Abstract
To analyze HIV-1 genetic variants in Kazakhstan, HIV-1 sequences were obtained from 205 antiretroviral-treated (ART) and naive patients in 2009–2013. Samples were collected in the most populous cities and provinces of Kazakhstan. On the basis of phylogenetic analyses of partial pol sequences, subtype A variant intravenous drug user (IDU)-A (which is dominant in the former Soviet Union) was found in 60.0% of the individuals, followed by CRF02_AG (34.6%); the rest of the samples were subtype B, CRF03_AB, CRF63_02A1, and CRF07_BC. The proportion of CRF02_AG has increased significantly since 2001–2003, when it was less than 5%. The majority of the CRF02_AG cases were found in Almaty, the former capital and the most populous city in Kazakhstan. The IDU-A variant dominated in the industrial regions of northern and central Kazakhstan and some other regions. Both dominant HIV-1 genetic variants were almost equally represented in the two main transmission groups: IDUs and heterosexuals. The analysis of drug-resistant mutations found a low prevalence of drug resistance in 165 therapy-naive individuals (3.0%). Thus, in the beginning of the second decade of the 2000s, the HIV epidemic in Kazakhstan is driven by two main genetic variants: IDU-A and CRF02_AG.
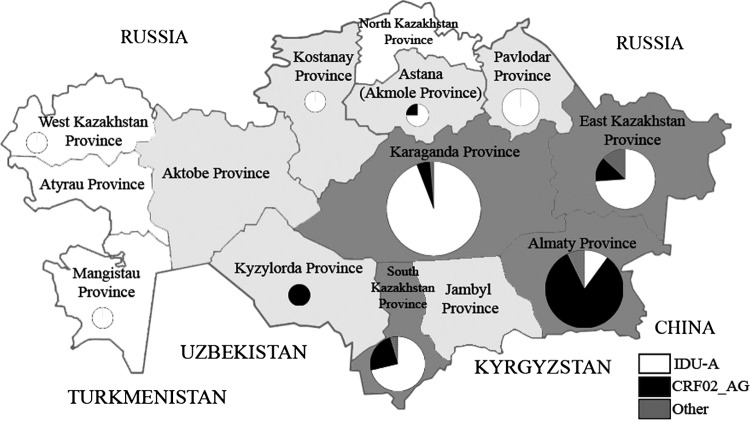
The epidemic of HIV infection in Kazakhstan, a former USSR (FSU) republic, started in 1997 when the first HIV case was registered in East Kazakhstan. At the time, massive HIV epidemics among intravenous drug users (IDUs) were already raging in Ukraine and Russia. As studies have shown,1,2 these epidemics were highly homogeneous and were caused by a monophyletic variant of subtype A1 that originated in Central Africa3,4 named IDU-A. After a short delay, HIV infection began spreading explosively among IDUs in Kazakhstan, and by December 2001, the number of HIV cases had reached 1,800, with IDUs constituting more than 90%.5 The most severely affected regions of the country were Karaganda, Pavlodar, and Shymkent (South Kazakhstan) (Fig. 1). The first study of HIV subtypes in Kazakhstan was conducted in these cities in 2001–2002,6 and the same IDU-A variant was present in all 43 samples studied. The detection of some genetic markers pointed to the possibility that two independent epidemic sources in Ukraine and Russia caused the explosions in Pavlodar, North Kazakhstan, and in Karaganda, Central Kazakhstan, respectively.
FIG. 1.
Map of Kazakhstan showing the distribution of the main HIV-1 genetic variants in the territories studied. The color corresponds to the number of people living in the region (white, <700,000; light gray, 700,000–1,200,000; dark gray, >1,200,000). The diameter of the circles corresponds to the number of sequences analyzed.
Since 2000, the HIV epidemic in Central Asia (including Kazakhstan, Uzbekistan, and other areas) has been accelerating. This growth is most likely attributable to escalating injection drug use, which, in turn, results from the intensification of drug trafficking in this region.7 Ten years later, most of the cumulative HIV cases were still associated with drug use, although infections were increasing rapidly in heterosexuals, who accounted for 50% or more of the proportion of new infections.8,9 A 2007 study7 involved mainly IDUs (98.1%) in different cities and provinces including Pavlodar, East Kazakhstan, Almaty, South Kazakhstan, Karaganda, and Mangistau.
The results confirmed the predominance of IDU-A in this group of patients (94.1%); additionally, four individuals infected with a CRF02_AG recombinant (4.7%) and one infected with subtype C (1.2%) were found. Among the CRF02_AG-infected individuals, three were from Almaty province and one was from South Kazakhstan; all were heterosexuals. The CRF02_AG variants were previously found in neighboring Uzbekistan,10 so this finding came as no surprise to the investigators; all of the Kazakh strains clustered with strains from Uzbekistan, reflecting the increasing distribution of CRF02_AG in Central Asia.
By 2011, the number of people living with HIV in Kazakhstan reached 17,760. The average prevalence was 12.5 per 100,000 of the total population.11 Among the new HIV cases in 2011 for which the mode of transmission was known (97%), 52.6% were infected heterosexually, 45.1% through injecting drug use, 1.4% through sex between men (MSM), and 0.9% through mother-to-child transmission (MTCT). The MSM group is traditionally hidden for diagnostic access, so the number and the proportion of HIV-infected MSMs are probably understated.
The medical authorities reported a decrease in incidence in some regions in 2012 compared with 2011; nevertheless, the prevalence of HIV is still the highest in Pavlodar and Karaganda, with 180 and 152 infections per 100,000, respectively. The epidemic in Almaty (the former capital and the largest city in Kazakhstan) is growing quickly as well. Antiretroviral treatment (ART) was introduced in the beginning of the 2000s, but the coverage (CD4<350) is still insufficient and comes to only approximately 35%.12
In this study, we present the current distribution of HIV-1 subtypes among the samples collected in 2009–2013 and compare them to the data reported previously. The aim of the work was to estimate the possible changes in the molecular landscape of the epidemic of HIV infection in Kazakhstan considering the growth of the epidemic and the increasing use of ART.
The total number of patients involved in this study was 205. Samples were collected in the cities of Astana, Karaganda, Kostanay, Kyzylorda, Mangistau, Pavlodar, and Almaty as well as in Almaty, East Kazakhstan, South Kazakhstan, and West Kazakhstan provinces (Table 1). The average age of the patients was 33 years (2 months to 53 years); 117 were male and 88 were female. All of the participants or their parents signed an informed consent form. Drug use was reported as the main risk of HIV acquisition by 88 persons (42.9%); 107 were infected via heterosexual transmission (52.2%) and the other cases were reported as MTCT (7), MSM (1), blood transfusion (1), or unknown (1).
Table 1.
The Distribution of HIV-1 Genotypes by Regions and Route of Transmission (2009–2013)
| Genotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IDU-A | CRF02_AG | Other genotypesa | ||||||||
| Region | IDUs | Hetero | Other | IDUs | Hetero | Other | IDUs | Hetero | Other | Total number |
| East Kazakhstan | 13 | 4 | 0 | 2 | 1 | 0 | 3 | 0 | 0 | 23 |
| Almaty | 1 | 4 | 2 | 25 | 31 | 2 | 1 | 4 | 0 | 70 |
| Pavlodar | 5 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 12 |
| Karaganda | 27 | 35 | 2 | 1 | 2 | 0 | 0 | 1 | 0 | 68 |
| South Kazakhstan | 4 | 11 | 0 | 0 | 5 | 0 | 0 | 1 | 0 | 21 |
| Other regions | 4 | 3 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 11 |
| Total | 54 | 63 | 6 | 28 | 41 | 3 | 4 | 6 | 0 | 205 |
Subtype B, CRF03_AB, CRF63_02A1, and CRF07_BC recombinants.
IDU, intravenous drug user; Hetero, heterosexual.
There were two sets of data obtained: the first set came from samples collected in 2009 from 50 confirmed HIV-infected naive patients only and was transported to the Moscow laboratory, and the second set of genotypes (155) was analyzed by local specialists in Almaty in 2012–2013 using both naive (115) and ART (40) patients. The proviral DNA samples collected in 2001–2002 (Pavlodar–5, Karaganda–3, and Shymkent–1) were kept in the laboratory at −70°C.
Total RNA and proviral DNA were extracted using the DNA/RNA Extractor Quicube (Qiagen, Germany) and the QIAamp DNA Blood Mini Kit (Qiagen, USA) according to the manufacturer's instructions. The DNA extracted from the peripheral blood mononuclear cells (PBMCs) of the samples collected in 2009 was analyzed using an in house set of primers13,14; as a result, sequences of the pol gene Pro (1–99 aa) (2253–2549 nucleotides with the HXB2 strain as the reference) and reverse transcriptase (RT) (45–241 aa) (2682–3272 nucleotides) regions were obtained. Sequences from Pro and RT individual primers were assembled using BioEdit, version 7.0.5.3. The 2012–2013 group of samples was studied by performing an RNA analysis using the AmpliSens HIV-Resist-Seq commercial test (InterLabService, Russia) for full Pro and partial RT region sequences, with 1302 nucleotides in total. All of the pol gene fragments were directly sequenced using the genetic analyzer ABI Prism 3130 (Applied Biosystems, USA). The alignment and phylogenetic analyses of the sequences were performed using MEGA5.015 and TreeMaker tool PhyML (www.hiv.lanl.gov). Genotyping and drug resistance mutation analyses were carried out using the online programs COMET HIV-1/2 and HCV v.0.2 (http://comet.retrovirology.lu/) and HIVdb Program v.6.2.0 (http://hivdb.stanford.edu/pages/algs/sierra_sequence.html), respectively.
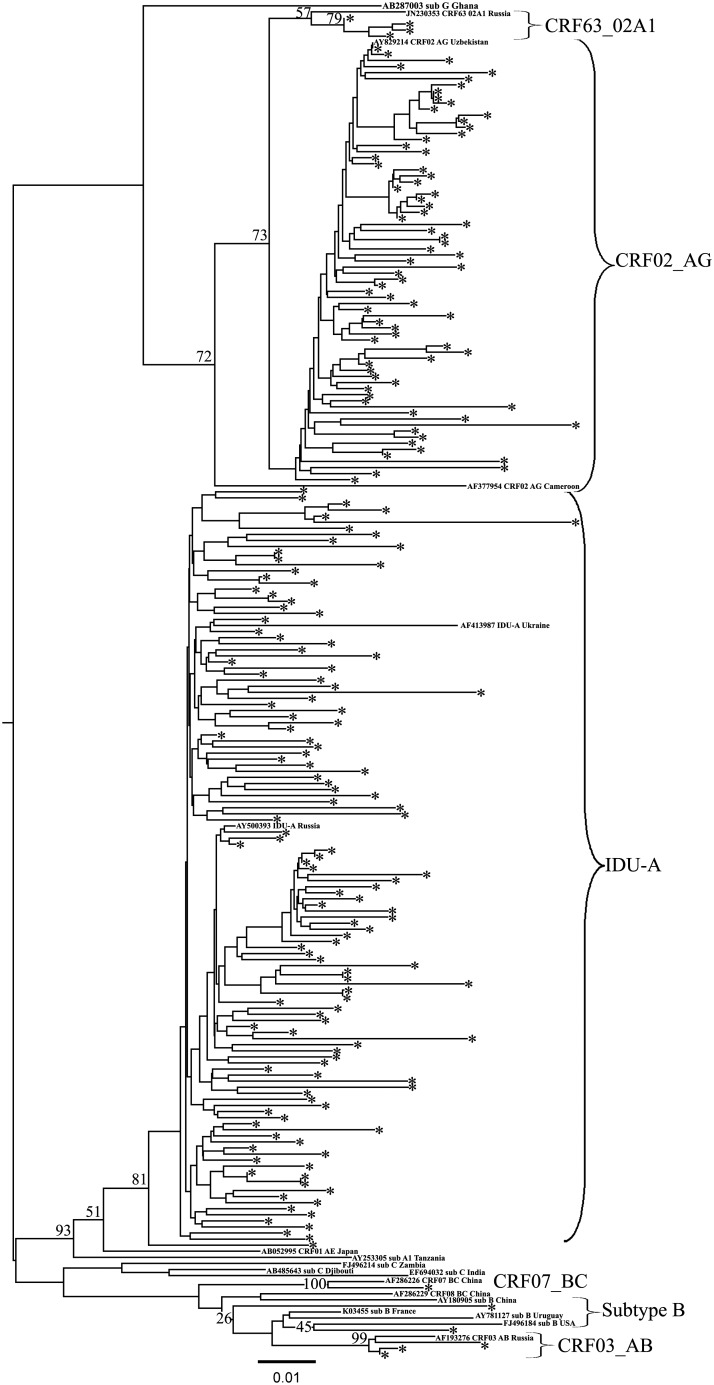
In the 2009 collection, 24 (48.0%) samples were subtyped by COMET as A1, two (4%) were CRF03_AB, and 24 (48.0%) were CRF02_AG. According to the phylogenetic analysis of the Pro-RT region, all of the A1 samples clustered with IDU-A variant samples from FSU countries (Fig. 2, AF413987 Ukraine and AY500393 Russia). Among the CRF02_AG samples 21 formed the common phylogenetic branch with recombinant variants collected in Uzbekistan in 1999–200016 (Fig. 2, AY829214). Three samples were found to belong to the newly described genetic variant CRF63_02A1.17 Two CRF03_AB samples were obtained from a male and a female who were possibly sexual partners; both reliably matched the CRF03_AB variant circulating in Eastern Europe and Kaliningrad.18 Most of the IDU-A-infected (21/24; 87.5%) and the CRF02_AG-infected patients (13/21; 61.9%) as well as all CRF63_02A1-infected patients reported drug use as the main route of infection.
FIG. 2.
Phylogenetic analysis of HIV-1 pol sequences (joined Pro-RT region) from 2009–2013 study individuals. The phylogenetic tree was constructed using the PhyML tool by the maximum likelihood method of phylogeny, the bootstrap method of branch support (100 replicates), and the GTR + gamma nucleotide substitution model. The optimization of the phylogenetic tree was carried out by FigTree v.1.3.1. Sequences obtained in this study are labeled as snowflakes. Bootstrap values as percentages are indicated at the nodes.
Among the samples analyzed in 2012–2013, 99 (63.9%) were identified as an IDU-A variant, 51 (32.9%) of the viruses were CRF02_AG, one was a CRF63_02A1 strain, and other genetic variants were identified in four (2.6%) samples. Of the four patients with other variants, two heterosexual men were infected with HIV-1 subtype B; a 42-year-old heterosexual man from Almaty harbored a CRF07_BC virus (confirmed by COMET HIV-1/2 and HCV v.0.2) and a female IDU, 51, from Almaty was infected with CRF03_AB in 2003 in Kaliningrad. In contrast to 2009, both IDU-A (60/99; 60.6%) and CRF02_AG patients (32/51; 62.7%) were infected mainly through sexual contact. Overall, from 2009 to 2013, both genotypes were almost equally distributed among IDUs and heterosexuals: drug use was the main route of transmission in 43.9% of IDU-A patients and 38.9% of CRF02_AG patients (Table 1).
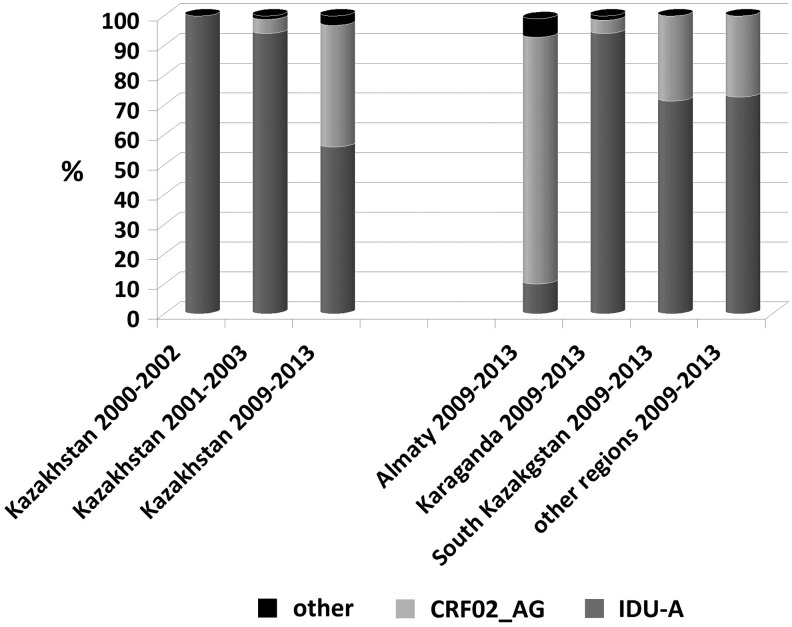
The distribution of the dominant HIV-1 genetic variants (IDU-A and CRF02_AG) among the administrative territories was highly unequal. In 2009, of the 22 samples collected in Almaty city and the province, 20 were the CRF02_AG variant (90.9%); in 2012–2013, the predominance was still pronounced (38/48; 77.1%). However, the IDU-A variant in Karaganda accounted for 100% (10/10) of the cases in 2009 and the vast majority (54/58; 93.1%) of the cases in 2012–2013. In South Kazakhstan as well in all other regions, IDU-A viruses were the most prevalent both in 2009 (14/18; 77.8%) and in 2012–2013 (38/49; 77.6%) (Fig. 3).
FIG. 3.
The comparative prevalence of HIV-1 genetic variants in Kazakhstan as reported in 2004,6 in 2007,7 and in the present study. The data published in 2004 resulted from the samples collected in Pavlodar, Karaganda, and Shymkent in 2000–2001. The publication of 2007 was based on the study of the strains found in different regions including Almaty in 2001–2003. The present study involved all the regions investigated before. The chart shows the ratio of the main HIV-1 variants—IDU-A and CRF02_AG—in the regions where the greatest numbers of samples were studied in 2009–2013.
The analysis of drug resistance found a low prevalence of drug-resistant mutations in 165 therapy-naive individuals. By applying the WHO TDRMs surveillance list 200919 we obtained a total value of 3.0%. It should be noted that not all of the patients were recently infected, so the analysis did not satisfy the WHO requirements, and this figure did not reflect the level of transmitted resistance correctly. As applied to the Pro region of the pol gene, there were two IDU-A strains each harboring single mutations: D30N associated with nelfinavir (NFV) resistance and M46L reducing susceptibility to many protease inhibitors. With regard to the RT region, three samples should be noted, each harboring M184I, K103N, and G190S single mutations. Some substitutions characteristic of the HIV-1 IDU-A variant were found in IDU-A samples, such as A62V (31.7%) in the RT region and V77I (34.2%) in the Pro region. In addition, 48/72 (66.7%) CRF02_AG samples had the K20I substitution characteristic of HIV-1 subtype A and G.
Among 40 ART patients most (33/40, 82.5%) were treated with the combination zidovudine/lamivudine (AZT/3TC); the third drug was nevirapine (NVP), efavirenz (EFV), or lopinavir/ritonavir (LPV/r). The rest of the patients received the alternative scheme tenofovir/emtricitabine/lopinavir/ritonavir (TDF/FTC/LPV/r). The analysis of HIV genotypes detected the major mutations in 15 patients only; the other 22 (62.5%) had the “wild” genotype, which is most likely attributed to the low level of adherence. The list of mutations was typical with regard to the combination used and included M41L, M184V, L210W, T215F/Y, K103N, K101E, V179D, G190A/S, and V106A (Table 2; the percentage of mutations was calculated with respect to the sequences with at least one major mutation).
Table 2.
The Major Drug Resistance Mutations in Antiretroviral-Treated Patients (n=15)
| Number of mutations | % | |
|---|---|---|
| NRTI | ||
| M41L | 3 | 20.0 |
| M184V | 11 | 73.3 |
| L210W | 1 | 6.7 |
| T215F/Y | 3 | 20.0 |
| NNRTI | ||
| K103N | 5 | 33.3 |
| K101E | 4 | 26.7 |
| V179D | 1 | 6.7 |
| G190A/S | 5 | 33.3 |
| V106A | 1 | 6.7 |
NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor.
Thus, the epidemic of HIV infection in Kazakhstan has been gradually changing. The first data on HIV-1 subtypes in Kazakhstan were obtained in 2001–2002.6 Forty-three samples were collected in Pavlodar, Karaganda, and Shymkent (South Kazakhstan province), where the HIV infection was the most prevalent, and were studied using the gag/env heteroduplex mobility assay and selective sequencing of the env gene.6 In this work, the RT genome region was reexamined in nine samples collected in 1999–2002 to be certain that they were not actually recombinant viruses that were misinterpreted as A1 subtypes. The sequences of the CRF02_AG and IDU-A variants circulating in Russia were used as references for the phylogenetic analysis. The results demonstrated that all samples with certainty were the IDU-A variant (data not shown). Thus, the epidemic of HIV infection started in the industrial regions in northern and central Kazakhstan, with the IDU-A variant dominant (Fig. 3). These regions bordered Russia, where this genetic variant was almost 100% prevalent at that time.
The next data on HIV infection molecular epidemiology in Kazakhstan came in 2007,7 when the results of sequencing the part of the pol gene in 85 HIV samples from Pavlodar, East Kazakhstan, Almaty, Karaganda, South Kazakhstan, Karaganda, and Mangistau (2001–2003) were described. Of the 80 individuals infected with subtype A, 73 (91.3%) reported injection drug use as the highest risk of infection, and for 5 of 80 subjects (6.3%) it was sexual transmission. By that time, CRF02_AG was already found in Uzbekistan,16 and in this study, CRF02_AG was first mentioned in the most populous regions of Kazakhstan. As shown, subtype A was the most common genetic variant (94.1%), followed by CRF02_AG (4.7%) and subtype C (1.2%). The four individuals infected with CRF02_AG were self-declared IDUs. It is worth noting that among them, three were from Almaty province and one was from South Kazakhstan. According to the authors' comments, “Kazakhstan subtype A is not genetically distinct from reference sequences” of the IDU-A strain. As to the CRF02_AG strains, although they “were only sequenced in part of pol, they clustered with similar strains identified in Uzbekistan, and it is most likely that they reflect the spread of that epidemic to Kazakhstan.”
Thus, in 2001–2003, the epidemic in Kazakhstan was an integral part of the expanding subtype A epidemic in the former USSR, and CRF02_AG was starting its spread in the country. These events coincided with a change in the transmission of HIV infection when the heterosexual route began to increase rapidly. Taken together, these facts most likely led to the consequences that are reflected in the results of the present study. The most notable finding is the rapid growth of CRF02_AG, which is currently catching up with IDU-A in the proportion of cases it causes.
The last variant still dominates in the regions where the epidemic started more than 10 years ago, but in some densely populated southeastern regions, including Almaty city and province, the HIV epidemic is driven mainly by the recombinant form of CRF02_AG (Fig. 3). This variant is almost equally distributed between IDUs and heterosexuals, with the proportion of women accounting for 40.1%. The rapid spread of CRF02_AG may be partially due to the probable increase in its pathogenic fitness,20 and prompts the question about strengthening prevention programs.
With regard to other HIV-1 genetic forms, none has become widely distributed in Kazakhstan. Subtype B and the recombinant CRF03_AB are occasionally found elsewhere in the former USSR, and their presence in Kazakhstan could be predicted. The only interesting exception is CRF07_BC, which was found in Almaty. The genetic variants commonly found in China have long been expected in Russia and other former USSR countries bordering China. Nevertheless, no traces of CRF07_BC have been observed until recently, for example, in the study of the Russian Far East region21 that has close contact with China. It is possible that this finding may foreshadow some upcoming events in the HIV epidemic in Central Asia and other FSU countries. Further investigations including full-length sequencing will provide new insights into the genetic diversity of HIV-1 in this “hotspot”8 region.
Sequence Data
The GenBank accession numbers for the sequences described in this article are HQ739036–HQ739040, HQ739042, HQ739045–HQ739050, JF682734–JF682738, JF682742–JF682746, JF682748–JF682751, JF682755, JF682756, JF682758–JF682764, JF718184–JF718209, JF718211–JF718213, JF718215–JF718226, KJ396596–KJ396604, KC123195–KC123197, KC156527–KC156531, KC215144–KC215157, KF194217, KF208426, KF498449–KF498527, KF512380–KF512470, KF554429–KF554452, KF678858–KF678860, and KF588572–KF588634.
Acknowledgments
We thank our colleagues from the Kazakh Republic Center for AIDS Prevention and Control, Almaty, for collecting the blood samples and epidemiological data. The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under the project “Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN)”—grant agreement N 223131. The study was supported in part by the International Science and Technology Center, project 3826, and the Ministry of Education and Science of the Russian Federation, projects 8148, 8154, and 8479.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bobkov A, Kazennova E, Selimova L, et al. : A sudden epidemic of HIV type 1 among injecting drug users in the former Soviet Union: Identification of subtype A, subtype B, and novel gagA/envB recombinants. AIDS Res Hum Retroviruses 1998;14(8):669–676 [DOI] [PubMed] [Google Scholar]
- 2.Bobkov AF, Kazennova EV, Selimova LM, et al. : Temporal trends in the HIV-1 epidemic in Russia: Predominance of subtype A. J Med Virol 2004;74(2):191–196 [DOI] [PubMed] [Google Scholar]
- 3.Riva C, Romano L, Saladini F, et al. : Identification of a possible ancestor of the subtype A1 HIV Type 1 variant circulating in the former Soviet Union. AIDS Res Hum Retroviruses 2008;24(10):1319–1325 [DOI] [PubMed] [Google Scholar]
- 4.Thomson MM, de Parga EV, Vinogradova A, et al. : New insights into the origin of the HIV type 1 subtype A epidemic in former Soviet Union's countries derived from sequence analyses of preepidemically transmitted viruses. AIDS Res Hum Retroviruses 2007;23(12):1599–1604 [DOI] [PubMed] [Google Scholar]
- 5.UNAIDS: Report on the global AIDS epidemic 2012. [cited August27, 2013]. Available from www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2012/gr2012/20121120_UNAIDS_Global_Report_2012_en.pdf
- 6.Bobkov AF, Kazennova EV, Sukhanova AL, et al. : An HIV type 1 subtype A outbreak among injecting drug users in Kazakhstan. AIDS Res Hum Retroviruses 2004;20(10):1134–1136 [DOI] [PubMed] [Google Scholar]
- 7.Eyzaguirre LM, Erasilova IB, Nadai Y, et al. : Genetic characterization of HIV-1 strains circulating in Kazakhstan. J Acquir Immune Defic Syndr 2007;46(1):19–23 [DOI] [PubMed] [Google Scholar]
- 8.Thorne C, Ferencic N, Malyuta R, et al. : Central Asia: Hotspot in the worldwide HIV epidemic. Lancet Infect Dis 2010;10(7):479–488 [DOI] [PubMed] [Google Scholar]
- 9.UNAIDS: Regional fact sheet 2012. Eastern Europe and Central Asia, Kazakhstan [cited August6, 2013]. Available from www.unaids.org/en/regionscountries/countries/kazakhstan/
- 10.Carr JK, Nadai Y, Eyzaguirre L, et al. : Outbreak of a West African recombinant of HIV-1 in Tashkent, Uzbekistan. J Acquir Immune Defic Syndr 2005;39(5):570–575 [PubMed] [Google Scholar]
- 11.World Health Organization: Key facts on HIV epidemic in Kazakhstan, 2011. Available from www.euro.who.int/__data/assets/pdf_file/0006/188754/Kazakhstan-HIVAIDS-Country-Profile-2011-revision-2012-final.pdf
- 12.UNAIDS: Report on the global AIDS epidemic 2012. [cited August27, 2013]. Available from www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2012/gr2012/20121120_UNAIDS_Global_Report_2012_en.pdf
- 13.Laga V, Kazennova EV, Vasil'ev AV, et al. : [Molecular-genetic characterization of the HIV-1 variants abundant in Kirghizia]. Vopr Virusol 2012;57(5):26–32 [PubMed] [Google Scholar]
- 14.Lapovok I, Laga V, Vasilyev A, et al. : [Molecular-genetic analysis of pol gene region coding HIV-1 integrase in patients from Russia and Ukraine]. HIV Infect Immunosuppress Disord 2012;4(2):73–81 [Google Scholar]
- 15.Tamura K, Peterson D, Peterson N, et al. : MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28(10):2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurbanov F, Kondo M, Tanaka Y, et al. : Human immunodeficiency virus in Uzbekistan: Epidemiological and genetic analyses. AIDS Res Hum Retroviruses 2003;19(9):731–738 [DOI] [PubMed] [Google Scholar]
- 17.Gashnikova NM, Totmenin AV, Bocharov EF, et al. : [New recombinant variant of human immunodeficiency virus of type 1, subtype envB/envA, isolated in Novosibirsk]. Zh Mikrobiol Epidemiol Immunobiol 2004;(5):53–58 [PubMed] [Google Scholar]
- 18.Liitsola K, Tashkinova I, Laukkanen T, et al. : HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS 1998;12(14):1907–1919 [DOI] [PubMed] [Google Scholar]
- 19.Bennett DE, Camacho RJ, Otelea D, et al. : Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009;4(3):e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tebit DM, Nankya I, Arts EJ, et al. : HIV diversity, recombination and disease progression: How does fitness “fit” into the puzzle? AIDS Rev 2007;9(2):75–87 [PubMed] [Google Scholar]
- 21.Laga M, Kazennova E, Vasilyev A, et al. : HIV-1 Genetic variants in Vladivostok, Russia. In 20th Conference on Retroviruses and Opportunistic Infections Atlanta, GA, March3–6, 2013 Abstract book, paper 503 [Google Scholar]