Abstract
Prefoldin is a cochaperone, present in all eukaryotes, that cooperates with the chaperonin CCT. It is known mainly for its functional relevance in the cytoplasmic folding of actin and tubulin monomers during cytoskeleton assembly. However, both canonical and prefoldin-like subunits of this heterohexameric complex have also been found in the nucleus, and are functionally connected with nuclear processes in yeast and metazoa. Plant prefoldin has also been detected in the nucleus and physically associated with a gene regulator. In this review, we summarize the information available on the involvement of prefoldin in nuclear phenomena, place special emphasis on gene transcription, and discuss the possibility of a global coordination between gene regulation and cytoplasmic dynamics mediated by prefoldin.
Keywords: prefoldin, gene transcription, cytoskeleton, chromatin, protein folding
2. Introduction
Misfolded proteins are detected shortly after their synthesis or after denaturation events, and are targeted to refolding. A large set of molecular chaperones and auxiliary factors are involved in this phenomenon (reviewed in [1]). Prefoldin was first described as a cochaperone [2], capable of capturing unfolded polypeptides and transferring them to the ATP-dependent class II chaperonin CCT [3], also known as c-cpn [4] or TriC [5].
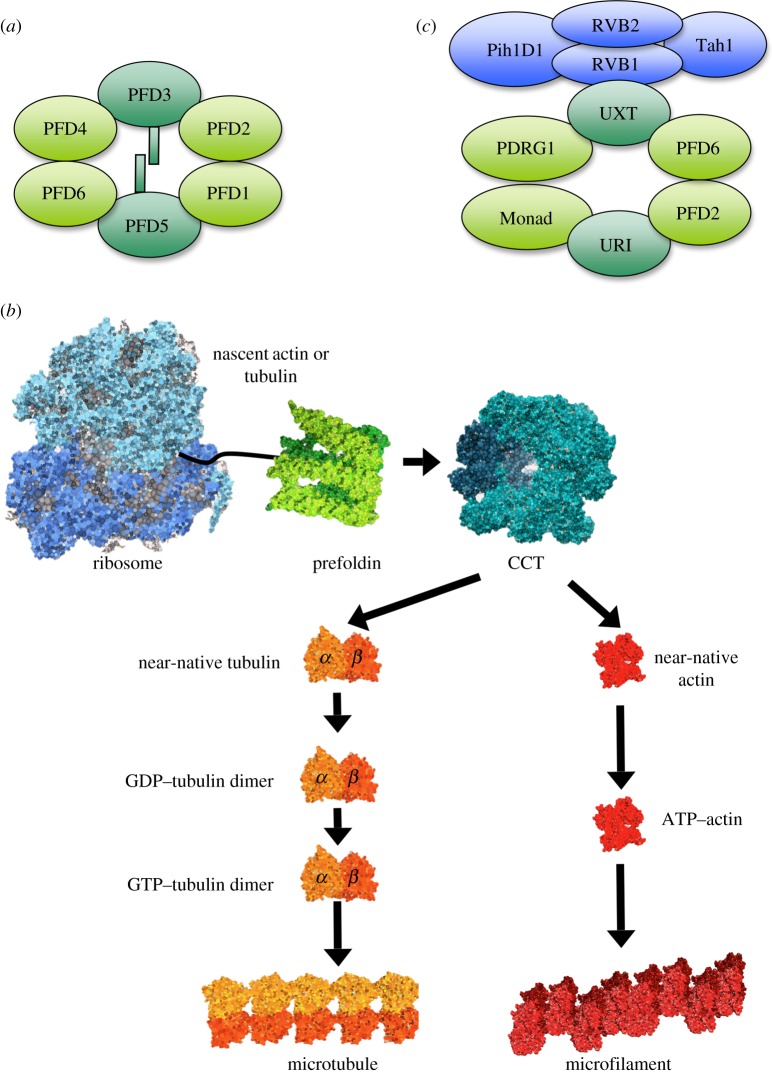
Prefoldin is not present in Eubacteria, but it is present in Archaea [6], suggesting that its ubiquitous presence in the eukaryotic kingdom is archaeal in origin [7]. Prefoldin is a heterohexameric complex. Whereas archaeal prefoldin is composed of two identical α and four identical β subunits, canonical eukaryotic prefoldin is composed of two different α and four different β subunits (figure 1a and table 1). All eukaryotic organisms, from yeast to human, share this heterohexameric structure, indicating an early differentiation of the prefoldin subunits during the evolution of eukaryotes. Eukaryotic prefoldin is evolutionarily conserved, because human and plant subunits functionally complement yeast prefoldin mutants [8,9]. In all cases, two subunits of the α class form a dimer, onto which four subunits of the beta class assemble and produce a jellyfish-like complex [10]. This complex consists of a double β barrel assembly with six long tentacle-like coiled coils protruding from it. The distal regions of these coiled coils expose hydrophobic patches, required for the binding of misfolded proteins. This feature situates prefoldin among the set of chaperone factors that use clamp-like structural features to grip substrate proteins [11].
Figure 1.
The prefoldin complex. (a) Canonical prefoldin is a heterohexameric complex composed of two α subunits (dark green), which play a central structural role, and four β subunits (light green). (b) The best-characterized function of prefoldin is the cotranslational folding of proteins. Prefoldin binds unfolded polypeptides and transfers them to the ATP-dependent chaperon CCT prior to its assembly into high-order protein structures, such as microtubules and actin filaments. (c) In addition to canonical complexes, which retain the structure of archaeal prefoldin, eukaryotes exhibit prefoldin-like complexes. In these, the two α and some of the β canonical subunits are replaced with alternative polypeptides. Prefoldin-like complexes interact and functionally cooperate with other cochaperones such as the R2TP complex (purple).
Table 1.
The canonical and prefoldin-like subunit nomenclature. Most prefoldin subunits have synonymic names. In order to facilitate comprehension, in this review, we have chosen the main nomenclature of metazoan prefoldin (PFDN1-6). Likewise, the mammalian prefoldin-like subunits have been named several times. We call them URI and UXT for the sake of simplicity.
| Archaea | S. cerevisiae | higher eukaryotes |
|---|---|---|
| β | Pfd1/Gim6 | PFDN1 |
| Pfd2/Gim4 | PFDN2 | |
| Pfd4/Gim3 | PFDN4 | |
| Pfd6/Gim1/Yke2 | PFDN6/HKE2 | |
| α | Pfd3/Gim2/Pac10/Rks2 | PFDN3/VBP1 |
| Pfd5/Gim5 | PFDN5/MM1 | |
| Bud27 | URI/RMP | |
| UXT/Art-27 |
Cytoskeleton components are the best-known targets of eukaryotic prefoldin [8]. This captures unfolded actin, α- and β-tubulin cotranslationally, and remains bound to the relatively unfolded polypeptides until their posttranslational delivery to CCT [12] (figure 1b). Recognition of actin and tubulin by prefoldin involves specific interactions between certain domains of the target proteins [13] and the distal ends of different, but overlapping, sets of prefoldin subunits [14]. The so-formed prefoldin-target binary complex is then able to interact with CCT. In this ternary complex, prefoldin transfers actin to CCT following a handoff mechanism [15]. Interestingly, the interaction between actin and eukaryotic prefoldin maps inside the cavity formed by its six subunits, whereas archaeal prefoldin stabilizes unfolded proteins by interacting with the distal regions of the chaperone tentacles. This suggests that the substrate interaction mechanism of prefoldin has diverged through evolution and potentially reflects a narrower range of substrates stabilized by prefoldin in eukaryotes [16]. Consistent with a role played by prefoldin in actin and tubulin folding, the deletion of prefoldin-encoding genes in Saccharomyces cerevisiae results in impaired cytoskeleton functions [2]. These cytoskeletal defects of the prefoldin mutants are even more marked in the absence of the phosducin-like protein 3, a factor that physically interacts with CCT and modulates its chaperonin ATPase activity in vitro [17].
While none of the prefoldin subunits is essential for yeast viability, the mutation of prefoldin genes or the depletion of their products in Caenorhabditis elegans and Drosophila results in embryonic lethality. In both cases, prefoldin-defective cells show low tubulin levels and several cytoskeleton abnormalities [18,19]. In the nematode, prefoldin depletion leads to a lower microtubule growth rate [18] and prevents pronuclear migration [20]. mgr flies, which are mutated in the gene encoding PFDN3, exhibit circular mitotic figures and loss of meiotic spindle integrity [19]. In Arabidopsis, PFDN3, PFDN5 and PFDN6 are required for normal microtubule dynamics and organization [9,21], whereas the genetic murine models affected in PFDN1 and PFDN5 display developmental defects that were interpreted in terms of cilia and cytoskeleton defects [22,23].
All these pieces of evidence have shaped the eukaryotic prefoldin concept as a highly specialized cochaperone for actin and tubulin folding. This specialization would be consistent with the more severe phenotype of CCT knockdown than the prefoldin one in C. elegans [18]. However, a more in-depth review of the scientific literature reveals that eukaryotic prefoldin has also been connected to phenomena that are not directly linked to the cytoskeleton, such as protein aggregation. Prefoldin prevents protein aggregation in brain cells under conditions where protein degradation is compromised [24]. This role of prefoldin explains its protective effect against polyglutamine toxicity and the accumulation of aggregated pathogenic huntingtin [25]. Human prefoldin also inhibits amyloid-β fibrillation and contributes to the formation of non-toxic amyloid-β aggregates in vitro, which is consistent with its upregulation in a murine model for Alzheimer's disease [26] and the genetic association of prefoldin variants with this pathology [27].
The involvement of prefoldin subunits in the cytoplasmic assembly of some non-cytoskeletal complexes has also been well established. In this case, mammalian PFDN2 and PFDN6 form a complex together with UXT, RPB5, WDR92/Monad, PDRG1 and URI [28]. In this complex, which is supposed to adopt a prefoldin-like structure, non-canonical prefoldin proteins substitute for the α subunits and some of the β subunits (figure 1c), although no evident sequence similarity exists between canonical and non-canonical prefoldin polypeptides. The prefoldin-like complex participates in the cytoplasmic assembly of RNA polymerase II [29], and in the stabilization and assembly of phosphatidylinositol-3 kinase-related protein kinase [30]. In both cases, it cooperates with another cochaperone, the R2TP complex, which is composed of four subunits: RVB1, RVB2, PihD1 and hSpagh [31]. The latter two subunits of R2TP interact with the Hsp90 chaperone, whereas RVB1 and RVB2 are AAA+ ATPases that participate in a set of additional cellular activities. They act in the context of R2TP, such as in the maturation of small nucleolar ribonucleoprotein complexes, or independently, contributing to histone acetylation, chromatin remodelling, telomere dynamics and mitotic spindle assembly (for a review, see [32]). Functional analyses in mammalian cells indicate that the URI subunit of this complex is a target of nutrient signalling and participates in TOR kinase-controlled gene expression [28]. URI is conserved through the evolution of eukaryotes, and its yeast homologue Bud27 also mediates TOR-controlled gene expression [28] and is involved in the cytoplasmic assembly of all three yeast nuclear RNA polymerases [33].
α-class prefoldin-like subunit UXT is also located in human centrosomes, associates with γ-tubulin and its overexpression disrupts the centrosome structure [34]. This finding indicates that the canonical prefoldin complex may share some of its cytoskeleton functions with non-canonical prefoldin subunits.
3. Prefoldin shuttles between the cytoplasm and the nucleus, and acts on DNA-binding proteins
All the phenomena related to prefoldin that we have examined so far in this review take place in the cytoplasm. However, both the canonical and prefoldin-like subunits have also been found in the nucleus. Proteomic analyses of URI nuclear interactors indicate that all the components of the R2TP/prefoldin-like complex, including the canonical prefoldin subunits PFDN2 and PFDN6, are found in the nucleus of prostate cells, and shuttle between the nucleus and the cytoplasm, probably together with RNA pol II [35]. The short isoform UXT-V2/Art-27, of UXT, which has 157 amino acids, is primarily present in the nucleus, whereas UXT-V1, which encompasses 169 amino acids, is predominantly found in the cytoplasm [36,37]. This nuclear localization preference of some splice variants also happens in the case of human PFDN5/MM-1. MM-1β and MM-1δ localize mainly to the cytoplasm, whereas MM-1α and MM-1γ are found in the nucleus [38].
Canonical plant prefoldin, at least its subunits PFDN5 and PFDN6, is also found in the nucleus of Nicotiana benthamiana and Arabidopsis thaliana leaf cells [39]. Its nuclear localization is not constitutive, but dependent on the physical interaction with the nuclear DELLA proteins. Plant prefoldin moves from the nucleus to the cytosol in response to environmental or endogenous cues that cause degradation of DELLA proteins [39]. Human PFDN3, also known as von Hippel–Lindau binding protein1 (VBP1), because of its physical interaction with VHL, localizes to the cytoplasm when it is solely expressed, but it translocates to the nucleus when it is co-expressed with VHL [40]. In turn, VHL localization to the nucleus depends on UXT [41]. Localization of URI in the nucleus also depends of its interaction with a partner, in this case with the DNA methyltransferase 1-associating protein (DMAP1), which inhibits the cytoplasmic localization signal of URI and favours its import into the nucleus [42].
In addition to their cytoplasmic localization, all yeast canonical prefoldin subunits can be detected in the nucleus. Moreover, they accumulate in the nucleus when the nuclear export systems based on Xpo1 and Mex67 become genetically inactivated [43]. So, from yeast to metazoa, prefoldin subunits are capable of migrating to the nucleus and of being translocated back to the cytoplasm by active export systems.
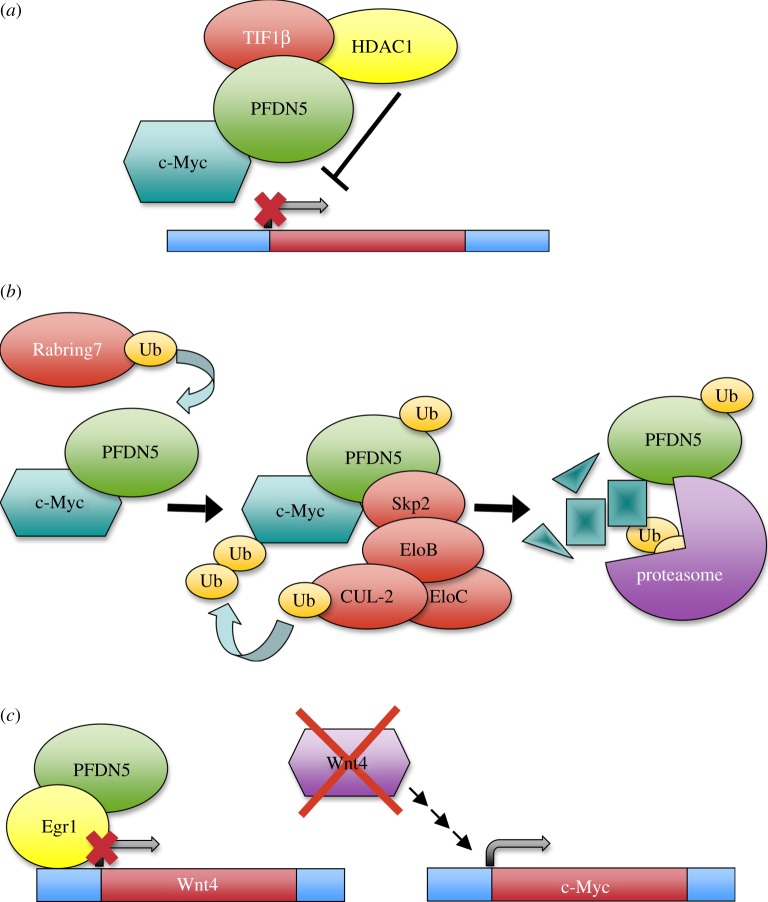
Localization of prefoldin in the nucleus may be the result of the regulation of its cytoplasmic functions by means of a cytoplasm-exclusion mechanism, as has been demonstrated in Arabidopsis [39]. In addition to this passive presence of prefoldin in the nucleus, several pieces of evidence indicate that the nucleus is not just a reservoir for stand-by prefoldin. URI, for instance, is required for DNA stability in C. elegans [44]. A clearer example of a nuclear function of prefoldin is its participation in the degradation of the HIV integrase after the provirus integration into the host genome, which is a strict requirement for HIV expression [45]. PFDN3 binds the integrase [46] and mediates its interaction with the cullin2-based VHL ubiquitin ligase, which is essential for its polyubiquitination and subsequent proteosome-mediated degradation [45] (figure 2). At least two other prefoldin subunits (PFDN1 and PFDN6) influence HIV expression at a post-integration step, suggesting that the whole prefoldin complex acts on the HIV integrase [45]. PFDN3 also facilitates the VHL-mediated degradation of other well-known DNA-interacting proteins, such as the DNA mismatch repair protein hMSH4 [47]. In this case, however, the co-localization of these two proteins is mainly perinuclear [48].
Figure 2.
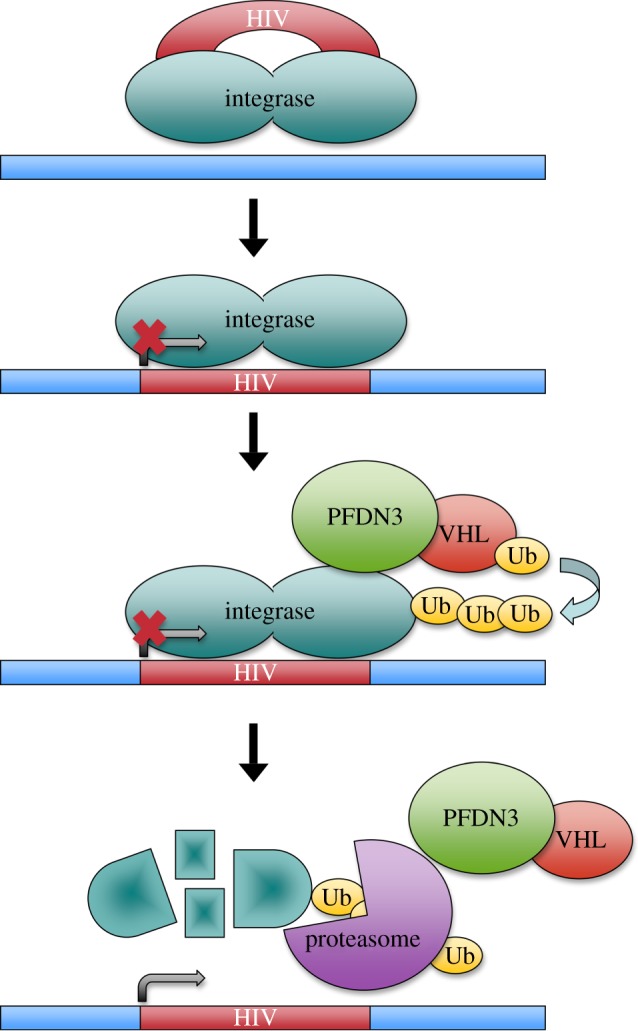
Human PFDN3 favours HIV transcription by driving its integrase into degradation. HIV integrase remains bound to proviral DNA after its integration into the host genome. PFDN3 is required for the in situ ubiquitination of the HIV integrase and its subsequent degradation by the proteasome, by virtue of its physical interaction with the E3-ubiquitin ligase VHL.
This nuclear role of mammalian PFDN3 in protein degradation is quite the opposite to that described for Drosophila PFDN3 in the microtubule dynamics context. In this case, PFDN3 promotes microtubule stabilization when tubulins are correctly folded by prefoldin, and tubulin destruction when they are not [19]. This double role of prefoldin in protein folding and degradation resembles that of protein segregases such as the VCP/p97/Cdc48 AAA-ATPase, which segregates ubiquitinated proteins from stable assemblies with proteins, membranes and chromatin [49]. Targets of VCP can either end in degradation, or can survive as free subunits and recycle [50]. No physical or functional connection between prefoldin and VCP has been established so far.
4. Prefoldin plays transcriptional roles
Among the nuclear proteins bound and influenced by prefoldin, transcription factors are the most frequent (table 2). Prefoldin-like subunit UXT binds the EVI1 transcriptional repressor and suppresses its cell transformation activity [59]. UXT is also an integral component of the NF-κB enhanceosome and is essential for its nuclear function [60]. Its knockdown leads to impaired NF-κB activity and dramatically attenuates the expression of NF-κB-dependent genes [60]. UXT is therefore an optimal target for NF-κB regulation. The Epstein–Barr virus BGLF4 kinase downregulates NF-κB transactivation by means of UXT phosphorylation, and the subsequent interference of this modification with the interaction between UXT and NF-κB [61]. Amyotrophic lateral sclerosis 2 protein has also been suggested to modulate the NF-κB pathway through its physical interaction with UXT [62].
Table 2.
Nuclear proteins that physically interact with canonical or prefoldin-like subunits. For detailed explanations, see the text.
| prefoldin subunit | interactor (biological process) | other factors involved | organism | reference |
|---|---|---|---|---|
| PFDN3 | HIV integrase | VHL | human | [45] |
| hMSH4 (DNA repair) | VHL, p97 | human | [47] | |
| NF-κB (transcription) | HBx | human | [51] | |
| PFDN5 | c-Myc (transcription) | HDAC1–mSin3, TIF1β | human | [52,53] |
| Skp2–ElonginB–ElonginC–Cullin2, Rabring7 | human | [54,55] | ||
| EGR1(transcription) | human | [56] | ||
| p73 (transcription) | human | [57] | ||
| PFDN3, PFDN5 | DELLA (gene regulation) | Arabidopsis thaliana | [39] | |
| URI | HBx (transcription) | Rpb5 | human | [58] |
| DMAP1 (transcription) | human | [42] | ||
| UXT | EVI1 (transcription) | human | [59] | |
| NF-κB (transcription) | human | [60] | ||
| HBV EGLF4 kinase (gene regulation) | human | [61] | ||
| ALS2 (gene regulation) | human | [62] | ||
| androgen receptor (transcription) | VHL | human | [36,41,63] | |
| URI | human | [35] | ||
| LRP16 (transcription) | human | [64] | ||
| TAF130 (transcription) | human | [36] | ||
| Sp1 (transcription) | human | [36] |
UXT is not only involved in NF-κB-dependent transcription regulation, but it also contributes to the regulation of androgen-dependent genes by binding the N-terminal domain of the androgen receptor (AR) and enhancing its transcriptional activation [36,63]. UXT also interacts with LRP16, a macrodomain-containing protein that functions as a coactivator of AR and other nuclear receptors [64]. The positive contribution of UXT- to AR-dependent transactivation seems to be mediated, at least partially, by the capacity of UXT to interact with VHL and to facilitate VHL-dependent ubiquitination of AR [41]. UXT interacts directly with VHL [41] and without the involvement of PFDN3, indicating that this ubiquitinating protein can interact with both canonical and prefoldin-like subunits. Two-hybrid assays suggest that UXT is also able to interact with the human transcription factor Sp1 and the TBP-interacting protein TAF130 [36].
A different prefoldin-like subunit, URI, can repress AR-mediated transcription by interacting physically with UXT in the chromatin context [35]. URI seems to inhibit AR recruitment to target genes because it is bound to chromatin prior to the hormonal activation of AR [35]. URI also inhibits transactivation by other gene-specific transcription factors, such as herpes simplex virus transactivator VP16 and hepatitis B virus protein X (HBx) [58]. In this case, URI inhibits HBx-activated transcription by competing with the activator to bind the RNA polymerase II subunit Rpb5 [58]. The co-repressor activity of URI is also related to its interaction with DMAP1, a partner of the histone deacetylase HDAC2 [42]. Interestingly, canonical prefoldin subunit PFDN3 also influences HBx transcriptional function, yet, in this case, it cooperates positively with this viral protein in the activation of the NF-κB transcription factor [51]. All these data indicate considerable plasticity in the ability of the different canonical and prefoldin-like subunits to interact with transcription factors, and suggest that they do not necessarily shape a single functional entity in the nucleus of mammalian cells.
In addition to the degradation of HIV integrase, the best-characterized contribution of a prefoldin subunit to transcription is the action of PFDN5/MM-1 as a co-repressor of the E-box-dependent transactivation activity of c-Myc [52]. The MM-1α and MM-1γ nuclear isoforms bind the N-proximal region of c-Myc, which accommodates one of its transactivation domains, and repress its transcriptional activity [38]. A point mutation in PFDN5, which is often observed in patients with leukaemia or lymphoma, abrogates all of its repressive activities towards c-Myc, indicating that PFDN5 behaves like a tumour suppressor [65]. The repressive function of PFDN5 is due to its ability to recruit the histone deacetylase HDAC1–mSin3 complex via transcriptional co-repressor TIF1β. This recruitment antagonizes the histone transacetylases bound to the N-terminal domain of c-Myc, thereby inhibiting chromatin remodelling [53] (figure 3a). This inhibitory effect of PFDN5 on c-Myc offers a chance to modulate this transcription factor; a good example of it is the ability of the hepatitis C virus ARFP/F protein to enhance the gene transactivation activity of c-Myc by interfering in its interaction with PFDN5 [66].
Figure 3.
Human PFDN5 is involved in three different control mechanisms of c-Myc. (a) PFDN5 binds the N-terminal region of c-Myc, and represses its transcriptional activity by recruiting the TIF1β correpressor and the histone deacetylase HDAC1–mSin3 complex. (b) PFDN5 drives c-Myc into proteasome-dependent degradation by recruiting the ubiquitin ligase Skp2–ElonginC–ElonginB–Cullin2 complex. PFDN5 mono-ubiquitination, which is induced by Rabring7, stimulates this process. (c) PFDN5 and Egr1 cooperate in the transcriptional repression of Wnt4, which is one of the elements of the Wnt-β-catenin pathway that positively controls the c-Myc gene.
The negative action of PFDN5 on the c-Myc function also takes place at two other levels. It favours c-Myc degradation by recruiting the ubiquitin ligase Skp2–ElonginC–ElonginB–Cullin2 complex, and driving it to the proteasome via the 26S subunit Rpt3 [54] (figure 3b). The monoubiquitination of PFDN5 by Rabring7, a Rab7-binding and RING finger-containing protein, stimulates this second role of PFDN5 in the control of c-Myc [55] (figure 3b). In addition, PFDN5 and the Egr-1 repressor bind and downregulate the promoter of the wnt4 gene. Because the c-Myc gene is the target of the Wnt–β-catenin pathway, PFDN5 also inhibits the expression of c-Myc by this indirect mechanism [56] (figure 3c).
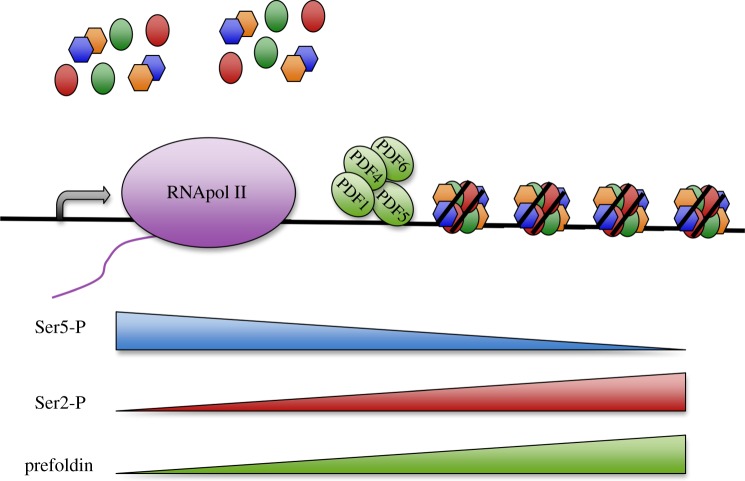
The above-described interactions indicate a role of prefoldin subunits in the regulation of the transactivation capacity or stability of gene-specific transcription factors. With the exception of its contribution to HIV expression, the transcriptional roles of prefoldin have been described for single subunits. Although the presence of other prefoldin subunits cannot be ruled out, the mechanisms described so far do not involve the action of prefoldin complexes. Saccharomyces prefoldin, however, seems to play a transcriptional role that involves at least the concerted action of PFDN1, PFDN2, PFDN5 and PFDN6 [43] (figure 4). These four canonical prefoldin subunits are recruited together to yeast transcribed genes in a transcription-dependent manner. The profile of recruited prefoldin parallels the phosphorylation of the Ser2 residues of Rpb1 CTD, a well-known marker of RNA polymerase II elongation activity. Accordingly, the deletion of any of the genes encoding these four prefoldin subunits provokes transcription elongation defects [43]. In agreement with this role of yeast prefoldin in transcription elongation, genes longer than 4 kpb are particularly affected. The absence of prefoldin increases the density of histones that remain bound to these genes under intensive transcription. However, nucleosome remodelling, reflected in the sensitivity of chromatin to micrococcal nuclease digestion, remains unaffected by the absence of prefoldin. Together, these pieces of evidence indicate a role of prefoldin in histone eviction after the cotranscriptional destabilization of nucleosomes [43]. This prefoldin–chromatin connection is fully consistent with the strong genetic interactions detected between prefoldin and a wide set of chromatin factors [67,68]. Interestingly, the yeast CCT mutants display the very same pattern of genetic interactions with chromatin factors [69]. Moreover, mouse CCT exhibits a nucleocytoplasmic distribution and associates to heterochromatin [70], and depletion of Cct2 interferes with HIV integration at a post-integration step, exactly as prefoldin does [45]. Taken altogether, the available experimental evidence suggests that prefoldin and CCT might also cooperate in the transcriptional dynamics of chromatin.
Figure 4.
Yeast prefoldin stimulates transcription elongation by favouring chromatin dynamics. A subset of canonical prefoldin subunits, including PFDN1, 4, 5 and 6, binds transcribed genes in a transcription-dependent manner and contributes to transcription elongation. This effect is maximal in long genes, where prefoldin stimulates cotranscriptional histone eviction. Prefoldin binding is absent in the promoter region and is maximal in the 3′ end of the gene body to correlate with the presence of Ser2-hyperphosphorylated RNA polymerase II.
5. Final perspective: prefoldin in global cell regulation
To date, we have shown that the scientific literature contains a significant amount of information on the presence of canonical and prefoldin-like subunits in the nucleus, and their functional involvement in gene transcription. The examples that we cite demonstrate that, from yeast to mammals, a given prefoldin subunit can play cytoplasmic and nuclear roles in the same organism. Moreover, a set of yeast canonical subunits plays concerted actions in both cytoplasmic cytoskeleton assembly and nuclear gene transcription. This fact might merely be the result of a functional divergence during the evolutionary transition from archaea to eukaryotes. Alternatively, the cytoplasmic and nuclear functions of prefoldin might be coupled in such a way as to favour a global coordination of gene expression and cytoskeleton dynamics. This hypothetical coordination is particularly expected in those situations where cells are challenged by stimuli that require a specific genomic response and, at the same time, the reorganization of cytoplasmic organelles. Although this speculative model is far from having been demonstrated, there are some experimental hints that are compatible with the notion of prefoldin as a global cell regulator.
The cytoskeletal function of prefoldin is not essential for the housekeeping assembly of microtubules or actin filaments. This is the reason why yeast prefoldin genes can be deleted without compromising cell viability [8]. Similarly, double knockout mice, lacking PFDN1, are also viable, at least until the fifth week of age, although they display a small size and cytoskeletal defects [22]. When β-tubulin is not expressed at high levels in Drosophila cells, its stabilization does not require prefoldin either, although it does when ectopic β-tubulin is expressed [19]. This indicates that prefoldin is rate-limiting only under strong cytoskeleton biogenesis conditions. Rate-limiting steps are the regulatory points in most biological pathways, and they are usually the most upstream stages. Accordingly, prefoldin plays its role in the very first step of microtubules and actin filaments biogenesis [18].
One of the most drastic cytoplasmic reorganizations takes place during B-lymphocyte activation by antigens, when they differentiate into highly secretory plasma cells. PFDN1 knockout mice are severely affected in this process. Interestingly, the mammalian PFDN6 gene is located in the centromeric portion of the class II region of the major histocompatibility complex [71], and its expression dramatically increases as a result of lymphocyte activation [72].
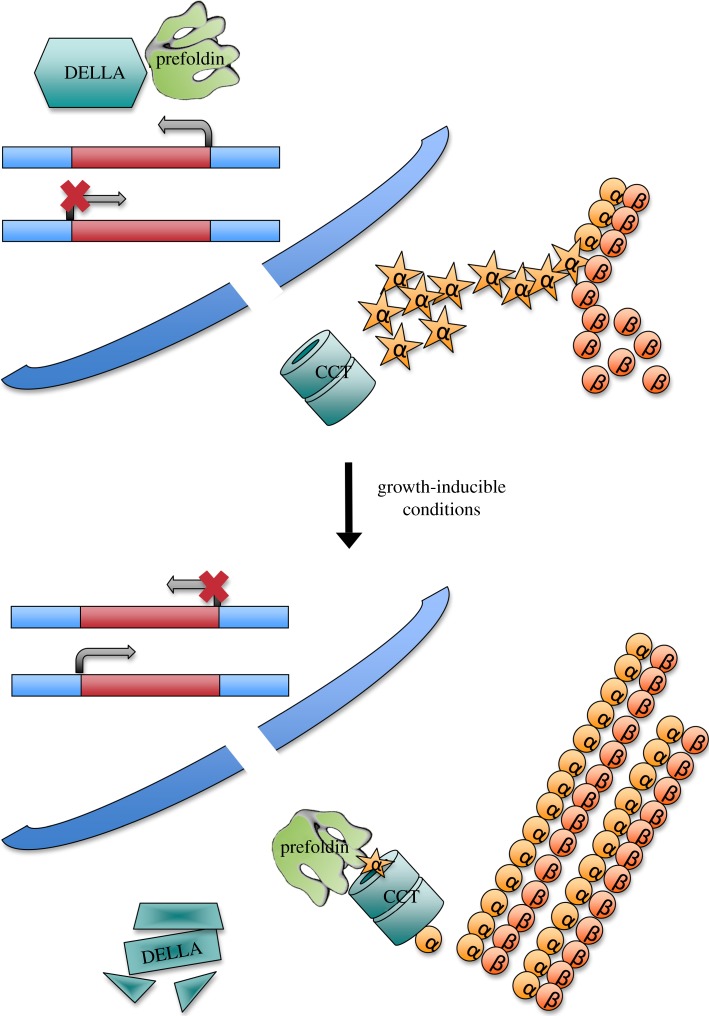
So far, the prefoldin role in lymphocyte activation has not been connected to any nuclear event, but there is one example where the cytoplasmic function of prefoldin is linked to the nucleus. This is the case of organ growth by cell expansion in the model plant Arabidopsis thaliana. The plant controls anisotropic cell elongation by a mechanism that finely coordinates the abundance of DELLA proteins in the nucleus with the subcellular localization of prefoldin [39]. When environmental conditions are not favourable for growth, DELLA proteins accumulate, and prefoldin is retained in the nucleus upon interaction, compromising its role in the cytoplasm. Thus, microtubule assembly is compromised, and anisotropic cell growth is prevented. On the contrary, environmental conditions that are favourable for growth promote degradation of DELLA proteins and prefoldin can move to the cytosol (figure 5) [39]. DELLAs are transcriptional regulators and, although no transcriptional function of prefoldin has been described to date in Arabidopsis, it is possible that prefoldin participates not only in the cytoplasmic part of this regulated process, but also in the transcriptional response.
Figure 5.
Plant prefoldin collaborates with the DELLA transcription factor in the regulation of cell expansion. When environmental conditions are not favourable for growth, prefoldin is imported to the nucleus by the DELLA transcription factor, which contributes to the regulation of a set of genes, either positively or negatively. Upon activation by growth-inducible conditions, DELLA is driven into degradation, allowing prefoldin to relocate to the cytoplasm and to participate in the cytoskeleton reorganization required for cell expansion.
The coordinated action in cytoskeleton dynamics and transcription would be facilitated if prefoldin acted on similar targets in these two processes. It is well known that the main cytoplasmic targets of prefoldin are actin and tubulin monomers. It is conceivable that actin, tubulin or both might also be nuclear targets of prefoldin. The transcriptional characterization of yeast prefoldin mutants, using chemical inhibitors of cytoskeleton assembly, indicates that the role of prefoldin in chromatin dynamics during transcription elongation is not mediated by actin or tubulin polymerization [43]. Yet the involvement of actin and tubulin monomers in the nuclear function of prefoldin is an open possibility. There is monomeric actin in the nucleus, and its contribution to gene transcription has already been reported (reviewed by [73]). The direct involvement of nuclear tubulin in gene transcription has also been described [74]. c-Myc, the best example of a transcription factor regulated by a prefoldin subunit (see above), also binds tubulin, and the region of the c-Myc protein that interacts with tubulin overlaps its PFDN5 interaction domain [75]. It has been proposed that microtubules might be involved in the migration of c-Myc to the cytoplasm, when cells exit the cell cycle [76,77]. Myc-nick, a cleavage product of c-Myc, promotes tubulin acetylation during cell differentiation [78], and MIZ-1, another c-Myc-interacting protein, is regulated by its association to microtubules and activate transcription in response to cytoskeleton changes [79]. All these data visualize physical and functional interactions between c-Myc and tubulin, which support functional coupling between c-Myc-dependent transcription and the cytoskeleton dynamics mediated by prefoldin.
The existence of such mechanisms of coordination between cytoplasmic dynamics and genome regulation, and the potential involvement of prefoldin and other protein-folding factors in them, is a challenging field for contemporary biology.
Acknowledgements
We thank all the members of S.C.'s laboratory for helpful discussion and Helen Warburton for English correction.
Funding statement
This work was supported by the Spanish Ministries of Economy and Competitiveness and Education (grants BFU-2010-21975-C03-03 to S.C., FPU fellowship to G.M.-Z.), FIPSE (grant 360946/10), the Andalusian Government (grants P08-CVI-03508 and P12-BIO-1938MO) and the European Union (Regional Development European Fund).
References
- 1.Saibil H. 2013. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 14, 630–642. (doi:10.1038/nrm3658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93, 863–873. (doi:10.1016/S0092-8674(00)81446-4) [DOI] [PubMed] [Google Scholar]
- 3.Kubota H, Hynes G, Carne A, Ashworth A, Willison K. 1994. Identification of six Tcp-1-related genes encoding divergent subunits of the TCP-1-containing chaperonin. Curr. Biol. 4, 89–99. (doi:10.1016/S0960-9822(94)00024-2) [DOI] [PubMed] [Google Scholar]
- 4.Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. 1992. A cytoplasmic chaperonin that catalyzes β-actin folding. Cell 69, 1043–1050. (doi:10.1016/0092-8674(92)90622-J) [DOI] [PubMed] [Google Scholar]
- 5.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. 1992. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 11, 4767–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leroux MR, Fandrich M, Klunker D, Siegers K, Lupas AN, Brown JR, Schiebel E, Dobson CM, Hartl FU. 1999. MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J. 18, 6730–6743. (doi:10.1093/emboj/18.23.6730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogumil D, Alvarez-Ponce D, Landan G, McInerney JO, Dagan T. 2014. Integration of two ancestral chaperone systems into one: the evolution of eukaryotic molecular chaperones in light of eukaryogenesis. Mol. Biol. Evol. 31, 410–418. (doi:10.1093/molbev/mst212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geissler S, Siegers K, Schiebel E. 1998. A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J. 17, 952–966. (doi:10.1093/emboj/17.4.952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Milla MA, Salinas J. 2009. Prefoldins 3 and 5 play an essential role in Arabidopsis tolerance to salt stress. Mol. Plant 2, 526–534. (doi:10.1093/mp/ssp016) [DOI] [PubMed] [Google Scholar]
- 10.Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. 2000. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 103, 621–632. (doi:10.1016/S0092-8674(00)00165-3) [DOI] [PubMed] [Google Scholar]
- 11.Stirling PC, Bakhoum SF, Feigl AB, Leroux MR. 2006. Convergent evolution of clamp-like binding sites in diverse chaperones. Nat. Struct. Mol. Biol. 13, 865–870. (doi:10.1038/nsmb1153) [DOI] [PubMed] [Google Scholar]
- 12.Hansen WJ, Cowan NJ, Welch WJ. 1999. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J. Cell Biol. 145, 265–277. (doi:10.1083/jcb.145.2.265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rommelaere H, De Neve M, Neirynck K, Peelaers D, Waterschoot D, Goethals M, Fraeyman N, Vandekerckhove J, Ampe C. 2001. Prefoldin recognition motifs in the nonhomologous proteins of the actin and tubulin families. J. Biol. Chem. 276, 41 023–41 028. (doi:10.1074/jbc.M106591200) [DOI] [PubMed] [Google Scholar]
- 14.Simons CT, Staes A, Rommelaere H, Ampe C, Lewis SA, Cowan NJ. 2004. Selective contribution of eukaryotic prefoldin subunits to actin and tubulin binding. J. Biol. Chem. 279, 4196–4203. (doi:10.1074/jbc.M306053200) [DOI] [PubMed] [Google Scholar]
- 15.Martin-Benito J, Boskovic J, Gomez-Puertas P, Carrascosa JL, Simons CT, Lewis SA, Bartolini F, Cowan NJ, Valpuesta JM. 2002. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 21, 6377–6386. (doi:10.1093/emboj/cdf640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Benito J, et al. 2007. Divergent substrate-binding mechanisms reveal an evolutionary specialization of eukaryotic prefoldin compared to its archaeal counterpart. Structure 15, 101–110. (doi:10.1016/j.str.2006.11.006) [DOI] [PubMed] [Google Scholar]
- 17.Stirling PC, Cuellar J, Alfaro GA, El Khadali F, Beh CT, Valpuesta JM, Melki R, Leroux MR. 2006. PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J. Biol. Chem. 281, 7012–7021. (doi:10.1074/jbc.M513235200) [DOI] [PubMed] [Google Scholar]
- 18.Lundin VF, Srayko M, Hyman AA, Leroux MR. 2008. Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Dev. Biol. 313, 320–334. (doi:10.1016/j.ydbio.2007.10.022) [DOI] [PubMed] [Google Scholar]
- 19.Delgehyr N, et al. 2012. Drosophila Mgr, a prefoldin subunit cooperating with von Hippel Lindau to regulate tubulin stability. Proc. Natl Acad. Sci. USA 109, 5729–5734. (doi:10.1073/pnas.1108537109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Bot N, Tsai MC, Andrews RK, Ahringer J. 2003. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13, 1499–1505. (doi:10.1016/S0960-9822(03)00577-3) [DOI] [PubMed] [Google Scholar]
- 21.Gu Y, Deng Z, Paredez AR, DeBolt S, Wang ZY, Somerville C. 2008. Prefoldin 6 is required for normal microtubule dynamics and organization in Arabidopsis. Proc. Natl Acad. Sci. USA 105, 18 064–18 069. (doi:10.1073/pnas.0808652105). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao S, Carlesso G, Osipovich AB, Llanes J, Lin Q, Hoek KL, Khan WN, Ruley HE. 2008. Subunit 1 of the prefoldin chaperone complex is required for lymphocyte development and function. J. Immunol. 181, 476–484. (doi:10.4049/jimmunol.181.1.476) [DOI] [PubMed] [Google Scholar]
- 23.Lee Y, Smith RS, Jordan W, King BL, Won J, Valpuesta JM, Naggert JK, Nishina PM. 2011. Prefoldin 5 is required for normal sensory and neuronal development in a murine model. J. Biol. Chem. 286, 726–736. (doi:10.1074/jbc.M110.177352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe A, Takahashi-Niki K, Takekoshi Y, Shimizu T, Kitaura H, Maita H, Iguchi-Ariga SM, Ariga H. 2013. Prefoldin plays a role as a clearance factor in preventing proteasome inhibitor-induced protein aggregation. J. Biol. Chem. 288, 27 764–27 776. (doi:10.1074/jbc.M113.476358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tashiro E, et al. 2013. Prefoldin protects neuronal cells from polyglutamine toxicity by preventing aggregation formation. J. Biol. Chem. 288, 19 958–19 972. (doi:10.1074/jbc.M113.477984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorgjerd KM, Zako T, Sakono M, Stirling PC, Leroux MR, Saito T, Nilsson P, Sekimoto M, Saido TC, Maeda M. 2013. Human prefoldin inhibits amyloid-β (Aβ) fibrillation and contributes to formation of nontoxic Aβ aggregates. Biochemistry 52, 3532–3542. (doi:10.1021/bi301705c) [DOI] [PubMed] [Google Scholar]
- 27.Broer L, et al. 2011. Association of HSP70 and its co-chaperones with Alzheimer's disease. J. Alzheimer‘s Dis. 25, 93–102. (doi:10.3233/JAD-2011-101560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302, 1208–1212. (doi:10.1126/science.1088401) [DOI] [PubMed] [Google Scholar]
- 29.Boulon S, et al. 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39, 912–924. (doi:10.1016/j.molcel.2010.08.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, Boulton SJ. 2010. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell 39, 839–850. (doi:10.1016/j.molcel.2010.08.037) [DOI] [PubMed] [Google Scholar]
- 31.Kakihara Y, Houry WA. 2012. The R2TP complex: discovery and functions. Biochim. Biophys. Acta 1823, 101–107. (doi:10.1016/j.bbamcr.2011.08.016) [DOI] [PubMed] [Google Scholar]
- 32.Nano N, Houry WA. 2013. Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Phil. Trans. R. Soc. B 368, 20110399 (doi:10.1098/rstb.2011.0399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miron-Garcia MC, Garrido-Godino AI, Garcia-Molinero V, Hernandez-Torres F, Rodriguez-Navarro S, Navarro F. 2013. The prefoldin bud27 mediates the assembly of the eukaryotic RNA polymerases in an rpb5-dependent manner. PLoS Genet. 9, e1003297 (doi:10.1371/journal.pgen.1003297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Wang Q, Zhang H, Liu Q, Du X, Richter M, Greene MI. 2005. UXT is a novel centrosomal protein essential for cell viability. Mol. Biol. Cell 16, 5857–5865. (doi:10.1091/mbc.E05-08-0705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mita P, Savas JN, Ha S, Djouder N, Yates JR, III, Logan SK. 2013. Analysis of URI nuclear interaction with RPB5 and components of the R2TP/prefoldin-like complex. PLoS ONE 8, e63879 (doi:10.1371/journal.pone.0063879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markus SM, Taneja SS, Logan SK, Li W, Ha S, Hittelman AB, Rogatsky I, Garabedian MJ. 2002. Identification and characterization of ART-27, a novel coactivator for the androgen receptor N terminus. Mol. Biol. Cell 13, 670–682. (doi:10.1091/mbc.01-10-0513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, Chen L, Zhou Y, Liu H, Yang J, Liu Z, Wang C. 2011. UXT-V1 protects cells against TNF-induced apoptosis through modulating complex II formation. Mol. Biol. Cell 22, 1389–1397. (doi:10.1091/mbc.E10-10-0827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagio Y, Kimura Y, Taira T, Fujioka Y, Iguchi-Ariga SM, Ariga H. 2006. Distinct localizations and repression activities of MM-1 isoforms toward c-Myc. J. Cell. Biochem. 97, 145–155. (doi:10.1002/jcb.20619) [DOI] [PubMed] [Google Scholar]
- 39.Locascio A, Blazquez MA, Alabadi D. 2013. Dynamic regulation of cortical microtubule organization through prefoldin–DELLA interaction. Curr. Biol. 23, 804–809. (doi:10.1016/j.cub.2013.03.053) [DOI] [PubMed] [Google Scholar]
- 40.Tsuchiya H, Iseda T, Hino O. 1996. Identification of a novel protein (VBP-1) binding to the von Hippel–Lindau (VHL) tumor suppressor gene product. Cancer Res. 56, 2881–2885. [PubMed] [Google Scholar]
- 41.Chen S, Chen K, Zhang Q, Cheng H, Zhou R. 2013. Regulation of the transcriptional activation of the androgen receptor by the UXT-binding protein VHL. Biochem. J. 456, 55–66. (doi:10.1042/BJ20121711) [DOI] [PubMed] [Google Scholar]
- 42.Delgermaa L, Hayashi N, Dorjsuren D, Nomura T, Thuy le TT, Murakami S. 2004. Subcellular localization of RPB5-mediating protein and its putative functional partner. Mol. Cell. Biol. 24, 8556–8566. (doi:10.1128/MCB.24.19.8556-8566.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millan-Zambrano G, Rodriguez-Gil A, Penate X, de Miguel-Jimenez L, Morillo-Huesca M, Krogan N, Chavez S. 2013. The prefoldin complex regulates chromatin dynamics during transcription elongation. PLoS Genet. 9, e1003776 (doi:10.1371/journal.pgen.1003776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M. 2006. URI-1 is required for DNA stability in C. elegans. Development 133, 621–629. (doi:10.1242/dev.02235) [DOI] [PubMed] [Google Scholar]
- 45.Mousnier A, Kubat N, Massias-Simon A, Segeral E, Rain JC, Benarous R, Emiliani S, Dargemont C. 2007. von Hippel–Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. Proc. Natl Acad. Sci. USA 104, 13 615–13 620. (doi:10.1073/pnas.0705162104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rain JC, Cribier A, Gerard A, Emiliani S, Benarous R. 2009. Yeast two-hybrid detection of integrase–host factor interactions. Methods 47, 291–297. (doi:10.1016/j.ymeth.2009.02.002) [DOI] [PubMed] [Google Scholar]
- 47.Xu Y, Her C. 2013. VBP1 facilitates proteasome and autophagy-mediated degradation of MutS homologue hMSH4. FASEB J. 27, 4799–4810. (doi:10.1096/fj.13-235127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Her C, Wu X, Griswold MD, Zhou F. 2003. Human MutS homologue MSH4 physically interacts with von Hippel–Lindau tumor suppressor-binding protein 1. Cancer Res. 63, 865–872. [PubMed] [Google Scholar]
- 49.Buchberger A. 2013. Roles of cdc48 in regulated protein degradation in yeast. Sub-cellular Biochem. 66, 195–222. (doi:10.1007/978-94-007-5940-4_8) [DOI] [PubMed] [Google Scholar]
- 50.Meyer H, Bug M, Bremer S. 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123. (doi:10.1038/ncb2407) [DOI] [PubMed] [Google Scholar]
- 51.Kim SY, Kim JC, Kim JK, Kim HJ, Lee HM, Choi MS, Maeng PJ, Ahn JK. 2008. Hepatitis B virus X protein enhances NFkappaB activity through cooperating with VBP1. BMB Rep. 41, 158–163. (doi:10.5483/BMBRep.2008.41.2.158) [DOI] [PubMed] [Google Scholar]
- 52.Mori K, Maeda Y, Kitaura H, Taira T, Iguchi-Ariga SM, Ariga H. 1998. MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J. Biol. Chem. 273, 29 794–29 800. (doi:10.1074/jbc.273.45.29794) [DOI] [PubMed] [Google Scholar]
- 53.Satou A, Taira T, Iguchi-Ariga SM, Ariga H. 2001. A novel transrepression pathway of c-Myc: recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J. Biol. Chem. 276, 46 562–46 567. (doi:10.1074/jbc.M104937200) [DOI] [PubMed] [Google Scholar]
- 54.Kimura Y, Nagao A, Fujioka Y, Satou A, Taira T, Iguchi-Ariga SM, Ariga H. 2007. MM-1 facilitates degradation of c-Myc by recruiting proteasome and a novel ubiquitin E3 ligase. Int. J. Oncol. 31, 829–836. [PubMed] [Google Scholar]
- 55.Narita R, Kitaura H, Torii A, Tashiro E, Miyazawa M, Ariga H, Iguchi-Ariga SM. 2012. Rabring7 degrades c-Myc through complex formation with MM-1. PLoS ONE 7, e41891 (doi:10.1371/journal.pone.0041891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida T, Kitaura H, Hagio Y, Sato T, Iguchi-Ariga SM, Ariga H. 2008. Negative regulation of the Wnt signal by MM-1 through inhibiting expression of the wnt4 gene. Exp. Cell Res. 314, 1217–1228. (doi:10.1016/j.yexcr.2008.01.002) [DOI] [PubMed] [Google Scholar]
- 57.Watanabe K, Ozaki T, Nakagawa T, Miyazaki K, Takahashi M, Hosoda M, Hayashi S, Todo S, Nakagawara A. 2002. Physical interaction of p73 with c-Myc and MM1, a c-Myc-binding protein, and modulation of the p73 function. J. Biol. Chem. 277, 15 113–15 123. (doi:10.1074/jbc.M111281200) [DOI] [PubMed] [Google Scholar]
- 58.Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N, Murakami S. 1998. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol. Cell. Biol. 18, 7546–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGilvray R, Walker M, Bartholomew C. 2007. UXT interacts with the transcriptional repressor protein EVI1 and suppresses cell transformation. FEBS J. 274, 3960–3971. (doi:10.1111/j.1742-4658.2007.05928.x) [DOI] [PubMed] [Google Scholar]
- 60.Sun S, Tang Y, Lou X, Zhu L, Yang K, Zhang B, Shi H, Wang C. 2007. UXT is a novel and essential cofactor in the NF-kappaB transcriptional enhanceosome. J. Cell Biol. 178, 231–244. (doi:10.1083/jcb.200611081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang LS, Wang JT, Doong SL, Lee CP, Chang CW, Tsai CH, Yeh SW, Hsieh CY, Chen MR. 2012. Epstein-Barr virus BGLF4 kinase downregulates NF-kappaB transactivation through phosphorylation of coactivator UXT. J. Virol. 86, 12 176–12 186. (doi:10.1128/JVI.01918-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enunlu I, Ozansoy M, Basak AN. 2011. Alfa-class prefoldin protein UXT is a novel interacting partner of amyotrophic lateral sclerosis 2 (Als2) protein. Biochem. Biophys. Res. Commun. 413, 471–475. (doi:10.1016/j.bbrc.2011.08.121) [DOI] [PubMed] [Google Scholar]
- 63.Taneja SS, et al. 2004. ART-27, an androgen receptor coactivator regulated in prostate development and cancer. J. Biol. Chem. 279, 13 944–13 952. (doi:10.1074/jbc.M306576200) [DOI] [PubMed] [Google Scholar]
- 64.Yang J, Zhao YL, Wu ZQ, Si YL, Meng YG, Fu XB, Mu YM, Han WD. 2009. The single-macro domain protein LRP16 is an essential cofactor of androgen receptor. Endocrine-Related Cancer 16, 139–153. (doi:10.1677/ERC-08-0150) [DOI] [PubMed] [Google Scholar]
- 65.Fujioka Y, Taira T, Maeda Y, Tanaka S, Nishihara H, Iguchi-Ariga SM, Nagashima K, Ariga H. 2001. MM-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer. J. Biol. Chem. 276, 45 137–45 144. (doi:10.1074/jbc.M106127200) [DOI] [PubMed] [Google Scholar]
- 66.Ma HC, Lin TW, Li H, Iguchi-Ariga SM, Ariga H, Chuang YL, Ou JH, Lo SY. 2008. Hepatitis C virus ARFP/F protein interacts with cellular MM-1 protein and enhances the gene trans-activation activity of c-Myc. J. Biomed. Sci. 15, 417–425. (doi:10.1007/s11373-008-9248-9) [DOI] [PubMed] [Google Scholar]
- 67.Collins SR, et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810. (doi:10.1038/nature05649) [DOI] [PubMed] [Google Scholar]
- 68.Costanzo M, et al. 2010. The genetic landscape of a cell. Science 327, 425–431. (doi:10.1126/science.1180823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dekker C, et al. 2008. The interaction network of the chaperonin CCT. EMBO J. 27, 1827–1839. (doi:10.1038/emboj.2008.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soues S, Kann ML, Fouquet JP, Melki R. 2003. The cytosolic chaperonin CCT associates to cytoplasmic microtubular structures during mammalian spermiogenesis and to heterochromatin in germline and somatic cells. Exp. Cell Res. 288, 363–373. (doi:10.1016/S0014-4827(03)00248-9) [DOI] [PubMed] [Google Scholar]
- 71.Herberg JA, Beck S, Trowsdale J. 1998. TAPASIN, DAXX, RGL2, HKE2 and four new genes (BING 1, 3 to 5) form a dense cluster at the centromeric end of the MHC. J. Mol. Biol. 277, 839–857. (doi:10.1006/jmbi.1998.1637) [DOI] [PubMed] [Google Scholar]
- 72.Ostrov DA, et al. 2007. Characterization of HKE2: an ancient antigen encoded in the major histocompatibility complex. Tissue Antigens 69, 181–188. (doi:10.1111/j.1399-0039.2006.00730.x) [DOI] [PubMed] [Google Scholar]
- 73.Grosse R, Vartiainen MK. 2013. To be or not to be assembled: progressing into nuclear actin filaments. Nat. Rev. Mol. Cell Biol. 14, 693–697. (doi:10.1038/nrm3681) [DOI] [PubMed] [Google Scholar]
- 74.Hoog G, Zarrizi R, von Stedingk K, Jonsson K, Alvarado-Kristensson M. 2011. Nuclear localization of γ-tubulin affects E2F transcriptional activity and S-phase progression. FASEB J. 25, 3815–3827. (doi:10.1096/fj.11-187484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexandrova N, Niklinski J, Bliskovsky V, Otterson GA, Blake M, Kaye FJ, Zajac-Kaye M. 1995. The N-terminal domain of c-Myc associates with α-tubulin and microtubules in vivo and in vitro. Mol. Cell. Biol. 15, 5188–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vriz S, Lemaitre JM, Leibovici M, Thierry N, Mechali M. 1992. Comparative analysis of the intracellular localization of c-Myc, c-Fos, and replicative proteins during cell cycle progression. Mol. Cell. Biol. 12, 3548–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemaitre JM, Buckle RS, Mechali M. 1996. c-Myc in the control of cell proliferation and embryonic development. Adv. Cancer Res. 70, 95–144. (doi:10.1016/S0065-230X(08)60873-8) [DOI] [PubMed] [Google Scholar]
- 78.Conacci-Sorrell M, Ngouenet C, Eisenman RN. 2010. Myc-nick: a cytoplasmic cleavage product of Myc that promotes α-tubulin acetylation and cell differentiation. Cell 142, 480–493. (doi:10.1016/j.cell.2010.06.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ziegelbauer J, Shan B, Yager D, Larabell C, Hoffmann B, Tjian R. 2001. Transcription factor MIZ-1 is regulated via microtubule association. Mol. Cell 8, 339–349. (doi:10.1016/S1097-2765(01)00313-6) [DOI] [PubMed] [Google Scholar]