Abstract
Patients coinfected with HIV and hepatitis C virus (HCV) have poor to modest rates of response with interferon-based therapies, which remain a backbone of the treatment in HIV/HCV-coinfected patients. The mechanisms responsible for poor responsiveness to interferon are not well described. In this study a targeted proteomic analysis of plasma from 42 patients infected with both HIV and HCV and undergoing therapy for HCV with peginterferon and ribavirin was performed. Higher baseline plasma levels of interleukin (IL)-23 were associated with sustained virologic response. Further investigation of how IL-23 facilitates interferon (IFN) responsiveness, as evidenced by a >2-fold increase in most interferon-stimulated genes (ISGs), revealed that IL-23 indirectly enhances IFN signaling in peripheral blood mononuclear cells and HCV continuous culture system by preventing the down-regulation of the IFNAR2 receptor after exposure to IFN-α. These findings suggest a unique role of the IL-23 pathway in enhancing host response to type I interferons, thereby facilitating eradication of HCV. Low levels of IL-23 present in plasma of nonresponders may reflect an impaired immune state that in the case of HIV/HCV-coinfected subjects could potentially lead to disruption of TH17 CD4+ T cells. This study suggests a major role for HIV-associated immune dysregulation present in HIV-infected subjects that subsequently determines the overall responsiveness to exogenous interferon-α-based HCV therapy.
Introduction
Hepatitis C virus (HCV) infection is a major public health problem with over 180 million people infected worldwide,1 and due to shared routes of transmission, approximately one-third of all human immunodeficiency virus (HIV)-infected patients are coinfected with HCV in the United States.2 In HIV coinfection, the rate of liver disease progression is accelerated, the response rate to standard of care therapy is diminished,3–5 and treatment with interferon (IFN)-based therapies results in poor sustained virologic response rates.6,7 The mechanisms by which IFN-α eradicates HCV are not fully understood,8 and thus a better understanding of molecular pathways, as well the influence of host factors is vital to improving treatment outcomes.
Given that interferon is an immune stimulatory therapy, there has been considerable interest in understanding the contribution of immune pathways involved in effecting treatment-induced viral clearance. This is particularly true in the context of HCV/HIV coinfection where additional adaptive immune dysregulation makes achieving a sustained virologic response (SVR) with IFN/ribavirin (RBV) therapy less likely. Interleukin-23 (IL-23) is a heterodimeric cytokine produced primarily by antigen-presenting cells that plays a role in the control of various viral infections, although the exact mechanisms are unclear.9–11 In the context of HCV infection, IL-23 has also been associated in vitro and in animal models with augmented HCV-specific CD8+ cytotoxic T cell function.9
In this study, we hypothesized that levels of cytokines in peripheral blood may influence treatment outcome in these subjects. A targeted proteomic approach identified pretreatment plasma cytokines associated with IFN-α-induced HCV clearance in an HIV/HCV-coinfected population. Initial analysis showed high levels of IL-23, a Th17 cytokine in the blood of subjects who achieved SVR when compared to nonresponders (NR). Therefore, we further investigated if levels of IL-23 could enhance IFN-α signaling and play an important role in determining HCV clearance.
Materials and Methods
Study design
Forty-two treatment-naive, genotype 1, HIV/HCV-coinfected subjects with known treatment outcomes were included in the study. Patients had been treated with pegylated IFN-α 2b at 1.5 μg/kg subcutaneously every week (Peg-Intron; Merck) or pegylated IFN-α 2a at 180 mg/week (Pegasys; Roche) and weight-based ribavirin (at 1,000 mg dose <75 kg, 1,200 mg dose for >75 kg) for a total of 48 weeks in two clinical trials conducted at the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NCT00018031 and NCT00085917). All subjects signed NIAID IRB-approved informed consent prior to enrollment in the study. Outcomes were defined as SVR (undetectable HCV RNA 24 weeks after the end of treatment) or NR (detectable HCV RNA through week 24 or a <2 log10 copies/ml reduction in HCV RNA at week 12 of treatment). HCV viral load was measured at frequent intervals using the Abbott HCV assay (Abbott Laboratories; lower limit of detection of 12 IU/ml). The clinical outcome data were previously reported.12,13
Proteomic analysis
Plasma from pretreatment samples was separated by centrifugation and stored at −80°C. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll Hypaque (GE Healthcare Biosciences Corp., Piscataway, NJ) density centrifugation and stored in liquid nitrogen. Plasma samples were thawed only once and aliquoted for further analysis. Then 200 μl plasma from 20 randomly selected treated subjects (10 SVR and 10 NR patients) and 6 normal volunteers (NV) was pooled and the samples run on a multiplex proteomic platform of 189 inflammatory mediators (Aushon Biosystems, Billerica, MA). We assessed 189 cytokines and chemokines in these 20 patients with differential clinical outcomes after IFN-α treatment. Differential cytokine expression relative to the two groups was estimated based on a 2-fold change and a one-way ANOVA test (p<0.01). IL-23 was quantitated in individual samples using a standard enzyme-linked immunoassay (EIA; R & D Biosystems Inc., Minneapolis, MN) with a lower limit of detection of 39.1 pg/ml. Following the initial pilot tests, IL-23 levels were analyzed in the entire cohort of 42 (21 NR and 21 SVR) patients.
IL-23 and IFN-α treatment of HCV J6/JF-infected Huh7.5 cell lines
A previously described, the J6/JFH-1/Huh7.5 cell (HCVcc) system was used to perform functional assays.14 Huh7.5 cells were maintained at 1×106 cell/ plate, incubated at 37°C in DMEM (Invitrogen Corporation Inc., Carlsbad, CA), and supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). Huh7.5 cells (HCV J6/JFH infected or uninfected) were incubated with either medium alone or recombinant human IL-23 (rhIL-23; 50 ng/ml, 100 ng/ml, or 200 ng/ml) for 48 h, at which time recombinant human IFN-α (rhIFN-α; 10 U/ml, 25 U/ml, or 50 U/ml) was added for an additional 48 h. Cell-free supernatant was collected at the end of incubation for proteomic analysis (R&D Biosystems Inc., Minneapolis, MN).
IL-23 and IFN-α treatment of PBMCs from SVR and NR patients
Frozen PBMCs from SVR and NR were thawed and plated in 96-well U-bottom plates at approximately 1 million cells/well in RPMI medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT), 100 U/ml penicillin/100 μg/ml streptomycin (Invitrogen, Carlsbad, CA), 1 mM HEPES (Invitrogen, Carlsbad, CA), and 2 mM l-glutamine (Invitrogen, Carlsbad, CA), and incubated for 54 h with medium alone or 100 ng/ml rhIL-23 at 37°C and 5% CO2, the last 6 h of which 10 U/ml rh IFN-α was added before cell lysis for RNA extraction.
Quantification of HCV RNA viral load and interferon-stimulated gene (ISG) expression via real-time PCR
RNA was extracted from infected Huh7.5 cells and PBMCs using the RNAqeous-4 PCR kit (Ambion Inc., Austin, TX). The extracted RNA was reverse transcribed using the High cDNA Archive Kit (Life Technologies Corp., Grand Island, NY) according to the manufacturer's instruction. For the cDNA from J6/JFH-infected Huh7.5 cells, HCV viral load was quantified by real-time polymerase chain reaction (PCR) (7500 PCR Real-Time PCR system, Life Technologies Corp., Grand Island, NY). PCR was performed in a volume of 20 μl containing 4 μl of PCR water, 2.5 μl of cDNA, 10 μl of Taqman Gene Expression MasterMix, 1.25 μl of primers (forward primer sequence: 5′-GCCATGGCGTTAGTATG AGTGT-3′; reverse primer sequence: 5′-CGCCCTATCAGGCAGTACCACAA-3′), and 1 μl of probe (6FAMTCTGCGGAACCGGTGAGTACACTAMRA; Life Technologies Corp., Grand Island, NY). ISG expression in rhIL-23 and rhIFN-α-treated PBMCs was assessed using Taqman Array 96-well plates (Life Technologies Corp., Grand Island, NY) containing primer/probe sets for ISGs previously found to be differentially expressed in HIV/HCV-coinfected PBMCs.14
Western blot analysis for IL-23 receptor and stat phosphorylation
The IL-23 receptor is a heterodimer consisting of an IL-12Rβ1 chain and an IL-23R chain, and the expression on Huh7.5 cells was assessed by western blot. Cells were grown until confluent as described above and then lysed in cold radioimmunoprecipitation assay (RIPA) buffer (ThermoScientific, Rockford, IL) supplemented with HALT protease inhibitor and HALT phosphatase cocktails (ThermoScientific, Rockford, IL) as per the manufacturer's instructions. Western blot analysis was performed and probed using polyclonal goat antihuman biotinylated IL-12RB1 (R&D Systems, Minneapolis, MN) and polyclonal goat antihuman biotinylated IL-23R (R&D Systems, Minneapolis, MN). To determine the effect of IL-23 and IFN-α on IFN-α-associated STAT signaling, cells were incubated in the presence or absence of 100 ng/ml rhIL-23 (rIL-23; R&D Systems) with or without 100 IU/ml rhIFN-α (rIFN-α; R&D Systems) for 15 min and lysed in RIPA buffer as previously described.15 Lysate was then run in a western blot as previously described and phosphorylated STAT1 (mouse antihuman; BD Biosciences) and phosphorylated STAT2 (rabbit antihuman; R&D Systems) were detected using appropriate antibodies.
Flow cytometry analysis of interferon receptors
Huh7.5 cells treated for 30 min with 100 ng/ml rhIL-23 and/or 100 IU/ml rhIFN-α were harvested by gentle mechanical dissociation and incubated in the dark at 4°C for 30 min with phycoerythrin (PE)-conjugated mouse antihuman IFNAR1 and fluorescein isothiocyanate (FITC)-conjugated mouse antihuman IFNAR2 (both from PBL Assay Science, Piscataway, NJ). Cells were fixed with Cytofix/Cytoperm solution as perthe manufacturer's guidelines (BD Biosciences, San Jose, CA) and analyzed on a FACSCanto II flow cytometer (BD Biosciences, San Jose, CA). A total of 50,000 events were collected per experimental condition, and receptor expression was analyzed using FlowJo software version 9 (Tree Star Inc., Ashland, OR).
Statistical analysis
The data are expressed as mean with standard error. Analysis of variance (ANOVA) with Tukey's multiple comparison test was used to compare means of the independent groups with p values<0.05 considered significant. The paired t test with Bonferroni adjustment for multiple testing was used to compare paired responses.
Results
Patient profile
Forty-two HIV/HCV-coinfected study subjects treated with interferon-based therapy were included in this study (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid). There were no significant differences between groups other than a higher proportion of African Americans in the NR group.
Elevated pretreatment IL-23 plasma levels in the SVR group compared to the NR group
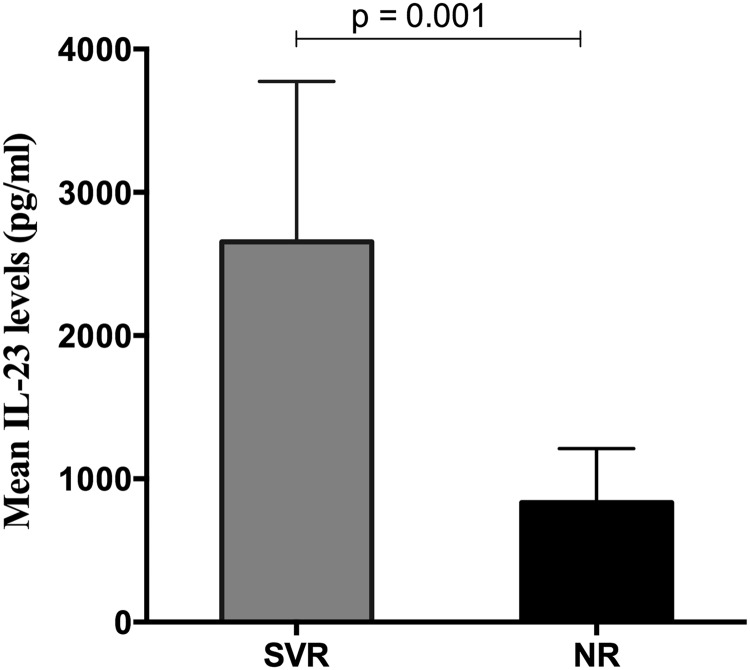
In the initial pilot analysis of 189 cytokines and chemokines in 20 patients, 10 proteins were differentially expressed in the NR versus SVR group (Table 1). All proinflammatory cytokines and chemokines were significantly higher in the SVR group than the NR group (p<0.05), and several (IL-1RA, CXCL9, CD40L, TRAIL) are known to be induced by IFN-α or IL-23 signaling. Pooled pretreatment plasma in the initial subset of 20 patients demonstrated differential IL-23 expression between the NR and SVR groups. NR subjects had lower mean IL-23 levels compared to SVR patients (Table 1, 37±9 versus 234±25 (pg/ml); p=0.001). These data were confirmed in all 42 individual plasma samples using a confirmatory immunoassay. SVR patients had higher mean IL-23 plasma levels compared to NR patients (Fig. 1: 2,654±1,120 vs. 835±376 (pg/ml), p=0.001). Notably, many NR had IL-23 levels below the level of quantification. These results suggest that IL-23 levels in pretreatment plasma could distinguish between patients who would subsequently achieve SVR or NR with IFN-α-based therapy.
Table 1.
Differentially Expressed Cytokines and Chemokines in Plasma from PEG-Interferon-α/Ribavirin Nonresponder and Sustained Virologic Response Patients
| Cytokine | NR (pg/ml) | SVR (pg/ml) | p value |
|---|---|---|---|
| Interleukin-23 | 37±9 | 234±25 | 0.001 |
| Interleukin-25 | 29±7 | 541±45 | 0.0001 |
| Beta-Defensin 2 | 153±26 | 493±54 | 0.007 |
| Visfatin | 585±21 | 79,368±298 | 0.00001 |
| Interleukin-1RA | 266±78 | 657±56 | 0.04 |
| Fibronectina | 34,765±7,650 | 79,838±5676 | 0.008 |
| CXCL-9 | 363±81 | 853±98 | 0.03 |
| sCD30 | 2,027±356 | 5,850±345 | 0.009 |
| CD40L | 39±10 | 152±10 | 0.007 |
| TRAIL | 44±35 | 112±54 | 0.044 |
ng/ml.
Levels of 189 proteins in pooled plasma of 10 NR and 10 SVR patients were measured as described in Materials and Methods. Those that were significantly different between the two groups are listed in the table.
NR, nonresponders; SVR, sustained virologic response.
FIG. 1.
Baseline interleukin (IL)-23 plasma concentration differs by clinical outcome in interferon (IFN)-α /ribavirin (RBV)-treated HIV/hepatitis C virus (HCV)-coinfected patients. Levels of IL-23 in plasma of nonresponders (NR) and sustained virologic response (SVR) were estimated using an enzyme immunoassay (EIA). As shown in the figure, subjects who achieved SVR had significantly higher levels of IL-23 (2,654±1,120 pg/ml SEM) than NR (835±376 pg/ml) (p=0.001).
Both chains of the IL-23R are expressed in Huh7.5 cells
An IL-23 receptor must be present for IL-23 to affect its function, and it has not been previously described on the Huh7.5 cell line used for in vitro HCV replication experiments in this study. Therefore, we used a western blot to confirm that Huh7.5 cells, both J6/JFH infected and uninfected, express both chains of the IL-23R. Supplementary Fig. S1 demonstrates the presence of IL-12RB1 and IL-23R, two components of the IL-23R heterodimer, in qualitatively similar amounts in Huh7.5 cells, regardless of J6/JFH infection, IL-23 treatment, or IFN-α exposure.
IL-23 does not directly affect HCV copy number in vitro but enhances IFN-α-mediated viral suppression
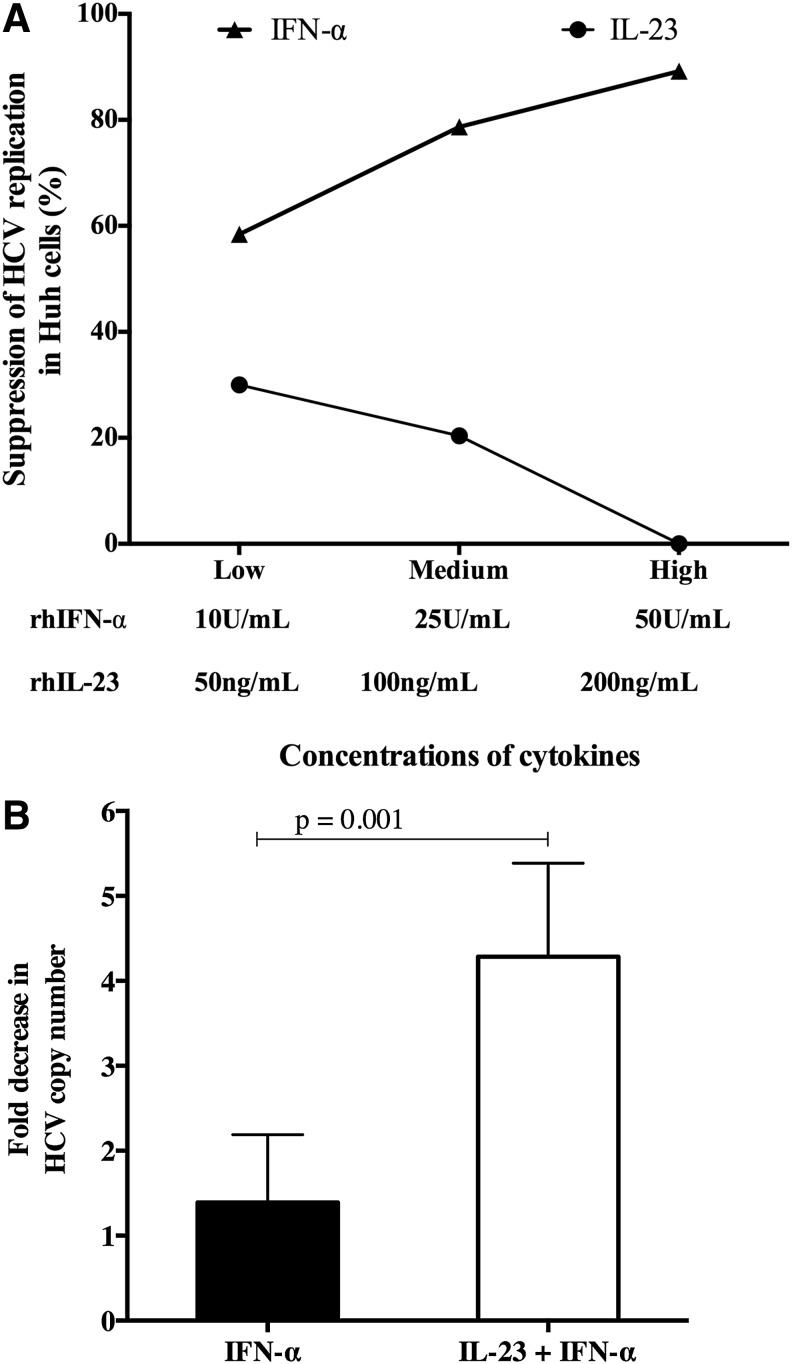
We tested whether higher SVR rates in patients with elevated baseline plasma IL-23 might be due to a direct antiviral effect of IL-23 by utilizing the in vitro HCVcc system. Huh7.5 cells were infected with chimeric J6/JFH as previously described14 and exposed to rhIL-23 and rhIFN-α, after which HCV viral suppression was measured. There was a direct relationship between the suppression of HCV replication and increasing rhIFN-α concentration demonstrating detectable in vitro viral replication and expected results with rhIFN-α treatment (Fig. 2A). In contrast, rhIL-23 treatment did not significantly inhibit HCV replication (Fig. 2A) regardless of concentration. Based on the Fig. 2A data, 100 ng/ml rhIL-23 and 10 IU/ml rhIFN-α were used in subsequent experiments to assess the effect of combination cytokine exposure on HCV copy number. When IL-23 was added to IFN-α, there was enhanced suppression of HCV replication in comparison to treatment of IFN-α alone (4.28±1.1 vs. 1.39±0.8-fold, p=0.001) (Fig. 2B).
FIG. 2.
IL-23 does not have a direct effect on HCV replication in vitro. (A) The effect of IL-23 on HCV copy number was determined using the HCVcc system as described. rhIL-23 treatment at a low (50 ng/ml), medium (100 ng/ml), or high (200 ng/ml) concentration had no significant effect on the HCV copy number. However, as expected, rhIFN-α at low (10 U/ml), medium (25 U/ml), and high (50 U/ml) concentrations demonstrated dose-dependent suppression of HCV replication. (B) IL-23 enhances IFN-α-mediated suppression of HCV replication. Consequently, 10 IU/ml rhIFN-α and 100 ng/ml rhIL-23 resulted in a significant decline in HCV copy number compared to IFN-α alone.
IL-23 does not induce ISGs in PBMCs, but enhances IFN-α-associated ISG expression
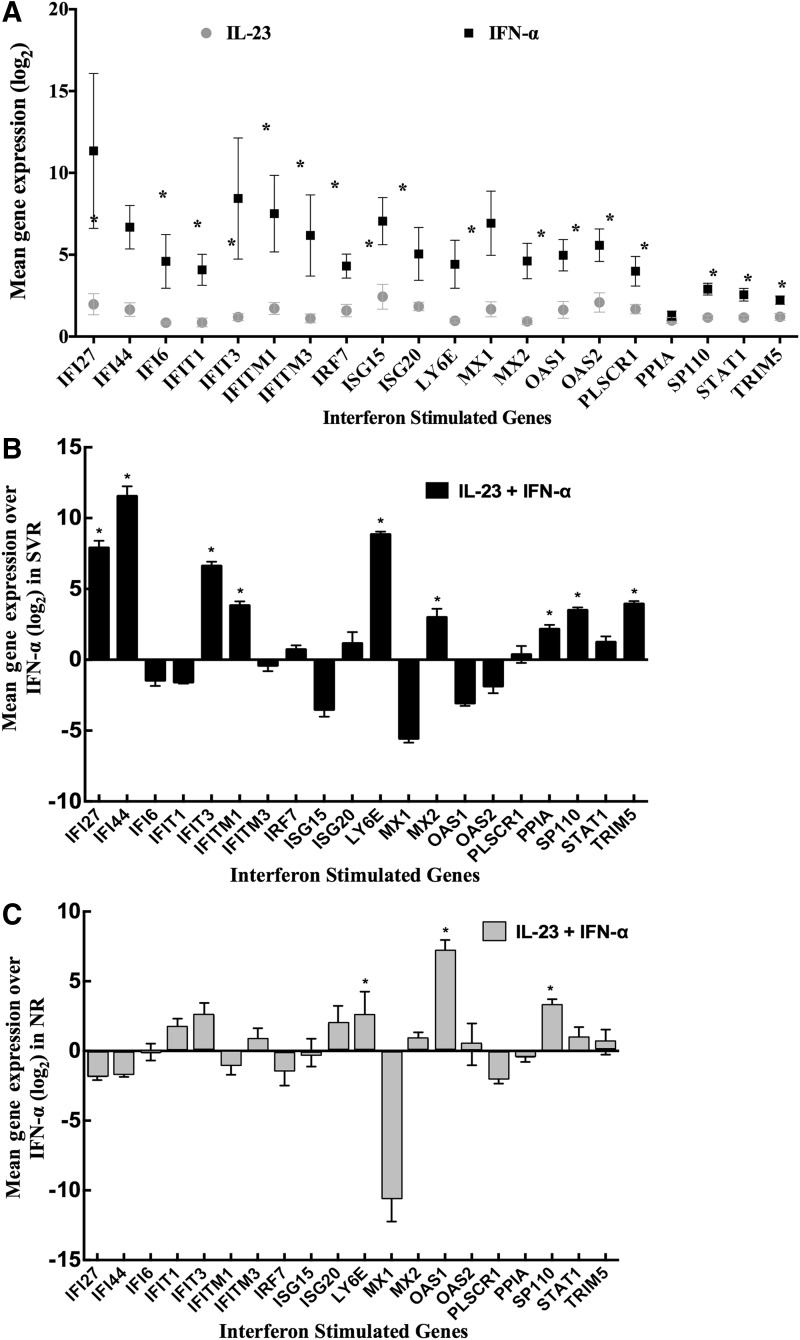
PBMCs from 22 patients, 11 SVR and 11 NR, were treated with IL-23, IFN-α, or both, and RNA was extracted for ISG expression analysis using TaqMan Array plates. IFN-α treatment induced expression of many ISGs, while IL-23 treatment of PBMCs did not significantly induce ISGs in normal volunteers (Fig. 3A). However, when IL-23 was combined with IFN-α, PBMCs from both SVR and NR groups showed significantly higher ISG expression than treatment with IFN-α alone. In PBMCs from SVR patients, IFI27, IFI44, IFIT3, IFITM1, LY6E, MX2, PPIA, SP110, and TRIM5 genes were highly induced with combination treatment of IL-23 and IFN-α (Fig. 3B). In the case of NR, the IL-23 and IFN-α treatment significantly induced LY6E, OASI, and SP110 genes compared to IFN-α alone (Fig. 3C).
FIG. 3.
IL-23 does not induce interferon-stimulated genes (ISGs) in peripheral blood mononuclear cells (PBMCs). (A) ISG expression was quantified in PBMCs from 22 patients after rhIL-23 and rhIFN-α treatment. (B) IL-23 and IFN-α induced significantly higher levels of ISGs than IFN-α treatment alone. (C) In the case of NR, treatment of IL-23 and IFN-α only significantly induced OASI genes compared to IFN-α alone.
IL-23 does not induce pSTAT1 in PBMCs
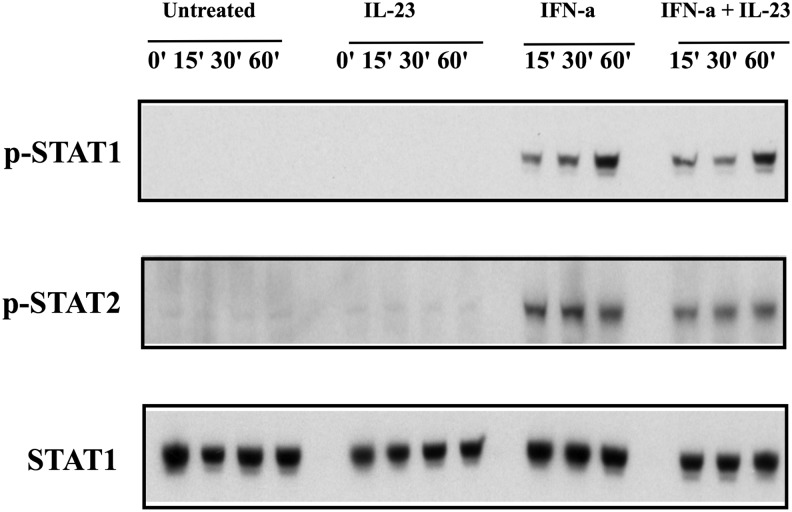
STAT1 phosphorylation is the initial step of IFN-α signal transduction and is critical in mediating the cellular response to IFN-α. STAT phosphorylation of Huh7.5 cells after IL-23 exposure was assessed to determine whether IL-23 may directly enhance IFN-α signaling. As shown in Fig. 4, there was no phosphorylation of STAT1 molecule with IL-23; however, exposure to IFN-α readily induced phosphorylation of STAT1. These results enforce that IL-23-mediated enhancement of IFN gene expression does not involve direct activation of early IFN-α signaling.
FIG. 4.
IL-23 does not induce pSTAT1 in PBMCs. pSTAT1 and pSTAT2 were induced in PBMCs by IFN-α but not IL-23.
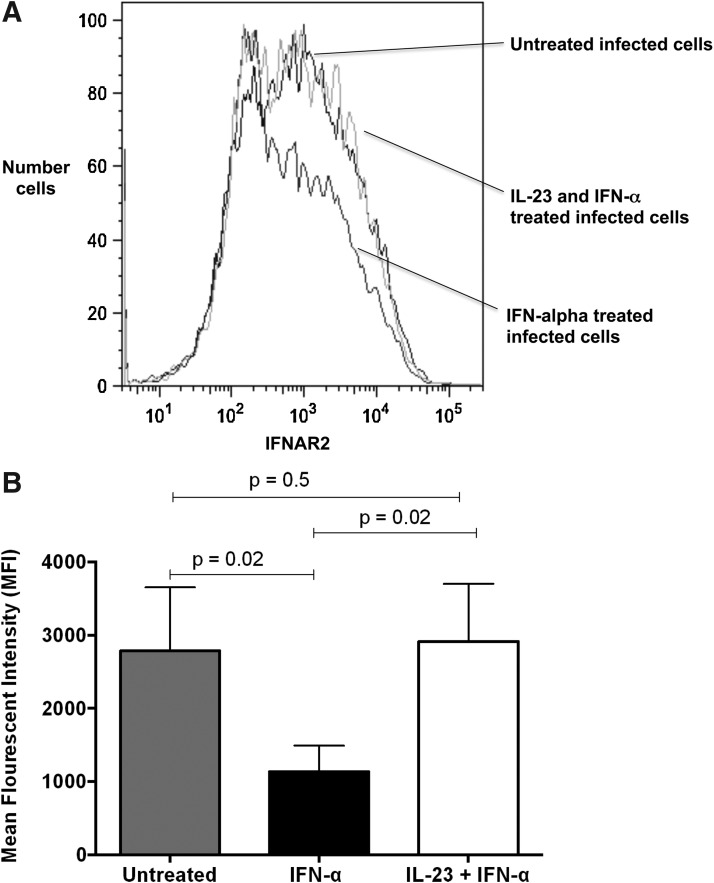
IL-23 treatment maintains IFNAR2 expression on Huh7.5 cells
Unlike IFN-αR1, IFN-αR2 pairs only with IFN-αR1 to mediate IFN-α signaling. Quantification of the IFNAR2 receptor on the surface of Huh7.5 cells before and after IL-23 and IFN-α treatment was performed. IL-23 exposure maintained a surface IFNAR2 receptor (Fig. 5A). Furthermore, IL-23 treatment of Huh7.5 cells restored the mean fluorescent intensity of the IFNAR2 receptor to the levels prior to treatment with IFN-α (2,912±786 for IFN-α+IL-23, 1,134±356 for IFN-α vs. 2,785±867 for untreated; Fig. 5B).
FIG. 5.
IL-23 treatment maintains IFNAR2 expression in Huh7.5 cells. The number of Huh7.5 cells expressing the IFNAR2 receptor was determined by flow cytometry. (A) IFN-α down-regulated IFNAR2 while IL-23 treatment maintained IFNAR2 expression. (B) The mean fluorescent intensity of IFNAR2 was significantly decreased in IFN-α but was maintained for the combination of IL-23 and IFN-α. (A representative sample from three separate experiments is shown.)
Discussion
Our study demonstrated that IL-23 specifically enhances interferon signaling pathways and is associated with HCV clearance in HIV/HCV-coinfected subjects. IL-23 levels were higher in the plasma of SVR subjects than those who were nonresponders, and in vitro studies demonstrate multiple possible mechanisms by which IL-23 is linked to improved HCV clearance. IL-23 augments IFN-α-mediated suppression of HCV replication in the HCVcc system, possibly via enhanced ISG expression and maintenance of surface IFNAR2 expression. Hence, we describe a novel mechanism for HCV clearance mediated by IFN-α-based therapy.
IL-23 is a cytokine secreted by antigen-presenting cells such as dendritic cells (DCs) and is a member of the IL-12 heterodimeric cytokine family.15 The heterodimer has two subunits, p40 and p19, that are secreted by activated DCs in response to endogenous or exogenous stress.16 IL-23 promotes T cell proliferation by inducing IL-17 secretion and has been implicated in several autoimmune disorders such as psoriasis, inflammatory bowel diseases, and experimental autoimmune encephalomyelitis.17 While the majority of studies have assessed IL-23 in the context of autoimmune disease, other studies in chronic viral infections such as HCV and HIV have also suggested a role for IL-23. Elevated plasma IL-23 levels have been associated with chronic hepatitis C virus genotype 4 infection,18 and monocytes from chronic HCV patients have exaggerated IL-23 production in vitro.19 However, these studies do not assess response to therapy, and no data exist on IL-23 levels with HCV therapy, or the potential role of IL-23 as a mechanism of viral clearance. In this study, we indicate for the first time that IL-23 concentration differs with clinical outcome in HIV/HCV-coinfected subjects. These findings extend previous associations between HCV and IL-23, and suggest a potential for mechanistic linkage between IL-23 and viral clearance in the context of IFN therapy.
We investigated the biological mechanisms that may underlie the association between elevated IL-23 and SVR using an in vitro HCVcc system. Based on the clinical data, IL-23 did not have any effect on HCV replication since SVR and NR patients had comparable levels of HCV VL at baseline. However, when used in combination with rIFN-α in vitro, IL-23 synergistically suppressed HCV replication, suggesting a role for IL-23 in augmenting the IFN signaling pathway. Hence, it is possible that high levels of IL-23 could enhance IFN signaling in target cells and enhance antiviral activity. However, it was not clear whether this was a universal IL-23 effect of IL-23 or was applicable only in individuals who may be more responsive to IFN therapy, such as those who achieve SVR. Thus, we treated PBMCs from SVR and NR individuals with IL-23 with or without IFN-α, and found that IL-23 specifically induced IFIG expression to similar levels in SVR and NR subjects; however, many more IFIGs were up-regulated in the SVR group compared to the NR group. These results demonstrate that IL-23 induction of IFN responsiveness is not a universal phenomenon, and the pathogenic mechanisms that trigger nonresponsiveness may include a lack of IL-23 induction.
Finally, when we explored specific mechanisms involved in IL-23-mediated enhancement of IFN signaling, we found that IL-23 maintained the cell surface expression of the IFNAR2 receptor, yet after treatment with IFN-α alone, this receptor was quickly down-regulated. Our data demonstrate that IL-23 treatment of Huh7.5 cells results in sustained expression of IFNAR and not an increase in receptor expression. This is possibly due to an effect of IL-23 in preventing or delaying the down-regulation of IFNAR, which is consistent with the findings that there is no phosphorylation of IFNAR with IL-23 treatment. This observation suggests a unique mechanism by which IL-23 is able to amplify the type I IFN gene signature. STAT1 phosphorylation is a pivotal part of IFN-α signaling, and IL-23 by itself had no effect on STAT1 phosphorylation, confirming the fact that IL-23 may not render direct effects on IFN signaling pathways. Instead, IL-23 seems to act indirectly via an as yet uncharacterized mechanism to maintain IFNA2 receptor surface expression. This phenomenon is known as receptor cross-modulation, and offers new possibilities in enhancing clinical responsiveness to IFN therapy not just in HCV infection but in other diseases treated with IFN-α.
Our findings suggest that there is a close relationship between high IL-23 levels and subsequent responsiveness to IFN-based therapy for HCV. It is possible that high levels of IL-23 were merely a marker of IFN responsiveness which has been reported in HCV-infected subjects previously.20 However, it is unclear why IL-23 levels are increased in subjects who go on to become SVR. One possibility is that in HIV-infected patients with more advanced liver disease and those who are HCV/HIV coinfected, there is increased disruption of TH17 cell induction in GALT21 and this may lead to reduced IL-23 levels in such individuals.14 Indeed, there were more subjects with advanced liver disease in the NR groups than SVR.12 This may lead to a selective down-regulation of IL-23 and Th17 pathways. Down-regulation of Th17 cells could be associated with advanced liver disease, which thereby renders immune cells less sensitive to IFN-α pathways. Further studies are required to delineate the interactions between Th17 cells and the type I IFN response. Another possibility could be the indirect effects of IL-23 on T-regulatory cells. IL-23 has been shown to reduce T-regulatory cells, which have been implicated in the pathogenesis of chronic hepatitis C.22 In the absence of an IL-23 response, as observed in NRs, there is sustained T regulatory activity, which could blunt the host response to HCV antigens, thereby reducing the ability to clear HCV. Although we did not specifically study T-regulatory cell activity in these patients due to lack of samples, a clear association between persistently elevated T-reg activity in patients with advanced liver disease, such as NR in this study, has been previously demonstrated.22
The main limitation in our study was a lack of samples, which prevented us from performing more functional assays that would help us to further understand the intricate interactions between the IL-23-driven Th17 response and type I interferon responses, culminating in enhanced HCV clearance from plasma. Furthermore, in this study, we did not perform or obtain gut-associated lymphoid tissue (GALT) or assess the correlation with IL-23 levels. There are very few studies that have performed gut biopsies to evaluate TH17 cells in HIV infection; however, collectively they have demonstrated a disruption of mucosal immunity and specifically disruption of TH17 cells in HIV-infected subjects.21 In summary, our study demonstrates a novel mechanism of IL-23-driven IFN responsiveness in subjects prior to initiation of therapy, enabling them to achieve an SVR.
Supplementary Material
Acknowledgments
This research was supported in whole by the Intramural Research Program of the NIH (National Institute of Allergy and Infectious Diseases and the Critical Care Medicine Department, Clinical Center). We acknowledge Apath LLC for providing us with J6/JFH-1 HCV clone and Huh7.5 cells.
This work was presented as an oral presentation at the AASLD Liver meeting in November 2011 in Boston, MA.
The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hanafiah KM, Groeger J, Flaxman AD, and Wiersma ST: Global epidemiology of hepatitis C virus infection: New estimates of age-specific antibody to HCV seroprevalence. Hepatology 2013;57:1333–1342 [DOI] [PubMed] [Google Scholar]
- 2.Funk EK, Shaffer A, Shivakumar B, et al. : Interferon/ribavirin treatment for HCV is associated with the development of hypophosphatemia in HIV/hepatitis C virus-coinfected patients. AIDS Res Hum Retroviruses 2013;29(9):1190–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman KE, Rouster SD, Chung RT, and Rajicic N: Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: A cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis 2002;34:831–837 [DOI] [PubMed] [Google Scholar]
- 4.Davis GL, Alter MJ, El-Serag H, Poynard T, and Jennings LW: Aging of hepatitis C virus (HCV)-infected persons in the United States: A multiple cohort model of HCV prevalence and disease progression. Gastroenterology 2010;138:513–521 [DOI] [PubMed] [Google Scholar]
- 5.Benhamou Y, Bochet M, Di Martino V, et al. : Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology 1999;30:1054–1058 [DOI] [PubMed] [Google Scholar]
- 6.Singal AK. and Anand BS: Management of hepatitis C virus infection in HIV/HCV co-infected patients: Clinical review. World J Gastroenterol 2009;15:3713–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pawlotsky JM: The results of phase III clinical trial with telaprevir and boceprevir presented at the Liver Meeting 2010: A new standard of care for hepatitis C virus genotype 1 infection, but with issues still pending. Gastroenterology 2011;140(3):746–754 [DOI] [PubMed] [Google Scholar]
- 8.Sadler AJ. and Williams BR: Interferon-inducible antiviral effectors. Nat Rev Immunol 2008;8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui M, Moriya O, Belladonna ML, et al. : Adjuvant activities of novel cytokines, interleukin-23 (IL-23) and IL-27, for induction of hepatitis C virus-specific cytotoxic T lymphocytes in HLA-A*0201 transgenic mice. J Virol 2004;78:9093–9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohs I, Windmann S, Wildner O, Dittmer U, and Bayer W: Interleukin-encoding adenoviral vectors as genetic adjuvant for vaccination against retroviral infection. PloS One 2013;8:e82528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Town T, Bai F, Wang T, et al. : Toll-like receptor 7 mitigates lethal West Nile encephalitis via interleukin 23-dependent immune cell infiltration and homing. Immunity 2009;30:242–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L, Kottilil S, Lempicki R, et al. : Hepatic histologic response (HR) to combination therapy among HCV/HIV-coinfected individuals: Interferon induces HR independent of sustained virologic response (SVR). AIDS Res Hum Retroviruses 2006;22:1091–1098 [DOI] [PubMed] [Google Scholar]
- 13.Osinusi A, Rasimas JJ, Bishop R, et al. : HIV/hepatitis C virus-coinfected virologic responders to pegylated interferon and ribavirin therapy more frequently incur interferon-related adverse events than nonresponders do. J Acquir Immune Defic Syndr 2010;53:357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kottilil S, Yan MY, Reitano KN, et al. : Human immunodeficiency virus and hepatitis C infections induce distinct immunologic imprints in peripheral mononuclear cells. Hepatology 2009;50:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank AC, Zhang X, Katsounas A, Bharucha JP, Kottilil S, and Imamichi T: Interleukin-27, an anti-HIV-1 cytokine, inhibits replication of hepatitis C virus. J Interferon Cytokine Res 2010;30:427–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlig HH, McKenzie BS, Hue S, et al. : Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity 2006;25:309–318 [DOI] [PubMed] [Google Scholar]
- 17.Cua DJ. and Tato CM: Innate IL-17-producing cells: The sentinels of the immune system. Nat Rev Immunol 2010;10:479–489 [DOI] [PubMed] [Google Scholar]
- 18.El Husseiny NM, Fahmy HM, Mohamed WA, and Amin HH: Relationship between vitamin D and IL-23, IL-17 and macrophage chemoattractant protein-1 as markers of fibrosis in hepatitis C virus Egyptians. World J Hepatol 2012;4:242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JM, Shi L, Ma CJ, et al. : Differential regulation of interleukin-12 (IL-12)/IL-23 by Tim-3 drives T(H)17 cell development during hepatitis C virus infection. J Virol 2013;87:4372–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lempicki RA, Polis MA, Yang J, et al. : Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: Class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. J Infect Dis 2006;193:1172–117 [DOI] [PubMed] [Google Scholar]
- 21.Sandler NG. and Douek DC: Microbial translocation in HIV infection: Causes, consequences and treatment opportunities. Nat Rev Microbiol 2012;10:655–666 [DOI] [PubMed] [Google Scholar]
- 22.Ferri S, Lalanne C, Lanzoni G, et al. : Redistribution of regulatory T-cells across the evolving stages of chronic hepatitis C. Dig Liver Dis 2011;43:807–813 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.