Abstract
Cyclic imine neurotoxins constitute an emergent family of neurotoxins of dinoflagellate origin that are potent antagonists of nicotinic acetylcholine receptors. We developed a target-directed functional method based on the mechanism of action of competitive agonists/antagonists of nicotinic acetylcholine receptors for the detection of marine cyclic imine neurotoxins. The key step for method development was the immobilization of Torpedo electrocyte membranes rich in nicotinic acetylcholine receptors on the surface of microplate wells and the use of biotinylated-α-bungarotoxin as tracer. Cyclic imine neurotoxins competitively inhibit biotinylated-α-bungarotoxin binding to Torpedo-nicotinic acetylcholine receptors in a concentration-dependent manner. The microplate-receptor binding assay allowed rapid detection of nanomolar concentrations of cyclic imine neurotoxins directly in shellfish samples. Although highly sensitive and specific for the detection of neurotoxins targeting nicotinic acetylcholine receptors as a class, the receptor binding assay cannot identify a given analyte. To address the low selectivity of the microplate-receptor binding assay, the cyclic imine neurotoxins tightly bound to the coated Torpedo nicotinic receptor were eluted with methanol, and the chemical nature of the eluted ligands was identified by mass spectrometry. The immobilization of Torpedo electrocyte membranes on the surface of microplate wells proved to be a high-throughput format for the survey of neurotoxins targeting nicotinic acetylcholine receptors directly in shellfish matrixes with high sensitivity and reproducibility.
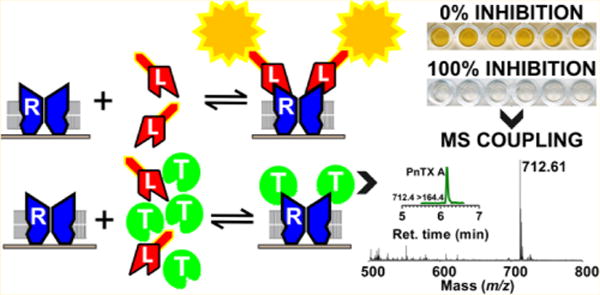
Cyclic imine neurotoxins constitute a large new family of emergent marine phycotoxins associated with harmful algal blooms and shellfish toxicity.1,2 The growing family of structurally related cyclic imines found in toxic dinoflagellates and/or contaminated shellfish includes 31 members: 4 gymnodimines,3,4 14 spirolides of which spirolides E and F are nontoxic,2 7 pinnatoxins,5 3 pteriatoxins, 2 prorocentrolides, and 1 spiro-prorocentrimine.1,2 In addition, a high number of fatty acid acyl esters derivatives of spirolides (21)6 and pinnatoxins (26)7 that are product of shellfish metabolism have been reported.
The dinoflagellate species producing cyclic imine toxins, namely, Karenia selliformis (gymnodimines),8 Alexandrium ostenfeldii (spirolides),9 Alexandrium peruvianum (spirolides and 12-methylgymnodimine),4 and Vulcanodinium rugosum (pinnatoxins),10,11 are globally distributed. Gymnodimines have been detected in New Zealand,8 Tunisia,12 and the United States.4 Spirolides have been reported in Canada,9 Norway,13 France,14 Spain,15 Italy,16 Chile,17 the United States, Denmark, and Scotland.2 Pinnatoxins have been found in Japan,18 China,18 Australia,5 New Zealand,5 Canada,7 and Norway.19
Cyclic imine toxins studied so far are competitive antagonists of nicotinic acetylcholine receptors (nAChRs) with high affinity and broad specificity toward muscle and neuronal cholinergic receptor subtypes from various animal species.12,20–22 A characteristic feature of cyclic imine toxins is the presence of a six- or seven-membered cyclic imine moiety (Figure S-1 Supporting Information), which was found to be the pharmacophore required for the interaction of the toxin with nAChRs.20,21,23
Although highly toxic to mice, cyclic imine toxins are not regulated because no human intoxications related to these toxins have been reported. However, mouse bioassay mortality following intraperitoneal injection with contaminated shellfish extracts forced the prophylactic closure of conchylicultural activities in Foveaux Strait (New Zealand)8 and in Arcachon (France).14 These events were associated with gymnodimine-A and 13-desmethyl spirolide-C, respectively.8,14 In 2011, the European Commission regulated the use of liquid chromatography coupled to mass spectrometry (LC/MS) and mouse bioassay for monitoring internationally regulated marine biotoxins. Whereas LC/MS will be the reference method for lipophilic toxin detection, mouse bioassay could be used until December 2014 for this purpose.24 However, mouse bioassay remain the reference method for paralytic shellfish poisoning toxins and will be periodically used in relaying areas for detecting unknown marine toxins. Novel functional assays are needed, however, to replace mouse bioassay for rapid detection of unknown and unanticipated neurotoxins related to harmful algal blooms and seafood safety that could facilitate LC/MS analysis.
In this study, we developed a target-directed functional method based on the mechanism of action of competitive agonists/antagonists of nAChRs. Key steps for method development were the immobilization of Torpedo electrocyte membranes rich in nAChRs on the surface of 96-well microplates, the use of biotinylated α-bungarotoxin (biotin-α-BgTx) as tracer, and the use of streptavidin coupled to horseradish peroxidase (streptavidin-HRP) for the detection of cyclic imine toxins directly on shellfish extracts. Our results showed that the microplate-receptor binding assay, although very sensitive and specific for the detection of neurotoxins targeting nAChRs as a class, is poorly selective. To address the low selectivity of the receptor binding assay, the analytes bound to the immobilized Torpedo-nAChRs were eluted from the wells and analyzed by mass spectrometry to determine the chemical nature of the contaminating cyclic imine neurotoxins in shellfish samples.
MATERIALS AND METHODS
Reagents
Maxisorp flat-bottomed microplates were obtained from NUNC (Kamstrupvej, Denmark). o-Phenylenediamine (OPD) tablets were obtained from DAKO (Glostrup, Denmark). Biotin-α-BgTx was obtained from Molecular Probes (Eugene, OR, USA). Streptavidin-HRP and α-BgTx were obtained from Sigma (St. Louis, MO, USA). Gymnodimine-A and 13-desmethyl spirolide-C were obtained from NRC–CNRC (Institute for Marine Biosciences, NRC, Halifax, NS, Canada). 13,19-didesmethyl spirolide-C and 20-methyl spirolide-G were obtained from CIFGA (Lugo, Spain). Pinnatoxin-A, pinnatoxin-G, and the amino ketone analogue of pinnatoxin-A were synthesized in the laboratory of Professor Armen Zakarian (University of California, Santa Barbara, California) as recently reported.21,25
Purification of Torpedo Electrocyte Membranes
Torpedo electrocyte membranes rich in nAChRs were purified from the electric organ of Torpedo marmorata as described previously.26,27
Extraction of Cyclic Imine Toxins from Shellfish Samples
One hundred grams of shellfish flesh from Glycymeris glycymeris (clams), Ostrea edulis (oysters), Mytilus galloprovincialis (mussels), and Pecten maximus (scallops) was homogenized five times with a Waring blender at full speed for 1 min at 4 °C. Then, 40 mL of acetone was added to 10 g of shellfish homogenate. The samples were vortexed, incubated for 15 min in a RollerMix at room temperature, and centrifuged (3500g, 15 min at 20 °C). The precipitates were re-extracted once with acetone as described above. The supernatants were recovered, and the solvent was evaporated under a stream of N2 at 35 °C. The resultant residues were resuspended in 4 mL of methanol, passed through 0.2-μm nylon filters, and ultrafiltered through 5000 molecular-weight cutoff centrifugal units. The extracts were stored at −80 °C.
Toxin Spiking Experiments
A mixed standard solution containing gymnodimine-A, 13,19-didesmethyl spirolide-C, pinnatoxin-A, 13-desmethyl spirolide-C, 20-methyl spirolide-G, and pinnatoxin-G at a concentration of 1 μM each was prepared in methanol. Ten grams of shellfish homogenate was spiked with 400 μL of the referred toxin mixture. The homogenate was vortexed and incubated for 2 h in a RollerMix at room temperature. Cyclic imine extraction proceeded as described above.
Microplate-Receptor Binding Assay
The method was optimized as described in the following subsections.
Coating
Torpedo electrocyte membrane (13.5 μg/mL total protein) was prepared in Tris-buffered saline (TBS) buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5). One hundred microliters of this membrane suspension was added to 96-well Maxisorp microplates. The plate was sealed and incubated overnight at 4 °C. Wells H1–H3 were filled with 100 μL of TBS to be used as control plate signals.
Blocking
The microplate was allowed to reach room temperature for 30 min. The membrane solution was discarded, and without washing, 250 μL of blocking buffer [TBS containing 0.5% bovine serum albumin (BSA)] was added to each well. The plate was sealed and incubated overnight at 4 °C.
Toxin/Extract Incubation
The blocking buffer was discarded, and without washing, the plates were loaded with 100 μL of toxin standards or shellfish extracts freshly prepared in blocking buffer. The methanol concentration in the reaction mixture was kept lower than 1% to avoid interference with the assay. The plate was sealed and incubated overnight at 4 °C for maximum sensitivity. Wells H4–H9 were filled with 100 μL of blocking buffer without toxin standards or extract samples for use as the 100% signal control. Wells H10–H12 were incubated with 100 μL of 1 μM α-BgTx in blocking buffer to be used as the 100% inhibition control.
Biotin-α-BgTx Competition
The microplate was allowed to reach room temperature for 30 min. Thereafter, 50 μL of 240 nM biotin-α-BgTx prepared in blocking buffer was added to the incubation mix. The whole mixture was incubated 30 min under shaking (50 rpm) at room temperature.
Washing
The wells were washed thrice with 250 μL of washing buffer (TBS, 0.1% Tween 20).
Streptavidin–Biotin-α-BgTx Reaction
One hundred microliters of streptavidin-HRP in TBS (220 ng/nL protein) was added to each well and incubated for 30 min at room temperature under shaking.
Washing
The wells were washed thrice with 250 μL of washing buffer.
Colorimetric Detection
One hundred microliters of the peroxidase substrate OPD, prepared as indicated by the supplier, was added to each well. After 3 min, the enzymatic reaction was stopped by addition of 100 μL of 0.5 M H2SO4. The optical density at 492 nm (OD492 nm) of the microplates was recorded with a microplate reader (GeniosPro, Männedorf, Switzerland).
Data Analysis
The OD492 nm data were transformed into inhibition percentages using the equation: percent inibition = 100 × [(100% signal − sample signal)/(100% signal − 100% inhibition)] where 100% signal is the maximum biotin-α-BgTx binding signal obtained from control wells in which Torpedo electrocyte membranes were incubated with biotin-α-BgTx in the absence of competitive agonists/antagonists of nAChR and 100% inhibition was obtained by incubating Torpedo-nAChR with α-BgTx that fully inhibited tracer binding. Background noise due to nonspecific retention of biotin-α-BgTx was controlled in signal plate wells. Typically, 100% inhibition and signal plate wells gave similar OD492 nm values. The binding assay data were analyzed using GraphPad Prism 5.02 (GraphPad Software Inc., San Diego, CA, USA).
Toxin Elution from Maxisorp Microplate Wells
Two columns of eight coated wells were incubated overnight with 100 μL of blocking buffer containing gymnodimine-A, 13,19-didesmethyl spirolide-C, pinnatoxin-A, 13-desmethyl spirolide-C, 20-methyl spirolide C, and pinnatoxin-G at a concentration of 1 μM each. The wells were washed three times with 250 μL of TBS. To elute the receptor-bound cyclic imine toxins, a volume of 50 μL of methanol was added to each well. After 5 min of incubation, the methanol was collected, pulled, and kept at −20 °C until analysis. Similarly, 16 microplate wells incubated with shellfish extracts were processed in the same manner.
Detection of Cyclic Imine Toxins with an Acquity Ultra-Performance Liquid Chromatograph Coupled to a Triple Quadrupole Detector (UPLC-TQD, Waters, MA)
Two microliters of a methanolic cyclic imine toxin standard mix or shellfish extract was chromatographed at a flow rate of 0.6 mL/min through a BEH C18 column at 40 °C (Waters, 2.1 × 100 mm; 1.8 μm particle size). Solvent A was water, 0.1% formic acid. Solvent B was acetonitrile, 0.1% formic acid. The gradient profile was as follows: 0–1 min, isocratic 15% B; 1–9 min, linear gradient 15–36.5% B; 9–9.2 min, linear gradient 36.5–100% B; 9.2–11 min, isocratic 100% B; 11–11.2 min, linear gradient 100–15% B; and 11.2–14 min, isocratic 15% B. All samples were run three times, and two blank runs (2 μL of methanol) were intercalated between different samples. The TQD mass spectrometer was operated in electrospray ionization positive mode. The capillary potential was 3.5 kV; desolvation temperature, 450 °C; source temperature, 150 °C; desolvation gas flow rate, 800 L/h and cone gas flow rate, 50 L/h. Data acquisition was performed in multiple-reaction-monitoring mode (MRM). Calibration curves were obtained using a toxin standard mix in the range from 1 pM to 1 μM and shellfish extracts samples spiked with toxin standards.
Detection of Cyclic Imine Toxins by MALDI-TOF MS
Mass spectra were obtained using a Voyager-DE-STR Workstation (AB Sciex, Les Ulis, France) equipped with a 337-nm pulsed nitrogen laser (20 Hz) operated in reflectron positiveion mode. The accelerating voltage was set at 20 kV, the grid voltage at 62% of the accelerating voltage, and the extraction delay time at 100 ns. The laser intensity was adjusted above the ion generation threshold to obtain peaks with the highest possible signal-to-noise ratio without significant peak broadening. Ten milligrams of 2,5-dihydroxybenzoic acid (DHB) was dissolved in 1 mL of water/acetonitrile (1:1) containing 0.3% trifluoroacetic acid (v/v). For MALDI-TOF analysis, 1 μL of 1 μM cyclic imine toxin standard in methanol was mixed with 1 μL of DHB directly on the stainless steel sample plate. Eluted toxin standards from the microplate wells were analyzed as described above. All data were processed with Data Explorer software (AB Sciex).
Safety Considerations
Live animals were maintained and treated according to the European standard protocols approved by the Animal Ethics Committee of CNRS. Experiments were performed in accordance with European Community guidelines for laboratory animal handling.
RESULTS AND DISCUSSION
Optimization of the Microplate-Receptor Binding Assay
We have developed a functional detection method based on the potent antagonism of cyclic imine toxins for Torpedo-nAChR. Important steps for method development were as follows: (i) immobilization of Torpedo electrocyte membranes on microplate plastic wells, (ii) use of biotin-α-BgTx as a nonradioactive tracer, and (iii) use of streptavidin-HRP for colorimetric detection of ligand–receptor binding. In the absence of competitive agonists/antagonists of nAChR, biotin-α-BgTx tightly binds to coated Torpedo-nAChR. The presence of cyclic imine toxins in the assay competitively inhibits biotin-α-BgTx binding to Torpedo-nAChRs in a concentration-dependent manner (Figure 1A,B).
Figure 1.
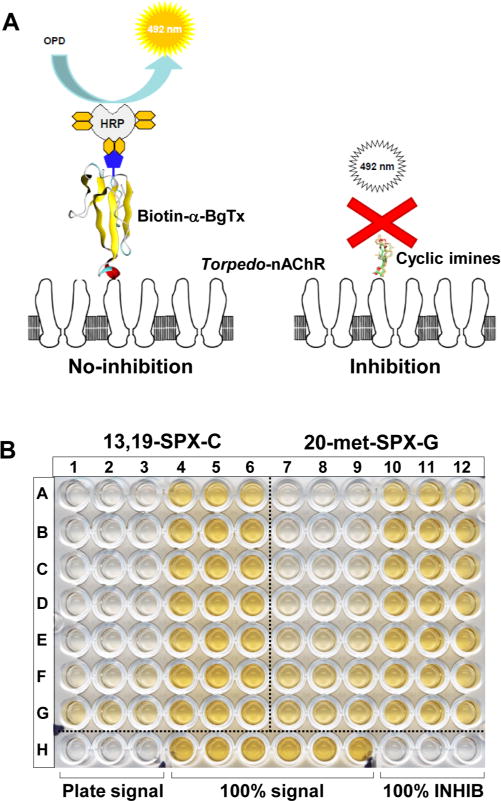
Microplate-receptor binding assay. (A) Schematic representation of the method. No inhibition: In the absence of competitive agonists/antagonists of nAChRs, biotin-α-BgTx binds to coated Torpedo-nAChRs. Subsequent reaction with streptavidin-HRP allows the colorimetric detection of bound biotin-α-BgTx. Inhibition: Competitive agonists/antagonists inhibit biotin-α-BgTx binding to coated Torpedo-nAChRs. Subsequent addition of streptavidin-HRP is washed out, and no color reaction takes place. (B) Comparative dose–response inhibition binding of biotin-α-BgTx to Torpedo-nAChR by 13,19-didesmethyl spirolide-C (13,19-SPX-C) and by 20-methyl spirolide-G (20-met-SPX-G). Each toxin dose was tested in triplicate. (Left) 13,19-Didesmethyl spirolide-C; concentration tested (wells): 2 μM (A1–A3), 667 nM (B1–B3), 222 nM (C1–C3), 74 nM (D1–D3), 24.7 nM (E1–E3), 8.2 nM (F1–F3), 2.7 nM (G1–G3), 913 pM (A4–A6), 304.4 pM (B4–B6), 101.5 pM (C4–C6), 33.8 pM (D4–D6), 11.3 pM (E4–E6), 3.7 pM (F4–F6), and 1.25 pM (G4–G6). (Right) 20-methyl spirolide-G; concentration tested (wells): 2 μM (A7–A9), 667 nM (B7–B9), 222 nM (C7–C9), 74 nM (D7–D9), 24.7 nM (E7–E9), 8.2 nM (F7–F9), 2.7 nM (G7–G9), 913 pM (A10–A12), 304.4 pM (B10–B12), 101.5 pM (C10–C12), 33.8 pM (D10–D12), 11.3 pM (E10–E12), 3.7 pM (F10–F12), and 1.25 pM (G10–G12). Plate signal: Control wells processed without membrane coating. 100% signal: Control wells in which Torpedo-nAChRs were processed in the absence of toxin or extract samples. 100% inhibition: Control wells where Torpedo-nAChRs were incubated with 1 μM α-BgTx.
Optimization of Torpedo electrocyte membrane coating was performed using MaxiSorp microplates designed for coating glycoproteins with mixed hydrophilic/hydrophobic domains.28 Torpedo electrocyte membranes are a mixture of lipoprotein vesicles, where the highly glycosilated nAChRs (7.5%) represent ~40% of total protein.29,30 TBS was selected as the membrane coating buffer because of its coating efficiency, pH, and ionic strength, as well as its known buffering capacity for the detection of cyanobacterial agonists of nAChR directly on complex matrixes.31 A Torpedo electrocyte membrane solution containing 13.5 μg/mL total protein was experimentally determined for optimal membrane coating considering that the receptor concentration should be kept to a minimum to avoid ligand depletion yet ensure a maximum signal-to-noise ratio.32 The coated microplates can be stored in blocking solution at 4 °C for up to 6 months and are ready for use or for transport. A tracer concentration of 80 nM biotin-α-BgTx (~3 times its Kd value) was determined for the microplate-receptor binding assay.
Detection of Cyclic Imine Toxin Standards by the Microplate-Receptor Binding Assay
Figure 1B shows the dose dependence and differential inhibition strength toward biotin-α-BgTx binding to Torpedo-nAChRs by 13,19-didesmethyl spirolide-C and 20-methyl spirolide-G in the concentration range from 1.25 pM to 2.0 μM.
IC50, concentration of inhibitor at which biotin-α-BgTx binding to Torpedo-nAChRs is inhibited by 50%, was calculated from the sigmoidal dose–response curves shown in Figure 2A. The snake toxin α-BgTx, which was used as a positive control, was the most potent competitive antagonist of nAChR, followed by 13,19-didesmethyl spirolide C, 13-desmethyl spirolide-C, pinnatoxin-G, 20-methyl spirolide-G, gymnodimine-A, and pinnatoxin-A (Table 1). As previously reported,21 the amino ketone analogue of pinnatoxin-A does not bind to Torpedo-nAChRs in the studied range (from 1.3 pM to 2.0 μM, Figure 2A).
Figure 2.
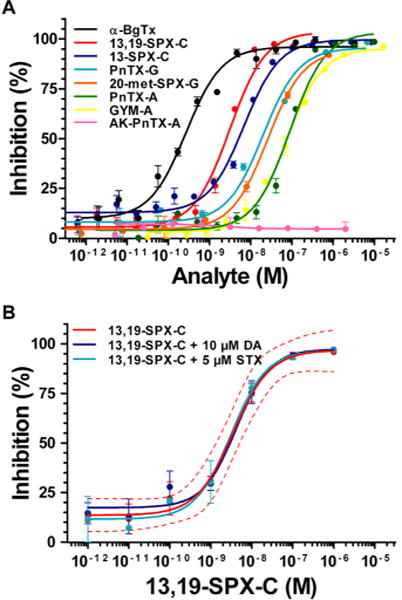
Detection of cyclic imine toxin by the microplate-receptor binding assay. (A) Inhibition of specific biotin-α-BgTx binding to Torpedo-nAChR by increasing concentrations of α-bungarotoxin (α-BgTx), 13,19-didesmethyl spirolide-C (13,19-SPX-C), 13-desmethyl spirolide-C (13-SPX-C), pinnatoxin-G (PnTX-G), 20-methyl spirolide-G (20-met-SPX-G), pinnatoxin-A, (PnTX-A), gymnodimine-A (GYM-A), and amino ketone pinnatoxin-A (AK-PnTX-A). Curve fitting was performed by nonlinear regression analysis using the Hill equation. Each data point is the mean value ± standard error of the mean (SEM) of at least three inhibition experiments. (B) Cross-reactivity: Inhibition of biotin-α-BgTx binding to Torpedo-nAChR by increasing concentrations of 13,19-SPX-C alone or in the presence of 10 μM domoic acid (DA) or 5 μM saxitoxin (STX). The dashed lines represent the 95% confidence band of the inhibition curve of 13,19-didesmethyl spirolide-C. Each data plot represents the mean value ± SEM of at least three independent experiments.
Table 1.
Detection of Cyclic Imine Toxins by the Microplate-Receptor Binding Assay: Binding Inhibition Parameters and Comparative Performance with Radioactive and Nonradioactive Assaysa
| 13,19-didesmethyl spirolide-C | 13-desmethyl spirolide-C | pinnatoxin-G | 20-methyl spirolide-G | gymnodimine-A | pinnatoxin-A | |
|---|---|---|---|---|---|---|
| IC50 (nM ± SEM) M- RBASTD-MeOH |
3.10 ± 1.23 | 6.84 ± 1.15 | 20.05 ± 13.32 | 25.57 ± 10.83 | 74.23 ± 10.80 | 75.82 ± 7.09 |
| IC50 (nM ± SEM) M- RBASTD-matrix |
2.57 ± 1.06 | 7.14 ± 1.07 | 24.74 ± 11.44 | 23.79 ± 11.51 | 70.40 ± 13.23 | 78.29 ± 15.27 |
| Kib (nM ± SEM) | 0.80 ± 1.22 | 1.77 ± 1.14 | 5.20 ± 1.33 | 6.63 ± 1.09 | 19.25 ± 8.28 | 23.00 ± 12.16 |
| LODc (nM/Inh%) | 0.8/15.8 | 1/21.5 | 2/15.5 | 2/10.2 | 2/5.2 | 5/9.8 |
| LOQd (nM/Inh%) | 2.0/34.5 | 4/42.1 | 8/32.5 | 6/21.7 | 20/28.5 | 24/23.4 |
| Hill slope | 1.33 ± 0.23 | 1.09 ± 0.14 | 0.65 ± 0.10 | 0.93 ± 0.06 | 0.82 ± 0.09 | 1.31 ± 0.21 |
| R2 | 0.9831 | 0.9915 | 0.9861 | 0.9988 | 0.9977 | 0.9826 |
| Ki (nM ± SEM) (RL-BA) | – | 0.080 ± 0.00220 | – | – | 0.23 ± 0.0820 | – |
| IC50 (nM ± SEM) (FP) | – | 108.2 ± 11.927 | – | – | 391.0 ± 55.327 | – |
| IC50 (nM ± SEM) (FP) | 63.60 ± 333 | – | – | – | – | |
| IC50 (nM ± SEM) (SP-RA) | – | 31.4 ± 4.934 | – | – | 526.0 ± 38.134 | – |
Abbreviations: M-RBASTD-MeOH, IC50 was determined using methanolic standards; M-RBASTD-matrix, IC50 was determined using methanolic standards in the presence of 25 mg/mL P. maximus extract; RL-BA, radioligand binding assay; FP, fluorescence polarization receptor assay; SP-RA, solid-phase receptor-based assay).
Ki was calculated by nonlinear regression analysis (biotin-α-BgTx = 80 nM; Kd of biotin-α-BgTx = 28.3 nM).
LOD was calculated from mean OD492 nm values at toxin concentrations lower than 100 pM ± 3 times the standard deviation (SD).
LOQ was calculated from mean OD492 nm values at toxin concentrations lower than 100 pM ± 10SD.
The microplate-receptor binding assay was less sensitive than the radioactive binding assay for the detection of 13-desmethyl spirolide-C, gymnodimine-A, and pinnatoxin-A (see Ki values in Table 1).20,21 The higher affinity of 125I-α-BgTx for Torpedo-nAChRs (Kd = 50 pM)20 compared to biotin-α-BgTx’s affinity (28 nM) might explain the differences in sensitivity. However, radioactive debris and radio decomposition (125I-α-BgTx halflife = 59.6 days) are serious drawbacks that hamper the use of radioactive ligand binding assays for routine monitoring of marine toxins. On the other hand, the microplate-receptor binding assay is more sensitive than the nonradioactive fluorescence polarization receptor assay27,33 or solid-phase receptor-based assay34 for the detection of cyclic imine toxins (Table 1).
To test for cross-reactivity, Torpedo-nAChRs were incubated with 13,19-didesmethyl spirolide-C in the range from 3.5 pM to 1 μM alone and in the presence of domoic acid or saxitoxin (Figure 2B). The 95% confidence band corresponding to the inhibition curve of 13,19-didesmethyl spirolide-C, represented by the red dashed lines in Figure 2B, indicates that neither 10 μM domoic acid nor 5 μM saxitoxin affected the inhibition kinetics of biotin-α-BgTx binding to Torpedo-nAChRs by 13,19-didesmethyl spirolide-C. Similar results were obtained for 13-desmethyl spirolide-C, gymnodimine-A, and pinnatoxin-A.
Further, Torpedo-nAChRs were incubated overnight with a fixed concentration of dinophysistoxin-1, azaspiracide-1, and okadaic acid (150 nM each). The latter lipophilic phycotoxins that can be coextracted with cyclic imine toxins from contaminated shellfish did not bind Torpedo-nAChRs. Thus, as was shown for okadaic acid, yessotoxin, and brevetoxin-2,27 bioactive molecules not interacting with nAChRs do not interfere with the detection of cyclic imine toxins by functional assays.
Detection of Cyclic Imine Toxins in Shellfish Extracts by the Microplate-Receptor Binding Assay
The aim of the present study was to develop a ligand binding assay to detect cyclic imine toxins directly on shellfish extracts. To this end, four different shellfish varieties were purchased from a seafood market and processed as described in the Materials and Methods section. Torpedo-nAChRs were incubated overnight with 25 mg/mL shellfish extracts. Unexpectedly, M. galloprovincialis strong inhibitory activity (39.9%) toward biotin-α-BgTx binding to Torpedo-nAChRs, followed by G. glycymeris (29.5%) and O. edulis (15.5%). In contrast, P. maximus extract did not inhibit biotin-α-BgTx binding to Torpedo-nAChRs (2.8%) and was used as a negative control.
To test the effect of negative shellfish extracts on the performance of microplate-receptor binding assay, we carried out dose–response inhibition experiments in which Torpedo electrocyte membranes were incubated with a given toxin standard in the presence of 25 mg/mL extract of P. maximus (Figure 3A). The 95% confidence bands calculated for pinnatoxin-G (light blue dashed lines) and pinnatoxin-A (green dashed lines) illustrate that the presence of P. maximus extract in the incubation mix did not interfere with the inhibitory activity of pinnatoxin-G or pinnatoxin-A toward biotin-α-BgTx binding to Torpedo-nAChRs (observe the inset in Figure 3A). Similar results were obtained for 13-desmethyl spirolide-C, 13,19-didesmethyl spirolide-C, 20-methyl spirolide-G, and gymnodimine-A (see the IC50 values in the presence of shellfish matrix in Table 1).
Figure 3.
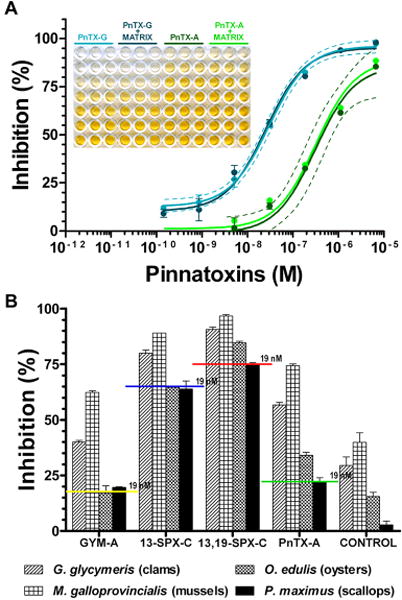
Shellfish matrix effect on the detection of cyclic imine toxin by the microplate-receptor binding assay. (A) Comparative inhibition binding of biotin-α-BgTx to Torpedo-nAChR by pinnatoxin-G and pinnatoxin-A in the concentration range from 350 pM to 10 μM in the absence and presence of 25 mg/mL P. maximus extract. The light blue dashed lines represent the 95% confidence band of the inhibition curve of pinnatoxin-G, and the green dashed lines represent the 95% confidence band of the inhibition curve of pinnatoxin-A in the absence of shellfish matrix. Each data plot represents the mean value ± SEM of at least three independent experiments. Inset: Representative microplate illustrating that P. maximus extract did not interfere with the detection of pinnatoxin-G and pinnatoxin-A. (B) Toxin spiking and shellfish matrix effect. Torpedo-nAChRs were incubated with 25 mg/ mL extract of G. glycymeris, O. edulis, M. galloprovincialis, or P. maximus previously spiked with 19 nM of gymnodimine-A (GYM-A), 13-desmethyl spirolide-C (13-SPX-C), 13,19-didesmethyl spirolide-C (13,19-SPX-C), or pinnatoxin-A (PnTX-A). The innate inhibitory activity of nonspiked shellfish extracts towards specific binding of biotin-α-BgTx to Torpedo-nAChRs is labeled as controls. The colored horizontal lines represent the inhibitory activity of the given cyclic imine toxin at a concentration of 19 nM extrapolated from the respective inhibition curve. Each bar represents the mean value ± SEM of at least three independent inhibition experiments.
To test how the positive shellfish extracts interfered with the method, Torpedo-nAChRs were incubated with different concentrations of cyclic imine toxins in the presence of extracts from M. galloprovincialis, G. glycymeris, O. edulis, or P. maximus (Figure S-2, Supporting Information). The analysis of shellfish extracts spiked with gymnodimine-A, 13-methyl spirolide-C, 13,19-didesmethyl spirolide-C, or pinnatoxin-A at a concentration of 19 nM each is shown in Figure 3B. The horizontal lines in this figure indicate the expected inhibitory activity of the given cyclic imine toxin extrapolated from the inhibition curves displayed in Figure 2A. P. maximus extract did not affect the inhibitory activity of any of the spiked toxins. In contrast, the increased inhibitory activity of the spiked cyclic imine toxins in the presence of extracts from M. galloprovincialis, G. glycymeris, or O. edulis indicated additive activity between these shellfish extracts and the spiked toxins. The potentiation of the inhibitory activity reflected the innate activity of each shellfish extract (see control bars in Figure 3B). Our results strongly suggest that M. galloprovincialis, G. glycymeris, and O. edulis samples, purchased in a fish market in France, were contaminated with cholinergic ligands.
Analysis of Shellfish Extracts by UPLC-MS/MS
Because the microplate-receptor binding assay cannot provide any information about the chemical nature of the putative contaminating ligand (s), the shellfish extracts were analyzed by UPLC-MS/MS for toxin identification. First, the chromatographic conditions were optimized for the simultaneous detection of the six cyclic imine toxin standards studied here (Figure 4A). MS/MS fragmentation conditions were optimized for each toxin to determine the MRM specific ion transitions (Table 2). The MRM response was toxin-dependent, with 13,19-didesmethyl spirolide-C giving a higher response (Figure S-3, Supporting Information). The calibration curves for the methanolic standards showed good linearity in the working range (from 1 pM to 1 μM) with correlation coefficients of >0.99 for all of the cyclic imine toxins studied and lower limits of detection (LOD) and limits of quantification (LOQ) that were consistent with previously published multitoxin LC/MS-based methods (Table 2).35–37
Figure 4.
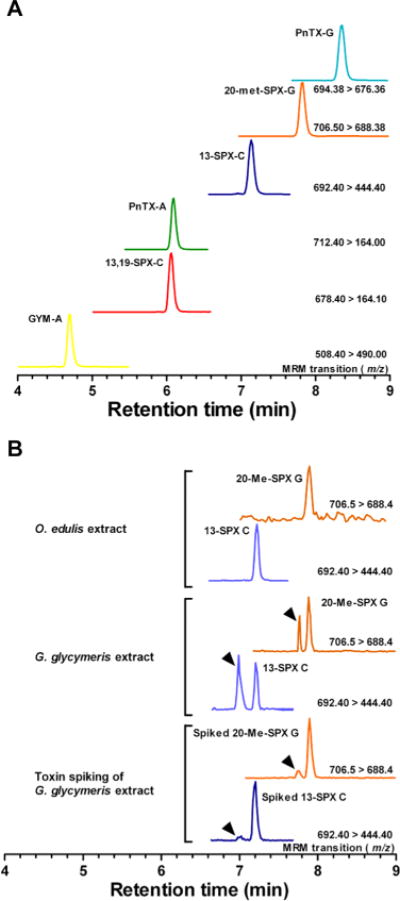
(A) UPLC-MS/MS simultaneous detection of pinnatoxin-G (PnTX-G), 20-methyl spirolide-G (20-met-SPX-G), 13-desmethyl spirolide-C (13-SPX-C), pinnatoxin-A, (PnTX-A), 13,19-didesmethyl spirolide-C (13,19-SPX-C), and gymnodimine-A (GYM-A). (B) Detection of 20-met-SPX-G and 13-SPX-C in contaminated extracts of O. edulis and G. glycymeris. Isomers of 20-met-SPX-G and 13-SPX-C were detected in G. glycymeris extract. Standard addition of 50 nM 20-met-SPX-G and 13-SPX-C in G. glycymeris extract confirmed the presence of the latter toxins in G. glycymeris extract.
Table 2.
UPLC-MS/MS Conditions for the Detection of Cyclic Imine Toxin Standards in Methanol and in Shellfish Matrixesa
| 13,19-didesmethyl spirolide-C |
13-desmethyl spirolide-C |
pinnatoxin-G | 20-methyl spirolide-G |
gymnodimine- A | pinnatoxin-A | |
|---|---|---|---|---|---|---|
| MRM transition (m/z) | 678.4 > 164.1 | 692.4 > 444.4 | 694.38 > 676.36 | 706.50 > 688.38 | 508.4 > 490.4 | 712.4 > 164.0 |
| cone voltage (V) | 50 | 60 | 75 | 65 | 50 | 65 |
| collision energy (eV) | 40 | 35 | 30 | 30 | 32 | 48 |
| retention time (min) | 6.12 | 7.20 | 8.42 | 7.89 | 4.71 | 6.17 |
| LOD per column (methanol) | 10 pM or 0.014 pg | 100 pM or 0.14 pg | 100 pM or 0.14 pg | 100 pM or 0.14 pg | 100 pM or 0.10 pg | 100 pM or 0.14 pg |
| LOQ per column (methanol) | 1 nM or 1.35 pg | 1 nM or 1.38 pg | 1 nM or 1.39 pg | 1 nM or 1.41 pg | 1 nM or 1.0 pg | 1 nM or 1.42 pg |
| R2 (methanol) | 0.9999 | 0.9999 | 0.9998 | 1.0000 | 0.9999 | 1.0000 |
| R2 (mussels) | 0.9632 | 0.9588 | 0.9565 | 0.9616 | 0.9605 | 0.9583 |
| LOD per column (bibliography) | – | 0.8 pg,37 0.2 pg36 | – | – | 3.7 pg,37 0.6 pg36 | – |
Abbreviations: MRM, multiple-reaction-monitoring mode; LOD, limit of detection (obtained from low-level standards with a signal-to-noise ratio equal to 3); LOQ, limit of quantification (obtained from low-level standards with a signal-to-noise ratio equal to 10).
Second, to quantify the contaminating cyclic imine toxins directly in the shellfish extracts, P. maximus extract was spiked with a mixture of cyclic imine toxins in the range from 100 pM to 10 μM. The standard addition of known toxin quantities resulted in linear calibration curves with correlation coefficients of >0.95 for all of the analytes (Table 2). By using the latter calibration curves, we detected 2.8 ng/g 13-desmethyl spirolide-C and 0.3 ng/g 20-methyl spirolide-G in O. edulis extracts (Figure 4B).
UPLC-MS/MS analysis of G. glycymeris extract showed two pairs of peaks with ion transitions similar to those of 13-desmethyl spirolide-C and 20-methyl spirolide-G (Figure 4B). Several cyclic imine toxins could have the same molecular mass but different structures; specifically, 13-desmethyl spirolide-C (C42H61NO7; MH+ m/z 692.4569)38 has a similar mass as spirolide-A (C42H61NO7, MH+ m/z 692.4503)38 and spirolide-G (C42H61NO7, MH+ m/z 692.4564)39 (Figure S-1, Supporting Information). UPLC-MS/MS analysis of G. glycymeris extract using the ion transitions m/z 692.4 > 164 (specific for 13-desmethyl spirolide-C), m/z 692.4 > 150 (specific for spirolide-A),38 and m/z 692 > 378 (specific for spirolide-G)39 confirmed the presence of 13-desmethyl spirolide-C in this extract, but failed to identify the first peak.
Similarly, 20-methyl spirolide-G (C43H63NO7, MH+ m/z 706.465)13 shares a similar molecular mass as spirolide-C (C43H63NO7, MH+ m/z 706.4698)38 and iso-spirolide-C of unknown structure.19 UPLC-MS/MS analysis of G. glycymeris extract using the ion transitions 706.5 > 458 (specific for spirolide-C),38 and 706.5 > 392 (specific for 20-methyl spirolide-G)13 confirmed the presence of 20-methyl spirolide-G. The first peak could not be identified.
Further, we added known amounts of 13-desmethyl spirolide-C and 20-methyl spirolide-G standards to G. glycymeris extracts. As a result, the MRM response of both toxins was potentiated at their characteristic retention times (Figure 4B). G. glycymeris samples contained 0.25 ng/g 20-methyl spirolide-G and 2.5 ng/g 13-desmethyl spirolide-C.
Despite the strong inhibitory activity of M. galloprovincialis extract (Figure S-2, Supporting Information), the nature of the cholinergic ligand (s) within the referred sample could not be identified by UPLC-MS/MS given the reduced number of cyclic imine toxin standards at our disposal.
Even if the microplate-receptor binding assay has higher LOD values than UPLC-MS/MS for the detection of cyclic imines (Tables 1 and 2), this functional assay is a high-throughput method for rapid detection of cyclic imines directly in shellfish extracts with minimal sample handling and reduced matrix effect and toxin cross-reactivity that could facilitate the use of mass-spectrometry-based methods for unequivocal identification of cyclic imine toxins.
Coupling the Microplate-Receptor Binding Assay with Mass Spectrometry
To answer the question of whether the microplate-receptor binding assay selectively captures cholinergic ligands, 16 wells containing coated Torpedo-nAChRs were incubated overnight with a 1 μM standard of a given cyclic imine toxin. Following a washing step, the bound toxins were eluted with methanol and analyzed by MALDI-TOF MS. Removing unbound molecules from the wells with a washing buffer containing 0.1% Tween 20 resulted in a complex background of low-molecular-weight molecules that masked the ionization signals of the eluted toxins. Washing the wells with only TBS resulted in high-quality MALDI spectra, as shown for pinnatoxin-A (Figure 5A) and 13,19-didesmethyl spirolide-C (Figure S-4A, Supporting Information) after elution from the microplate wells. The insets in these figures show the MRM profiles of the eluted cyclic imine toxin standards. Similar results were obtained for all of the cyclic imine toxins studied here.
Figure 5.
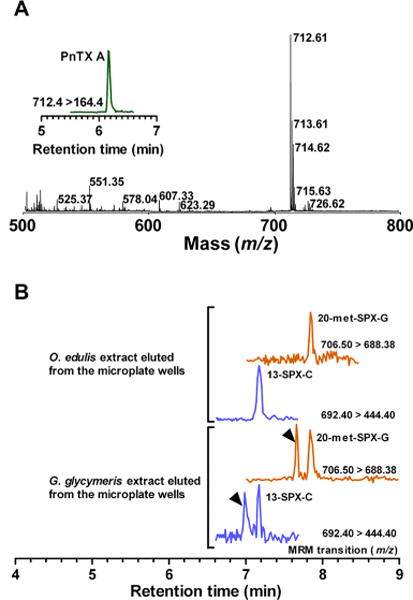
Coupling of the microplate-receptor binding assay with mass spectrometry methods. (A) Torpedo-electrocyte membranes were incubated with cyclic imine toxin standards. Following a wash step, bound toxins were eluted from the wells with methanol and analyzed by MALDI-TOF MS. Full scan, m/z 500–800, showing the protonated molecular ion of pinnatoxin-A (m/z 712.61) eluted from microplate wells. Inset: MRM analysis of pinnatoxin-A eluted from microplate wells. (B) UPLC-MS/MS analysis of O. edulis and G. glycymeris extracts eluted from microplate wells. Torpedo-electrocyte membranes were incubated with O. edulis and G. glycymeris extracts. Following a wash step, bound toxins were eluted from the wells with methanol and analyzed by UPLC-MS/MS: 20-Methyl spirolide-G and 13-desmethyl spirolide-C were detected in both shellfish samples. Note that isomers of 20-methyl spirolide-G and 13-desmethyl spirolide-C were also retrieved in eluted samples from G. glycymeris extract.
Once the proof-of-principle of toxin elution from Torpedo-nAChRs was validated (Figures 5A and S-4 A,B, Supporting Information), we applied the same procedure to the analysis of shellfish extracts. The basic idea of receptor affinity purification of cholinergic ligands present in the shellfish extracts and fast determination of their molecular ion masses by MALDI-TOF MS hit the low sensitivity of this method. Under our experimental conditions, the LOD of MALDI-TOF MS for any of the six cyclic imine toxin standards was 50 nM. The low concentration of the eluted toxins in the shellfish extracts hampered their subsequent analysis by MALDI-TOF.
In contrast, the results of the analysis of the eluates by UPLC-MS/MS were consistent with the direct analysis of shellfish extract samples using the same technique. That is, 13-desmethyl spirolide-C and 20-methyl spirolide-G were detected in eluted samples from G. glycymeris and O. edulis extracts. Moreover, the double peaks found using the MRM transitions for 13-desmethyl spirolide-C and 20-methyl spirolide-G were obtained in the eluted samples from G. glycymeris (Figure 5B), showing the feasibility of receptor affinity purification of ligands directed against Torpedo-nAChR by using the microplate-receptor binding assay.
CONCLUDING REMARKS
Cyclic imine toxins are not internationally regulated because no human fatalities associated with this family of neurotoxins have been reported. However, the risk of long-term exposure to sublethal doses of cyclic imine toxins is of concern given their oral toxicity,40,41 their capacity to cross the blood–brain barrier,42 and their nanomolar to picomolar affinities for human neuronal nAChRs in vitro.12,20,21 Mouse bioassay is being replaced by LC/MS to monitor internationally regulated marine lipophilic biotoxins.24 Novel functional assays are needed, however, to replace mouse bioassay for the rapid detection of unknown and unanticipated neurotoxins from shellfish samples that could facilitate the use of LC/MS methods. The microplate-receptor binding assay is based on the immobilization of Torpedo electrocyte membranes on the surface of plastic microplate wells providing a high-throughput format for routine detection of neurotoxins targeting nAChRs from environmental samples. Although highly sensitive and specific for the detection of neurotoxins acting on nAChRs, the microplate-receptor binding assay is poorly selective. To overcome the lack of selectivity of the method, the microplate-receptor binding assay was coupled with mass spectrometry, enabling its use for the discovery and receptor-affinity purification of novel nAChR ligands from marine organisms.
Supplementary Material
Acknowledgments
We thank D. Ladant for fruitful discussions. The technical assistance of P. Villeneuve is gratefully acknowledged. This work was supported in part by research grants from the Agence Nationale de la Recherche (ANR CES 2008-Aristocya) and National Institutes of Health (USA, NIGMS R01 GM077379 to A.Z.), and by Grant 2009-1/117 PHARMATLANTIC (to L.M.B. and J.M.).
Footnotes
Supporting Information Chemical structures of the investigated neurotoxins, map of a microplate designed to assess the competitive inhibition of biotin-α-BgTx binding to Torpedo nAChR by shellfish extracts as well as spiking experiments on M. galloprovincialis extracts, row data and their respective analysis, UPLC-MS/MS calibration curves of cyclic imine toxin standards, and coupling of the microplate-receptor binding assay with mass spectrometry methods. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
R.A., L.M.B., and J.M. designed research; R.A., S.R., F.P., N.V., and V.G. performed experiments; R.A., E.B., A.Z., and J.M. analyzed data; R.A., L.M.B., A.Z., and J.M. wrote the article. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Molgó J, Girard E, Benoit E. In: Phycotoxins: Chemistry and Biochemistry. Botana LM, editor. Blackwell Publishing Ltd; Oxford, UK: 2007. pp. 319–335. [Google Scholar]
- 2.Gueret SM, Brimble MA. Nat Prod Rep. 2010;27:1350–1366. doi: 10.1039/c005400n. [DOI] [PubMed] [Google Scholar]
- 3.Miles CO, Wilkins AL, Stirling DJ, MacKenzie AL. J Agric Food Chem. 2003;51:4838–4840. doi: 10.1021/jf030101r. [DOI] [PubMed] [Google Scholar]
- 4.Van Wagoner RM, Misner I, Tomas CR, Wright JLC. Tetrahedron Lett. 2011;52:4243–4246. [Google Scholar]
- 5.Selwood AI, Miles CO, Wilkins AL, van Ginkel R, Munday R, Rise F, McNabb P. J Agric Food Chem. 2010;58:6532–6542. doi: 10.1021/jf100267a. [DOI] [PubMed] [Google Scholar]
- 6.Aasen JA, Hardstaff W, Aune T, Quilliam MA. Rapid Commun Mass Spectrom. 2006;20:1531–1537. doi: 10.1002/rcm.2501. [DOI] [PubMed] [Google Scholar]
- 7.McCarron P, Rourke WA, Hardstaff W, Pooley B, Quilliam MA. J Agric Food Chem. 2012;60:1437–1446. doi: 10.1021/jf204824s. [DOI] [PubMed] [Google Scholar]
- 8.Seki T, Satake M, Mackenzie L, Kaspar HF, Yasumoto T. Tetrahedron Lett. 1995;36:7093–7096. [Google Scholar]
- 9.Cembella AD, Lewis NI, Quilliam MA. Phycologia. 2000;39:67–74. [Google Scholar]
- 10.Nézan E, Chomerat N. Cryptogam: Algol. 2011;32:3–18. [Google Scholar]
- 11.Rhodes L, Smith K, Selwood A, McNabb P, Munday R, Suda S, Molenaar S, Hallegraeff G. Phycologia. 2011;50:624–628. [Google Scholar]
- 12.Kharrat R, Servent D, Girard E, Ouanounou G, Amar M, Marrouchi R, Benoit E, Molgó J. J Neurochem. 2008;107:952–963. doi: 10.1111/j.1471-4159.2008.05677.x. [DOI] [PubMed] [Google Scholar]
- 13.Aasen J, MacKinnon SL, LeBlanc P, Walter JA, Hovgaard P, Aune T, Quilliam MA. Chem Res Toxicol. 2005;18:509–515. doi: 10.1021/tx049706n. [DOI] [PubMed] [Google Scholar]
- 14.Amzil Z, Sibat M, Royer F, Masson N, Abadie E. Mar Drugs. 2007;5:168–179. doi: 10.3390/md504168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez AV, Rodriguez-Velasco ML, Ben-Gigirey B, Botana LM. Toxicon. 2006;48:1068–1074. doi: 10.1016/j.toxicon.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Ciminiello P, Dell’Aversano C, Fattorusso E, Magno S, Tartaglione L, Cangini M, Pompei M, Guerrini F, Boni L, Pistocchi R. Toxicon. 2006;47:597–604. doi: 10.1016/j.toxicon.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez G, Uribe E, Avalos P, Mariño C, Blanco J. Toxicon. 2010;55:638–641. doi: 10.1016/j.toxicon.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Uemura D, Chou T, Haino T, Nagatsu A, Fukuzawa S, Zheng SZ, Chen HS. J Am Chem Soc. 1995;117:1155–1156. [Google Scholar]
- 19.Rundberget T, Aasen JAB, Selwood AI, Miles CO. Toxicon. 2011;58:700–711. doi: 10.1016/j.toxicon.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Bourne Y, Radic Z, Aráoz R, Talley TT, Benoit E, Servent D, Taylor P, Molgó J, Marchot P. Proc Natl Acad Sci USA. 2010;107:6076–6081. doi: 10.1073/pnas.0912372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aráoz R, Servent D, Molgó J, Iorga BI, Fruchart-Gaillard C, Benoit E, Gu Z, Stivala C, Zakarian A. J Am Chem Soc. 2011;133:10499–10511. doi: 10.1021/ja201254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellyer SD, Selwood AI, Rhodes L, Kerr DS. Toxicon. 2011;58:693–699. doi: 10.1016/j.toxicon.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Duroure L, Jousseaume T, Aráoz R, Barré E, Retailleau P, Chabaud L, Molgó J, Guillou C. Org Biomol Chem. 2011;9:8112–8118. doi: 10.1039/c1ob06257c. [DOI] [PubMed] [Google Scholar]
- 24.Commission Regulation. Off J Eur Union. 2011;54:3–7. [Google Scholar]
- 25.Stivala CE, Zakarian A. J Am Chem Soc. 2008;130:3774–3776. doi: 10.1021/ja800435j. [DOI] [PubMed] [Google Scholar]
- 26.Hill JA, Nghiem HO, Changeux JP. Biochemistry. 1991;30:5579–5585. doi: 10.1021/bi00236a034. [DOI] [PubMed] [Google Scholar]
- 27.Vilariño N, Fonfría ES, Molgó J, Aráoz R, Botana LM. Anal Chem. 2009;81:2708–2714. doi: 10.1021/ac900144r. [DOI] [PubMed] [Google Scholar]
- 28.Esser P. Principles in Adsorption to Polystyrene. Thermo Fisher Scientific Inc; Waltham, MA: 2010. (Technical Bulletin: 06a). [Google Scholar]
- 29.da Costa CJB, Kaiser DEE, Baenziger JE. Biophys J. 2005;88:1755–1764. doi: 10.1529/biophysj.104.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albuquerque EX, Pereira EFR, Alkondon M, Rogers SW. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aráoz R, Herdman M, Rippka R, Ledreux A, Molgó J, Changeux JP, Tandeau de Marsac NT, Nghiêm HO. Toxicon. 2008;52:163–174. doi: 10.1016/j.toxicon.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Y, Prusoff WH. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 33.Fonfría ES, Vilariño N, Molgó J, Aráoz R, Otero P, Espiña B, Louzao MC, Alvarez M, Botana LM. Anal Biochem. 2010;403:102–107. doi: 10.1016/j.ab.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez LP, Vilariño N, Molgó J, Aráoz R, Antelo A, Vieytes MR, Botana LM. Anal Chem. 2011;133:10499–10511. doi: 10.1021/ac200423s. [DOI] [PubMed] [Google Scholar]
- 35.Sleno L, Volmer DA. Anal Chem. 2005;77:1509–1517. doi: 10.1021/ac0486600. [DOI] [PubMed] [Google Scholar]
- 36.Fux E, McMillan D, Bire R, Hess P. J Chromatogr A. 2007;1157:273–280. doi: 10.1016/j.chroma.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Gerssen A, Mulder PPJ, McElhinney MA, de Boer J. J Chromatogr A. 2009;1216:1421–1430. doi: 10.1016/j.chroma.2008.12.099. [DOI] [PubMed] [Google Scholar]
- 38.Hu TM, Burton IW, Cembella AD, Curtis JM, Quilliam MA, Walter JA, Wright JLC. J Nat Prod. 2001;64:308–312. doi: 10.1021/np000416q. [DOI] [PubMed] [Google Scholar]
- 39.MacKinnon SL, Walter JA, Quilliam MA, Cembella AD, LeBlanc P, Burton IW, Hardstaff WR, Lewis NI. J Nat Prod. 2006;69:983–987. doi: 10.1021/np050220w. [DOI] [PubMed] [Google Scholar]
- 40.Munday R, Quilliam MA, Le Blanc P, Lewis N, Gallant P, Sperker SA, Ewart HS, MacKinnon SL. Toxins. 2012;4:1–14. doi: 10.3390/toxins4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Authority EFS. EFSA J. 2010;8:1628–1667. [Google Scholar]
- 42.Gill S, Murphy M, Clausen J, Richard D, Quilliam M, MacKinnon S, LaBlanc P, Mueller R, Pulido O. Neurotoxicology. 2003;24:593–604. doi: 10.1016/S0161-813X(03)00014-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


