Abstract
Ratiometric fluorescent reporters have recently emerged a new technique to non-invasively measure aspects of cell physiology such as redox status, calcium levels, energy production, and NADH levels. These reporters consist of either a single or pair of fluorophores along with specific modifications, such as the addition of a protein domain which binds to a metabolite of interest, thereby producing gradual alterations in fluorescence in response to changes in the measured parameter. Measurement of the changes in fluorescence produces a quantitative read-out of the cellular environment. While these reporters were initially developed to easily visualize and track changes in cultured cells, several groups have adapted these reporters to use in Caenorhabditis elegans which opens a new avenue through which to explore cell physiology during development or aging, in response to changes in external environment, or in response to genetic manipulation. These reporters have the advantage of being easily targeted to any part of the worm, and because C. elegans is transparent both the reporters and changes in their fluorescence can be clearly observed in vivo. Here we discuss the application of ratiometric reporters to C. elegans, and outline a method to quantitatively measure changes in intracellular peroxide levels using the HyPer ratiometric reporter. However, these principles can be applied to alternate ratiometric reporters which are designed to measure either other chemical species or other cellular parameters.
Keywords: fluorescent proteins, transgene, Caenorhabditis elegans, metabolism, digital imaging, ratiometric reporters
1. Introduction
An area of long-standing interest in biochemistry and cell biology has been the connection between changes in cell behavior and changes in cell physiology. The changes in cell status produced processes such as hormone signaling, medications, stressors, and aging have been thought to be linked to changes in metabolism and many other cellular parameters, however the techniques available to examine these changes both in vivo and in real time have been limited. For example, the use of in vitro biochemical assays to measure metabolites of interest have been valuable, but these techniques are limited by the need to prepare cell lysates which require both time and sufficient biomass to permit their detection. Further, the biochemical data is drawn from a population of cells so the study of individual variation is lost via this approach, and also the need to prepare cell lysates makes serial study of the same cell population not possible.
The development of fluorescent proteins, such as green fluorescent protein (GFP), as research tools has produced multiple benefits in the understanding in the areas of cell biology, developmental biology, and gene expression, because these processes could now be easily seen in a non-destructive manner through the use of fluorescence microscopy [1]. Further these fluorescent proteins could be modified using the tools of molecular biology to create novel fusion proteins that can be targeted to specific locations, come in different colors, or have other new properties. Among these fusion proteins are a significant number of ratiometric reporters which demonstrate changes in fluorescence in response to changes in a parameter of interest [2–12] (Table 1). These ratiometric reporters generally fall into two categories with one category consisting of a single fluorescent protein inserted or attached to a parameter responsive protein domain [2, 8, 9, 11, 13] and the second category consisting of two fluorescent proteins, which exhibit fluorescence resonance energy transfer (FRET) [14–17], flanking a parameter responsive protein domain. These domains are often drawn from prokaryotic genes and the encoded protein often binds or produces other conformational changes in response to interactions with a metabolite or other chemical. Alternatively, the domain could be smaller and have functional groups which are responsive to changes in a cellular parameter, such as pH or redox status, or the domain could have high affinity phosphorylation sites for a specific protein kinase. The resulting conformational changes serve to either alter the structure of the single fluorophore or to change the FRET coupling between the pair of fluorophores (Figure 1). The net effect is that physiological changes in the concentration of the relevant parameter result in reproducible and measurable changes in the excitation and emission spectra of the reporter. These changes can be easily measured via the use of fluorescent microscopy with changes in the observed fluorescence between control and intervention cells providing a measure of changes in the parameter of interest. The use of a transgene-encoded fluorescent protein carries certain advantages such as the ability to limit measurements to specific cell-types or to specific compartments within the cell.
Table 1.
Genetically encoded fluorescent ratiometric reporters:
| Category | Name | Type | Localization | Spectral properties | references |
|---|---|---|---|---|---|
| pH sensors | phluorin | GFP | Cytosol | Ex 410/470 Em 535 |
[47–49] |
| Pt-GFP | GFP | Cytosol | Ex 390/475 Em 508 |
[50] | |
| Redox Sensors | HyPer | cpYFP | Cytosol | Ex 420/500 Em 516 |
[2, 9, 10] |
| roGFP1 roGFP2 |
GFP | Cytosol, Mitochondrial | Ex 400/490 Em 500 |
[10, 51–54] | |
| ATP sensors | ATeam | FRET based | Cytosol | Ex 435 Em 475/527 |
[14, 16, 17] |
| IP(3) sensors | LIBRA | FRET based | Cytosol | Ex 425 Em 480/535 |
[55] |
| cAMP sensors | ---------------- | FRET based | Cytosol | Ex 430 Em 480/545 |
[56] |
| Glucose sensors | FLIPglu | FRET based | Cytosol | Ex 433 Em 485/502 |
[57] |
| NADH sensors | NADH Sensor2 RexYFP |
cpYFP | Cytosol, Mitochondrial | Ex 420 Em 500/518 |
[12, 18, 58] |
| Calcium sensor | Cameleon YC2.1, YC3.1, YC6.1 |
FRET based | Cytosol, Nucleus | Ex 434 Em 477/514 |
[59] |
| Calcium sensor | Cameleon YC3.6 |
cpYFP | Cytosol | Ex 440 Em 535 |
[60, 61] |
GFP: green fluorescent protein
YFP: yellow fluorescent protein
CFP: cyan fluorescent protein
pt-GFP: Ptilosarcus gurneyi
cpYFP: circularly permuted yellow fluorescent protein
roGFP: reduction–oxidation-sensitive GFP
cAMP: Cyclic adenosine monophosphate
IP(3): Inositol 1,4,5-trisphosphate
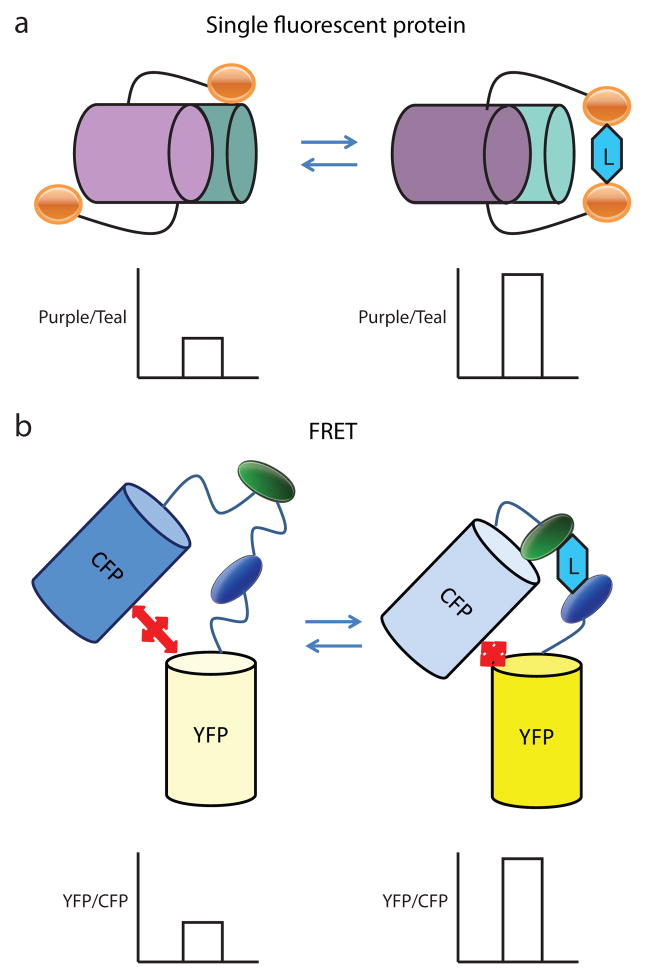
Figure 1.
Cartoon comparing the structure of the two types of ratiometric reporters. (a) The single fluorescent protein reporters consist of a single fluorophore with two excitation maxima, shown in purple and teal, linked to a protein domain, shown in orange, which is sensitive to changes in a particular metabolite which is the ligand (L) for the domain. Changes in the metabolite concentration thereby produces changes in the confirmation of this domain (right panel). This often results in changes in the spectral properties of the fluorescent protein, such as a change in the relative strength of two excitation maxima in the liganded and unliganded forms of the fusion protein, which in this example is indicated by changes in the purple and teal colors that represent each of the maxima. The change in the ratio of the fluorescence produced by each maxima is also shown in the graph below each form. The reproducible change in the excitation maxima can be measured via fluorescent imaging and then used to provide a quantitative readout of the cellular environment. (b) The FRET-coupled fluorescence reporters contain two fluorescent proteins, which exhibit fluorescence resonance energy transfer between the fluorophores. These fluorescent proteins flank a protein domain which is sensitive to changes in a metabolite of interest. Conformational changes in this protein domain then change the spatial arrangement of the fluorophores and alter the FRET efficiency between them, which is shown in this example by changes in CFP, shown in blue, and YFP, shown in yellow. The reproducible change in FRET efficiency can be measured via fluorescent imaging and similarly be used to provide a readout of the cellular environment as shown in the graphs below the liganded and unliganded forms of the fusion protein.
The quantitative analysis on the ratiometric reporters can be done in two major ways: imaging based or microplate based. Microplate based measurement allows rapid, kinetic measurement of intracellular fluorescence in a large population without taking individual images, but requires a specific fluorometric imaging plate reader. In this paper, we will focus on the imaging approach with along with a discussion on the choice of appropriate fluorescence reporters, microscopes, filters, and cameras for imaging intracellular biological changes in C. elegans.
2. Methods
2.1 Choice of ratiometric reporters
There are several reasons why fluorescence-based ratiometric reporters are an attractive and reliable way to assess cellular physiology in vivo. First, these biosensors are protein-based and encoded by a specific transgene. As a result, the reporter is relatively stable both over time and between animals, and also does not involve the treatment of animals with exogenous products. Second, through the use of transgene modifications, the reporter can be targeted to specific cell types, developmental stages, or even subcellular compartments. Finally, often extensive work has been conducted during the creation of individual reporters to produce a biosensor that is both sensitive to physiological changes in the metabolite of interest while at the same time being selective for this metabolite compared to even closely related metabolites. For example, the HyPer ratiometric reporter is able to detect small changes in cellular H2O2 levels while at the same time, this reporter is insensitive to other oxidants tested, such as superoxide, oxidized glutathione, and nitric oxide [8].
Due to these advantages, in the past ten years ratiometric reporters have been developed to monitor a variety of parameters in cell physiology including cellular pH changes, Ca2+ fluxes, changes in intracellular signaling, and changes in cellular metabolism (Table 1). These reporters consist either of a single fluorescent protein attached to a domain which changes conformation in response to a change in a specific cellular parameter, or the reporter consists of two FRET-coupled fluorophores with a responsive insert sandwiched between the two fluorophores. In the former type of reporter, changes in the conformation of the attached domain produce small changes in the three dimensional structure of the fluorescent protein and hence alter the spectral properties of the fluorophore. Importantly, many fluorescent proteins, such as yellow fluorescent protein (YFP), have more than one excitation peak resulting in light emission at a single emission wavelength. The changes in the fluorophore conformation typically result in reproducible changes in the ratio of these excitation maxima [2, 8, 9, 11, 13]. In contrast, the reporters consisting of two FRET-coupled fluorophores utilize non-radiative energy transfer between a donor fluorophore, cyan fluorescent protein (CFP), and an acceptor fluorophore (YFP) [14–17]. This transfer of energy depletes the excitation energy of the donor and produces a decrease in the fluorescence emission from the donor, while the transferred energy excites the acceptor and results in increased emission from this fluorophore (Figure 1). The changes in the conformation of the insert change the geometry of the two fluorophores in the fusion protein and hence produce reproducible changes in the amount of FRET occurring between the fluorophores. Both types of reporters can be successfully used to measure changes in cell physiology though one needs to be aware of the types and properties of the fluorophores to select the proper imaging equipment and conditions.
Important factors to consider in selecting a reporter for use include the sensitivity and selectivity for a ratiometric reporter for a parameter of interest; the resistance of the reporter to potential confounding variables such as pH; the availability of published worm strains with the reporter; the availability of the reporter cDNA from a company, repository, or the lab of origin; and whether targeting the reporter to a specific cellular compartment, tissue, or developmental stage is important for experimental design. Information on the reporter and its sensitivity and selectivity can usually be obtained from prior publications. These same properties may not be seen in worms due to differences between worms and mammalian cells grown in culture, such as the temperature used for growth [14]. Another important consideration is that some fluorophores also show changes in fluorescence in response to changes in pH so if changes in pH could be a factor in the experimental design, i.e. examining changes in energy production, it will be important to determine if this factor has been examined in prior publications [12, 18]. Most of the ratiometric reporters have not been used in worms, but strains are reported for reporters measuring pH, oxidative stress, calcium fluxes, and adenosine triphosphate (ATP) levels [2, 14, 19, 20]. As a result, most applications will involve creating new transgenic animals for use, and hence the availability of the reporter cDNA is an important consideration. Fortunately, a number of reporters are available from either Addgene (Cambridge, MA) or Evrogen (Moscow, Russia) which can streamline the process of requesting and receiving the desired reporter. If a new transgene and transgenic animal will be constructed, this step will also permit the use of targeting sequences or specific promoters to control when and where the reporter is expressed.
2.2 Construction of Transgenes
A few C. elegans strains expressing ratiometric reporters have been created, and a limited number, such as worms expressing the calcium sensors D3cpv and GCaMP or the pH sensor pHluorin, are available at the Caenorhabditis Genetics Center (CGC) [2, 14, 19–25]. Because of this limited availability, the most common way to utilize a ratiometric reporter is by creating a transgene containing the reporter of interest and then using this transgene to produce transgenic worms either via microinjection, mos1-mediated single copy transgene insertion (mosSCI), or microparticle bombardment [26–30]. This approach, while somewhat time-consuming, does carry the advantage of being able to create a transgene with features that are optimized for a specific experiment. For example, sequences could be inserted to localize the reporter to specific intracellular structures, such as the nucleus or mitochondria, or one could use tissue- or stage-specific promoters to limit expression of the reporter to the desired tissues or developmental stages.
These transgenes are often produced through the use of standard cloning or alternate approaches such as Gateway cloning (Life Technologies, Grand Island, NY) can be used especially if multiple transgenes are being produced at one time. Most of the reporters have been constructed for use in mammalian cell culture, so issues such as differences in codon usage between mammals and worm and the lack of internal introns, which can lead to reduced transgene expression in worms, are possible difficulties involved in utilizing a reporter cDNA directly to generate a transgene for use in C. elegans [30]. However, we have found that replacing the GFP sequence in a Fire lab vector, such as pPD93.97, with the reporter cDNA and placing a promoter of interest upstream is often sufficient to get moderate to high level expression in C. elegans without modification (H. Wang and A. Fisher, unpublished). The pPD93.97 vector is available from Addgene (Cambridge, MA), and the vector has synthetic introns both upstream and downstream of the GFP sequence, which may influence the expression of the cDNA. To replace the GFP sequence in the vector, we generate a PCR fragment containing the reporter sequence flanked by the sequence 5′-GGTACCGGTAGAAAAAATG-3′ on the 5′ side and placing an EcoRI restriction site (5′-GAATTC-3′) on the 3′ side of the cDNA. The 5′ sequence represents the immediate 5′ UTR sequence from many Fire lab vectors and ends with the initiating ATG. The use of these specific oligos for PCR allows the GFP sequence to be replaced with an Acc65I - EcoRI fragment encoding the protein of interest. The PCR step can also be used to add targeting sequences to cDNA, such as a nuclear localization signal, mitochondrial targeting sequence, or myristoylation sequence, to localize the reporter to a specific cellular compartment.
If desired, the resulting plasmid can be modified to carry the unc-119+ gene via the use of homologous recombination in E. coli [31, 32]. This modification allows the plasmid to be used to generate transgenic animals with rescue of the unc-119 mutation being used as a selectable marker. Alternately, selectable markers, such as the rol-6 marker or antibiotic resistance, can be used to identify transgenic animals [29, 33, 34]. Fluorescent markers, such as GFP or mCherry, are also an option as long as care is taken to express either a fluorescent protein that will not conflict with the selected reporter or to express the marker in a tissue distinct from those of experimental interest.
2.3 Generation of Transgenic Animals
Once the desired transgene has been created, several approaches, including microinjection, microparticle bombardment, or mosSCI, can be used to generate transgenic animals [27–29]. Each approach has trade-offs with regards to the effort involved in getting a transgenic animal and the type of transgene produced. We have favored the use of bombardment to generate transgenic animals due to the ability to obtain a mix of animals with a range of extrachromasomal arrays and integrated transgenes [27]. We feel this produces animals with a range of transgene expression levels which might make identifying a transgenic line with the optimal expression level of the transgene easier. However, the bombardment procedure can damage the genomic DNA especially when integrated transgenes are produced. In addition to bombardment, both microinjection and mosSCI can be successful approaches to produce transgenic animals. The microinjection technique usually produces transgenic animals with extrachromosomal arrays and requires some practice to become skilled at the injection technique, but this approach can be carried out quickly to obtain the desired animals. Recently, mosSCI has emerged as a powerful technique to obtain single copy transgenes that are inserted in a defined genomic locus [28]. This approach gives the investigator a great deal of control over the type of transgenic animal created, but this approach can be more time consuming and can produce transgenic animals with a more limited range of transgene expression levels.
Microparticle bombardment utilizes a specialized apparatus to propel DNA-coated gold microparticles driven by high-pressure helium into a large population of animals. A small percentage of the animals will have the microparticle penetrate the gonad and transfer the associated DNA into the germline to produce transgenic progeny with mainly extrachromosomal arrays and a small number with integrated transgenes. Several detailed protocols for the use of bombardment have been described including some from our lab [27, 35, 36].
Regardless of the procedure used to generate transgenic animals, the resulting strain will need to be out-crossed to remove any genetic differences related to the starting strains needed to select for transgenic animals or the transformation process. The transgene can also be moved into other genetic backgrounds via the use of standard crosses.
2.4 Use of Control Experiments to Validate Reporter Function
An important first step in the use of a new ratiometric reporter is to determine whether changes in the fluorescence of the reporter occur when the cellular parameter of interest is changed. Seeing such an effect both validates the successful expression and activity of the reporter, and also confirms that physiologically relevant changes in the metabolite or other parameter can be detected by the reporter. These types of control experiments can be performed by using an exogenous treatment that would be expected to change the parameter of interest in a predictable manner, such as inhibiting mitochondrial function to change ATP levels, NADH levels, or pH. Alternately, either a genetic mutant or RNAi treatment could be an option to produce changes that can be visualized by the reporter.
For example, we treated transgenic worms carrying a rpl-17p::HyPer transgene that widely expresses the HyPer ratiometric reporter, which consists of circularly permuted YFP inserted into the regulatory domain of the E. coli OxyR protein, with tert-butyl hydroperoxide (tBHP) to generate peroxide [2, 8]. We chose to use tBHP due to its greater stability and higher membrane permeability while retaining the ability to oxidize amino acids including those in OxyR [37–40]. The HyPer reporter shows changes in the 420 nm and 500 nm excitation maxima of YFP in response to changes in peroxide levels, with the 420 nm maxima decreasing and the 500 nm maxima increasing following exposure to increasing peroxide levels [8]. To perform this experiment we used the following protocol:
Grow several 6-cm. plates of mixed stage transgenic worms.
Wash worms from plates with S-Basal and collect in a 15 ml centrifuge tube.
Pellet worms by centrifugation 30–60 seconds at 3000 rpm and remove supernatant.
Wash once with an equal volume of S-basal.
Add 10 mL of fresh hypochlorite solution to worms (4ml bleach, 2.5ml 10M KOH, and 43.5ml dH2O). Mix well by inversion and monitor for the disappearance of adults and liberation of eggs using a dissecting microscope.
Collect the eggs by centrifugation at 3000 rpm for 30–60 seconds.
Add ~200 eggs to fresh 6-cm NGA plates and incubate at 20°C for three days to generate a synchronized population of day 1 adult transgenic worms.
Add an aliquot of tBHP dissolved in water to the surface of a 6-cm OP50-spotted NGA plate and allow the plate to dry for at least 1 hour. As a control, add a similar amount of water to an additional plate and similarly allow the plate to dry.
Move adult transgenic worms to the tBHP or control spotted NGA plate by hand and incubate the plates at 20°C for one hour.
2.5. Mounting of Worms for Imaging
To image the worms, we mount them on standard agarose pads [41]. However, an important concern is the choice of anesthetic agent because the biochemical effects of some of the agents may alter the parameter of interest. For example, the commonly used anesthetic sodium azide impairs mitochondrial function and can produce both decreases in ATP levels and oxidative stress [42, 43]. To circumvent the limitations of a specific anesthetic, it is possible to switch anesthetics or to immobilize worms via another approach such as gluing [44]. To image the animals with the rpl-17p::HyPer transgene, we chose to use the alternative anesthetic levamisole, which acts as a acetylcholine receptor agonist in C. elegans [45]. Beyond levamisole, other anesthetics and immobilization techniques are available for use with imaging. The online resources Wormbook and Worm Breeder’s Gazette can be useful resources for identifying alternate techniques. To prepare slides and mount the worms with levamisole, we used the following protocol:
Using laboratory label tape, tape two flat microscope slides and lie them parallel to each other on the laboratory bench.
Place a third blank slide between the two taped slides.
Boil 2% agarose in distilled water.
Using a plastic pipette, place 2–3 drops of 2% agarose onto the middle slide.
Immediately lay a fourth slide perpendicular to the other three slides on top of the agar.
Once the agar has set, carefully slide apart the taped slides, so that the agar pad is left in the center of the middle slide.
Pipet 20 μL 10 mM levamisole in M9 or S-basal onto the agar pad.
Transfer worms to the agar pad using a pick or piece of nylon monofilament fishing line.
Set the edge of a coverslip at the side of the agar pad opposite to the levamisole and slowly drop it to avoid air bubbles.
2.6 Selection of Microscope and Filters
Fluorescence emitted by the ratiometric reporter is visualized and measured by fluorescence microscopy and digital imaging. In this section, we discuss some of the options for microscopes and cameras along with important considerations for choosing microscopy equipment.
Both confocal and widefield epifluorescence microscopes can be used for imaging the reporters. Confocal microscopes capture images with high z-resolution by rejecting light from outside of the imaging plane of interest. This approach is particularly attractive if the reporter is expressed in multiple tissues in the worm as the microscope could be focused to only visualize specific cells in the animal while excluding unwanted signal from other cells or tissues above or below the region of interest. This is less of a concern if a tissue-specific reporter transgene is constructed as the chosen promoter would serve to limit fluorescence only to the cell types of interest. A trade off associated with the use of confocal is the need to use either brighter transgenes or longer acquisition times during imaging because only the fluorescence from a thin section is visualized. In contrast to confocal imaging, a widefield epifluorescence microscope captures fluorescence from a thicker region of the animal which could permit either dimmer reporters or shorter acquisition times to be used. Another advantage for widefield microscope use is that they are often more common and therefore more accessible on a university campus. This may be due to their lower cost to purchase or the greater ease with reserving time to use the microscope in an imaging core. Also when using a core facility instrument, the increased availability also lowers competition for time with other users. Lastly, the widefield microscope is often familiar to researchers so there is less of a learning curve compared to the use of confocal microscopy. Despite these differences, both approaches can be successful.
Another major difference between standard widefield microscopes and confocal microscopes is the illumination source. Most confocal systems use a laser light source with only selected, monochromatic light wavelengths available on a specific system. Most university-owned systems have been purchased with multiple lasers that provide light sources compatible with standard fluorophores, such as fluorescent proteins or FRET-based ratiometric reporters. However, it is important to investigate the system during the planning phase of reporter use as a microscope system lacking the needed laser will be of little use experimentally. Furthermore, adding a new laser to an existing system is expensive and not always possible. Most widefield fluorescent microscope systems instead use a single broad-spectrum light source which utilizes filters to generate light of the desired excitation wavelength. While almost any light source can be used, often filters need to be purchased as most of the ratiometric reporters need excitation wavelengths that do not match the standard DAPI, FITC, or TRITC filters included in most standard microscope configurations. These filters can be purchased from multiple suppliers and are reasonably inexpensive.
In addition to the light source, the emitted fluorescent light from the sample also needs to be filtered to limit imaging to specific wavelengths of interest. Some confocal microscopes utilize spectral emission pathways, which use a monochromator to separate out desired wavelengths. This can provide great flexibility with regards to the emission wavelengths that can be chosen. On filter-based confocal microscopes or widefield microscopes lacking a monochromator, an emission filter is instead used to remove light that is not of the desired wavelength. The use of an emission filter allows most fluorescent microscopes to be used for visualizing the ratiometric reporters but often these filters need to be specially purchased for this purpose. Filters are typically quite easy to add to a widefield microscope, whereas filter modification on a confocal microscope can be tricky.
Most fluorescent widefield microscopes install the excitation and emission filters together in a cube with a dichroic mirror which permits the excitation and emission light to share the same path through the microscope objective. The dichroic mirror is selected to reflect the excitation wavelength and to transmit the emission wavelength. This allows the faint signal, or emitted light, to be detected by removing it from the excitation light which is brighter but of a lower wavelength. Filters are selected for a defined range of wavelengths, typically considered as a range around a wavelength, instead of a single wavelength. The excitation and emission filters are selected to approximate the excitation and emission maxima of the reporter fluorophores. However, properly selected filters often do not correspond solely to the single peak wavelength as the entire excitation and emission spectra and the interaction with other signals, such as autofluorescence or other fluorophores, must be taken into consideration. Often a narrow-band filter that has been carefully selected to optimize the coverage of the excitation and emission peaks can be used to enhance imaging of the reporters while minimizing autofluorescence or other unwanted signals. For example, to image the HyPer and HyPer2 peroxide-sensing reporters, which have excitation maxima at 420 and 500 nm and an emission maxima at 516 nm, we use an excitation filter that is centered at 414 nm with a bandwidth of 46 nm, a excitation filter centered at 494 nm with a bandwidth of 20 nm, and a 529 nm emission filter with a bandwidth of 24 nm [8, 9]. With these filters, each of the maxima fall within the bandwidth of one of the filters. Once purchased the emission and excitation filters and dichroic mirror are assembled into a microscope manufacturer specific cube for use and installed in the microscope. For example, for HyPer and HyPer2, we paired the 414 nm excitation filter and 529 nm emission filter to visualize the 420 nm maxima, and we paired the 494 nm excitation filter and a second 529 nm emission filter to visualize the 500 nm maxima. Both cubes contain a 506 nm long pass dichroic mirror. Many filter manufacturers will both supply the cube housing and insert the filters and mirror for an additional charge.
The microscope objective choice is another important factor to consider in setting up a microscope for use with ratiometric reporters. Objectives with the appropriate magnification should be chosen depending on whether large regions of the animal are being imaged, which would favor a lower magnification, versus a limited focus on a small region of the animal, such as a specific neuron, which would favor the use of higher magnification. Importantly, an achromatic objective should be selected to limit the effects of chromatic aberration, which could distort the image data especially at higher magnifications. Finally, objectives with higher numerical aperture values tend to be more useful for imaging because the higher numerical aperture produces a brighter image, which decreases the exposure time required, and reduces potential for animal movement or photobleaching of reporter.
2.7 Selection of the Microscope Camera
Another important part of the microscope set-up is the camera used for capturing the images. Beyond simply representing a picture of the worm, the image also contains the intensity data which is utilized to calculate the status of the reporter. The ideal camera should show low read noise, high sensitivity, a high speed of acquisition and a wide dynamic range. Most commercially available cameras do well at capturing images at a range of resolutions which are suitable for use in analyzing reporters in C. elegans. To optimize the dynamic range, one should consider the use of a monochrome camera so the brightness of the image is directly related to the intensity of the light transmitted by the emission filter. Furthermore, these cameras have higher quantum efficiencies, which allow for a brighter picture and shorter exposure time.
An additional concern is the number of bits used by the camera. Many color cameras are 8 bit so only 28 or 256 shades are captured by the camera. Higher quality monochrome cameras can provide high resolution, similar to that of color scientific cameras, as well as 12 bit depth which captures 212 or 4096 shades. The advantage of the 12 bit camera is greater sensitivity, a wider dynamic range, and greater sensitivity to small differences in fluorescence. Some cameras are available that even function at 16 bits or 216 or 65,536 shades.
2.8 Imaging of Nematodes
Once the ratiometric reporter expressing worms are mounted on a slide and positioned in a suitable microscope for imaging, digital images at each of the relevant excitation and emission wavelength pairs need to be captured for subsequent use. In preparing to acquire the images, three issues need to be kept in mind to obtain images suitable for the downstream ratiometric analysis. First, the animals should be absolutely paralyzed because the slightest movement of the animal between capturing each of the two images may result in large deviations in the ratiometric image, especially at high magnifications. Second, the selected acquisition time for the digital camera must result in pixel fluorescence intensities that lie within the dynamic range of the camera and specifically avoids saturated pixels. Saturated pixels have a fluorescence level which lies outside of the range that can be measured by the camera (Figure 2). Using images with saturated pixels in the subsequent analysis will produce erroneous results as these pixels are assigned the maximum value measured by the camera while the true intensity of the pixel could be much higher (see below for further discussion). This underestimates the differences between the compared images. Third, the camera acquisition times for each of the excitation and emission wavelength pairs, which correspond to the excitation maxima, do not need to be the same, but these times do need to be same between experimental groups. Ratiometric reporters are typically used to compare the effects of an intervention or genetic mutation on a parameter of interest, so a typical experiment will consist of a control group and one or more treatment or mutant groups which are then compared. To facilitate this comparison, distinct camera acquisition settings can be used for each of the individual excitation and emission pairs imaged, but these settings cannot vary between experimental groups. Also, the images will ideally be captured at the same time, such as by arranging the worms on the slide so that both the experimental and control animals are seen in the same field. In this section, we will discuss the concerns about acquisition time in more detail.
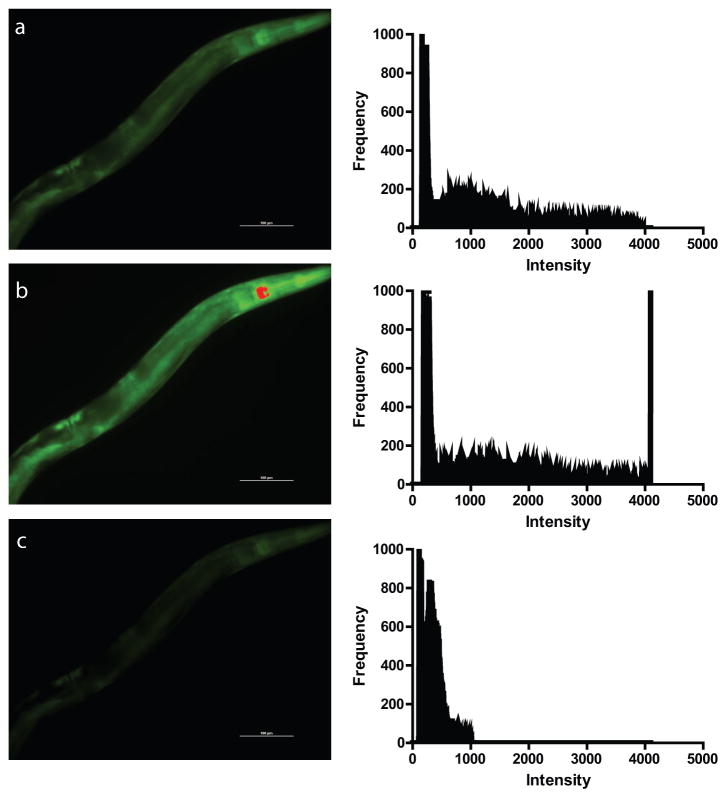
Figure 2.
Sample images from HyPer expressing transgenic C. elegans showing optimal versus over- and under-exposure. The panels on the left show digital images of rpl-17p::HyPer transgenic worms were the indicator function in the microscope software has been used to highlight saturated pixels in red. Note the changes saturated pixels between images captured using (a) optimal acquisition settings, (b) over-exposed settings, and (c) under-exposed settings. The right panels show the corresponding histogram for each image which shows the image pixel distribution with each acquisition setting. In each histogram, the X-axis indicates intensity and the Y-axis indicates frequency of this intensity value. Note the wide distribution of the pixels along the X-axis with no pixels exceeding the 4095 maxima of the 12-bit camera for the (a) optimal acquisition setting. In contrast, the over-exposed image shows a sharp spike of saturated pixels at this 4095 maxima at the right side of the histogram (b) whereas the under-exposed image (c) shows clustering of the pixels along the left side of the histogram where the intensities are under 1000. In each graph, the large peak at the left edge represents the background signal from the black background of the image.
To avoid using an acquisition time which produces saturated pixels, several approaches can be used. Many of the imaging software packages, such as the NIS Elements software from Nikon, have indicators which will highlight saturated pixels in a live image, often through the use of a distinctive color, to provide real-time feedback to the user. If saturated pixels are observed, the acquisition time can be reduced so that the highlighted pixels disappear. An alternate approach is to use a histogram function, if available, which shows the range of pixel intensities as well as the relative abundance of each pixel intensity (Figure 2). The advantage of the histogram function is that the acquisition time can be adjusted based on feedback from the histogram to both avoid saturated pixels and to shift the bulk of the non-background pixels into the middle of the histogram and avoid an under-exposed image (Figure 2). By favoring the middle of the range, the capability of the camera to identify increases or decreases in the fluorescence of the reporter is greater (Figure 3).
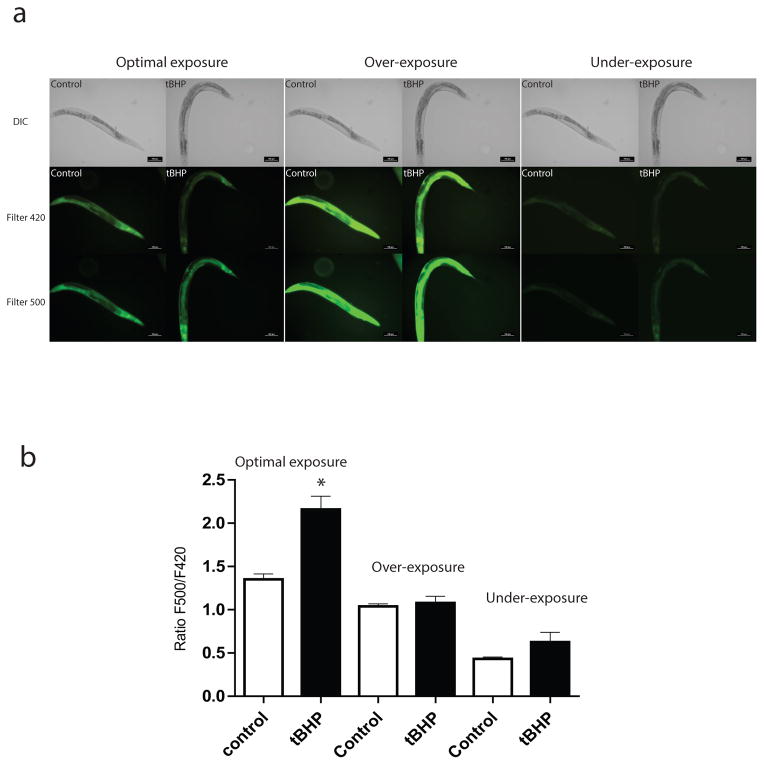
Figure 3.
Response of HyPer to exogenous H2O2 treatment. We transferred rpl-17p::HyPer transgenic worms either to tert-butyl hydroperoxide (tBHP) containing plates for 1 hour or to a control plate without tBHP. The worms were then mounted and imaged using two filter cubes. The first consisted of an excitation filter centered at 414 nm paired with a 529 nm emission filter (Filter 420), and the second consisting 494 nm excitation filter paired with an identical 529 nm emission filter (Filter 500). (a) Representative epifluorescence images of HyPer transgenic C. elegans exposed to 12mM tBHP or control plates using the optimal, over- and under-exposed camera acquisition times. The top row shows a brightfield image of the same worm for reference (b) Graph showing the ratio of the fluorescence resulting from excitation at 500nm divided by the fluorescence resulting from excitation at 420nm (F500/F420) for the same group of 10 animals from each treatment group imaged with each setting. The one-hour exposure to tBHP caused a significant increase in F500/F420 ratio consistent with the increase in intracellular peroxide levels, but the difference was largest and only statistically significant in the images acquired using the optimal exposure time. * P<0.05.
Obtaining images without saturated pixels is fairly straightforward when capturing a single or small number of images in a single experimental group, however to be useful the reporter will need to be imaged in multiple treatment groups or genetic mutants where the spectral properties of the reporter are expected to be different due to changes in the parameter being measured. As a result, imaging conditions that would work well with the control animals, may result in either excessively dim or saturated images in another group. To avoid these concerns, we use an initial group of control and experimental animals to establish constant imaging conditions for each of the relevant excitation and emission pairs that provide pixel intensities that are clearly above background and not saturated in each group of animals. Investing this effort initially is important as obtaining suboptimal images from any of the experimental groups can jeopardize an entire experiment.
Once the camera settings and acquisition times are optimized, imaging the animals in each of the groups is fairly straightforward. We have found that imaging as few as 10 animals in each group can demonstrate a clear change in the reporter measures. More animals are likely needed if the magnitude of the expected change in cellular physiology is smaller or if there is greater animal-to-animal variability in either the reporter measure or the physiological change. Often a pilot experiment can be very informative for determining the numbers of animals needed to obtain a robust dataset.
2.9 Image Analysis
Ratiometric reporter images are often analyzed at a time after being captured, but some imaging software packages allow the images to be processed in real-time. This live-analysis allows the researcher to identify the regions of interest while performing the imaging to make changes to the imaging protocol or to verify whether changes in reporter fluorescence seen in an animal are then reliably found in other animals. The ability to perform live-analysis can be performed, for example, using the newer versions of the Nikon imaging software. This software also offers a feature where the ratio image is normalized for the intensity of the underlying pixels, which results in dim regions of the images being effectively excluded from the analysis without a region of interest being selected. The post-analysis approach is more commonly used because it requires less sophisticated imaging software, and also minimizes the amount of user time spent on a microscope, which can be important for heavily used instruments. Here we will focus on the post-analysis using the free ImageJ software [46].
To extract the ratiometric data from the images, the investigator is essentially dividing the pixel intensities in an image captured for one excitation-emission pair by the pixel intensities in an image captured using the same animal with another excitation-emission pair. This resulting ratio provides a measure of the parameter measured by the reporter and differences in the ratio between experimental groups indicates a change in the underlying parameter. In this section we will discuss how this ratio can be calculated from the obtained images. Fortunately, a number of software packages, such as software provided by a microscope manufacturer or the freely available ImageJ, can be used to facilitate this analysis [46].
The initial step in analyzing the image involves determining the background- subtracted pixel intensities for the images corresponding to each relevant excitation-emission pair for a given animal. This involves using software to calculate the level of background fluorescence for the entire image, and the subtracting this value from all of the pixel intensities in the image. Failing to remove these background values can decrease the differences observed by artificially inflating the values from the dimmer image. Within ImageJ, this function can be carried out by using the “subtract background” option within the “process” menu. Most other software packages are also able to perform this operation.
After background subtraction is performed for the pair of images, the pixel intensities for the pair of images are then mathematically divided to obtain a new “image” consisting of the resulting ratio. Ideally the calculation is performed using pixel-by-pixel data so that the effects of noisy pixels are minimized. Within ImageJ the “image calculator” option within the “process” menu is used for this purpose. Often in C. elegans experiments the reporter is expressed in only a subset of the image so regions of interest, often based on the reporter expression pattern, can then be identified to measure the ratio of the two images only in these areas. Within ImageJ, the selection tool buttons can be used to define the region of interest, and then the “measure” option within the “analyze” menu can be used to obtain the desired ratio data. With many of the software packages, the regions of interest can be specified either manually or by using the software, and many of the steps can be automated to reduce the hands-on manipulation time involved.
After performing these steps for each animal imaged within each group, the resulting ratio data can then be plotted and compared using scientific graphing software such as Prism (Graphpad Software). We are typically comparing two groups so calculating a mean and comparing the groups through the use of the t-test is adequate to determine if differences between the groups exist. More complicated experimental designs would require the use of different statistical approaches.
3. Results and Discussion
To illustrate the ratiometric behavior of the HyPer ratiometric reporter which detects hydrogen peroxide, we treated worms expressing HyPer in most tissues by transferring 10 rpl-17p::HyPer transgenic worms to a plate containing tert-butyl hydroperoxide (tBHP) for 1 hour [2, 8]. Because of its high membrane permeability, tBHP has been used as an alternative to H2O2 and similarly to H2O2, it can oxidize and activate OxyR [38–40]. We then mounted the worms using levamisole as an anesthetic to avoid exposure to sodium azide which could independently produce oxidative stress in the animals. HyPer has two excitation peaks with maxima at 420 nm and 500 nm, and one emission peak with maximum at 516 nm which we measured via digital fluorescent imaging. First we imaged these animals using a filter cube consisting of an excitation filter centered at 414 nm paired with a 529 nm emission filter to measure fluorescence related to the 420 nm excitation maxima (Figure 3). Immediately after acquiring the first image, we then imaged the same animal using a second filter cube consisting of a 494 nm excitation filter paired with an identical 529 nm emission filter to measure fluorescence related to the 500 nm excitation maxima (Figure 3).
Upon exposure to H2O2, the excitation peak of HyPer at 420 nm decreases proportionally to the increase in the peak at 500 nm [2, 8]. Consistently, we observe an increase in the fluorescence in the images measuring this 500 nm peak, while we see less fluorescence in the images measuring the 420 nm peak (Figure 3A). As a result, this HyPer transgenic line is showing the expected ratiometric reporter properties in this control experiment. The next step in this experiment is to image additional animals and to then analyze the resulting images as described above. We find that imaging additional worms shows a clear difference between control and tBHP treated worms when the images are analyzed and the resulting ratio data are plotted (Figure 3B). To demonstrate the pitfalls of under- and over-exposed images, we imaged the same animals with acquisition times that are either too short or too long (Figure 3A). Analysis of these images fails to show the same difference in HyPer fluorescence as seen in the optimally exposed animals (Figure 3B).
In summary, ratiometric reporters have emerged as a powerful new tool to non-invasively monitor cell physiology. The transparent nature of C. elegans has allowed these reporters to be applied to measuring a wide-range of cellular parameters (Table 1) inside of intact, living animals. The coming years will likely see growing use of these reporters for real-time monitoring of an increasing number of metabolites, signaling pathways, and chemical conditions through development, aging, and environmental changes and to determine how genetic pathways can alter these parameters. These reporters offer a compelling blend of sensitivity, high spatial and temporal resolution, and non-invasive measurements using readily available lab equipment. As a result, the ratiometric reporter approach may greatly expand our understanding of the cellular biology of living animals.
Acknowledgments
This work was supported by NIH grants (R01 ES017761 and R01 AG044768) to A.L.F., as well as support from the San Antonio GRECC of the South Texas VA Healthcare system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Back P, De Vos WH, Depuydt GG, Matthijssens F, Vanfleteren JR, Braeckman BP. Free Radic Biol Med. 2012;52:850–859. doi: 10.1016/j.freeradbiomed.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 3.Ting AY, Kain KH, Klemke RL, Tsien RY. Proc Natl Acad Sci U S A. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Ma Y, Taylor SS, Tsien RY. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truong K, Sawano A, Mizuno H, Hama H, Tong KI, Mal TK, Miyawaki A, Ikura M. Nature structural biology. 2001;8:1069–1073. doi: 10.1038/nsb728. [DOI] [PubMed] [Google Scholar]
- 6.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 7.Miyawaki A, Tsien RY. Methods in enzymology. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 8.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 9.Markvicheva KN, Bilan DS, Mishina NM, Gorokhovatsky AY, Vinokurov LM, Lukyanov S, Belousov VV. Bioorg Med Chem. 2011;19:1079–1084. doi: 10.1016/j.bmc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 11.Bilan DS, Matlashov ME, Gorokhovatsky AY, Schultz C, Enikolopov G, Belousov VV. Biochimica et biophysica acta. 2013;1840:951–957. doi: 10.1016/j.bbagen.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung YP, Albeck JG, Tantama M, Yellen G. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer AJ, Dick TP. Antioxidants & redox signaling. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 14.Tsuyama T, Kishikawa J, Han YW, Harada Y, Tsubouchi A, Noji H, Kakizuka A, Yokoyama K, Uemura T, Imamura H. Analytical chemistry. 2013;85:7889–7896. doi: 10.1021/ac4015325. [DOI] [PubMed] [Google Scholar]
- 15.Tsou P, Zheng B, Hsu CH, Sasaki AT, Cantley LC. Cell Metab. 2011;13:476–486. doi: 10.1016/j.cmet.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Proc Natl Acad Sci U S A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liemburg-Apers DC, Imamura H, Forkink M, Nooteboom M, Swarts HG, Brock R, Smeitink JA, Willems PH, Koopman WJ. Pharmaceutical research. 2011;28:2745–2757. doi: 10.1007/s11095-011-0492-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, Yu Z, Xu L, Wang X, Yang Y, Loscalzo J. Cell Metab. 2011;14:555–566. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allman E, Johnson D, Nehrke K. Am J Physiol Cell Physiol. 2009;297:C1071–1081. doi: 10.1152/ajpcell.00284.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nehrke K. J Biol Chem. 2003;278:44657–44666. doi: 10.1074/jbc.M307351200. [DOI] [PubMed] [Google Scholar]
- 21.Nakai J, Ohkura M, Imoto K. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 22.Wagner J, Allman E, Taylor A, Ulmschneider K, Kovanda T, Ulmschneider B, Nehrke K, Peters MA. Am J Physiol Cell Physiol. 2011;301:C1389–1403. doi: 10.1152/ajpcell.00139.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coburn C, Allman E, Mahanti P, Benedetto A, Cabreiro F, Pincus Z, Matthijssens F, Araiz C, Mandel A, Vlachos M, Edwards SA, Fischer G, Davidson A, Pryor RE, Stevens A, Slack FJ, Tavernarakis N, Braeckman BP, Schroeder FC, Nehrke K, Gems D. PLoS Biol. 2013;11:e1001613. doi: 10.1371/journal.pbio.1001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoefler D, Thamsen M, Koniczek M, Niemuth NJ, Diederich AK, Jakob U. Mol Cell. 2012;47:767–776. doi: 10.1016/j.molcel.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stout RF, Jr, Parpura V. Cell calcium. 2011;50:98–108. doi: 10.1016/j.ceca.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Mol Cell Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Praitis V, Casey E, Collar D, Austin J. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mello CC, Kramer JM, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mello C, Fire A. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 31.Ferguson AA, Fisher AL. Plasmid. 2009 doi: 10.1016/j.plasmid.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson AA, Cai L, Kashyap L, Fisher AL. Methods Mol Biol. 2013;940:87–102. doi: 10.1007/978-1-62703-110-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semple JI, Garcia-Verdugo R, Lehner B. Nat Methods. 2010;7:725–727. doi: 10.1038/nmeth.1495. [DOI] [PubMed] [Google Scholar]
- 34.Giordano-Santini R, Milstein S, Svrzikapa N, Tu D, Johnsen R, Baillie D, Vidal M, Dupuy D. Nat Methods. 2010;7:721–723. doi: 10.1038/nmeth.1494. [DOI] [PubMed] [Google Scholar]
- 35.Hochbaum D, Ferguson AA, Fisher AL. J Vis Exp. 2010 doi: 10.3791/2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilm T, Demel P, Koop HU, Schnabel H, Schnabel R. Gene. 1999;229:31–35. doi: 10.1016/s0378-1119(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 37.Fuangthong M, Helmann JD. Proc Natl Acad Sci U S A. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charoenlap N, Eiamphungporn W, Chauvatcharin N, Utamapongchai S, Vattanaviboon P, Mongkolsuk S. FEMS microbiology letters. 2005;249:73–78. doi: 10.1016/j.femsle.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. Journal of bacteriology. 2000;182:4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolbe K, Schonherr R, Gessner G, Sahoo N, Hoshi T, Heinemann SH. J Physiol. 2010;588:2999–3009. doi: 10.1113/jphysiol.2010.192468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porta-de-la-Riva M, Fontrodona L, Villanueva A. Cer, J n, JoVE. 2012:e4019. doi: 10.3791/4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagido C, Pettitt J, Flett A, Glover LA. BMC physiology. 2008;8:7. doi: 10.1186/1472-6793-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kell A, Ventura N, Kahn N, Johnson TE. Free Radic Biol Med. 2007;43:1560–1566. doi: 10.1016/j.freeradbiomed.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerr RA. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.113.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarpey MM, Fridovich I. Circulation research. 2001;89:224–236. doi: 10.1161/hh1501.094365. [DOI] [PubMed] [Google Scholar]
- 46.Abramoff MDMPJ, Ram SJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 47.Beg AA, Ernstrom GG, Nix P, Davis MW, Jorgensen EM. Cell. 2008;132:149–160. doi: 10.1016/j.cell.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miesenbock G, Kevrekidis IG. Annual review of neuroscience. 2005;28:533–563. doi: 10.1146/annurev.neuro.28.051804.101610. [DOI] [PubMed] [Google Scholar]
- 49.Miesenbock G, De Angelis DA, Rothman JE. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 50.Schulte A, Lorenzen I, Bottcher M, Plieth C. Plant methods. 2006;2:7. doi: 10.1186/1746-4811-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 52.Meyer AJ. Journal of plant physiology. 2008;165:1390–1403. doi: 10.1016/j.jplph.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 53.Schwarzlander M, Fricker MD, Sweetlove LJ. Biochimica et biophysica acta. 2009;1787:468–475. doi: 10.1016/j.bbabio.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 54.Schwarzlander M, Fricker MD, Muller C, Marty L, Brach T, Novak J, Sweetlove LJ, Hell R, Meyer AJ. Journal of microscopy. 2008;231:299–316. doi: 10.1111/j.1365-2818.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- 55.Tanimura A, Nezu A, Morita T, Turner RJ, Tojyo Y. J Biol Chem. 2004;279:38095–38098. doi: 10.1074/jbc.C400312200. [DOI] [PubMed] [Google Scholar]
- 56.Zaccolo M. Circulation research. 2004;94:866–873. doi: 10.1161/01.RES.0000123825.83803.CD. [DOI] [PubMed] [Google Scholar]
- 57.Deuschle K, Okumoto S, Fehr M, Looger LL, Kozhukh L, Frommer WB. Protein science: a publication of the Protein Society. 2005;14:2304–2314. doi: 10.1110/ps.051508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bilan DS, Matlashov ME, Gorokhovatsky AY, Schultz C, Enikolopov G, Belousov VV. Biochimica et biophysica acta. 2013 doi: 10.1016/j.bbagen.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyawaki A, Griesbeck O, Heim R, Tsien RY. Proc Natl Acad Sci U S A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagai T, Sawano A, Park ES, Miyawaki A. Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]