOI] [PubMed] [Google Scholar]
- 23.Colaneri A, Wang T, Pagadala V, Kittur J, Staffa NG, Jr, Peddada SD, Isganaitis E, Patti ME, Birnbaumer L. 2013. A minimal set of tissue-specific hypomethylated CpGs constitute epigenetic signatures of developmental programming. PLoS ONE 8, e72670 (doi:10.1371/journal.pone.0072670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartley I, Elkhoury FF, Heon Shin J, Xie B, Gu X, Gao Y, Zhou D, Haddad GG. 2013. Long-lasting changes in DNA methylation following short-term hypoxic exposure in primary hippocampal neuronal cultures. PLoS. One 8, e77859 (doi:10.1371/journal.pone.0077859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi T, et al. 2013. Estimation of the fraction of cancer cells in a tumor DNA sample using DNA methylation. PLoS ONE 8, e82302 (doi:10.1371/journal.pone.0082302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YK, Jin S, Duan S, Lim YC, Ng DP, Lin XM, Yeo GSH, Ding C. 2014. Improved reduced representation bisulfite sequencing for epigenomic profiling of clinical samples. Biol. Proced. Online 16, 1 (doi:10.1186/1480-9222-16-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho YK, Chung JW, Kim JK, Ahn YS, Kim MY, Park YO, Kim WT, Byun JH. 2008. Comparison of 7 staging systems for patients with hepatocellular carcinoma undergoing transarterial chemoembolization. Cancer 112, 352–361. (doi:10.1002/cncr.23185) [DOI] [PubMed] [Google Scholar]
- 28.Smith DD, Schwarz RR, Schwartz RE. 2005. Impact of total lymph node count on staging and survival after gastrectomy for gastric cancer: data from a large US-population database. J. Clin. Oncol. 23, 7114–7124. (doi:10.1200/JCO.2005.14.621) [DOI] [PubMed] [Google Scholar]