Abstract
Background
Sterol 14α-demethylase (cytochrome P450 51, CYP51, P45014DM) is a microsomal enzyme that in eukaryotes catalyzes formation of sterols essential for cell membrane function and as precursors in biosynthesis of steroid hormones. Functional properties of CYP51s are unknown in non-mammalian deuterostomes.
Methods
PCR-cloning and sequencing and computational analyses (homology modeling and docking) addressed CYP51 in zebrafish Danio rerio, the reef fish sergeant major Abudefduf saxatilis, and the sea urchin Strongylocentrotus purpuratus. Following N-terminal amino acid modification, zebrafish CYP51 was expressed in Escherichia coli, and lanosterol 14α-demethylase activity and azole inhibition of CYP51 activity were characterized using GC/MS.
Results
Molecular phylogeny positioned S. purpuratus CYP51 at the base of the deuterostome clade. In zebrafish, CYP51 is expressed in all organs examined, most strongly in intestine. The recombinant protein bound lanosterol and catalyzed 14α-demethylase activity, at 3.2 nmol/min/nmol CYP51. The binding of azoles to zebrafish CYP51 gave KS (dissociation constant) values of 0.26 μM for ketoconazole and 0.64 μM for propiconazole. Displacement of carbon monoxide also indicated zebrafish CYP51 has greater affinity for ketoconazole. Docking to homology models showed that lanosterol docks in fish and sea urchin CYP51s with an orientation essentially the same as in mammalian CYP51. Docking of ketoconazole indicates it would inhibit fish and sea urchin CYP51s.
Conclusions
Biochemical and computational analyses are consistent with lanosterol being a substrate for early deuterostome CYP51s.
General Significance
The results expand the phylogenetic view of animal CYP51, with evolutionary, environmental and therapeutic implications.
Keywords: Sterol 14α-demethylase, CYP51, zebrafish, deuterostomes, sterol biosynthesis, azole inhibition
Introduction
The major sterol in vertebrates is cholesterol, the starting precursor molecule in the biosynthesis of hormones and other derivatives (e.g. androgens, estrogens, corticosteroids, vitamin D and bile acids) that play important reproductive, developmental, and physiological roles. Cholesterol is also an essential component of cell membranes, contributing to optimal membrane fluidity and permeability. The initial steps of sterol biosynthesis are conserved in the eukaryotes and derive from acetyl CoA, through to the biosynthesis of isopentenyl pyrophosphate and the condensation of six such molecules to form squalene. The cyclization of oxidosqualene results in formation of the first parental sterol molecule, which is enzymatically tailored by up to 20 biosynthetic steps to yield cholesterol. The third step in the post-squalene part of the pathway is catalyzed by sterol-14α-demethylase (cytochrome P450 family 51; CYP51; P45014DM; EC 1.14.13.70). CYP51 has been described in mammals, as well as in fungi, plants and microbes [1]. We address CYP51 in non-mammalian deuterostomes.
In mammals CYP51 catalyzes the oxidative removal of the methyl group at the C14α-position of lanosterol and dihydrolanosterol. This monooxygenase reaction consists of three sequential steps converting the methyl group to the hydroxymethyl- and then carboxyaldehyde- intermediates, followed by the elimination of formic acid and concomitant formation of the ⊗14,15 double bond in the resulting sterol product, 4, 4-dimethyl-5〈-cholesta-8,14,24-triene-3ßol (Figure 1). This product is converted in several additional steps to cholesterol. The two intermediate products of the CYP51 reactions, 4, 4-dimethyl-5〈-cholesta-8,14,24-triene-3ß-ol (follicular fluid meiosis activating sterol; FF-MAS), and a reductase product of FF-MAS, termed testis-meiosis activating sterol (T-MAS), have been suggested to stimulate the resumption of meiosis in mouse oocytes [2-4].
Figure 1.
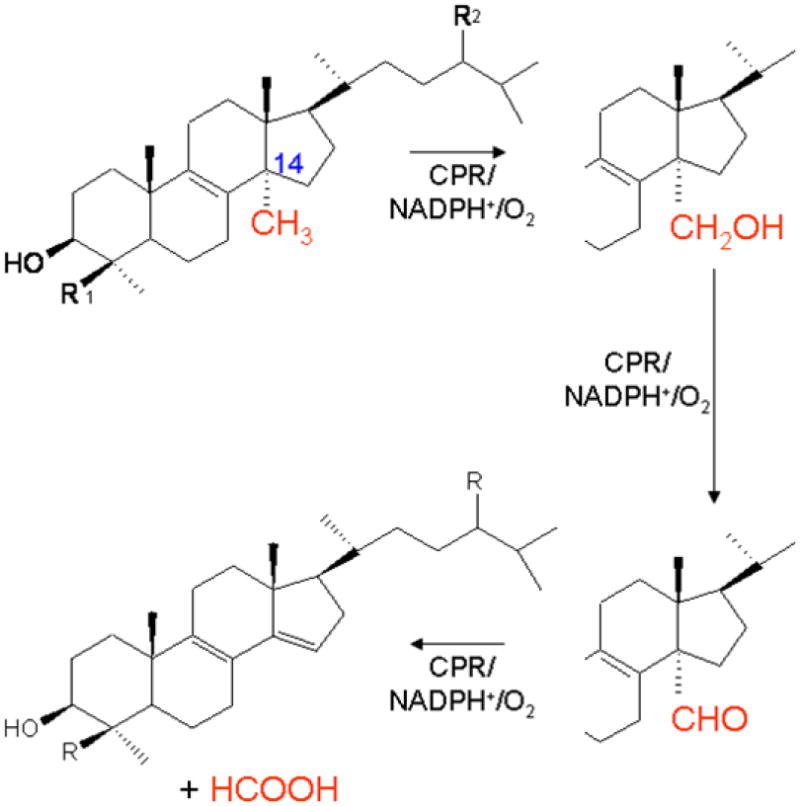
The ubiquitous sterol 14α-demethylase (CYP51) biochemical reaction, an essential step in sterol biosynthesis.
CYP51 is an essential enzyme; deletion of CYP51 is embryo-lethal in mice [5], and disruption of cholesterol biosynthesis can have adverse consequences for reproduction, digestion, growth, and cell maintenance [2-4, 6]. Given the essential nature of sterols and the pivotal role of CYP51 in their biosynthesis, this enzyme is an important drug target. Azole drugs that inhibit CYP51 are agents of choice to treat fungal and protozoal infections of humans, animals and plants, and have potential to be used to treat fungal pathogens of fish. Azole N-heterocycle nitrogen binding to the P450 heme Fe blocks enzyme activity, leading to the accumulation of 14α-methyl sterol precursors and preventing pathogen growth. Antifungal agents, however, can adversely affect the host, or non-target species, through inhibition of host/non-target CYP51 or other CYP enzymes [7, 8].
In this paper we report cloning of cDNAs for CYP51s from three non-mammalian deuterostomes, the biomedical model zebrafish (Danio rerio), a globally distributed coral reef fish sergeant major (Abudefduf saxatilis), and the sea urchin (Strongylocentrotus purpuratus), an echinoderm in the earliest deuterostome lineage. The phylogeny of CYP51 was re-assessed with emphasis on the relationship of CYP51s among animals. The zebrafish CYP51 cDNA sequence was expressed in Escherichia coli, and the recombinant protein used to investigate catalytic activity. Given azole interaction with other CYP51s, we explored the affinity of recombinant zebrafish CYP51 to a therapeutic imidazole (ketoconazole) used to treat fungal infections in the clinic, and an agricultural triazole (propiconazole), which occurs as an environmental contaminant. Molecular modeling and docking studies were employed to assess similarity in function between echinoderm and early vertebrate CYP51s, and the mammalian enzymes.
Materials and Methods
Chemicals
Lanosterol was purchased from Sigma Chemicals (St. Louis, MO). BSTFA was obtained from Suppelco (Bellefonte, PA). Propiconazole was generously provided by Syngenta Crop Protection, Inc. (Greensboro, NC). Authentic FF-MAS standard was generously provided by Drs. Mogens Baltson and Anne Grete Byskov. All other chemicals were of the highest quality and were purchased from Fisher Scientific.
Animals and RNA
Zebrafish, (D. rerio) Tupfel Long-fin, were obtained as before [9], and liver was obtained from adult male fish. Sergeant major (A. saxatilis), were collected in Bermuda, and liver and gonad tissues were obtained from two male and one female fish. Fish were placed on ice and then killed by cervical section. The protocols were approved by the WHOI Institutional Animal Care and Use Committee. Tissues were weighed and immediately placed in RNAlater (Ambion). Pacific purple sea urchins (S. purpuratus) were obtained from Marine Research and Educational Products (Escondido, CA). All sea urchins were female. The ovary and viscera were removed and placed in RNAlater. Messenger RNA was prepared from zebrafish liver tissue using the MicroPolyA Pure kit (Ambion). Total RNA was prepared from A. saxatilis liver and testes and from sea urchin tissues using RNA STAT-60 (Tel-Test).
cDNA cloning and sequencing
RACE and PCR specific primers used for all three species are shown in Supplemental Table S1. RACE-ready cDNA was prepared using Powerscript Reverse Transcriptase (Clontech) according to the manufacturer's recommendations. For zebrafish CYP51, specific PCR primers, 51F345 and 51R1059, were designed based on CYP51 fragments from the zebrafish EST database (NCBI). For sergeant major, degenerate primers were designed based on highly conserved regions of known mammalian CYP51 sequences and the zebrafish EST partial CYP51 sequences (see Supplemental Table S1) yielding a PCR fragment of approximately 170 bp. Sequences of the PCR fragments were confirmed as encoding CYP51 from BLAST results against the NCBI database. For zebrafish and sergeant major, the initial CYP51 fragments generated by PCR were used to design nucleotide primers for use with the SMART RACE kit (Clontech). For sea urchin, specific primers for PCR and SMART RACE were designed from EST fragments of CYP51 obtained from the NCBI website (http://www.ncbi.nlm.nih.gov/).
A full-length cDNA sequence was obtained from zebrafish liver, using the forward primer 51F20dr and the reverse primer 51R1681dr (Supplemental Table S1). A full-length cDNA sequence was obtained from sergeant major liver using the forward and reverse primers indicated in Table S1. Sea urchin cDNA was generated from viscera total RNA, using the Powerscript Reverse Transcription kit, following the recommended protocol. A full-length cDNA CYP51 sequence was obtained using the forward primer, 51F175sp, and the reverse primer, 51R1721sp (Supplemental Table S1) with Deep Vent polymerase according to the manufacturer's recommendations, with the addition of 5% (v/v) DMSO.
The PCR and RACE products were cloned into the pGEM-T Easy Vector (Promega) and sequenced. Sequencher software (Gene Codes Corporation) was used for sequence analysis.
Sampling of tissues for quantitative real time PCR
Adult zebrafish were anaesthetized by MS222 and killed by decapitation, and multiple tissues were obtained following a dissection protocol similar to Gupta and Mullins [10]. Three replicates were collected for both males and females, resulting from four individuals pooled per replicate for each organ. The dissected organs were flash frozen in liquid nitrogen and were stored at -70 °C until RNA isolation.
Quantitative real time PCR
Quantitative real time PCR was performed using the iQ SYBR Green Supermix (BioRad) on a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad), according to the manufacturer's instructions. A CYP51 primer pair for real time PCR (forward, 5′-AAGTTTGCTTACATCCCGTTTGG-3′; reverse, 5′-GATGGTCTTGATCTGGACGTAGG-3′) was synthesized by Eurofins MWG Operon (Huntsville, AL, USA). A melt curve analysis was performed at the end of each PCR run to ensure that a single product was amplified. Sequencing of the amplicon confirmed that the right product was amplified by qPCR. Relative mRNA expression of the CYP51 gene was normalized to that of arnt2 and ef1α and comparisons of tissue transcript abundance were then conducted, as previously published (E−ΔΔCt, as before [11]). PCR efficiencies (E) for within-experiment amplicon groups were determined by the LinRegPCR program [12, 13].
Heterologous expression of zebrafish CYP51 in E. coli: plasmid preparation
The bacterium, Escherichia coli, was selected for expression of zebrafish CYP51. Unlike mammalian and yeast cell lines, E. coli do not have any CYPs, including CYP51. Therefore P450 expression in E. coli would necessarily be the result of induced heterologous expression of the recombinant CYP51. Expression of eukaryotic genes in a prokaryotic organism requires an appropriate plasmid and modifications of the gene such that the bacterial transcription and translation machinery function properly [14, 15]. Following the recommendations of Barnes [14], modifications were made to the first eight codons of the zebrafish CYP51 cDNA (Table 1) before cloning it into the pCWOri+ plasmid vector (a gift of Dr. Michael Waterman).
Table 1.
N-terminus nucleotide modifications to the first 8 amino acids in the zebrafish cDNA sequence to enhance expression in E. coli.
| Modification | Sequence (5′ → 3′) | |||||||
|---|---|---|---|---|---|---|---|---|
| Native | M | T | I | L | E | V | G | S |
| ATG | ACG | ATC | CTG | GAG | GTG | GGC | AGT | |
| CYP51ZF | M | A | I | L | E | V | G | S |
| ATG | GCT | ATT | TTA | GAA | GTA | GGA | AGT | |
| CYP51ZF17 | M | A | L | L | L | A | V | F |
| ATG | GCT | CTG | TTA | TTA | GCA | GTT | TTT | |
| CYP51ZFT | Replacement of first 35 amino acids with start codon (ATG) | |||||||
Three zebrafish CYP51 cDNA sequences (CYP51ZF, CYP51ZF17, and CYP51ZFT) were prepared, with slight modifications for expression in E. coli. CYP51ZF modifications included substitution of the second codon to encode the amino acid alanine and A-T enrichment of the next six codons. The 3′-terminus of CYP51ZF (but not CYP51ZF17 or CYP51ZFT) was also modified, with the addition of four codons for histidine immediately preceding the stop codon. (The histidine tag was inserted to facilitate purification by Ni+ affinity columns although eventually purification was accomplished without Ni+ affinity columns). CYP51ZF17 contained a modification that replaced the first eight amino acid codons with those of modified bovine CYP17. The final CYP51, CYP51ZFT, encoded a truncated CYP51 sequence produced by removing the first 35 amino acids of the 5′-terminus, which correspond to the membrane anchor sequence of the CYP51 protein.
Several full-length zebrafish CYP51 clones were sequenced and one with no PCR errors was chosen for expression in E. coli. Modified cDNA sequences were constructed by PCR using this plasmid as template. The primers for each modification are shown in Supplemental Table S2. PCR cycling conditions are shown in the Supplemental Material.
Resulting PCR products were purified with GENECLEAN II (BIO 101). The purified products were A-tailed using Taq Gold polymerase (Perkin Elmer), and cloned into the pGEM T Easy vector (Clontech). Plasmids were prepared by alkaline lysis and ethanol precipitation using the Rev Prep Orbit (Genemachines) and sequenced by capillary sequencing on an ABI 3700 at the Josephine Bay Paul Center (Marine Biological Laboratory, Woods Hole, MA). Clones without PCR errors were selected and restriction digested with Hind III and Nde I (Fisher Scientific). The digested fragment was ligated into the same restriction sites in the pCWOri+ plasmid.
Heterologous expression of zebrafish CYP51 in E. coli: protein isolation
E. coli cultures for zebrafish CYP51 expression were developed based on the methods of Stromstedt et al. [16]. Expression cultures were grown in Terrific Broth and incubated at 37°C and 250 rpm until the OD 550 of the culture was at least 0.5 and less than 1.2. Delta-aminolevulinic acid (ALA, 0.5 mM final concentration) was added to the culture to promote bacterial heme synthesis, and isopropyl β-D-thiogalactoside (IPTG, 1 mM final concentration) was added to initiate transcription. Cultures were incubated at 28-30°C for 20-24 hours at a rotation speed of 180 rpm. Cultures showing predominantly cytochrome P420 in reduced CO spectra, required the addition of the antibiotic chloramphenicol, which shifted P420 expression to P450 expression [17]. A range of chloramphenicol concentrations was tested to identify a concentration allowing optimal bacterial growth and that yielded spectral P450 (10 μg/mL). Cells were isolated by centrifugation and pellets resuspended in 12.5 mL digestion buffer (10 mM potassium phosphate, pH 7.4, 20% (v/v) glycerol, 0.5 mg/mL lysozyme). Solubilized pellets were incubated on ice for 20-30 minutes, then 0.5% (w/v) CHAPS and 100 mM PMSF were added, and the mixture was sonicated on ice for 6 × 20 seconds. After sonication, the mixture was centrifuged at 100,000 × g, and the resulting supernatant contained the solubilized recombinant heterologously expressed zebrafish CYP51. Cytochrome P450 content was determined by reduced CO difference spectra, measured on a Shimadzu UV 2401PC spectrophotometer according to Omura and Sato [18], using an extinction coefficient of 91.1 mM−1cm−1. Aliquots of the solubilized P450 were stored at −80°C. Additional details are provided in the Supplemental Material.
Sterol 14α-demethylase reconstitution assays
The sterol 14α-demethylase activity of CYP51ZFT was assayed using 0.1 nmol CYP51ZFT, 0.2 nmol recombinant human cytochrome P450 reductase (Panvera), 30 μg dilauryl phosphatidylcholine (DLPC) in 100 mM potassium phosphate, pH 7.4, and lanosterol dissolved in 30% (v/v) Tween 80 and 100 mM potassium phosphate, pH 7.4. NADPH (0.3 mM) was added to initiate the reaction to a final volume of 0.2 mL. Reaction mixtures were held in a 25°C water bath with constant shaking for 60 minutes, and the reaction was stopped by addition of 1 mL of 10% (w/v) potassium hydroxide in methanol, and saponified for 1 hour at 80°C. Sterols were extracted by the addition of 1 mL of a 1:1 mixture of diethyl ether and pentane and analyzed by GC-MS analysis on a Hewlett Packard 6890 Gas Chromatograph coupled to a Hewlett Packard 5973 Mass Selective Detector (MSD) using Electron Impact. The column used was a J&W DB-5ms, 60 m long, 320μm I.D., with a 0.25μm film thickness. The GC effluent was split between the MSD and a flame ionization detector (FID). The temperature program was an initial 50°C, hold for 1 minute, ramp 20°C /min to 80°C, hold for 7 minutes, ramp at 4°C /minute to 320°C, and a final hold for 5 minutes at 320°C. The activity of zebrafish CYP51 as a lanosterol 14α-demethylase was determined from the conversion ratio of lanosterol to the metabolite 4,4-dimethyl-5α-cholesta-8,14-dien-3ß-ol (FF-MAS).
Lanosterol and azole binding to CYP51ZFT
Spectral change due to substrate binding to CYP51ZT (Type I spectral change) was recorded on a Shimadzu UV 2401PC spectrophotometer. Lanosterol was added to CYP51ZT at 35 μM, in 30% (v/v) Tween 80 and 100 mM potassium phosphate, pH 7.4. To investigate the binding of ketoconazole and propiconazole to zebrafish CYP51, 0.5 μM CYP51ZFT in 100 mM potassium phosphate, pH 7.4, 20% (v/v) glycerol was added to the reference and sample cuvettes. Increasing concentrations of azole dissolved in DMSO were titrated into the sample cuvette, and a corresponding volume of DMSO was added to the reference cuvette. After 3 minutes, the spectrum was recorded from 460 to 380 nm. Titrations were continued until azole saturation of the enzyme was observed. The amount of DMSO after the final titration did not exceed 1% (v/v) of the sample volume.
Carbon monoxide displacement by azole antifungals
Ketoconazole and propiconazole binding to zebrafish CYP51 was further investigated by examining the displacement of carbon monoxide from the heme iron of CYP51. 0.5 μM CYP51ZFT in 100 mM potassium phosphate, pH 7.4, 20% (v/v) glycerol was mixed with azole dissolved in DMSO. The mixture was incubated for approximately 2-5 minutes and bubbled with carbon monoxide. The mixture was split between the sample and reference cuvettes, and the baseline between 490-400 nm was recorded. Several grains of sodium dithionite were added to the sample cuvette and mixed. The absorbance at 447 nm was monitored until stable, at which time the spectrum was recorded. The baseline assay contained DMSO alone. The concentration of cytochrome P450 was determined from the difference spectrum.
Phylogenetic analysis of CYP51
CYP51 amino acid sequences were obtained from publicly available databases and websites (Supplemental Table S3) and from translation of the cDNA sequences described above. Choanoflagellate CYP51 was used as the outgroup. Sequence comparisons and alignments were carried out using ClustalW [19] and BioEdit (Ibis Biosciences, Carlsbad, CA). Phylogenetic trees were constructed by analyzing inferred or confirmed amino acid sequences under maximum likelihood using RAxML (v 7.2.6 [20], with the PROT-CAT-WAG model of amino acid substitution [21], and with Bayesian methods using MrBayes (v 3.2.1) and a WAG model, uninformative priors, and running 1e6 generations of Markov chain Monte Carlo with the relative burnin set to 0.25 [22].
Molecular modeling of fish and sea urchin CYP51
Homology models of fish and sea urchin CYP51s were generated using Modeller (v9.8 [23]). Multiple models were generated based on three templates of human CYP51 (PDB:3LD6, 3JUS, 3JUV [24]). The best model from the generated structures was selected based on the Discrete Optimized Protein Energy (DOPE) score [25, 26]. Models were further evaluated using Procheck [27]. Side chain positions were optimized in a solvated model by molecular dynamics minimization and equilibration using NAMD (v2.8) [28]. The P450 heme-cysteine bonds were explicitly patched, and models were solvated in a cube of water to allow periodic boundary condition, charge-neutralized, and minimized for 100 steps prior to relaxation for 2500 steps (5ps). Models and crystal structures were prepared for docking using AutoDockTools (v1.5.4) [29], with the addition of polar hydrogens and the assignment of Gasteiger-Marsili partial charges. Ligand structure and charge minimization was performed with semiempirical methods (PM6 Hamiltonian in Mopac2009 [30, 31]). Gasteiger-Marsili partial charges were used in the final docking runs. High throughput docking was performed using Autodock Vina (v1.1.2) [32]. Flexible ligands were docked into models with rigid protein backbones and rigid or flexible side chains, performing 100 replicate dockings and retaining a broad range of calculated energies (6 kcal/mol). For the evaluation of azole binding modes, docking was performed with Autodock (v4.2.5.1), and the azole nitrogen was constrained to a position 2.0 Å above the heme based on the crystal structure of ketoconazole with human CYP51 (PDB:3LD6 [24]).
Results
Sequence analysis of cloned CYP51s
Using PCR approaches, cDNAs representing CYP51 full-length open reading frames from zebrafish liver, from sergeant major liver, and from sea urchin tissues, were successfully cloned and sequenced (GenBank ID AY496939, GenBank ID AY496940 and GenBank ID AY496941, respectively). The percent amino acid identity between the newly cloned sequences and several known CYP51 sequences is presented in Table 2. The zebrafish and sergeant major CYP51 amino acid sequences are between 75-79% identical to mammalian CYP51 sequences and 32-36% identical to plant, yeast, and bacterial CYP51s. The sea urchin CYP51 amino acid sequence is approximately 65% identical to both the mammalian and fish sequences, suggesting a more basal evolutionary position to these groups. Notably, the zebrafish CYP51 sequence is only as identical to the CYP51 sequences inferred for other fish (Takafugu rubripes, Fundulus heteroclitus) and cloned from sergeant major, as it is to CYP51 from mammals.
Table 2.
Percent amino acid identity between cloned CYP51 and selected reference sequences. Human CYP51 identities are included for reference.
| Human | Mouse | Rat | Killifish | |
|---|---|---|---|---|
| S. purpuratus | 65.2% | 63.6% | 64.6% | 66.3% |
| D. rerio | 75.1% | 73.7% | 74.1% | 76.9% |
| A. saxatilis | 77.5% | 75.5% | 75.9% | 88.3% |
| H. sapiens | 100% | 91.0% | 93.4% | 77.1% |
An alignment of the fish and sea urchin CYP51 amino acid sequences with those from human and chicken illustrates identity and similarity among the sequences from disparate animal groups (Supplementary Figures S1-S2). Maximum likelihood was used to elucidate the evolutionary relationship among CYP51 sequences within animals (Figure 2). Bootstrap support values associated with each clade and taxon are shown at each node on the tree, and horizontal branch lengths are proportional to the evolutionary change along that branch. The resolution of CYP51 phylogeny is high, based on the high bootstrap support values.
Figure 2. Molecular phylogenetic tree of CYP51 evolution determined by analysis of CYP51 amino acid sequences.
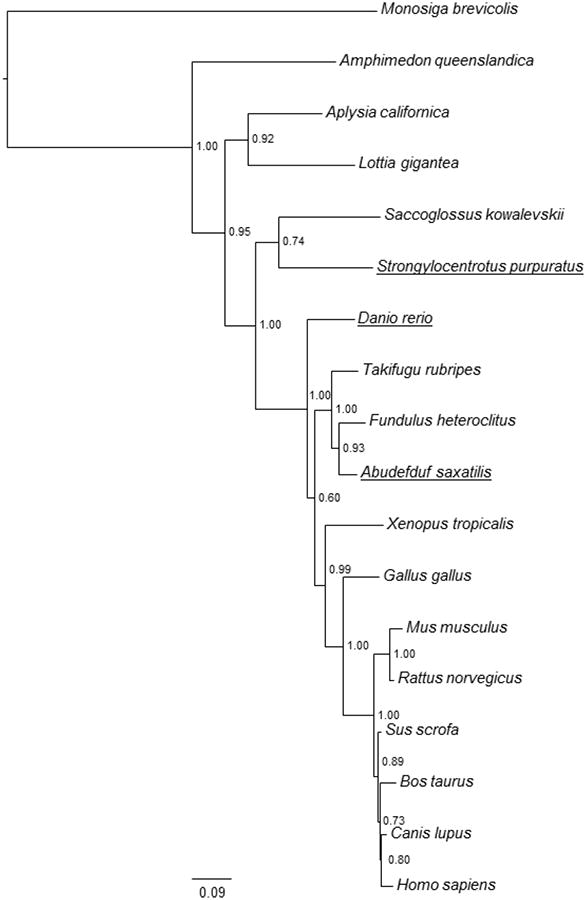
Selected animal CYP51 sequences are shown, including sequences from genome projects. Source and database identifiers are in the supplemental information. Values at branch points are posterior probabilities obtained from Bayesian analysis and maximum likelihood bootstrap support percentages resulting from 500 bootstrap replicates.
Tissue distribution of CYP51 in zebrafish
The expression of CYP51 was assessed in multiple organs of both male and female zebrafish. The highest levels of expression were found in intestine in both sexes, followed by liver, especially in female, and then brain (Figure 3). There were lower levels of CYP51 expression in ovary, testis and heart.
Figure 3. Tissue distribution of CYP51 in adult zebrafish.
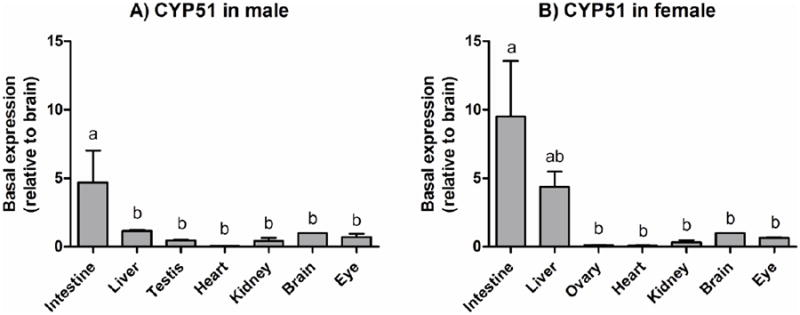
Transcript levels of CYP51 were determined by qPCR in seven tissues from males (A) and females (B). Data were normalized by geometric mean of two reference genes, EF1α and ARNT2. Results are shown as values in each tissue relative to the average value in brain (mean + SD; n = 3 for each sex), as this organ showed one of the least inter-individual variability and sex-differences in the CYP51 expression levels. Statistical differences in transcript levels among tissues were determined by one-way ANOVA followed by Tukey's multiple comparisons test and are shown by different letters (p < 0.05).
Heterologous expression of D. rerio CYP51
Three zebrafish CYP51 cDNA sequences (CYP51ZF, CYP51ZFT and CYP51ZF17) were generated through PCR, each with slight codon modifications that could potentially facilitate heterologous expression of the protein in E. coli (see Table 2). The CYP51ZF and CYP51ZFT cDNA sequences were successfully expressed, as indicated by the presence of reduced CO difference spectra typical of native cytochrome P450 in the solubilized membrane fractions from E. coli. The reduced CO difference spectrum for CYP51ZFT is shown in Figure 4A. With some CYP genes addition of chloramphenicol to cultures has been observed to enhance the expression of native P450 in E. coli, possibly linked to a cold-stress response [17]. Thus, expression of CYP51ZF cytochrome P450 was dependent on the addition of 10 μg/ml chloramphenicol to the culture broth (see Supplementary Figure S3). However, with chloramphenicol, bacterial growth was substantially reduced, decreasing CYP51ZF yield. Cultures expressing CYP51ZF and CYP51ZFT produced approximately 22 nmol/L and 81 – 250 nmol/L of CYP51 protein, respectively. No spectral cytochrome P420 or cytochrome P450 was observed in the solubilized membrane fraction of E. coli cultures expressing CYP51ZF17, with or without chloramphenicol. Solubilized membrane fractions from cultures expressing the pCWOri+ vector without a CYP51 cDNA insert also showed no absorbance at or near 450 nm.
Figure 4. Spectral analysis of recombinant CYP51ZFT.
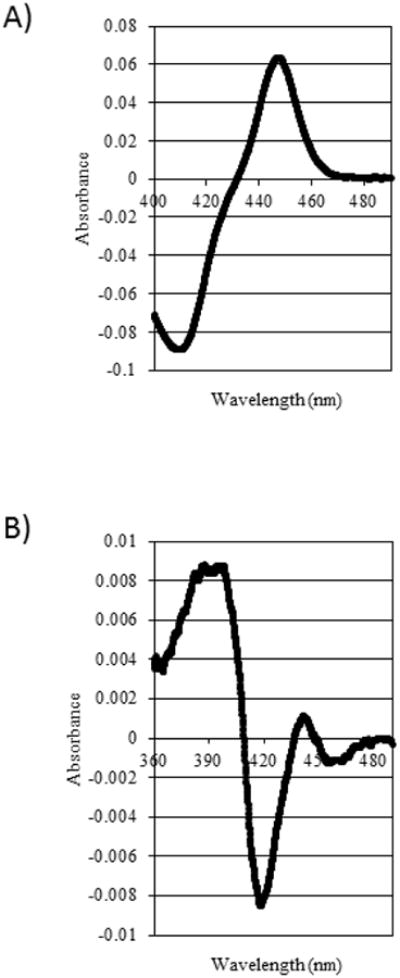
(A) Reduced CO difference spectrum of CYP51ZFT. (B) Type I binding spectrum of CYP51ZFT for lanosterol.
In addition to the zebrafish CYP51, attempts were made to express the sergeant major and the sea urchin CYP51s. However, neither the truncated nor full-length sequences in expression plasmids yielded any spectral cytochrome P450 or cytochrome P420. Additional efforts are warranted.
D. rerio CYP51 sterol binding and enzymatic reconstitution
CYP51ZFT demonstrated a Type I spectrum when mixed with 35μM lanosterol (Figure 4B), reflecting a heme Fe spin-state change indicative of substrate binding to the active site. Sterol 14α-demethylase activity was catalyzed in the reconstituted assay with both CYP51ZF and CYP51ZFT, determined by GC-MS detection of production of 4, 4-dimethyl-5α-cholesta-8,14,24-triene-3ß-ol (FF-MAS) (illustrated in Figure 5 for CYP51ZFT). The expressed CYP51ZFT had substantially greater activity than CYP51ZF; the conversion of 50 μM lanosterol to the 4, 4-dimethyl-5α-cholesta-8,14,24-triene-3β-ol metabolite was measured as 3.20 nmol/min/nmol CYP51ZFT compared to 0.29 nmol/min/nmol CYP51ZF. The reaction demonstrated NADPH dependence, ketoconazole sensitivity, and potassium cyanide insensitivity. Solubilized proteins from the E. coli culture expressing the empty pCWOri+ vector did not yield any sterol 14α-demethylase activity.
Figure 5. Gas chromatogram of the extracted sterols following reconstitution of CYP51ZFT monooxygenase.
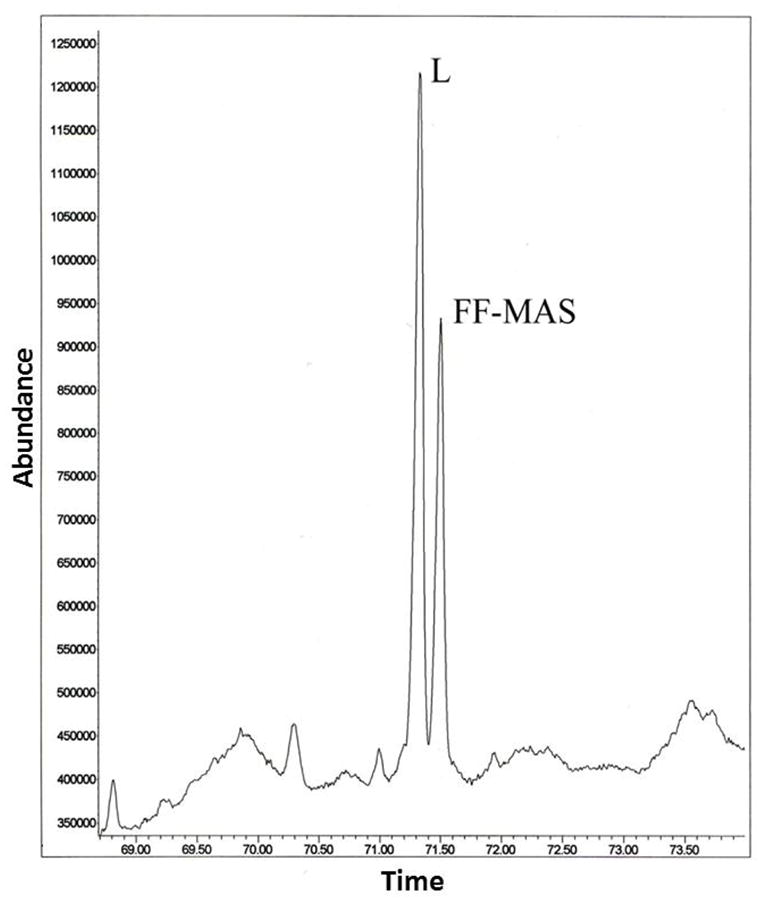
Peaks L and FF-MAS correspond to lanosterol and its 14α-demethylated product 4, 4-dimethyl-5α-cholesta-8,14,24-triene-3β-ol. GC-MS analysis was carried out using an Electron Impact Mass Selective Detector as described in the Methods and Supplemental Materials.
Azole binding and CO displacement studies
In other species, binding of antifungal azole compounds to CYP51 produces a Type II spectral shift, as the inhibitor imidazole N3 or triazole N4 binds directly with the heme iron, causing an increase in the low spin state [33]. Both ketoconazole and propiconazole produced Type II spectra with CYP51ZFT (Figure 6A). The difference between the peak and trough of the spectra increased with increasing concentration of azole, until saturation occurred. Ketoconazole induced a spectrum peak at approximately 432 nm and a trough at 412 nm (Figure 6B). The peak for propiconazole was at 427 nm, and the trough was observed at 409 nm (Figure 6C). Saturation occurred at approximately equimolar concentrations of CYP51ZFT and ketoconazole (0.5 μM), indicating a 1:1 stoichiometry between the enzyme and the azole, but propiconazole saturation did not occur until approximately 0.75 μM propiconazole (Figure 7A). Replicate assays of different preparations of CYP51ZFT gave consistent results for both azole additions. The KS values were 0.26 μM and 0.64 μM for ketoconazole and propiconazole, respectively, indicating that ketoconazole has a higher affinity for CYP51ZFT than propiconazole.
Figure 6. Type II spectral analysis of CYP51ZFT for ketoconazole and propiconazole.
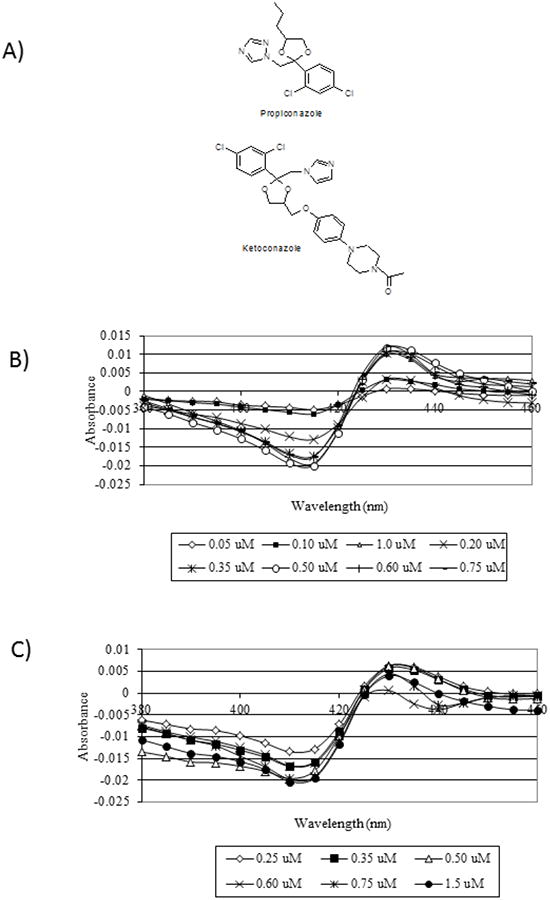
(A) Chemical structures of ketoconazole and propiconazole. (B) Induced type II spectra of CYP51ZT for ketoconazole. (C) Induced type II spectra of CYP51ZT for propiconazole
Figure 7. Azole binding and CO displacement analysis to CYP51ZFT.
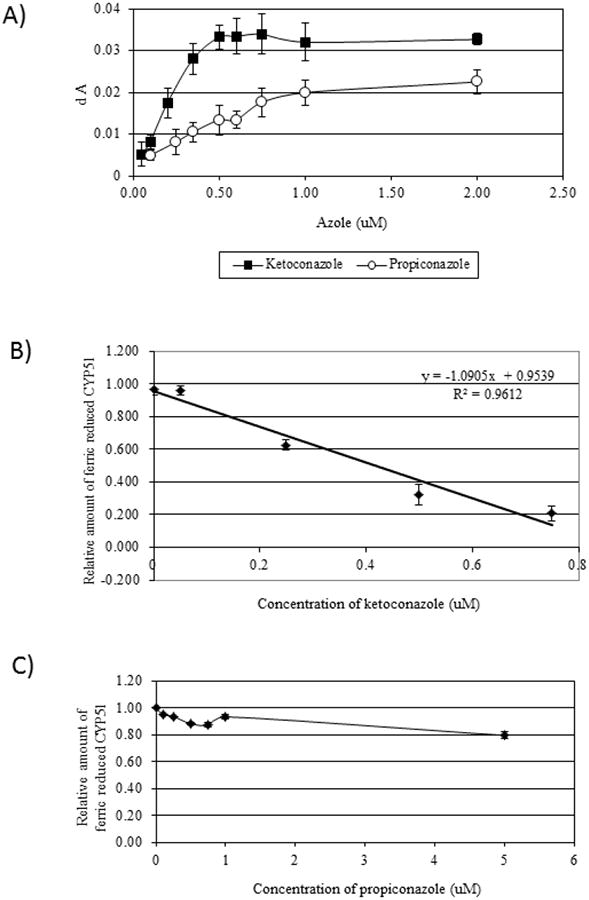
(A). Comparative analysis of azole saturation of CYP51ZFT with ketoconazole (▪) and propiconazole (○). Each data point is the average of 3 to 9 measurements for propiconazole and 2 to 5 measurements for ketoconazole. Error bars represent the standard deviation of the mean absorbance difference for each azole concentration. (B). Displacement of CO binding to CYP51ZFT recombinant expressed protein by ketoconazole. (C) Displacement of CO binding to CYP51ZFT recombinant expressed protein by propiconazole. Error bars indicate standard error.
The displacement of carbon monoxide binding to the cytochrome P450 heme Fe by an azole is indicative of the binding affinity of the inhibitor. Ketoconazole demonstrated a strong affinity for CYP51ZFT and a linear response to increasing ketoconazole concentrations, with 50% inhibition of CO binding occurring at approximately 0.42 μM (Figure 7B). The displacement of CO binding by propiconazole demonstrated non-linear rates of inhibition of CO binding to the CYP51ZFT heme upon increasing concentration of propiconazole (Figure 7C).
Modeling and docking with CYP51s
Homology models of the cloned CYP51s were based on human CYP51 crystal structures. Models for all three exhibited very good optimization and Procheck scores, as expected based on the high sequence identities with human CYP51 (65-79%) as noted above (Table 4). The high degree of similarity of the sea urchin CYP51 three-dimensional model to that of the human crystal structures (RMSD average 1.6 Å) supports the functional assignment of this CYP.
Docking of lanosterol to the zebrafish, sargeant major, and sea urchin CYP51 homology models showed that the 14α-methyl group is found in closest proximity to the heme iron in the lowest energy (best) docking position for all three species (Figure 8). Lanosterol is in a productive orientation, located 4.1-5 Å from the heme iron (and thus merely 2-2.5 Å from the computed position of a transient iron-bound oxo moiety). The docking position of lanosterol in all of the models overlays that of lanosterol docked into the human crystal structure (PDB:3LD6 [24]). Flexible docking of lanosterol to zebrafish CYP51 (Supplementary Figure S4) in which amino acid side chains were allowed to move during the docking calculations, revealed that F226, H228, and W235 (corresponding to human F234, H236, and W239) play important roles in ligand positioning in the zebrafish enzyme, as noted by Strushkevich et al (2010) for human CYP51 [24].
Figure 8. Molecular models of zebrafish, sea urchin, and sergeant major CYP51 overlaid with the human CYP51 showing docked lanosterol.
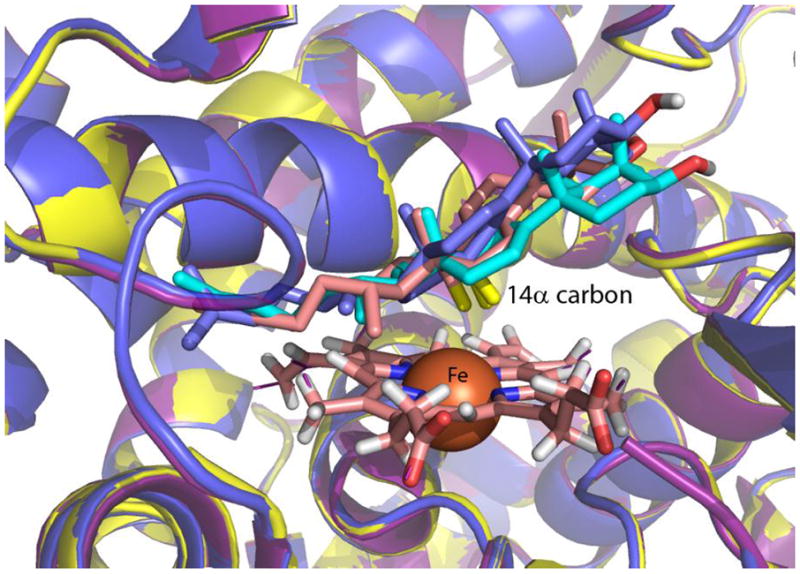
CYP51s from zebrafish (yellow), sea urchin (purple), and sergeant major (dark blue) are overlaid with the human (blue) CYP51 showing lanosterol docked. Ligand docking of lanosterol into the models showed that the 14α-methyl group oxidized and removed by mammalian CYP51 is positioned in an productive conformation in all the modeled proteins, including sea urchin, in the same way as in the crystallized human protein.
Docking of azole inhibitors into CYP structures or models is complicated by the strong bonds formed between the heme iron and the azole nitrogen atom. Docking of flexible ligands into rigid proteins allows for the simplest calculation of binding affinities for direct comparisons between ligands, but does not necessarily provide accurate ligand positions. Despite these limitations, in a re-docking experiment the position of ketoconazole docked into the human CYP51 crystal structure (PDB:3LD6) overlays the structure of the co-crystalized ketoconazole, but does not reflect the proper orientation of the azole nitrogen towards the heme (Supplementary Figure S5). Docking experiments in which the azole nitrogen was constrained to a position obtained from the crystal structure do not adequately reproduce the ligand orientation found in the crystal structure (Supplementary Figure S6).
Binding constants calculated for the three ligands docking into zebrafish CYP51 were 14 nM, 76 nM, and 3.7 μM for lanosterol, ketoconazole, and propiconazole, respectively (corresponding to -10.7, -9.7, and -7.4 kcal/mol). The relative order of modeled binding constants (lanosterol > ketoconazole > propiconazole) is the same as determined biochemically in substrate binding experiments to the heterologously expressed CYP51ZFT. The Autodock calculated affinity constants are within an order of magnitude of the observed constants, not ideal but acceptable given the proper ranking of substrates.
Discussion
The data presented here establish that CYP51 is expressed in early deuterostomes, including invertebrate echinoderms as well as fish. A number of studies have suggested that CYP51 is the only P450 that is broadly distributed in bacteria, plants, fungi and metazoans [1, 34, 35]. A recent analysis points to CYP51 as having occupied the “P450 Genesis Locus,” a genomic locus in the last common animal ancestor that may have given rise to all animal CYPs [36]. However, until now, functional studies have only been carried out with fungal, protozoal, plant and mammalian CYP51. The results here indicate that early deuterostome CYP51s are functionally similar to the mammalian CYP51 orthologs. Earlier studies suggested that echinoderms are capable of cholesterogenesis [37].
Characterization of recombinant zebrafish CYP51 showed that properties of lanosterol metabolism by the zebrafish enzyme are similar to the mammalian enzymes. As with mammalian enzymes, the product of the CYP51 reaction was identified as 4,4-dimethyl-5α-cholesta-8,14,24-triene-3β-ol (FF-MAS). The 14α-demethylase activity of CYP51ZF and of CYP51ZFT were of a similar order of magnitude compared to that of sterol 14α-demethylase of purified rat liver CYP51 [38], recombinant heterologously expressed rat liver CYP51 [39], recombinant heterologously expressed human CYP51 [40] as well as yeast sterol 14α-demethylases [40, 41] (Table 3). This emphasizes the high level of activity of the expressed CYP51ZFT.
Table 3.
Selected examples of reported CYP51 activities values for reconstituted 14α-sterol demethylase assays with the substrates lanosterol (L), a mixture of lanosterol (L) and 24,25 – dihydrolanosterol (DHL), obtusifoliol (O), 3β-hydroxylanost-7-en-32-ol (HL) or 24-methylene dihydrolanosterol. The underlying reasons for differences between enzymes purified from tissue and the same species expressed and purified are not known but could involve architectural differences between native membrane and bacterial expressed proteins or differences in native membrane vs. reconstituted systems.
| Species | CYP51 source | Activity (nmol/m in/nmol P450) | Substrate concentration | Reference |
|---|---|---|---|---|
| Zebrafish | Heterologously expressed, CYP51ZF | 0.29 | 50 μM L | This study |
| Zebrafish | Heterologously expressed, CYP51ZFT | 3.2 | 50 μM L | This study |
| Rat | Liver, purified | 4.1 | 50 μM DHL | [36] |
| Rat | Heterologously expressed, purified | KM 10.5 μM VMax 13.9 | Various L | [37] |
| Human | Heterologously expressed, purified | KM 29.4 μM VMax 0.47 | Various HL | [38] |
| Human | Heterologously expressed, purified | ≈30.0 | Various L, DHL, O | [65] |
| Saccharomyces cerevisiae | Purified, cells | 9.0 | 13 nmol L | [66] |
| C. albicans | Heterologously expressed, purified | KM 20.8 μM VMax 0.15 | Various HL | [38] |
| C. albicans | Heterologously expressed, purified, ATCC -1 | 0.338 | 15 nmol L | [39] |
| C. albicans | Heterologously expressed, purified, ATCC -2 | 0.324 | 15 nmol L | [39] |
| C. albicans | Heterologously expressed, purified, DUMC | 0.388 | 15 nmol L | [39] |
| C. albicans | Heterologously expressed, purified | ≈28.0 | Various L, O, DHL | [65] |
| Sorghum bicolor | Heterologously expressed, purified | 5.0 | 500 μM O | [67] |
| Trypanosoma brucei | Heterologously expressed, purified | 6.2 | Various O | [68] |
| Trypanosoma cruzi | Heterologously expressed, purified | 2.4 | Various M | [68] |
| Leishmania infantum | Heterologously expressed, purified | ≈9.0 | Various O | [65] |
The KS value for ketoconazole binding to zebrafish CYP51 (0.26 μM) is 20-fold higher than that obtained when ketoconazole was titrated against Candida albicans CYP51 (KS ∼ 0.012 μM), and approximately 4 fold higher when titrated against human CYP51 (Ks ∼ 0.061 μM) [42]. Furthermore, when comparing propiconazole binding to zebrafish CYP51, the KS value is 15-fold higher than that obtained when propiconazole was titrated against Candida albicans CYP51 (KS ∼ 0.038 μM), and approximately 2-fold higher when titrated against truncated human CYP51 (KS ∼ 0.335 μM). These results indicate an azole binding similarity of the zebrafish enzyme and the mammalian forms, and suggest that selective azole antifungal treatment of fish fungal infections is feasible.
Lamb and colleagues [40] found that 50% inhibition of CO binding to 0.3 nmol Candida albicans CYP51 occurred at 0.15 nmol ketoconazole; however, 0.9 nmol ketoconazole was required to inhibit 50% of CO binding to 0.3 nmol human CYP51. Our results on CO displacement indicate that ketoconazole has a stronger affinity for zebrafish CYP51ZFT than human CYP51 but less affinity compared to yeast CYP51. The displacement of CO binding by propiconazole showed non-linear binding to the CYP51ZFT. Guardiola-Diaz et al. [43] observed interesting spectral binding characteristics of Mycobacterium tuberculosis (MT) CYP51 and azaconazole, econazole and ketoconazole. Azaconazole had two dissociation constants for binding to MTCYP51, which could result from two distinct binding sites within the heme pocket of the MT CYP51 or suggest that the azole binds in different conformations, producing different affinities observed by spectral measurement [43]. In the present study, we found approximately a 1:1 stoichiometry between ketoconazole and CYP51ZFT in saturation studies, and a 1.5:1 stoichiometry between propiconazole and CYP51ZFT. These observations might suggest that, similarly to azaconazole, propiconazole (but not ketoconazole) is able to bind multiple sites or adopt various conformations within the active site of CYP51ZFT. This would be made possible by the drastically smaller size of propiconazole and azaconazole compared to ketoconazole. Clearly, more work is needed to understand the dynamics of the interaction of zebrafish CYP51 and propiconazole.
The modeling and docking data support results with the expressed enzyme showing that interaction of lanosterol and ketoconazole with zebrafish CYP51 is similar to the mammalian enzymes. Calculated azole affinities reproduced the rank order of the observed binding affinities. Docking of lanosterol also strongly suggests that sea urchin (echinoderm) CYP51 is a lanosterol 14α-demethylase, susceptible to azole inhibition. An important consideration in molecular modeling is sequence identity between the template (crystal structure) and the target, which ideally should be more than 30%. The CYP51 sequences here share 65% or more identity, and in the case of highly conserved proteins such as CYP51, homology modeling is likely to provide excellent structural predictions even for distantly related species. The results support the use of ligand docking to homology models as a screen for ligand binding to these enzymes.
Sterol synthesis
A CYP51 role in sterol synthesis provides a rationale for its proposed position as a “founder” CYP [1, 36]. Sterols are components of nearly all eukaryotes and principally play a key role in controlling cell membrane flexibility and fluidity. Furthermore, in multicellular eukaryotes sterol products are the precursor molecules from which hormones are biosynthesized such as steroids in mammals, brassinosteroids in plants, and ecdysteriods in insects, crustaceans and other ecdysozoans. Eukaryotic post-squalene sterol biosynthesis involves over 20 proteins, including a number of other cytochrome P450 enzymes.
The studies here show that CYP51 and presumably sterol synthesis occur in early deuterostomes. However, not all animals, nor even all deuterostomes, are capable of de novo cholesterol biosynthesis, nor do all animals possess a CYP51 gene (Figure 9). The hexactinellid sponges appear not to be capable of de novo cholesterol biosynthesis [44], but other sponges do appear capable of this [45, 46], and the demosponge Amphimedon queenslandica does contain a CYP51 gene. This suggests that the common ancestor of animals possessed the ability to synthesize cholesterol implying the presence of a CYP51 gene. Indeed, CYP51 genes are found in the genomes of the choanoflagellates Monosiga brevicolis, M. ovata, and Salpingoeca rosetta ([36] and unpublished data), although the functions have not been confirmed.
Figure 9. Schematic diagram of the phylogenetic relationship of the animal kingdom based on 18S ribosomal RNA data [68] and data from the Tree of Life project [69].
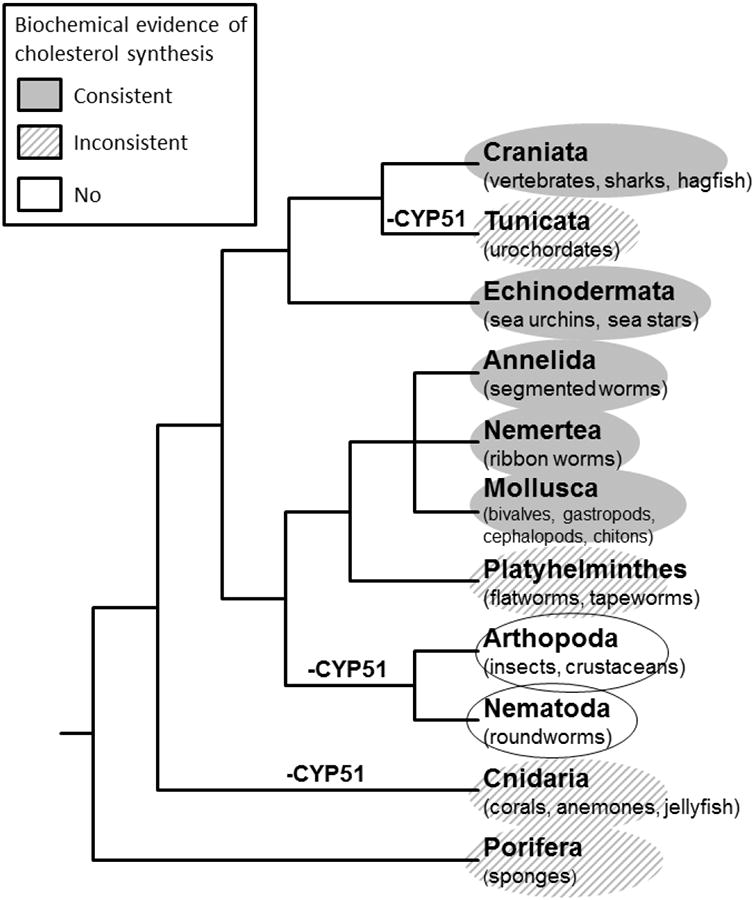
Classes of organisms capable of de novo cholesterol biosynthesis determined by radiolabeled acetate or mevalonate incorporation are shaded [70]. Groups partially shaded have produced conflicting data regarding de novo cholesterol biosynthesis [44, 45, 70-72]. No shading indicates that there is no biochemical evidence supporting de novo cholesterol biosynthesis. The loss of a CYP51 gene is indicated on the figure, where the supporting genomic data are available [47, 73].
Despite the suggestion of early evolutionary appearance of CYP51, analysis of available animal genome sequences indicates that CYP51 is absent from a number of groups [36], including all the ecdysozoans investigated to date. We find it intriguing that a deuterostome phylum, the Tunicata (Urochordata), also lacks CYP51; a CYP51 gene was not detected in genomes of the ascidians Ciona intestinalis or Ciona savignyi. Tunicates are evolutionarily positioned between the echinoderms and the vertebrates, both of which we show here do express CYP51. Notably, 17β-hydroxysteroid dehydrogenase type 7, also involved in the post-squalene portion of cholesterol biosynthesis, is absent from the C. intestinalis genome as well [47]. Without sterol-14α-demethylase and 17β-hydroxysteroid dehydrogenase type 7, tunicates would be incapable of de novo cholesterol biosynthesis. Despite this, de novo sterol synthesis has been reported in ascidian tunicates [48]. This observation may be explained by the discovery that a cyanobacterial symbiont of Didemnid ascidian tunicates appears to synthesize sterols for its host, indicating dependence of these tunicates on their symbiotic bacteria, and suggesting that other tunicates could have similar obligate symbioses [49]. Thus, CYP51 and cholesterol biosynthesis have been selectively retained and lost throughout animal evolution, and different strategies have evolved to allow animals to survive without CYP51.
The expression and function of CYP51 can be involved in reproduction in some animals. However, while the role of CYP51 in cholesterol synthesis is clear and essential, the role in other processes is less so. The immediate product of the CYP51 reaction, FF-MAS and its metabolite T-MAS, have been suggested to stimulate the resumption of meiosis in mammalian oocytes [2]. However, while FF-MAS is formed by zebrafish CYP51, it is unlikely to have any role in resumption of meiosis in fish. Rather, this is accomplished by the maturation-inducing steroids 17,20 β-dihydroxy-4-pregnen-3-one (DHP) and 17,20 β,21-trihydroxy-4-pregnen-3-one (20 β-S), the latter synthesized from 11-deoxycortisol [50, 51]. CYP51 mRNA was expressed in sea urchin ovary, but there is no indication that sterols are involved in the release from meiotic arrest in echinoderms [52, 53]. The absence of a CYP51 gene in Ciona spp. argues against any role for the sterol 14α-demethylase products as MAS in tunicates. Recent results with germ cell-specific CYP51 knockout mice cast doubt on the role of MAS in mammals [54]. Involvement in release from meiotic arrest, if this is a function of CYP51, must differ among deuterostomes [55].
Environmental and therapeutic implications
The susceptibility of zebrafish CYP51 to azole inhibition was similar to that seen with human CYP51 but less than that seen with yeast and fungal CYP51. Azoles that inhibit fungal CYP51 are teratogenic in humans, potentially leading to symptoms of Antley-Bixler syndrome [56, 57]. Defects in or inhibition of cholesterol synthesis in fish are as yet poorly known, and there is opportunity for such studies in this model system, for biomedical and environmental consequences of azole exposure, including evaluation of new drugs.
Our results indicate that it may be possible to selectively inhibit CYP51 of fish fungal pathogens in an analogous fashion seen in the clinic where fungal pathogens such as C. albicans are targeted through selective inhibition of CYP51 over the human enzyme. Fungal infections (mycosis) frequently occur in wild and farmed salmonid broodstock with most infections attributed to the genus Saproleggnia, which can cause mortality in all salmonid lifestages [58]. Other problematic fungal infections of farmed fish include Exophiala salmonis, Phialophora, Ichthyophonus hoeferi, Dermocystidium and Paecilomyces farinosus. Determining the azole sensitivity of CYP51 of various fish species relative to that of fish fungal pathogens will be important in developing selective inhibitors for treatment of fungal infections in aquaculture.
Although CYP51 is the target of many azole anti-fungal drugs and pesticides, these chemicals have been shown to interact with other cytochrome P450 enzymes including those in families 1, 2, 3, and 4 [59-63]. Azole fungicides are often applied in conjunction with other pesticides, such as organophosphate pesticides. Studies indicate that exposure of non-target species to azole fungicides can affect the ability of an organism to metabolize pesticides, increasing the toxicity of the chemicals [64-67]. Levine and Oris [65] observed that exposure of fathead minnows to propiconazole prior to exposure to the organophosphate parathion increased the acute toxicity of parathion. Such complexities would need to be considered when assessing the use of azoles for treatment of fish fungal pathogens.
In summary, the studies here demonstrate expression of CYP51 in fish and echinoderms, and provide initial characterization of zebrafish CYP51 expressed in E. coli. The characteristics of lanosterol metabolism as well as susceptibility to inhibition by azoles were largely similar to those of mammalian enzymes. Computational analyses (homology modeling and docking) suggest that other fish and also echinoderm CYP51s will catalyze the 14α-demethylation of lanosterol, and be similar in susceptibility to azoles. The results provide a foundation for further studies of CYP51, with implication for the evolution of sterol synthesis, effects of chemicals in the environment, and the design of possible therapeutic treatments for infection by fungal pathogens in finfish aquaculture.
Supplementary Material
Highlights.
- CYP51s were cloned from fish and sea urchin, the first from non-mammalian metazoans.
- Zebrafish CYP51 expressed in E. coli catalyzed lanosterol 14α-demethylase activity.
- Zebrafish CYP51 was inhibited by the azoles ketoconazole and propiconazole.
- Docking studies indicate sea urchin CYP51 is a sterol 14α-demethylase inhibited by azoles.
- The phylogeny of CYP51 informs our assessment of the evolution of sterol biosynthesis.
Acknowledgments
This work was supported by NIH Toxicology Training Grant T32EF07155, and by Superfund Research Program grants P42ES059471 (AMM) and P42ES007381 (JJS, JVG, AK and BL) from the National Institute of Environmental Health Sciences, and by a Wellcome Trust Travel Award to DCL. Japan Society for the Promotion of Science Postdoctoral Fellowship and Postdoctoral Fellowship for Research Abroad to AK (nos. 4313 and 820, respectively) are acknowledged. We would like to thank Sean Sylva and Chris Reddy (Woods Hole Oceanographic Institution) for guidance with the GC-MS. Thanks also to Drs. M. Waterman and N. Kagawa for the gift of the pCW expression vector. We would like to thank Syngenta Crop Protection, Inc. for the generous gift of the propiconazole, and Mogens Baltson and Anne Grete Byskov for the generous gift of the FF-MAS standard.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lepesheva GI, Waterman MR. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim Biophys Acta. 2007;1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byskov AG, Andersen CY, Nordholm L, Thogersen H, Guoliang X, Wassman O, Guddal JVAE, Roed T. Chemical structure of sterols that activate oocyte meiosis. Nature. 1995;374:559–562. doi: 10.1038/374559a0. [DOI] [PubMed] [Google Scholar]
- 3.Byskov AG, Andersen CY, Leonardsen L, Baltsen M. Meiosis activating sterols (MAS) and fertility in mammals and man. J Exp Zool. 1999;285:237–242. [PubMed] [Google Scholar]
- 4.Byskov AG, Andersen CY, Leonardsen L. Role of meiosis activating sterols, MAS, in induced oocyte maturation. Mol Cell Endocrinol. 2002;187:189–196. doi: 10.1016/s0303-7207(01)00707-9. [DOI] [PubMed] [Google Scholar]
- 5.Keber R, Motaln H, Wagner KD, Debeljak N, Rassoulzadegan M, Acimovic J, Rozman D, Horvat S. Mouse Knockout of the Cholesterogenic Cytochrome P450 Lanosterol 14{alpha}-Demethylase (Cyp51) Resembles Antley-Bixler Syndrome. J Biol Chem. 2011;286:29086–29097. doi: 10.1074/jbc.M111.253245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson HC. Disorders of post-squalene cholesterol biosynthesis leading to human dysmorphogenesis. Cell Mol Biol (Noisy-le-grand) 2002;48:173–177. [PubMed] [Google Scholar]
- 7.Akiyoshi T, Saito T, Murase S, Miyazaki M, Murayama N, Yamazaki H, Guengerich FP, Nakamura K, Yamamoto K, Ohtani H. Comparison of the inhibitory profiles of itraconazole and cimetidine in cytochrome P450 3A4 genetic variants. Drug Metab Dispos. 2011;39:724–728. doi: 10.1124/dmd.110.036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mast N, Zheng W, Stout CD, Pikuleva IA. Antifungal Azoles: Structural Insights into Undesired Tight Binding to Cholesterol-Metabolizing CYP46A1. Mol Pharmacol. 2013;84:86–94. doi: 10.1124/mol.113.085902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handley-Goldstone HM, Grow MW, Stegeman JJ. Cardiovascular gene expression profiles of dioxin exposure in zebrafish embryos. Toxicol Sci. 2005;85:683–693. doi: 10.1093/toxsci/kfi116. [DOI] [PubMed] [Google Scholar]
- 10.Gupta T, Mullins MC. Dissection of organs from the adult zebrafish. J Vis Exp. 2010 doi: 10.3791/1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubota A, Bainy AC, Woodin BR, Goldstone JV, Stegeman JJ. The cytochrome P450 2AA gene cluster in zebrafish (Danio rerio): Expression of CYP2AA1 and CYP2AA2 and response to phenobarbital-type inducers. Toxicol Appl Pharmacol. 2013;272:172–179. doi: 10.1016/j.taap.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 13.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnes HJ. Maximizing Expression of Eukaryotic Cytochrome P450s in Escherichia coli. In: Johnson EF, Waterman MR, editors. Methods in Enzymology. Vol. 272. Academic Press; San Diego: 1996. pp. 3–14. Cytochrome P450 (Part B) [DOI] [PubMed] [Google Scholar]
- 15.Waterman MR, Jenkins CM, Pikuleva I. Genetically engineered bacterial cells and applications. Toxicol Lett. 1995;82-83:807–813. doi: 10.1016/0378-4274(95)03597-4. [DOI] [PubMed] [Google Scholar]
- 16.Stromstedt M, Rozman D, Waterman MR. The ubiquitously expressed human CYP51 encodes lanosterol 14 alpha- demethylase, a cytochrome P450 whose expression is regulated by oxysterols. Arch Biochem Biophys. 1996;329:73–81. doi: 10.1006/abbi.1996.0193. [DOI] [PubMed] [Google Scholar]
- 17.Kusano K, Waterman MR, Sakaguchi M, Omura T, Kagawa N. Protein synthesis inhibitors and ethanol selectively enhance heterologous expression of P450s and related proteins in Escherichia coli. Arch Biochem Biophys. 1999;367:129–136. doi: 10.1006/abbi.1999.1248. [DOI] [PubMed] [Google Scholar]
- 18.Omura T, Sato R. The carbon monoxide binding pigment of liver microsomes. I. Evidence for its haemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 21.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 22.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5 doi: 10.1002/0471250953.bi0506s15. Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strushkevich N, Usanov SA, Park HW. Structural basis of human CYP51 inhibition by antifungal azoles. J Mol Biol. 2010;397:1067–1078. doi: 10.1016/j.jmb.2010.01.075. [DOI] [PubMed] [Google Scholar]
- 25.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eramian D, Shen MY, Devos D, Melo F, Sali A, Marti-Renom MA. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 28.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosconati S, Forli S, Perryman AL, Harris R, Goodsell DS, Olson AJ. Virtual Screening with AutoDock: Theory and Practice. Expert Opin Drug Discov. 2011;5:597–607. doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart JJ. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J Mol Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart JJ. MOPAC2009, Stewart Computational Chemistry. Colorado Springs, CO; 2008. MOPAC2009. [Google Scholar]
- 32.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida Y, Aoyama Y. Interaction of azole antifungal agents with cytochrome P-45014DM purified from Saccharomyces cerevisiae microsomes. Biochem Pharmacol. 1987;36:229–235. doi: 10.1016/0006-2952(87)90694-0. [DOI] [PubMed] [Google Scholar]
- 34.Lepesheva GI, Waterman MR. CYP51–the omnipotent P450. Mol Cell Endocrinol. 2004;215:165–170. doi: 10.1016/j.mce.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem Biophys Res Commun. 2000;273:799–804. doi: 10.1006/bbrc.2000.3030. [DOI] [PubMed] [Google Scholar]
- 36.Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120474. doi: 10.1098/rstb.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith AG, Goad LJ. Sterol biosynthesis by the sea urchin Echinus esculentus. Biochem J. 1974;142:421–427. doi: 10.1042/bj1420421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trzaskos J, Kawata S, Gaylor JL. Microsomal enzymes of cholesterol biosynthesis. Purification of lanosterol 14 alpha-methyl demethylase cytochrome P-450 from hepatic microsomes. J Biol Chem. 1986;261:14651–14657. [PubMed] [Google Scholar]
- 39.Nitahara Y, Aoyama Y, Horiuchi T, Noshiro M, Yoshida Y. Purification and characterization of rat sterol 14-demethylase P450 (CYP51) expressed in Escherichia coli. J Biochem (Tokyo) 1999;126:927–933. doi: 10.1093/oxfordjournals.jbchem.a022536. [DOI] [PubMed] [Google Scholar]
- 40.Lamb DC, Kelly DE, Waterman MR, Stromstedt M, Rozman D, Kelly SL. Characteristics of the heterologously expressed human lanosterol 14alpha-demethylase (other names: P45014DM, CYP51, P45051) and inhibition of the purified human and Candida albicans CYP51 with azole antifungal agents. Yeast. 1999;15:755–763. doi: 10.1002/(SICI)1097-0061(19990630)15:9<755::AID-YEA417>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Aoyama Y, Kudo M, Asai K, Okonogi K, Horiuchi T, Gotoh O, Yoshida Y. Emergence of fluconazole-resistant sterol 14-demethylase P450 (CYP51) in Candida albicans is a model demonstrating the diversification mechanism of P450. Arch Biochem Biophys. 2000;379:170–171. doi: 10.1006/abbi.2000.1852. [DOI] [PubMed] [Google Scholar]
- 42.Warrilow AG, Parker JE, Kelly DE, Kelly SL. Azole affinity of sterol 14alpha-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob Agents Chemother. 2013;57:1352–1360. doi: 10.1128/AAC.02067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guardiola-Diaz HM, Foster LA, Mushrush D, Vaz AD. Azole-antifungal binding to a novel cytochrome P450 from Mycobacterium tuberculosis: implications for treatment of tuberculosis. Biochem Pharmacol. 2001;61:1463–1470. doi: 10.1016/s0006-2952(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 44.Blumberg M, Thiel V, Pape T, Michaelis W. The steroids of hexactinellid sponges. Naturwissenschaften. 2002;89:415–419. doi: 10.1007/s00114-002-0343-x. [DOI] [PubMed] [Google Scholar]
- 45.Djerassi C, Silva C. Sponge sterols: origin and biosynthesis. Acc Chem Res. 1991;24:371–378. [Google Scholar]
- 46.Silva CJ, Wunsche L, Djerassi C. Biosynthetic studies of marine lipids. 35. The demonstration of de novo sterol biosynthesis in sponges using radiolabeled isoprenoid precursors. Comp Biochem Physiol B. 1991;99:763–773. doi: 10.1016/0305-0491(91)90140-9. [DOI] [PubMed] [Google Scholar]
- 47.Marijanovic Z, Laubner D, Moller G, Gege C, Husen B, Adamski J, Breitling R. Closing the gap: identification of human 3-ketosteroid reductase, the last unknown enzyme of mammalian cholesterol biosynthesis. Mol Endocrinol. 2003;17:1715–1725. doi: 10.1210/me.2002-0436. [DOI] [PubMed] [Google Scholar]
- 48.Voogt PA, van Rheenen JW. On the sterols of some ascidians. Arch Int Physiol Biochim. 1975;83:563–572. doi: 10.3109/13813457509071400. [DOI] [PubMed] [Google Scholar]
- 49.Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR, Phillippy AM, Schatz MC, Piel J, Haygood MG, Ravel J, Schmidt EW. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc Natl Acad Sci U S A. 2011;108:E1423–1432. doi: 10.1073/pnas.1111712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trant JM, Thomas P. Isolation of a novel maturation-inducing steroid produced in vitro by ovaries of Atlantic croaker. Gen Comp Endocrinol. 1989;75:397–404. doi: 10.1016/0016-6480(89)90174-3. [DOI] [PubMed] [Google Scholar]
- 51.Tubbs C, Tan W, Shi B, Thomas P. Identification of 17,20beta,21-trihydroxy-4-pregnen-3-one (20beta-S) receptor binding and membrane progestin receptor alpha on southern flounder sperm (Paralichthys lethostigma) and their likely role in 20beta-S stimulation of sperm hypermotility. Gen Comp Endocrinol. 2011;170:629–639. doi: 10.1016/j.ygcen.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Dale B, Marino M, Wilding M. The ins and outs of meiosis. J Exp Zool. 1999;285:226–236. [PubMed] [Google Scholar]
- 53.Sagata N. Introduction: meiotic maturation and arrest in animal oocytes. Semin Cell Dev Biol. 1998;9:535–537. doi: 10.1006/scdb.1998.0247. [DOI] [PubMed] [Google Scholar]
- 54.Keber R, Acimovic J, Majdic G, Motaln H, Rozman D, Horvat S. Male germ cell-specific knockout of cholesterogenic cytochrome P450 lanosterol 14alpha-demethylase (Cyp51) J Lipid Res. 2013;54:1653–1661. doi: 10.1194/jlr.M035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo GL, Wilding M, Marino M, Dale B. Ins and outs of meiosis in ascidians. Semin Cell Dev Biol. 1998;9:559–567. doi: 10.1006/scdb.1998.0250. [DOI] [PubMed] [Google Scholar]
- 56.Pursley TJ, Blomquist IK, Abraham J, Andersen HF, Bartley JA. Fluconazole-induced congenital anomalies in three infants. Clin Infect Dis. 1996;22:336–340. doi: 10.1093/clinids/22.2.336. [DOI] [PubMed] [Google Scholar]
- 57.Horvat S, McWhir J, Rozman D. Defects in cholesterol synthesis genes in mouse and in humans: lessons for drug development and safer treatments. Drug Metab Rev. 2011;43:69–90. doi: 10.3109/03602532.2010.540580. [DOI] [PubMed] [Google Scholar]
- 58.Hatai K, Hoshiai G. Mass mortality in cultured coho salmon (Oncorhynchus kisutch) due to Saprolegnia parasitica coker. J Wildl Dis. 1992;28:532–536. doi: 10.7589/0090-3558-28.4.532. [DOI] [PubMed] [Google Scholar]
- 59.Ronis MJ, Ingelman-Sundberg M, Badger TM. Induction, suppression and inhibition of multiple hepatic cytochrome P450 isozymes in the male rat and bobwhite quail (Colinus virginianus) by ergosterol biosynthesis inhibiting fungicides (EBIFs) Biochem Pharmacol. 1994;48:1953–1965. doi: 10.1016/0006-2952(94)90594-0. [DOI] [PubMed] [Google Scholar]
- 60.Ronis MJ, Badger TM. Toxic interactions between fungicides that inhibit ergosterol biosynthesis and phosphorothioate insecticides in the male rat and bobwhite quail (Colinus virginianus) Toxicol Appl Pharmacol. 1995;130:221–228. doi: 10.1006/taap.1995.1027. [DOI] [PubMed] [Google Scholar]
- 61.Ronis MJ, Celander M, Badger TM. Cytochrome P450 enzymes in the kidney of the bobwhite quail (Colinus virginianus): induction and inhibition by ergosterol biosynthesis inhibiting fungicides. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;121:221–229. doi: 10.1016/s0742-8413(98)10043-9. [DOI] [PubMed] [Google Scholar]
- 62.Rodrigues AD, Lewis DF, Ioannides C, Parke DV. Spectral and kinetic studies of the interaction of imidazole anti-fungal agents with microsomal cytochromes P-450. Xenobiotica. 1987;17:1315–1327. doi: 10.3109/00498258709047162. [DOI] [PubMed] [Google Scholar]
- 63.Hegelund T, Ottosson K, Radinger M, Tomberg P, Celander MC. Effects of the antifungal imidazole ketoconazole on CYP1A and CYP3A in rainbow trout and killifish. Environ Toxicol Chem. 2004;23:1326–1334. doi: 10.1897/03-155. [DOI] [PubMed] [Google Scholar]
- 64.Levine SL, Czosnyka H, Oris JT. Effect of the fungicide clotrimazole on the bioconcentration of benzo[a]pyrene in gizzard shad (Dorosoma cepedianum): In vivo and in vitro inhibition of cytochrome P4501A activity. Environmental Toxicology and Chemistry [Environ Toxicol Chem] 1997;16:306–311. [Google Scholar]
- 65.Levine SL, Oris JT. Enhancement of Acute Parathion Toxicity to Fathead Minnows Following Pre-exposure to Propiconazole. Pestic Biochem Physiol. 1999;65:102–109. [Google Scholar]
- 66.Levine SL, Oris JT, Denison MS. Modulation of CYP1A expression in rainbow trout by a technical grade formulation of propiconazole. Environ Toxicol Chem. 1999;18:2565–2573. [Google Scholar]
- 67.Levine SL, Oris JT. Noncompetitive mixed-type inhibition of rainbow trout CYP1A catalytic activity by clotrimazole. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1999;122:205–210. doi: 10.1016/s0742-8413(98)10108-1. [DOI] [PubMed] [Google Scholar]
- 68.Knoll AH, Carroll SB. Early animal evolution: emerging views from comparative biology and geology. Science. 1999;284:2129–2137. doi: 10.1126/science.284.5423.2129. [DOI] [PubMed] [Google Scholar]
- 69.Maddison DRe. The Tree of Life Project. In: Maddison DR, editor. The Tree of Life Project. 2004. [Google Scholar]
- 70.Goad LJ. The sterols of marine invertebrates: composition, biosynthesis, and metabolites. In: Scheuer PJ, editor. Marine Natural Products: Chemical and Biological Perspectives. Vol. 2. Academic Press; New York: 1978. pp. 76–172. [Google Scholar]
- 71.Awharitoma AO, Opute FI, Ali SN, Obiamiwe BA. Lipid biosynthesis in Paramphistomum microbothrium (Trematoda) Angew Parasitol. 1990;31:51–53. [PubMed] [Google Scholar]
- 72.Meyer H, Provasoli L, Meyer F. Lipid biosynthesis in the marine flatworm Convoluta roscoffensis and its algal symbiont Platymonas convoluta. Biochim Biophys Acta. 1979;573:464–480. doi: 10.1016/0005-2760(79)90221-2. [DOI] [PubMed] [Google Scholar]
- 73.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


