Summary
Many biological processes involve gene-expression regulation by alternative splicing. Here, we identify the splicing factor SRSF6 as a regulator of wound healing and tissue homeostasis in skin. We show that SRSF6 is a proto-oncogene that is frequently overexpressed in human skin cancer. Overexpressing it in transgenic mice induces hyperplasia of sensitized skin and promotes aberrant alternative splicing. We identify 139 target genes of SRSF6 in skin, and show that this SR protein binds to alternative exons of the extracellular-matrix protein tenascin C pre-mRNA, promoting the expression of isoforms characteristic of invasive and metastatic cancer in a cell-type-independent manner. SRSF6 overexpression additionally results in depletion of Lgr6+ stem cells, and excessive keratinocyte proliferation and response to injury. Furthermore, the effects of SRSF6 in wound healing assayed in vitro depend on the TNC isoforms. Thus, abnormal SR-protein expression can perturb tissue homeostasis.
Most mammalian genes utilize alternative splicing (AS) to express multiple mRNA isoforms; thus, AS is a major contributor to proteome complexity. The serine/arginine-rich (SR) protein SRSF6 (SRp55) belongs to a family of highly conserved RNA-binding splicing-factor proteins1,2, with one or two RNA-recognition motifs (RRMs), and a carboxy-terminal arginine/serine-rich domain (RS domain)3. The RRMs mediate binding to specific exonic splicing enhancer (ESE) motifs, whereas the RS domain engages in protein-protein interactions, modulated by serine phosphorylation and dephosphorylation.
Given the central role of SR proteins in splicing, their deregulation could be causally related to or influence disease. In the context of cancer, many mutations affect splicing of oncogenes, tumor-suppressors, and other cancer-associated genes; however, many splicing abnormalities found in cancer are not associated with mutations in the affected genes4. Instead, they may arise from aberrant expression of splicing factors5. Indeed, certain SR proteins are over-expressed in human cancers, notably SRSF16,7, SRSF66, and SRSF38. Moreover, SRSF1 is oncogenic in certain contexts6,7, e.g. by regulating AS of the proto-oncogene MSTR1 (RON), resulting in increased cell motility9.
Cancer resembles a state of chronic wound healing, in which the latent wound-healing program is erroneously activated and perpetuated10. Notably, cancer can result from chronic inflammation11. Gene-expression profiling of wounded skin and tumors revealed striking similarities12. Normally, tissue injury triggers a pathway that attempts to repair damaged tissue and restore homeostasis through three distinct but overlapping phases: inflammation, tissue formation, and tissue remodeling13. Stem cells throughout the skin are a source of keratinocyte/progenitor cells that can be rapidly generated to repair tissue injury. AS was strongly tied to this pathway when the fibronectin gene (FN) was found to be alternatively spliced during wound healing. During wound healing, FN AS generates embryonic isoforms with distinct functions associated with cell migration and proliferation14.
AS is an intrinsic mechanism to introduce proteome complexity and exert temporal and spatial regulation. Because AS regulation is complex and incompletely understood, models that reproduce in vivo conditions are clearly needed. Here we set out to establish a transgenic mouse model with conditional overexpression of SRSF6 to study AS regulation in a natural context, and to characterize the functional consequences of aberrant SRSF6 expression in tissues. Surprisingly, SRSF6-overexpressing mice developed pronounced skin hyperplasia, accompanied by stem-cell depletion and aberrant splicing. We identify SRSF6 as a master-regulator of tenascin C AS. This is the first evidence of a causal role of AS misregulation by an SR protein in wound healing and hyperplasia.
Results
SRSF6 overexpression induces epithelial hyperplasia
We generated a mouse transgenic for human SRSF6 cDNA and IRES-EGFP under the control of a tetracycline-responsive promoter (TRE-tight) at the ColA1 locus (ColA1-SRSF6). The reverse tetracycline transactivator (rtTA) was expressed from the Rosa26 locus (R26-rtTA)(Supplementary Fig. 1a). Upon doxycycline-treatment (DOX) of adult mice, RT-PCR and immunoblotting showed high SRSF6 transgene expression in skin and small intestine, and low expression in spleen, liver, kidney, and heart (Fig. 1a, Supplementary Fig. 1b). The transgene expression pattern was consistent with previous use of the same system to express shRNA15. We used the TREtight promoter to avoid potential deleterious effects of ectopic SRSF6 expression during embryogenesis.
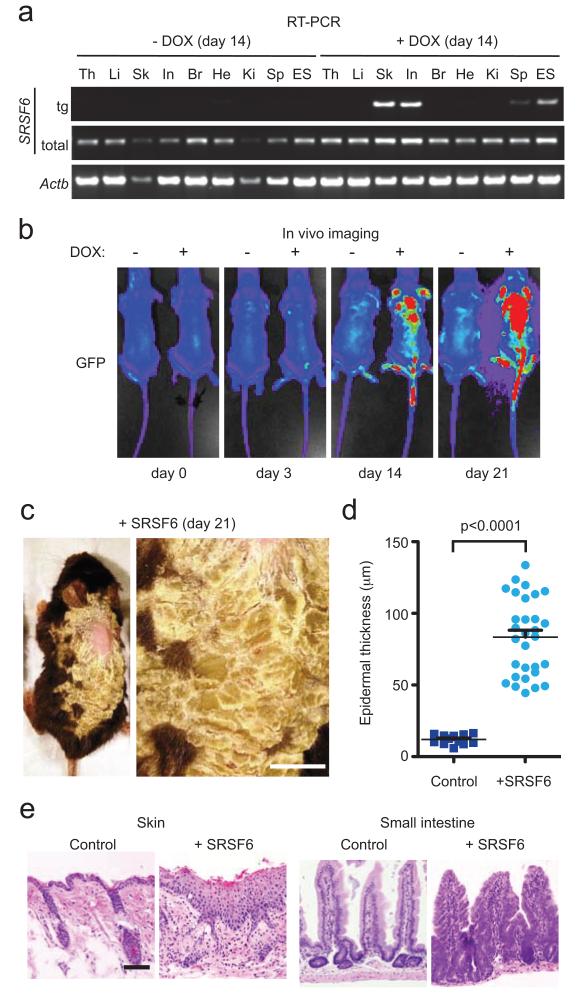
Figure 1.
SRSF6 overexpression induces skin and intestinal hyperplasia in mice. (a) RT-PCR showing expression of transgenic (tg) and total SRSF6 in DOX-treated R26-rtTA/ColA1-SRSF6-transgenic mice, and Actb mRNA as a loading control, in RNA extracted from thymus (th), liver (li), skin (sk), small intestine (in), brain (br), heart (he), kidney (ki), spleen (sp), and parental SRSF6-targeted ES cells. (b) In vivo imaging showing DOX induction of GFP after 0, 3, 14, or 21 d, in transgenic versus control mice. (c) Left panel: Abnormal skin phenotype of transgenic R26-rtTA/ColA1 mouse treated for 21 d with DOX. Right panel: Magnification of region with abnormal back skin. Bar = 0.5 cm. (d) Analysis of epidermal thickness: control skin (solid square) and SRSF6-expressing skin (solid circle), with thickness in μm (Mann-Whitney test, p<0.0001, skin samples from 3 treated and 3 control mice, n=21 each). Data are represented as mean +/− s.e.m. (e) Histopathology (H&E staining) of transgenic tissues from skin and small intestine of mice induced with DOX for 14 d, showing severe hyperplasia.
Using live-imaging of shaved, DOX-treated animals, we analyzed GFP induction for 21 days (Fig. 1b). We observed a gradual increase in GFP expression in the skin, and severe epithelial hyperplasia in skin and small intestine (Fig. 1c-e, Supplementary Table 1a). Hyperplasia is the first stage of cancer development.
The initial lesion was an abnormal increase in keratinocyte number in the interfollicular epithelium, and infundibulum and distal isthmus of the hair follicles. There was mild to moderate achanthosis, hypergranulosis, and hyperkeratosis (Supplementary Fig. 2a-b vs. 2c-d). The severity of the hyperplasia increased during the DOX-induction time course. In the more severe lesions, the progression of morphologic differentiation generally followed the normal pattern, but was severely hyperplastic and mildly to moderately dysplastic, with thickening and loss of organization of the stratum spinosum, irregularities in the stratum granulosum, and severe parakeratotic hyperkeratosis (Supplementary Fig. 2e-f). The epithelium of the hair follicles was hyperplastic and abnormal hair formation was often seen. The adnexa were also hyperplastic with squamous metaplasia. The lesions were clearly demarcated (Supplementary Fig. 2a-b). In advanced lesions, other abnormalities included intraepidermal microabscesses, dyskeratotic keratinocytes, spongiosis, apoptotic basal cells (Civatte bodies), and increased apoptotic bodies (Supplementary Fig. 3). There were areas of epidermal thickening, with an ~8-fold increase in thickness, compared to normal skin (p<0.0001)(Fig.1d). Depilation by plucking gave a more severe phenotype (Supplementary Fig. 4).
In the most severe cases, the skin was remarkably thickened, with thick verrucose proliferative lesions adjacent to papules with severe epidermal hyperplasia, especially in the mice with plucked hair. In the severe verrucose lesions, the parakeratotic hyperkeratotic stratum corneum was up to 1-mm thick and it surrounded and trapped the hair shafts. The parakeratosis was often severe, with retention of large nuclear fragments and many apoptotic bodies. The surface was covered with crusts. The underlying epidermis was markedly thickened and formed folds reminiscent of papillary development. The epidermis was irregular, with marked variation in cellular differentiation and orientation. The granular cell layer was prominent, often thickened, and displayed variation in keratohyalin granules. There was severe dyskeratosis, many apoptotic bodies, and mitoses above the basal layer. Keratin pearls were present in the epidermis, as a result of trapping in misoriented interpapillary areas (Supplementary Fig. 4). In the middle of the most severely affected areas, the hair bulb, stem, and glands of the follicles were often absent. This loss of intact follicles is typical of hyperplastic epidermis and wound healing.
In small intestine, we observed segmental areas of severe crypt-epithelial hyperplasia, with blunting of the villi. The lower penetrance of intestinal hyperplasia (5/9) compared with skin (8/8) may be due to the intestinal lesions being segmental and occurring in random portions of the intestine that we did not always sample. Control skin from double-transgenic animals in the absence of DOX, or from DOX-treated wild-type or R26-rtTA mice showed no signs of hyperplasia (Supplementary Table 1a). We conclude that SRSF6 overexpression in mouse skin and small intestine induces hyperplasia.
To further characterize SRSF6-expressing cells, we analyzed the epidermis for co-localization with skin-differentiation markers in mild-to-moderate lesions. Ectopic SRSF6 expression was strongly associated with keratins 5 and 14 (Fig. 2a,c). These markers of proliferating basal cells were expressed throughout the epidermis, suggesting a failure to differentiate and mature. We also detected a high level of proliferation, measured by KI67 and also by keratin 6, which is strongly expressed in activated keratinocytes and is normally observed during epidermal injury16 (Fig. 2a). In contrast, epithelial hyperplasia coincided with reduced levels of loricrin (Fig. 2c), a keratinocyte-differentiation marker. Many of these gene-expression changes are characteristic of hyperproliferative skin due to various causes. The abnormal expression of differentiation markers preceded the gross morphological changes, suggesting that abnormal molecular changes occurred early in the hyperplastic epidermis. We infer that enforced expression of SRSF6 maintains cells in an aberrant proliferating state, suppressing the normal epithelial differentiation program.
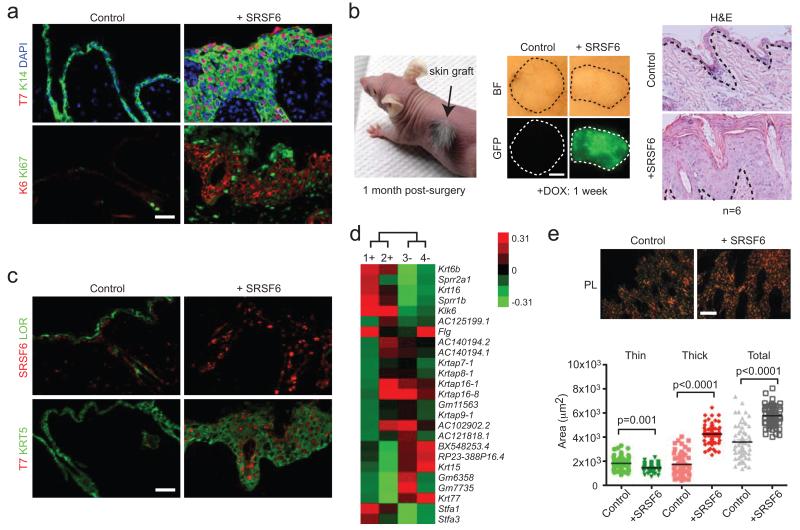
Figure 2.
SRSF6 overexpression results in a wound-healing expression signature. (a) Two-color immunofluorescence of transgenic SRSF6 (T7), keratin 14 (KRT14), keratin 6 (KRT6) and KI67 in transgenic skin (DOX: +SRSF6). (b) Left panel: nude mouse with transplanted skin graft, 1 month after surgery. Middle panel: bright field (BF) or GFP images of allogenic skin transplants induced by DOX for 1 week (Control versus R26-rtTA/ColA1-SRSF6). Right panel: Histopathology showing skin hyperplasia in SRSF6-expressing transplants (H&E staining); n=6. (c) Expression of an epidermal differentiation marker, loricrin (LOR), a basal cell marker, keratin 5 (KRT5), and SRSF6. (d) Unsupervised hierarchical clustering of SRSF6-induced versus control skin (n=4) based on the 25 most variable genes; DOX-treated (1+ and 2+) and control (3− and 4−) samples represent biological replicates. (e) Top: Analysis of fibrillar type I+III collagen levels by Picrosirius-red staining of skin obtained from DOX-induced (+SRSF6) or control transgenic mice. Images are for bright field (BF) or polarized light (PL) conditions. Bottom: Image quantification of thin (green), thick (red), or total (gray) collagen (type I and III) (Mann-Whitney test, p<0.0001, n=60 fields per condition). Bar = 50 μm (a, c), 2 mm (b), and 100 μm (e). Data are represented with the mean for each group.
SRSF6-driven skin phenotype is cell-autonomous
Immunofluorescence microscopy showed that ectopic SRSF6 (T7-tagged) protein was exclusively expressed in the hyperplastic epithelium and hair follicles (Fig. 2a). There was no transgene expression in the stroma adjacent to the affected epithelium. We tested whether skin hyperplasia was cell-autonomous or depended, e.g., on cells originating in the hematopoietic system in the spleen or bone marrow, which showed sporadic SRSF6 expression (Supplementary Fig. 1c). Allogenic transplants made from double-transgenic or control skin were grafted onto recipient nude mice; upon DOX-treatment, only mice with double-transgenic transplants developed hyperplasia (Supplementary Table 1b), indicating that skin hyperplasia occurs cell-autonomously upon induction of SRSF6 expression within the epithelium (Fig. 2b).
SRSF6 induces a wound-healing expression signature
To identify genes affected by SRSF6 overexpression, we used genome-wide expression microarrays, which yielded sets of differentially expressed genes in epidermal hyperplasia: 989 genes were upregulated and 1201 were downregulated >2-fold (Supplementary Table 2). Clustering analysis clearly separated the skin samples from the DOX-treated versus the normal mice, indicating inherent changes in their transcriptional profiles (Fig. 2d). Gene Ontology (GO) analysis of the upregulated genes showed significant enrichment in terms associated with the mitotic cell cycle (p<2.6×10−23), response to wounding (p<8.3×10−12), epidermal differentiation (p<4.7×10−7), and cell proliferation (p<1.3×10−3) (Supplementary Table 2).
RT-PCR validation (Supplementary Fig. 5, Supplementary Note 1 for primer sequence information) confirmed the strong induction of keratin 6 (154-fold) and keratin 16 (13-fold)—two markers of tissue injury16. Similarly, many pro-inflammatory cytokines important for wound healing were upregulated, including Tnf (6-fold), Il1f (24-fold), Cxcl2 (97-fold), and Ccl3 (43-fold). Importantly, tenascin C (Tnc), an extracellular-matrix protein commonly expressed at sites of epithelial-mesenchymal-transition (EMT) in embryos17 and at sites of inflammation18, increased 6-fold.
A hallmark of wound healing is collagen deposition at injury sites and adjacent stroma during the proliferative phase13. Despite the lack of ectopic SRSF6 expression in stroma, we detected increased levels of thick collagen fibrils and total collagen by quantitative polarized-light microscopy of Picrosirius-red-stained tissues (p<0.0001)(Fig. 2e). Collectively, these results indicate that epidermal SRSF6 overexpression activates genes directly involved in the wound-healing response, and results in excessive collagen deposition.
A novel role of SRSF6 in wound healing
We next searched for conditions that modulate the observed phenotype. Intriguingly, simply shaving the hair greatly enhanced SRSF6-induced epidermal hyperplasia (Fig. 3a). This implicates stem-cell activation in the phenotype, as depilation or injury can activate stem cells within the hair-follicle bulge19,20. Supporting this interpretation, hair-follicle areas of unshaved skin occasionally harbored a few GFP-positive cells, upon DOX-induction (unpublished), and a few focal areas of local hyperplasia appeared without shaving (Fig. 3a). Similarly, full-thickness longitudinal incisions to the tails of transgenic animals resulted in progressive development of epidermal hyperplasia proximal to the wound, within 1 week after DOX-induction; in contrast, no GFP-positive epidermal cells were present in regions distal to the wound (Fig. 3b). Cross-sections confirmed that wound-induced hyperplasia was closely restricted to the wound site and surrounding tissue. Consistent with this observation, epidermal thickness increased ~3-fold at sites distal to the wound, compared to control epidermis (Fig. 3c, right). Epidermal thickness at wound-proximal sites likewise increased significantly (Fig. 3c, right). Although SRSF6 had no effect on the wound-closure rate, it resulted in significantly wider wounds (unpublished).
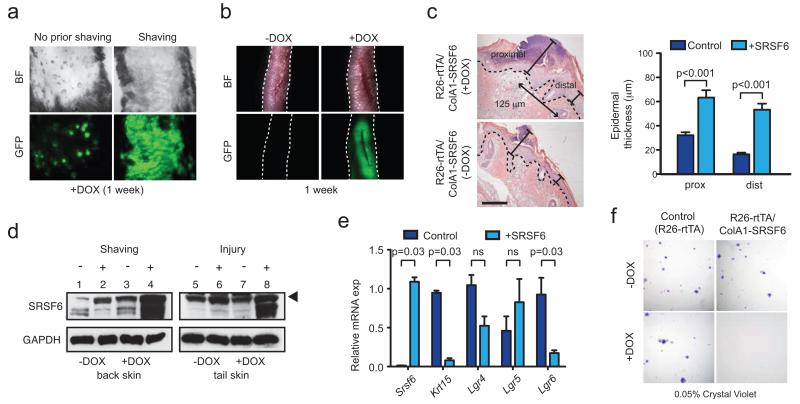
Figure 3.
SRSF6 is involved in wound healing. (a) Effect of hair shaving on SRSF6-induced skin hyperplasia. (b) Full-thickness incision wound healing of mouse tails from transgenic mice, induced by DOX or controls (1 week). (c) Measurement of wound-induced hyperplasia upon SRSF6 induction. Left: Representative tail-skin wounds in the presence or absence of SRSF6 induction (1 week post-injury), H&E staining; bar = 100 μm. Right: Quantification of epidermal thickness at sites proximal (center) or distal (125 μm from the center) from the wounds (Mann-Whitney test, p<0.001, n=26). (d) Immunoblotting for endogenous SRSF6 expression in wounded or shaved skin from DOX-induced or control mice. Arrow indicates fully phosphorylated SRSF6. (e) qPCR of stem-cell markers in total skin extracts, showing depletion of skin stem cells upon SRSF6-induced hyperplasia: Keratin 15 (Krt15), Lgr4, Lgr5, and Lgr6 (Mann-Whitney test, n=4 samples per condition). (f) Clonogenic assay of primary keratinocytes isolated from R26-rtTA/ColA1-SRSF6 (double-transgenic) or R26-rtTA (control) mice Representative images of Crystal-Violet-stained colonies, showing that SRSF6 overexpression strongly reduced colony formation (plating efficiency %: Fisher’s exact test, p<0.05, n=3 per condition). Data are represented as mean +/− s.e.m.
We next investigated whether SRSF6 is normally involved in wound healing. We extracted protein from wounded tail skin or shaved back skin of double-transgenic mice for immunoblot analysis. Indeed, one week after wounding, endogenous SRSF6 increased (Fig. 3d; lanes 5,6). Moreover, wounding was required to maximally elevate total SRSF6 protein in the context of DOX induction (Fig. 3d; lanes 7,8). We observed a very similar pattern in shaved skin: shaving increased endogenous SRSF6 protein, and was required to maximally increase total SRSF6 upon DOX induction (Fig. 3d; lane 1 vs. 2, lane 3 vs. 4). These results suggest that SRSF6 is important for normal wound healing; however, under SRSF6-overexpression conditions, the normal tissue-repair process is misregulated, resulting in skin hyperplasia, most likely due to aberrant stem-cell activation.
To further investigate skin-stem-cell dynamics during SRSF6-induced skin hyperplasia, we analyzed the expression of the hair-follicle stem-cell markers Krt15, Lgr4, Lgr5, and Lgr621. Interestingly, qPCR analysis showed specific depletion of Lgr6, which marks an upper hair-follicle stem-cell subtype, in contrast to the lower hair-follicle LGR5+ stem cells, which did not change significantly in skin samples with SRSF6-induced hyperplasia (Fig. 3e). The hair-follicle-bulge marker Krt15 was also reduced upon hyperplasia, as expected. In certain situations, broad stem-cell activation can lead to stem-cell depletion, e.g., upon ectopic c-Myc overexpression in mouse skin22. These results further support the notion that hair-follicle stem cells are directly activated, and may be required for initiating SRSF6-induced epidermal hyperplasia.
To test if SRSF6 can affect stem-cell self-renewal or differentiation in vitro, we purified and cultured primary keratinocytes from double-transgenic or control mice, and plated them to determine the effect of SRSF6 overexpression on the number of colonies formed—reflecting the original number of stem cells seeded23. After 10 days in culture, we did not detect any colonies from DOX-induced SRSF6-transgenic keratinocytes (Fig. 3f), consistent with specific stem-cell depletion upon SRSF6 overexpression. We also showed by qPCR and immunoblotting that induction of SRSF6 promotes differentiation of cultured primary keratinocytes (Supplementary Fig. 6a, b). Importantly, markers of differentiation were strongly induced (Krt1: +743-fold; Flg: +32-fold), whereas markers of undifferentiated cells changed only slightly (Krt5: +3-fold; Krt14: +2-fold). These results suggest that SRSF6 can impair stem-cell renewal and/or promote differentiation of epidermal stem cells, both in vivo and in vitro. In vivo, SRSF6 overexpression in skin prevents basal keratinocytes from undergoing complete terminal differentiation.
SRSF6 promotes cassette-exon inclusion in hyperplastic skin
To investigate the impact of SRSF6 overexpression on AS, we analyzed the raw microarray expression data using the exon-splicing-index method24. Pairwise comparisons identified common AS events (ASEs), yielding a set of 154 alternatively spliced exons from 139 unique genes (Fig. 4a). Illustrating SRSF6’s inherent function as a splicing activator, we observed almost exclusively increased exon inclusion (153 of 154 events) upon SRSF6 induction in skin (Fig. 4a, Supplementary Table 2). These changes were not attributable to gene-expression changes, as the latter did not correlate with exon splicing-index (SI) scores.
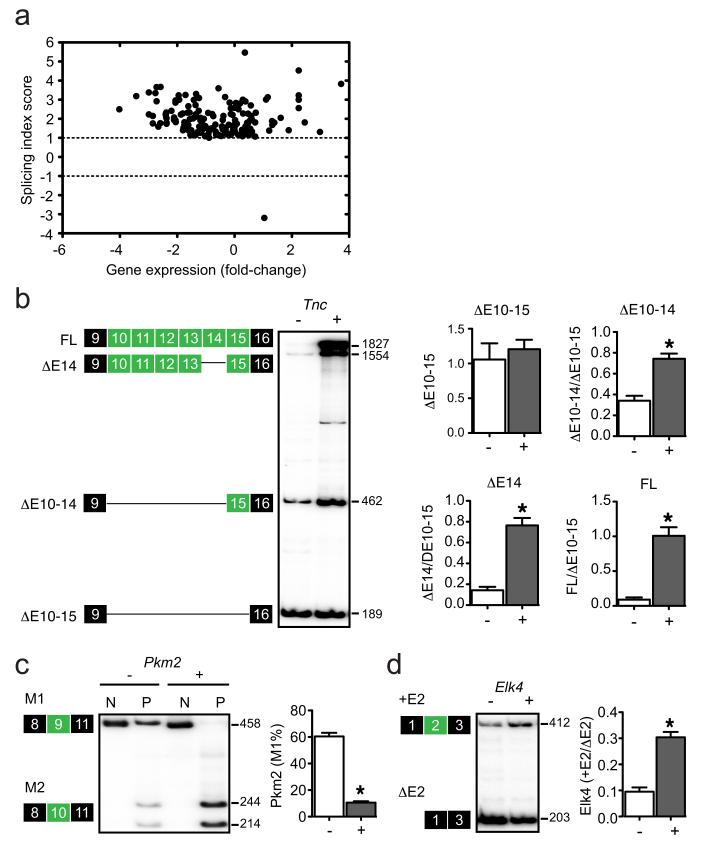
Figure 4.
SRSF6 regulates AS in skin hyperplasia. (a) Microarray analysis of AS events (ASEs) showing 154 changes identified by two independent pairwise comparisons between SRSF6-induced and control skin samples. (b-d) Radioactive RT-PCR validations of SRSF6-responsive ASEs: tenascin C (Tnc), pyruvate kinase 2 (Pkm2), and ETS-domain protein (SRF accessory protein 1) (Elk4) (Mann-Whitney test; *p<0.05, n=10). Data are represented as mean +/− s.e.m.
SR proteins, including SRSF6, bind to degenerate ESEs in pre-mRNA, from where they activate splicing25. To further address the mechanisms of SRSF6-induced hyperplasia, we performed binding-motif prediction using MEME26. Omitting 5′- and 3′-terminal exons from the set of 154 alternatively spliced exons yielded 84 internal exons that we used in combination with a random set of cassette exons (as background reference) to predict de novo a consensus SRSF6 motif. We identified a 9-mer motif with an e-value of 2.5×108 (‘SRSF6 in vivo motif’). This motif differed only slightly from the 6-mer motif found by in vitro functional SELEX27, consistent with its presumed functionality in vivo (Supplementary Fig. 7e). Furthermore, the SRSF6 motif did not resemble the SELEX motifs of other SR proteins28.
Next, we validated the AS changes using RT-PCR with an independent set of skin samples—either SRSF6-overexpressing or controls (n=10). We successfully validated 16 of 22 cassette exons—an overall success rate of 73% (Supplementary Fig. 4a-c, Supplementary Table 2-3a). Prominent validated examples include: pyruvate kinase 2 (Pkm2) exon 9 (−3.2); tenascin C (Tnc) exons 10 (+3.0), 11 (+2.6), 12 (+3.2), 13 (+4.5), and 15 (+3.8); and transcription factor Elk4 exon 2 (+1.7)(Fig. 4b-d). hnRNPA1/A2, PTB, and SRSF3 contribute to regulating PKM2 AS29-31, and our present data also implicate SRSF6 in regulating murine Pkm2 AS. The extracellular-matrix protein TNC is a particularly intriguing new target of SRSF6, because it is specifically expressed at sites of tissue injury and inflammation, in chronic wounds and cancer18,32. Alternatively spliced exons of Tnc scored among the highest changes in our analysis, consistent with this gene’s potential role as a functionally relevant target of SRSF6.
Tenascin C is a target gene of SRSF6
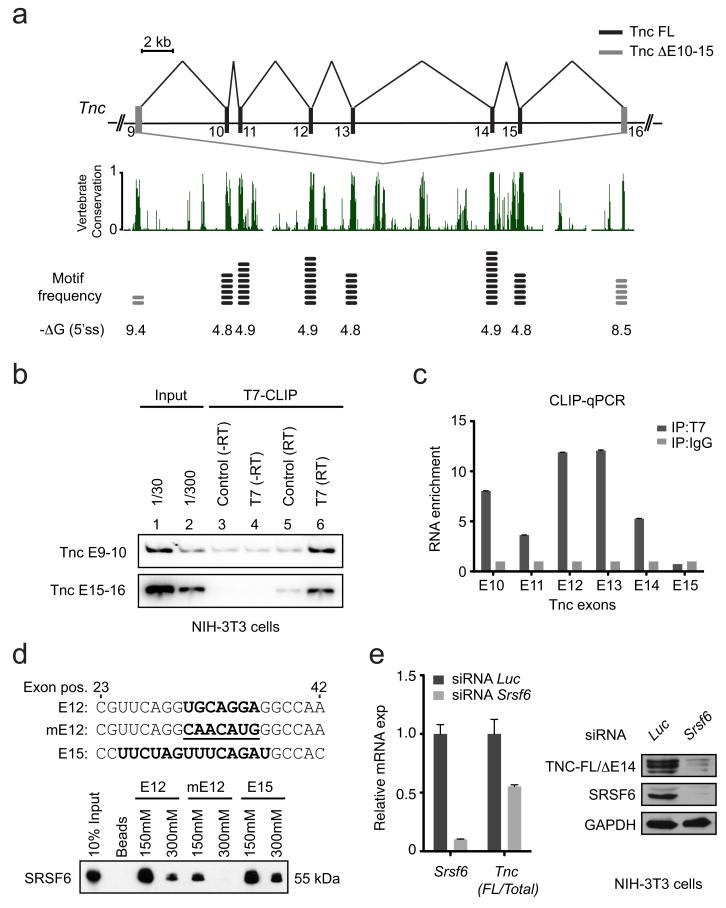
To determine how SRSF6 controls Tnc AS, we searched for putative SRSF6 motifs within exons at or near the alternatively spliced region of Tnc, using SFmap33. The predicted SRSF6 motif was enriched in the highly conserved alternative exons of Tnc, compared to the flanking constitutive exons, although these are smaller (Fig. 5a, Supplementary Table 3b). Furthermore, the presence of SRSF6 motifs in alternative exons inversely correlated with the strength of their 5′ splice sites (Fig. 5a, Supplementary Table 3b), suggesting that SRSF6 binding to these ESE motifs is required to promote more efficient alternative-exon inclusion.
Figure 5.
Tenascin C AS is regulated by SRSF6 in skin. (a) Top: schematic representation of exons 9 to 16 of the mouse Tnc gene. Black: alternative cassette exons (exons 10 to 15) regulated by SRSF6; gray: constitutive exons 9 and 16. Middle: the exon 9 to 16 region of Tnc is highly conserved in vertebrates. Bottom: inverse correlation between putative SRSF6 in vivo binding motif (5-mer core motif; WKSWG) frequency and 5′-splice-site-strength predictions. (b) RNA CLIP from NIH-3T3 cells with primer pairs complementary to mouse Tnc exons 9 and 10, or 15 and 16, respectively. (c) RNA CLIP-qPCR analysis with MNase digestion. (d) RNA-affinity pulldown using immobilized 21-mer RNA oligonucleotides comprising the wild-type SRSF6 binding motif in exon 12 (E12; UGCAGGA), exon 15 (E15; UUCUAGUUUCAGAU) or a mutated motif (mutE12; CAACAUG). (e) Left: qPCR analysis of siRNA knockdown of SRSF6 and luciferase control, showing effects on Tnc AS (Mann-Whitney test, p<0.05, n=6). Right: immunoblotting after SRSF6 knockdown showing effect on TNC protein isoform levels. Data are represented as mean +/− s.e.m.
To directly measure whether SRSF6 binds alternatively spliced Tnc mRNA, we transiently over-expressed T7-tagged SRSF6 cDNA or empty-vector control in NIH-3T3 cells. After UV-irradiating the cells, we isolated cross-linked adducts by immunoprecipitation with immobilized anti-T7-tag antibody. We then purified the bound mRNA and analyzed it by RT-PCR using primers specific for either Tnc exons 9 and 10 or exons 15 and 16. In both cases, SRSF6 specifically bound to alternatively spliced Tnc mRNA, compared to empty-vector control (Fig. 5b; lanes 5,6). As expected, there was no product when we omitted reverse transcriptase (lanes 3,4).
To investigate where along Tnc mRNA SRSF6 binds in cells, we repeated the experiment, but with micrococcal-nuclease (MNase) digestion before immunoprecipitation. We then measured by qPCR the relative RNA enrichment for each alternative Tnc exon (10 to 15)(Fig. 5c). SRSF6 associated with alternative exons 10 to 14, but not with exon 15. However, the latter could be due to weak nuclease protection or inefficient crosslinking, as exon 15 has 6 SRSF6 motifs. To test the functionality of the predicted SRSF6 binding sites, we performed RNA pulldowns with three 21-mer oligonucleotides comprising nucleotide positions 23 to 42 of mouse Tnc exons: one from exon 12; another with a mutated binding site; and the last with two SRSF6 binding sites from exon 15. Intriguingly, endogenous SRSF6 strongly bound mouse wild-type E15 and E12, but not mutE12, under splicing conditions in nuclear extract from HeLa cells (Fig. 5d).
We then asked whether SRSF6 knockdown affects Tnc AS. We transiently transfected NIH-3T3 cells with short interfering RNA (siRNA) against luciferase or Srsf6. We quantitated Tnc AS as the ratio between long isoforms and total Tnc, using specific qPCR primers for exons 8/9 (total Tnc) and 15/16 (long isoforms). Srsf6 knockdown resulted in downregulation of the long Tnc isoforms, compared to total Tnc (Fig. 5e, left, and Supplementary Note 2 for target sequence information). We confirmed this result by RT-PCR, and importantly, the short Tnc isoform, ΔE10-15 (E=exon), was unaffected by Srsf6 knockdown (Supplementary Fig. 7d). This is consistent with the Tnc splicing changes observed upon SRSF6 overexpression, in that the ΔE10-15 isoform was likewise unaffected (Fig. 4c). We confirmed the decrease in the long TNC protein isoform by immunoblotting using isoform-specific antibody (Fig. 5e, right).
Tenascin C AS correlates with phenotypic severity
To study the spatial and temporal effects of SRSF6 expression on Tnc splicing, and its effects on skin pathology, we performed a time-course experiment, feeding mice DOX for 0-7 days. We observed a delay in the Tnc switch towards the long isoform after SRSF6 induction (Fig. 6a). This fits with the expectation that SRSF6 expression needs to reach a certain level before it can affect splicing of its targets. Furthermore, the Tnc splicing change correlated with the appearance in hyperplastic skin of TNC-FL protein, which was undetectable in normal uninduced skin (Fig. 6b). Moreover, histopathology of induced skin tissues confirmed that the Tnc splicing switch was associated with induction of hyperplasia (Fig. 6c). However, the timing and duration of SRSF6 induction was critical. During normal tissue wound healing, the onset of keratinocyte proliferation starts after ~1-2 d following injury12,13. Here, we observed that if SRSF6 was persistently expressed on days 0-7 following injury, severe skin hyperplasia was induced. In contrast, if SRSF6 enforced expression was stopped by removing DOX on day 2 following injury, skin hyperplasia was no longer present on day 7 (Fig. 6c, lower right). Hence, sustained SRSF6 expression is required during the proliferative phase of keratinocytes to drive hyperplasia.
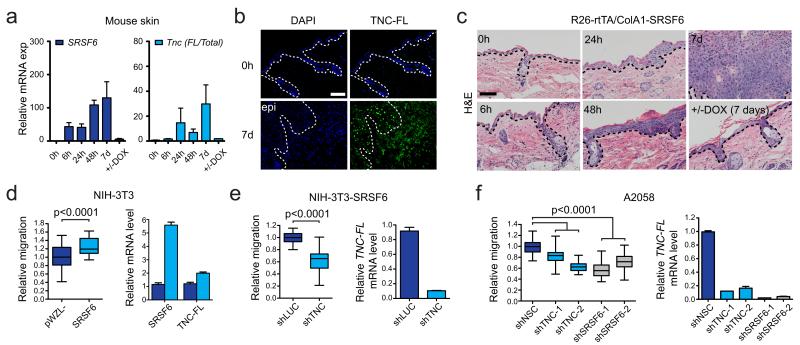
Figure 6.
Tnc-FL expression is linked to hyperplasia severity. (a) qPCR analysis of skin from time-course experiment. Left: total SRSF6; right: Tnc isoform ratio (Tnc-FL/Total Tnc) (n=3). (b) Immunofluorescence of TNC-FL protein isoform expression in skin upon SRSF6 induction for 0 h and 7 d. Epidermis=epi. (c) Correlation between skin hyperplasia, SRSF6 induction, and splicing switch towards the Tnc-FL isoform; 0 h to 7 d. Bottom right: Reversion of skin hyperplasia upon DOX withdrawal (5 d) after SRSF6 induction (2 d). (d) In vitro wound-healing assay (18 h), showing enhancement of cell migration by SRSF6 overexpression in NIH-3T3 cells (n=24, p<0.0001) (left), and qPCR analysis of concomitant increases in relative Tnc-FL mRNA levels (right). (e) Cell migration assay showing that shRNA knockdown of Tnc suppresses the effect of SRSF6 overexpression (n=24, p<0.0001) (left); qPCR analysis of relative Tnc-FL mRNA levels (right). (f) In vitro wound-healing assay; inducible shRNA knockdown of either TNC or SRSF6 (two hairpins each) in A2058 human melanoma cells, showing reduced cell migration; +DOX (10 μg/ml) for 4 d (n=24, p<0.0001) (left). qPCR analysis of relative TNC-FL mRNA levels after knockdown of TNC or SRSF6 (right). Bar = 100 μm (b, c). Data are represented as mean +/− s.e.m. All p-values were calculated by Mann-Whitney tests.
To further explore the role of SRSF6 and its specific target isoform, Tnc-FL, in wound healing, we used in vitro assays. Consistent with its role in hyperplasia, SRSF6 significantly increased cell migration when overexpressed in NIH-3T3 cells (Fig. 6d). Importantly, this stimulatory effect of SRSF6 was suppressed by shRNA knockdown of Tnc, which resulted in strong down-regulation of the Tnc-FL isoform (Fig. 6e). In addition, we tested whether SRSF6 controls cell migration through splicing regulation of TNC in A2058 human melanoma cells, which express high levels of TNC-FL34; SRSF6 knockdown using two independent inducible shRNAs resulted in reduced levels of TNC-FL and decreased cell migration. We obtained similar results by inducible shRNA knockdown of TNC-FL (Fig. 6f, Supplementary Fig. 8). These findings are consistent with a previous report showing that TNC promotes cell migration mainly through its long isoform (TNC-FL), whereas the short isoform lacks this ability35. Based on these data, we conclude that regulation of TNC AS by SRSF6 is cell-type-independent, and plays a causal role in promoting wound healing in vitro.
Overexpression of both SRSF6 and TNC-FL in skin cancer
SRSF1 is a proto-oncogene that can transform rodent fibroblasts and mammary epithelial cells36,37. As SRSF1 and SRSF6 are closely related orthologs, we investigated whether SRSF6 is likewise an oncogenic splicing factor. We transduced MEFs (p53−/−, c-Myc+) with either SRSF6 cDNA or empty vector, injected them subcutaneously into nude mice, and allowed tumors to develop (Supplementary Table 4). SRSF6 strongly promoted sarcomagenesis after only 4 weeks (Fig. 7a).
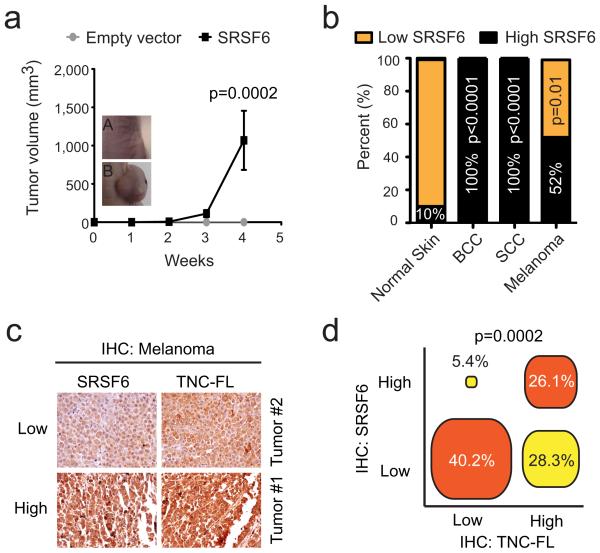
Figure 7.
SRSF6 is a proto-oncogene overexpressed in human skin cancer. (a) Analysis of tumor growth in nude mice injected with 2×106 MEF +c-Myc, p53−/− cells in each flank (n=8). Control (A), SRSF6 (B). (b) Tissue microarray analysis of SRSF6 protein expression in human tumors, showing SRSF6 overexpression (High SRSF6) in BCC (100%, p<0.0001, n=20), SCC (100%, p<0.0001, n=18), and malignant melanoma (52%, p=0.01, n=17), compared to normal skin (10%, n=20). (c) Serial tissue microarrays of malignant melanoma tissues stained with either anti-SRSF6 or anti-TNC-FL antibodies for expression-correlation analysis (n=92). Tumor #1-2: representative tissues. (d) Diagram showing strong correlation between levels of SRSF6 and TNC-FL proteins in most of the tissues analyzed (66.3%). Data in a are represented as mean +/− s.e.m. All p-values were calculated by Fisher’s exact test.
Considering the skin hyperplasia caused by SRSF6 overexpression, and its overexpression in different human cancers6, we measured its levels in skin cancers in particular. To assess SRSF6 protein expression, we stained tissue microarrays comprising multiple tumor specimens, with a monoclonal antibody. Remarkably, we found significantly elevated levels of SRSF6 protein in basal-cell carcinomas (BCC; n=20, p<0.0001), squamous-cell carcinomas (SCC; n=18, p<0.0001), and malignant melanomas (n=27, p=0.01)(Fig. 7b). These data are consistent with SRSF6’s potential role in human skin cancer, and with SRSF6 being a proto-oncogene. We also stained serial tissue microarrays containing 92 human melanoma specimens with either SRSF6 or TNC-FL antibodies. SRSF6 and TNC-FL protein isoform levels were strongly correlated (66.3% of the cases, p=0.0002) (Fig. 7c-d). High SRSF6 levels were only infrequently linked to low levels of TNC-FL protein (5.4%). These results suggest that in addition to its role in normal skin, SRSF6 controls TNC-FL protein levels in the context of skin cancer. However, our data suggest that additional factors also regulate TNC/FL levels, as some tumors have high TNC-FL levels without SRFS6 overexpression.
In summary, SRSF6 regulates multiple AS events in vivo. When its normal level is perturbed—as it is in many cancers—it gives rise to skin hyperplasia, and we identified tenascin C as one of its prominent target genes, whose AS pattern is strongly associated with skin cancer. These observations reinforce the importance of AS regulation for cancer progression. Furthermore, our study identifies SRSF6 and/or the genes it controls as potential targets for therapeutic intervention in skin cancer.
Discussion
We demonstrated that overexpression of the SR protein SRSF6 causes skin hyperplasia in mice by deregulating tissue homeostasis. We also found statistically significant overexpression of SRSF6 in human BCC, SCC, and malignant melanoma. Moreover, we showed that enforced expression of SRSF6 is sufficient to transform p53-null, Myc-expressing primary fibroblasts in allograft assays. The latter observation resembles earlier findings in other contexts for the SRSF6 paralogs SRSF16,7 and SRSF38, both of which are proto-oncogenes, thus extending the evidence of oncogenic SR proteins.
Our results suggest that SRSF6 has a role in normal wound healing, potentially related to its role in cancer. Indeed, we found that endogenous SRSF6 is elevated upon skin injury. We hypothesize that under physiological conditions reminiscent of tissue injury, SRSF6 is induced to regulate certain target effector genes at the level of AS, allowing an efficient and appropriate wound-healing response. Our data suggest a possible mechanism for how excessive SRSF6 expression might instead result in abnormal hyperplasia (Fig. 8). Normally, keratinocytes respond to injury by becoming activated—a state characterized by strong expression of Krt6 and Krt16. Activated keratinocytes also release the cytokine Il-1—the first signal upon wounding—to further increase keratinocyte activation and alert surrounding tissues38. Indeed, using microarray expression analysis of skin tissues, we found that SRSF6 induction resulted in strong upregulation of Krt6, Krt16, Il-1, and many other wound-healing markers.
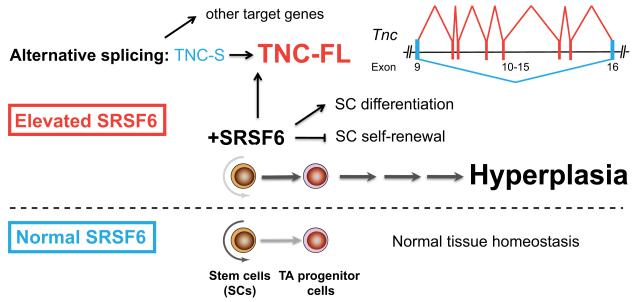
Figure 8.
Model for the dynamic role of SRSF6 in skin hyperplasia and wound healing. Bottom (blue): Normal tissue homeostasis in skin. Top (red): During tissue injury, expression of SRSF6 is induced (and perhaps required) to regulate or promote unknown processes that are part of normal wound healing, through AS of target genes, e.g., Tnc and others. When SRSF6 expression is elevated, skin stem cells are aberrantly activated upon stimulation (by shaving or wounding) causing skin hyperplasia. SRFS6 likely suppresses stem-cell self-renewal signals and/or stimulates stem-cell differentiation to keratinocyte progenitors that are maintained in a proliferating state. Aberrant levels of the SRSF6 splicing target Tnc-FL protein isoform may have a disruptive role for maintenance of the stem-cell niche microenvironment.
One of the main pathological features of SRSF6-induced hyperplasia is the dependence on somatic stem-cell activation to achieve the full phenotype. Thus, the onset of hyperplasia was strongly enhanced by either hair shaving or tissue wounding—conditions in which skin stem cells are specifically activated21,39. We did observe focal hyperplastic lesions in unshaved mice, which likely reflect scratching or hair pulling.
Intriguingly, skin hyperplasia was associated with depletion of LGR6+ cells. These skin stem cells reside in the upper part of the hair-follicle, and participate in wound healing upon tissue injury21. We demonstrated that the effects of SRSF6 are cell-autonomous, and therefore consistent with a leading role of stem-cell activation, as transplanted skin grafts recapitulated the phenotype upon shaving/DOX-treatment. Supporting evidence with cultured primary keratinocytes showed that SRSF6 expression repressed their colony-formation capacity and promoted differentiation. These findings suggest a role of SRSF6 in repressing self-renewal signals and promoting differentiation. The reasons why SRSF6 promotes proliferation and the initial steps of differentiation, but represses terminal differentiation, remain unknown. Overexpression of SRSF6 may maintain keratinocytes in an abnormal activation state that otherwise would be temporary during normal tissue repair. Additional studies are needed to understand this phenomenon in detail.
Strikingly, for the genes that showed changes in AS in skin upon SRSF6 overexpression, nearly all the changes consisted of increased exon inclusion, confirming SRSF6’s role as a strong splicing activator. A consensus 9-mer binding motif was present in all 84 internal alternative exons. Moreover, we demonstrated specific binding of SRSF6 to alternatively spliced Tnc exons with the binding motifs.
Due to the complex nature of AS regulation in vivo, and the global effects of individual SR proteins, such as SRSF6, it is difficult to demonstrate a direct causal relationship between expression of individual isoforms and the observed hyperplastic phenotype.
Intriguingly, however, some of the isoforms promoted by SRSF6 represent the reappearance of an embryonic splicing pattern. In particular, the M2 isoform of Pkm2 is specifically expressed in embryonic tissues and in cancers40. Similarly, the long Tnc isoform (+E10-15 or +E10-18 (human)) is expressed in neuronal stem cells and embryonic tissue41, and in many types of invasive and metastatic cancer42-47. In breast-cancer cells, the two long TNC isoforms, +E10-15 and ΔE14, drive increased cell migration and proliferation in vitro46. Also, the plasma level of the long TNC isoform is a putative predictive marker for metastatic colon cancer48,49. We showed here that SRSF6 specifically promotes AS of the +E10-15 and ΔE14 Tnc isoforms, without changing the levels of the constitutive ΔE10-15 isoform, even though we observed an increase in total Tnc expression upon induction of hyperplasia.
Consistent with SRSF6 being a likely regulator of Tnc AS, we observed a direct relationship between the magnitude of the change in Tnc AS, the SRSF6-expression level, and the extent of hyperplasia over time. Moreover, when SRSF6 induction was withdrawn, Tnc AS reverted, as did the hyperplastic phenotype. The relevance of SRSF6 as a regulator of Tnc AS was not limited to cell lines and mouse skin, but was also evident in human melanomas, which showed a strong correlation between SRSF6 and TNC-FL protein levels. We have thus discovered an important connection between clinically proven, important isoforms of Tnc and a member of the SR-protein family of splicing regulators.
Future work will be required to confirm that Tnc AS plays a key role in driving the development and maintenance of skin hyperplasia, and possibly other related pathological conditions. It will also be important to understand how SRSF6 is naturally regulated during wound healing, and whether other splicing factors contribute to these processes as well.
Methods
ES-cell targeting and production of transgenic mice
Details of the targeting construct, pBS31’-RBGpA TREtight ColA1, are in the Supplementary Information. A DOX-responsive element controlling SRSF6 expression was targeted downstream of the Cola1 locus by Flp/FRT recombinase-mediated site-specific integration in KH2 ES cells, as described50,51. Injection of targeted ES cells into tetraploid blastocysts to produce fully ES-cell-derived transgenic mice was performed by the CSHL Gene Targeting Shared Resource. DOX was administered to adult mice via food pellets (625 mg/kg) (Harlan Teklad). Hair removal was performed either by shaving, plucking, or applying NAIR lotion for 3 min (Church & Dwight Co) immediately before DOX administration. In vivo imaging of skin GFP expression was measured using a Xenogen IVIS imaging system (Perkin Elmer). No experiments were blinded; each group consisted of gender- and age-matched mice (6 to 52 weeks old C57/BL6/mixed background). As the skin phenotype was not anticipated, we did not perform a power calculation to deduce an adequate group size; instead we used a small group size of 8 treated and 8 control animals, which proved adequate for statistical-significance tests. All mouse experiments were approved by the CSHL Animal Care and Use Committee.
RNA extraction, RT-PCR and quantitative PCR
Total RNA from cells or tissues was extracted using Trizol (Life Technologies). Digestion with RQ1 RNase-Free DNase (Promega) for 30 min was used to eliminate genomic DNA contamination. cDNA synthesis was performed using 1 μg of total RNA, random primers (Sigma-Aldrich), and Improm-II reverse transcriptase (Promega). cDNA was amplified by PCR using AmpliTaq DNA polymerase (Applied Biosystems). For quantitative measurement of mRNA levels, cDNA was amplified by PCR with Power SYBR Green PCR Master Mix on a 7900HT Fast Real-Time System (Applied Biosystems). For details of primer sequences, see Supplementary Information.
Splicing analysis
32P-radiolabeled PCR products were separated by 6% native PAGE, followed by phosphorimage analysis to quantify band intensities, as described52. Splicing changes were validated using an independent set of samples, consisting of 4 controls and 6 SRSF6-expressing skin samples.
Tissue-microarray evaluation
Immunohistochemical analysis of SRSF6 expression in human tumors was performed using skin-cancer tissue microarrays (US Biomax) as described53. Staining intensities were scored by attributing score values proportional to the staining level, ranging from 0 to 4 for each tissue specimen (n=75 and n=92). Two independent evaluation rounds were performed for each tissue microarray, and any scoring discrepancy was re-evaluated by J.E.W. Specimens were divided into two groups: low staining (0-2) and high staining (3-4) according to the scores.
Cell transduction
Two different primary MEF cell lines (p53−/−) were generated by retroviral transduction: MSCV-c-Myc-dsRED/MSCV-SRSF6-IRES-EGFP and MSCV-c-Myc-dsRED/MSCV-IRES-EGFP (control). Stable double-labeled cells were sorted by flow cytometry using a FACSAria II instrument (BD). All other cell lines were generated as described previously54 or obtained from CSHL’s mammalian cell repository. All cell lines are routinely tested for mycoplasma contamination.
Expression and splicing microarray analysis
Microarray experiments were carried out at the CSHL Microarray Shared Resource. Skin tissues were homogenized using liquid nitrogen, and total RNA was isolated using an RNeasy Mini Kit (Qiagen). RNA quality was determined on an Agilent 2100 Bioanalyzer; samples with a RIN score of ≥7.0 passed the quality control. RNA was amplified, and converted into cDNA using a WT Expression kit (Ambion). This was followed by cDNA fragmentation and terminal biotin-labeling, using a GeneChip WT Terminal Labeling kit (Affymetrix). Samples were then hybridized, washed, and scanned according to the manufacturer’s instructions, using Mouse Gene ST 1.0 GeneChips (Affymetrix). Raw data were normalized, and analyzed for gene expression and AS in parallel, using AltAnalyze software55. The splicing index method was used to calculate AS for pairwise comparisons. Putative AS events were identified as common identities from two independent pairwise comparisons. Unsupervised hierarchical clustering was performed with the 25 most variable genes across all microarrays, using Cluster 3.0 (average linkage method) (Stanford University), and visualized by Java TreeView56. Gene Ontology annotation analysis was performed using DAVID Bioinformatic Resources 6.757.
Primary keratinocytes
Primary keratinocytes were isolated from control (R26-rtTA) or double-transgenic (R26-rtTA/ColA1-SRSF6) neonates, and cultured in CnT-57 media, according to the manufacturer’s instructions (CELLnTEC Advanced Cell Systems AG). Clonogenic assays were performed by seeding freshly isolated cells (2500 cells/well in a 6-well plate) in CnT-02 media, in the presence or absence of DOX (1 μg/ml). Colonies were fixed and stained for 30 min with 6% (v/v) glutaraldehyde/0.5% (w/v) Crystal Violet after incubation for 10 d, as described58. Differentiation was induced in CnT-02 media/+1.2 mM CaCl2 for 5 d, in the presence or absence of DOX (1 μg/ml).
Wound-healing assay
Mice expressing R26-rtTA/ColA1-SRSF6 were anesthetized, and longitudinal full-thickness incision wounds (1 cm) were made with a razor blade. Half the mice were fed DOX for 7 d. On day 7, tail-wound regions were analyzed by light microscopy. Wound width and epidermal thickness were measured at the wound site (proximal) or 125 μm from the wound center (distal) using ImageJ software (n=26). Scratch wound-healing assays were performed as described previously59; A2058 cell lines were treated with 10 μg/ml DOX for 3 d prior to assaying.
Allogenic transplantations
Back skin from R26-rtTA/ColA1-SRSF6 transgenic or wild-type/R26-rtTA control animals was transplanted onto the back of nude recipient mice, as described60. Skin grafts were allowed to re-grow hair before they were shaved and animals induced by DOX for 1 week.
Picrosirius-red staining of collagen
For quantitative measurement of fibrillar type I+III collagen levels, skin tissue sections from either DOX-treated or control animals were stained with Weigert’s hematoxylin and then 0.1% Picrosirius red for 1 hour. 16-bit images were obtained using circular polarized light microscopy, and analyzed with Volocity software (PerkinElmer). RGB image settings used were: R: 3660-15781, G: 981-4246, B: 859-2389 (red-yellow colored thick collagen); R: 1920-4563, G: 869-7012, B: 1087-7744 (green colored thin collagen). We measured a total of 60 representative regions (100 × 100 μm) per condition.
Immunofluorescence microscopy
Paraffin-embedded mouse skin sections were analyzed by immunofluorescence microscopy using fluorescence-labeled anti-mouse Alexa Fluor 568 goat anti-mouse IgG (A11031) and Alexa Fluor 647 goat anti-rabbit IgG (A21245) secondary antibodies (Life Technologies). Images were acquired on a Zeiss Axiovert 200M microscope and further processed using AxioVision software (Zeiss).
Subcutaneous tumor growth in nude mice
For subcutaneous injections, 2 × 106 cells (see above) were resuspended into 150 μl of 1× PBS and injected into each flank of γ-irradiated (400 rad) NIH nu/nu nude mice using a 26-gauge needle (females, 6 to 8 weeks of age). Eight flank injections were performed per cell line, and tumor development was measured by weekly palpation for 4 weeks. We chose a small group size that would be sufficient for statistical significance, assuming strong tumorigenic activity.
RNA-binding motif and 5′-splice-site prediction
The MEME Motif Discovery Tool61 for RNA-binding-motif prediction was used to derive a de novo consensus motif for SRSF6 in mouse skin. The analysis was performed using full-length sequences of the 84 internal exons identified as alternatively spliced by microarray analysis. A random set of 500 mouse cassette exon sequences was used as a background reference. WebLogo software62 was used to generate a representative consensus frequency plot. Calculations of 5′-splice-site strength were based on the algorithm published by Shapiro and Senapathy63 or based on the free energy, ΔG, of base pairing to U1 snRNA64 using the Splice-Site Analyzer Tool (ibis.tau.ac.il/ssat).
RNA CLIP
RNA immunoprecipitation of Tnc mRNA bound to SRSF6 protein was performed as described65, with slight modifications. 30 μg pcDNA3.1-T7-SRSF6 or control plasmid was transiently transfected into NIH-3T3 cells in a 15-cm plate, and UV-crosslinking was performed at 400 mJ/cm2 using a Stratalinker UV Crosslinker (Stratagene). Immunoprecipitation was done using anti-T7 antibody (CSHL; mAb 42 1-87) coupled to Protein G-Dynabeads (Life Technologies); RNA was extracted from the immunoprecipitate, and cDNA was synthesized using random primers (Sigma-Aldrich). RT-PCR was performed using primers specific for each alternatively spliced Tnc isoform, i.e., to amplify across exons 9 and 10, or exons 15 and 16, respectively (for details of primer sequences, see Supplementary Information), and 32P-radioactive PCR products were separated as described above. RNA CLIP qPCR was performed essentially as above, except that extracts were treated with micrococcal nuclease before immunoprecipitation using Protein G-dynabeads coupled with either anti-T7 (CSHL; mAb 42 1-87) or control IgG1 (CSHL; AK105, against E. coli MBP) antibody. RNA enrichment was measured by RT-qPCR using exon-specific primers, and calculated as the relative levels in T7 antibody versus control IgG1 immunoprecipitates.
RNA interference
For knockdown of Srsf6, NIH-3T3 cells were transfected with either 25 nM control siRNA (Luciferase) or mouse Srsf6 siRNA using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions. Cells were incubated for 72 h before lysis, and protein and RNA were then extracted as described54. Details of siRNA/shRNA target sequences are given above in Supplementary Information.
Antibodies
For immunofluorescence microscopy, the following primary antibodies were used: monoclonal anti-T7 (Covance; 9115), anti-SRSF6 (CSHL; mAb 9 1-56), anti-keratin 6 (Abcam; SPM269) and polyclonal anti-keratin 5 (Covance; PRB-160P), anti-keratin 14 (Covance; PRB-155P), anti-loricrin (Covance; PRB-14P), anti-KI67 (Epitomics; 15580), anti-c-myc (Abcam; 39688), anti-β-catenin (Cell Signaling; 9587). For immunoblotting, the antibodies used were: monoclonal anti-SRSF6 (CSHL; mAb 9 1-56), anti-GAPDH (Abcam; 6C5), anti-TNC (4C8MS; Novus Biologicals; 4C8MS) and polyclonal anti-keratin 5 (Covance; PRB-160P). For tissue microarray analysis, monoclonal anti-SRSF6 (CSHL; mAb 9 1-56) was used. Typical antibody dilutions were 1: 25 for IHC and 1:1000 for immunoblotting.
Statistical analysis
For RT-PCR-based splicing analysis, we applied the Mann-Whitney statistical test to identify significant splicing changes between normal and SRSF6-induced skin samples (n=10). For tissue-microarray analysis of SRSF6 and TNC-FL protein expression, statistically significant score differences between normal and skin cancer subtypes, or co-expression of SRSF6 and TNC-FL were calculated using Fisher’s exact test. Significant differences in epidermal thickness after tail injury (n=26) and in collagen deposition measured by Picrosirius-red staining (n=60) were both calculated using the Mann-Whitney test.
Original images of gels, autoradiographs and blots used in this study can be found in Supplementary Figure 9.
Supplementary Material
Acknowledgements
We thank R. Jaenisch (Whitehead Institute) for KH2 ES cells; C. Miething (Memorial Sloan-Kettering Cancer Center) for the targeting construct; P. Premsrirut for useful discussions and help with production of stable ES-cell lines; S.Y. Kim from the Gene Targeting Shared Resource (CSHL) for help with tetraploid complementation procedures; C. Johns for support with microarray analysis; M. Motley for assistance with RT-PCR analysis; and L. Chartarifsky for tissue microarray staining. We thank L. Bianco for help with animal procedures. We are grateful to B. Boettner, M. Egeblad, and A. Mills for critical comments on the manuscript. This study was supported by grant CA13107 from the U.S. National Cancer Institute, and was performed with assistance from CSHL Shared Resources funded in part by Cancer Center Support grant 5P30CA045508. M.A. Jensen was supported by a Danish Cancer Society postdoctoral fellowship.
Footnotes
Author Contributions
M.A.J and A.R.K designed the study and wrote the paper. M.A.J. and J.E.W. carried out the experiments and analyzed the data. All authors read the manuscript.
Accession codes
All raw microarray data files are available from the Gene Expression Omnibus (GSE52458).
References
- 1.Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. (1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 2.Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. (1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. (1993;21:5803–16. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. (2006;119:2635–41. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 5.Ritchie W, Granjeaud S, Puthier D, Gautheret D. Entropy measures quantify global splicing disorders in cancer. PLoS Comput Biol. 2008;4:e1000011. doi: 10.1371/journal.pcbi.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. (2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anczukow O, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. (2010;6:806–26. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghigna C, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. (2005;20:881–90. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. (1986;315:1650–9. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 11.Dunham LJ. Cancer in man at site of prior benign lesion of skin or mucous membrane: a review. Cancer Res. (1972;32:1359–74. [PubMed] [Google Scholar]
- 12.Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. (2008;9:628–38. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- 13.Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. (1999;341:738–46. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 14.Ffrench-Constant C, Van de Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J Cell Biol. (1989;109:903–14. doi: 10.1083/jcb.109.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McJunkin K, et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci U S A. (2011;108:7113–8. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paladini RD, Takahashi K, Bravo NS, Coulombe PA. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J Cell Biol. (1996;132:381–97. doi: 10.1083/jcb.132.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiquet-Ehrismann R, Mackie EJ, Pearson CA, Sakakura T. Tenascin: an extracellular matrix protein involved in tissue interactions during fetal development and oncogenesis. Cell. (1986;47:131–9. doi: 10.1016/0092-8674(86)90374-0. [DOI] [PubMed] [Google Scholar]
- 18.Midwood KS, Orend G. The role of tenascin-C in tissue injury and tumorigenesis. J Cell Commun Signal. (2009;3:287–310. doi: 10.1007/s12079-009-0075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. (2009;137:811–9. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenn KS, Paus R. Controls of hair follicle cycling. Physiol Rev. (2001;81:449–494. doi: 10.1152/physrev.2001.81.1.449. [DOI] [PubMed] [Google Scholar]
- 21.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. (2010;327:1385–9. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 22.Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. (2001;28:165–8. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- 23.Kaur P, Li A, Redvers R, Bertoncello I. Keratinocyte stem cell assays: an evolving science. J Investig Dermatol Symp Proc. (2004;9:238–47. doi: 10.1111/j.1087-0024.2004.09306.x. [DOI] [PubMed] [Google Scholar]
- 24.Salomonis N, et al. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol. 2009;5:e1000553. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. (2002;3:285–98. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 26.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. (1994;2:28–36. [PubMed] [Google Scholar]
- 27.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. (1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & development. (1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clower CV, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. (2010;107:1894–9. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. J Mol Cell Biol. 2011 doi: 10.1093/jmcb/mjr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. (2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orend G, Chiquet-Ehrismann R. Tenascin-C induced signaling in cancer. Cancer Lett. (2006;244:143–63. doi: 10.1016/j.canlet.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009;10:R30. doi: 10.1186/gb-2009-10-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukunaga-Kalabis M, et al. Tenascin-C promotes melanoma progression by maintaining the ABCB5-positive side population. Oncogene. (2010;29:6115–24. doi: 10.1038/onc.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsunoda T, et al. Involvement of large tenascin-C splice variants in breast cancer progression. The American journal of pathology. (2003;162:1857–67. doi: 10.1016/S0002-9440(10)64320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anczukow O, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nature structural & molecular biology. (2012;19:220–8. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature structural & molecular biology. (2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morasso MI, Tomic-Canic M. Epidermal stem cells: the cradle of epidermal determination, differentiation and wound healing. Biol Cell. (2005;97:173–83. doi: 10.1042/BC20040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasper M, et al. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci U S A. (2011;108:4099–104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. (2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 41.Yagi H, et al. HNK-1 epitope-carrying tenascin-C spliced variant regulates the proliferation of mouse embryonic neural stem cells. J Biol Chem. (2010;285:37293–301. doi: 10.1074/jbc.M110.157081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang C, et al. Tenascin-C patterns and splice variants in actinic keratosis and cutaneous squamous cell carcinoma. Br J Dermatol. (2006;155:763–70. doi: 10.1111/j.1365-2133.2006.07401.x. [DOI] [PubMed] [Google Scholar]
- 43.Guttery DS, et al. Association of invasion-promoting tenascin-C additional domains with breast cancers in young women. Breast Cancer Res. 2010;12:R57. doi: 10.1186/bcr2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esposito I, et al. Tenascin C and annexin II expression in the process of pancreatic carcinogenesis. J Pathol. (2006;208:673–85. doi: 10.1002/path.1935. [DOI] [PubMed] [Google Scholar]
- 45.Schliemann C, et al. Three clinical-stage tumor targeting antibodies reveal differential expression of oncofetal fibronectin and tenascin-C isoforms in human lymphoma. Leuk Res. (2009;33:1718–22. doi: 10.1016/j.leukres.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Tsunoda T, et al. Involvement of large tenascin-C splice variants in breast cancer progression. Am J Pathol. (2003;162:1857–67. doi: 10.1016/S0002-9440(10)64320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frey K, et al. Different patterns of fibronectin and tenascin-C splice variants expression in primary and metastatic melanoma lesions. Exp Dermatol. (2011;20:685–8. doi: 10.1111/j.1600-0625.2011.01314.x. [DOI] [PubMed] [Google Scholar]
- 48.Takeda A, et al. Plasma large Tenascin-C spliced variant as a possible biomarker for the prediction of hepatic recurrence in colorectal cancer. Surgery. (2007;141:124–5. doi: 10.1016/j.surg.2006.07.044. [DOI] [PubMed] [Google Scholar]
- 49.Takeda A, et al. Clinical significance of large tenascin-C spliced variant as a potential biomarker for colorectal cancer. World J Surg. (2007;31:388–94. doi: 10.1007/s00268-006-0328-6. [DOI] [PubMed] [Google Scholar]
- 50.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. (2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 51.Premsrirut PK, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. (2011;145:145–58. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anczukow O, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aaboe M, et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. (2006;66:3434–42. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 54.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. (2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salomonis N, et al. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput Biol. 2009;5:e1000553. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. (2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 58.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. (2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 59.Lampugnani MG. Cell migration into a wounded area in vitro. Methods Mol Biol. (1999;96:177–82. doi: 10.1385/1-59259-258-9:177. [DOI] [PubMed] [Google Scholar]
- 60.McFarland HI, Rosenberg AS. Skin allograft rejection. Curr Protoc Immunol. 2009 doi: 10.1002/0471142735.im0404s84. Chapter 4, Unit 4 4. [DOI] [PubMed] [Google Scholar]
- 61.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. (1994;2:28–36. [PubMed] [Google Scholar]
- 62.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. (2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. (1987;15:7155–74. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. (2004;10:828–40. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanford JR, et al. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.