Abstract
Green fluorescent proteins (GFPs) and their derivatives are widely used as markers to visualize cells, protein localizations in in vitro and in vivo studies. The use of GFP fusion protein for visualization is generally thought to have negligible effects on cellular function. However, a number of reports suggest that the use of GFP may impact the biological activity of these proteins. Heparin is a glycosaminoglycan (GAG) that interacts with a number of proteins mediating diverse patho-physiological processes. In the heparin-based interactome studies, heparin-binding proteins are often prepared as GFP fusion proteins. In this report, we use surface plasmon resonance (SPR) spectroscopy to study the impact of the GFP tagging on the binding interaction between heparin and a heparin-binding protein, the Roundabout homolog 1 (Robo1). SPR reveals that heparin binds with higher affinity to Robo1 than GFP-tagged Robo1 and through a different kinetic mechanism. A conformational change is observed in the heparin-Robo1 interaction, but not in the heparin-Robo1-GFP interaction. Furthermore the GFP-tagged Robo1 requires a shorter (hexasaccharide) than the tag-free Robo1 (octadecasaccharide). These data demonstrate that GFP tagging can reduce the binding affinity of Robo1 to heparin and hinder heparin binding-induced Robo1 conformation change.
Keywords: heparin, Robo1, green fluorescent protein, interaction, surface plasmon resonance, fusion protein
Introduction
Green fluorescent proteins (GFPs) and their derivatives are widely used as markers in molecular biology and cell biology. GFP was first isolated from the jellyfish, Aequorea victoria, and has been subsequently modified to produce brighter and more stable variants with emission of a wide array of colors [1,2]. Although fluorescent proteins from different species have different protein sequences, they are ~25 kD in size and share a common three-dimensional structure consisting of an 11-stranded β-barrel motif surrounding a central chromophore that is responsible for their fluorescent properties [3-6]. Due to their ease of expression in vitro and in vivo and their lack of required exogenous substrates and cofactors for fluorescence, GFPs are often used to monitor gene expression and protein localization in living organisms [7,8]. For example, the GFP gene can be co-transfected with a gene of interest to serve as a marker of co-transfection. The use of GFPs as endogenously produced protein markers is generally thought to have negligible effects on cellular function [9]. However, a number of reports argue that the use of GFP may impact the biological activity of the fusion proteins and that these tags may not be as innocuous in all systems as previously believed [9-16]. For example, it was demonstrated that expression of GFP in muscle impairs its contractile properties due to GFP impacts actin-myosin interactions by binding to the actin-binding site of myosin [9,10]. It was also reported that transgenic expression of GFP caused a dilated cardiomyopathy in two independent transgenic mouse lines [15] and dose-dependent co-expression of eGFP and galactosidase in the cytoplasm of forebrain neurons resulted in growth retardation, weakness, and premature lethality [16]. More recently, Koelsch and co-workers reported that GFP affects T-cell activation, leading to defects in clustering, up-regulation of the activation marker CD25 and IL-2 cytokine production [14].
Heparin/heparan sulfate (HS) are glycosaminoglycans (GAGs), anionic and often highly sulfated, complex, polydisperse linear polysaccharides. GAGs are ubiquitous molecules exhibiting a wide range of biological functions. HS consists predominantly of repeating disaccharide motif comprised of β-D-glucuronic acid and N-acetyl-α-D-glucosamine residues connected through 1→4 glycosidic linkages. Each HS disaccharide unit can be differentially substituted with 2-O-sulfo groups in the uronic acid residue and 6-O-, 3-O- and N-sulfo groups in the glucosamine residue [17,18]. Since each biosynthetic modification is incomplete, sequence heterogeneity results and is thought to serve as an important mechanism in the regulation of the specificity of HS interaction with cellular proteins including various growth and differentiation factors and morphogens, extracellular matrix components, protease inhibitors, protease, lipoprotein lipase, and various pathogens [19-22]. Interactions between heparin/HS and proteins mediate diverse patho-physiological processes including: blood coagulation, cell growth and differentiation, host defense and viral infection, lipid transport and metabolism, cell-to-cell and cell-to-matrix signaling, inflammation, angiogenesis and cancer [17,19-23]. Thus, an understanding of heparin/ HS-protein interactions at the molecular level is of fundamental importance to biology and will aid in the development of highly specific glycan-based therapeutic agents [17,21].
Since the popular application of GFP involving both in vitro and in vivo experiments, many heparin-binding proteins have been expressed as GFP fusion proteins for the heparin-protein interaction studies. Thus, it is important to understand whether the GFP tagging impacts protein-heparin interaction. In the present study, we used Robo1 (Roundabout homolog 1, with or without GFP tagging) in a heparin binding study. Robo1 is the receptor for SLIT1-3 which are thought to act as molecular guidance cue in cellular migration, including axonal navigation at the ventral midline of the neural tube and projection of axons to different regions during neuronal development, and in neovascularization [24-27]. We used surface plasmon resonance (SPR) spectroscopy to study the effect of the GFP tagging on the heparin-binding interactions with Robo1.
Materials and Methods
Materials
Porcine intestinal heparin (16 kDa) and low molecular weight (LMW) heparin (4.8 kDa) were provided by Celsus Laboratories (Cincinnati, OH). Heparin oligosaccharides included disaccharide (degree of polymerization (dp)2) tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide (dp10), dodecasaccharide (dp12), tetradecasaccharide (dp14), hexadecasaccharide (dp16) and octadecasaccharide (dp18) were prepared from controlled partial heparin lyase 1 treatment of bovine lung heparin (Sigma) followed by size fractionation. Sensor SA chips were from GE Healthcare (Uppsala, Sweden). SPR measurements were performed on a BIAcore 3000 (GE Healthcare, Uppsala, Sweden) operated using BIAcore 3000 control and BIAevaluation software (version 4.0.1).
Protein expression and purification
Human Robo1 (immunoglobulin domains 1 and 2) was expressed as a soluble secreted fusion protein in HEK293 by transient transfection of HEK293 suspension cultures. The coding region of the human roundabout homolog 1 precursor (NP_002932, Uniprot Q9Y6N7) immunoglobulin domains 1 and 2 (residues 58-266) was chemically synthesized by GeneArt AG (Regensburg, Germany) with an additional NH2-terminal fusion of the 7 amino acid recognition sequence of the tobacco etch virus (TEV) protease and a termination codon at the 3’ end of the coding region. The synthetic DNA contained a flanking 5’ EcoRI site and a 3’ HindIII site and was subcloned into a similarly digested chemically synthesized vector containing additional NH2-terminal fusion protein sequences and a vector backbone containing a CMV promoter, intron, post-regulatory element, termination, and terminal repeat sequences as previously described (pGEn2 vector [28]. The final fusion protein sequence was comprised of a 25 amino acid NH2-terminal signal sequence from the T. cruzi lysosomal α-mannosidase [29] followed by an 8xHis tag, 17 amino acid AviTag [30] “superfolder” GFP [31], the TEV protease recognition site, and the immunoglobulin 1-2 domains of Robo1. This expression vector was designated Robo1-pGEn2 and the recombinant product termed Robo1-GFP.
Suspension culture HEK293f cells (Life Technologies, Grand Island, NY) were transfected and recombinant products were purified essentially as described [28] by chromatography on Ni-NTA superflow (Qiagen, Valencia, CA) followed by elution in 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 300 mM NaCl, 300 mM imidazole, pH 7.0. Fractions containing fluorescence were pooled and concentrated to ~1 mg/ml using an ultrafiltration pressure cell membrane (Millipore, Billerica, MA) with a 10 kDa molecular weight cutoff. Robo1-GFP was further purified by Superdex 75 chromatography (GE Healthcare Life Sciences, Pittsburgh, PA) in a buffer containing 20 mM HEPES, 250 mM NaCl, 60 mM imidazole, pH 7.5. Fractions containing fluorescence were concentrated by ultrafiltration and buffer exchanged into 100 mM ammonium bicarbonate, pH 7.4. For removal of the GFP fusion tags, the concentrated Robo1-GFP preparation following Ni-NTA chromatography was treated with recombinant His-tagged GFP-TEV protease (1:20 ratio) at 4°C overnight followed by 15-fold dilution in 25 mM HEPES, 300 mM NaCl, pH 7.0. The sample was then passed through a Ni-NTA column to remove the cleaved fusion tag and His-tagged GFP-TEV and further purified by Superdex 75 chromatography. The final protein preparation was concentrated by ultrafiltration and buffer exchanged into 100 mM ammonium bicarbonate, pH 7.4. The final preparation of Robo1-GFP was 2 mg/ml and Robo1 was 0.6 mg/ml.
Preparation of heparin biochip
Biotinylated heparin was prepared by reaction of sulfo-N-hydroxysuccinimide long-chain biotin (Piece, Rockford, IL) with free amino groups of unsubstituted glucosamine residues in the polysaccharide chain following a published procedure [32]. Biotinylated heparin was immobilized to streptavidin chip based on the manufacturer's protocol. In brief, 20- μl solution of the heparin-biotin conjugate (0.1 mg/ml) in HBS-EP running buffer was injected over flow cell 2 of the streptavidin chip at a flow rate of 10 μl/min. The successful immobilization of heparin was confirmed by the observation of a ~250 resonance unit (RU) increase in the sensor chip. The control flow cell was prepared by 1 min injection with saturated biotin.
Measurement of interaction between heparin and Robo1 and Robo1-GFP using BIAcore
The protein samples were diluted in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4,). Different dilutions of protein samples were injected at a flow rate of 30 μl/min. At the end of the sample injection, the same buffer was flowed over the sensor surface to facilitate dissociation. After a 3 min dissociation time, the sensor surface was regenerated by injecting 30 μl of 2 M NaCl to get fully regenerated surface. The response was monitored as a function of time (sensorgram) at 25 °C.
Variable contact time of the interaction of Robo1/Robo1-GFP with heparin
Robo1/Robo1-GFP (500 nM) in HBS-EP running buffer was injected at different injection times (30, 60, 90 and 120 s) over flow cell 1 (control) and flow cell 2 the SA sensor chip. A kinetic injection mode was used, flowing the protein for (30, 60, 90 and 120 s) and monitoring the dissociation for 3 min. The response was monitored as a function of time, and after each analyte injection the surface was regenerated with 1min injection of 2 M NaCl. The conformational change that occurs in the complex was examined by comparison of the dissociation phase of the normalized sensorgrams.
Solution competition study between heparin on chip surface and heparin-derived oligosaccharides in solution using SPR
Protein (500 nM) mixed with 1000 nM of heparin oligosaccharides, including disaccharide (degree of polymerization (dp) 2) tetrasaccharide (dp4), hexasaccharide (dp6), octasaccharide (dp8), decasaccharide (dp10), dodecasaccharide (dp12), tetradecasaccharide (dp14), hexadecasaccharide (dp16) and octadecasaccharide (dp18) in HBS-EP buffer were injected over heparin chip at a flow rate of 30 μl/min, respectively. After each run, the dissociation and the regeneration were performed as described above. For each set of competition experiments on SPR, a control experiment (only protein without any heparin or oligosaccharides) was performed to make sure the surface was completely regenerated and that the results obtained between runs were comparable.
Computation Modeling
A BLAST-P [36, 37] search of the PDB database for homologous structures to both the Robo1 and GFP portions of the constructs using the default settings. PDB 2V9Q [38] and 4J88 [39] were selected for modeling the Robo1 and GFP portions, respectively, having over 90% homology. Homology model for Robo1 were generated using the homology modeling tool of MOE [40]. Due to the presence of a modified chromophore, the GFP portion was hand-modeled. Structures were then minimized using the AMBER99 forcefield. Structures were then prepared using MOE's partial charge and Protonate3D utilities. Electrostatic surfaces were then generated.
Results
Kinetics measurement of heparin-Robo1 (with and without GFP tagging) interactions
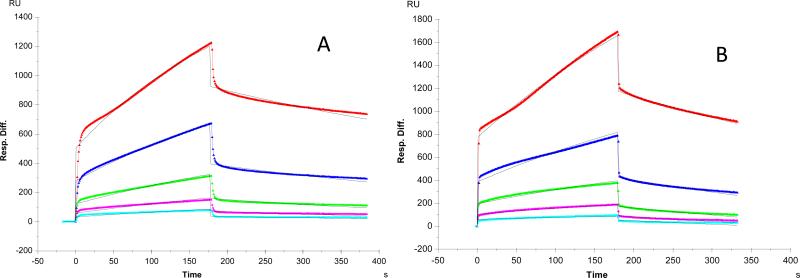
It was reported that Robo1 interacts with heparin/HS, and the interaction is important for Slit2-Robo1 signaling since heparin/HS binds to both Slit2 and Robo1 facilitating the ligand-receptor interaction [24-27]. In our previous report, we used SPR to characterize the binding of Robo1 to heparin immobilized on SA sensor chips [35]. In order to dissect the impact of GFP tagging on the interaction between heparin and Robo1 in this report, two versions of Robo1 (with or without GFP tagging) were used for the binding kinetics measurements. Sensorgrams of Robo1-heparin and GFP-tagged Robo1-heparin interactions are shown in Figure 1. The sensorgrams fit well to a Langmuir 1:1 binding model and the binding kinetics are presented in Table 1. The binding kinetic data show different binding profiles of Robo1 to heparin with different binding affinity (KD for Robo1-heparin interaction: 0.65 μM; GFP-tagged Robo1-heparin interaction: 5.5 μM). The results suggest that the fusion of GFP decreases the binding affinity of Robo1 to heparin.
Figure 1.
SPR sensorgrams. A: Robo1-heparin interaction [35]. Concentrations of Robo1 (from top to bottom): 1000, 500, 250, 125 and 63 nM, respectively. B: Robo1-GFP-heparin interaction. Concentrations of Robo1-GFP (from top to bottom): 1000, 500, 250, 125, and 63 nM, respectively. The black curves are the fitting curves using models from BIAevaluate 4.0.1.
Table 1.
Summary of kinetic data of protein- heparin interactions*
| Interaction | ka (1/MS) | kd (1/S) | Kd (M) |
|---|---|---|---|
| Robol/Heparin [35] | 3.3 x 103 | 2.0 x 10-3 | 6.5 x10-7 |
| (± 1.3 x103) | (± 0.4 x10-3) | (±1.3 x10-7) | |
| Robol-GFP/Heparin | 5.6 x 102 | 2.7x 10-3 | 5.5 x 10-6 |
| (± 2.2 x 102) | (± 0.2 x10-3) | (± 2.6 x10-6) |
The data with (±) in parentheses are the standard deviations (SD) from triplicate binding experiments.
Variable contact time of the interaction of Robo1/Robo1-GFP with heparin
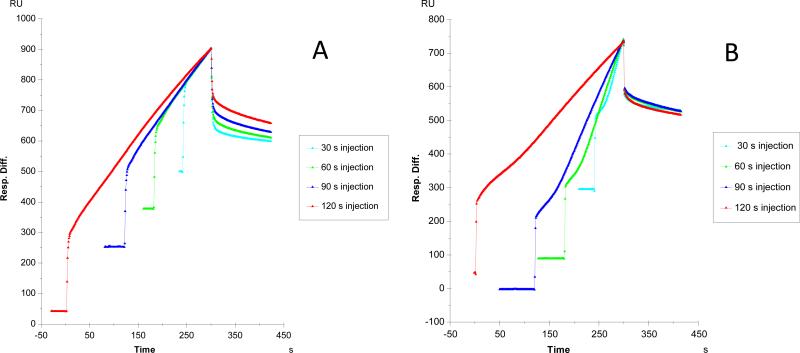
In our previous report [35], we found a much better fitting profile was obtained when fitting with the two-state (conformational change) binding model on the binding of heparin to Robo1 suggesting the interaction may induce a conformation change in Robo1. When the sensorgrams of GFP-tagged Robo1-heparin interaction were fit with the same two-state binding model, no improvement of fitting was obtained (data not shown).
Different contact times of the interaction of Robo1/Robo1-GFP with heparin were performed to test if the protein underwent a conformational change on binding heparin. Longer association times allow the conformational change to take place to a greater extent, generating a more stable conformation, which is reflected by a slower dissociation rate. Sensorgrams of Robo1-heparin, Robo1-GFP-heparin interactions, with variable contact times, are shown in Figure 2. The conformational change that occurs in the Robo1 heparin complex was examined by comparison of the dissociation phase of the normalized sensorgrams. A conformational change is observed on Robo1 binding to heparin as the different contact times of heparin-Robo1 interaction show different profiles in the dissociation phase (Fig. 2A). In contrast, in the Robo1-GFP-heparin interaction, different contact times showed similar dissociation phase profiles (Fig. 2B), suggesting no conformational change takes place on Robo1-GFP binding to heparin. Solution competition study on the interaction between heparin (on surface) with protein to heparin-derived oligosaccharides (in solution) using SPR
Figure 2.
Sensorgrams of Robo1-heparin interaction with variable contact times, A: Robo1 [35]; B: Robo1-GFP
Solution/surface competition experiments were performed by SPR to examine the effect of saccharide chain size of heparin on the heparin-protein interaction. Different size heparin-derived oligosaccharides (from dp2 to dp18) were used in the competition study. The same concentration (1000 nM) of heparin oligosaccharides were present in the protein/heparin interaction solution.
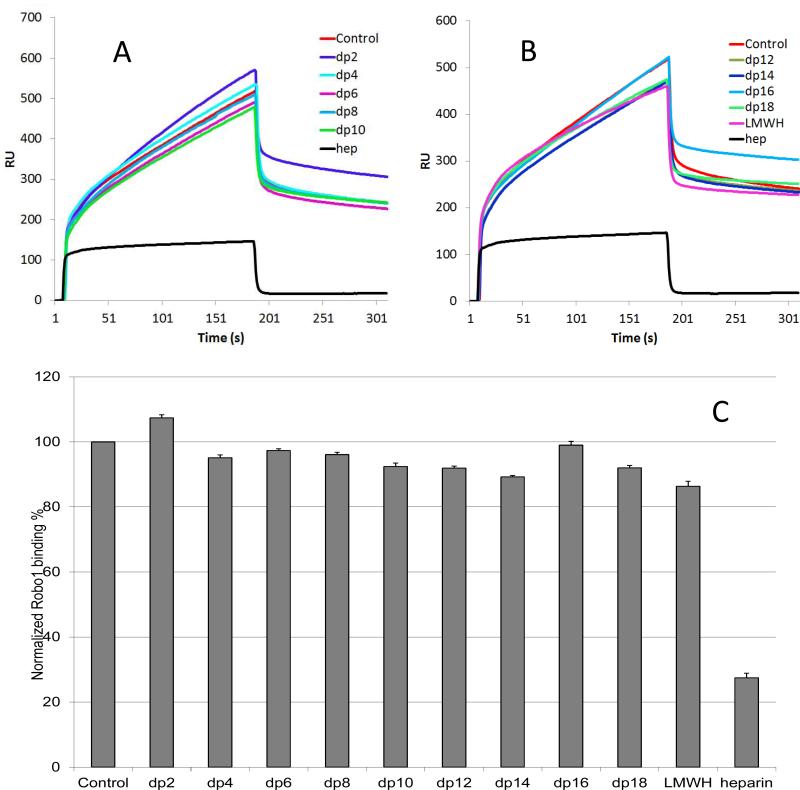
In the case of Robo1-heparin interaction, no competition effect was observed when 1000 nM of disaccharide present in the protein solution. Oligosaccharides (dp 4 to dp18) were next tested and increasing the size of the oligosaccharides, slightly decreased the observed binding of Robo1 to the surface heparin (Fig. 3). The variation of the Robo1 binding observed suggests that the interaction between Robo1 and heparin is chain-length dependent and Robo1 prefers to bind a long chain heparin.
Figure 3.
Oligosaccharide competition experiments for Robo1-heparin binding [35]. A and B: Sensorgrams of solution heparin oligosaccharides - surface heparin competition. Robo1 concentration was 500 nM and concentrations of heparin oligosaccharides in solution were 1000 nM. C: Normalized Robo1 binding to surface immobilized heparin in the presence of different size heparin oligosaccharides in solution in competitive SPR analysis. The data was summarized from triplicate experiments and are presented as mean with standard deviation.
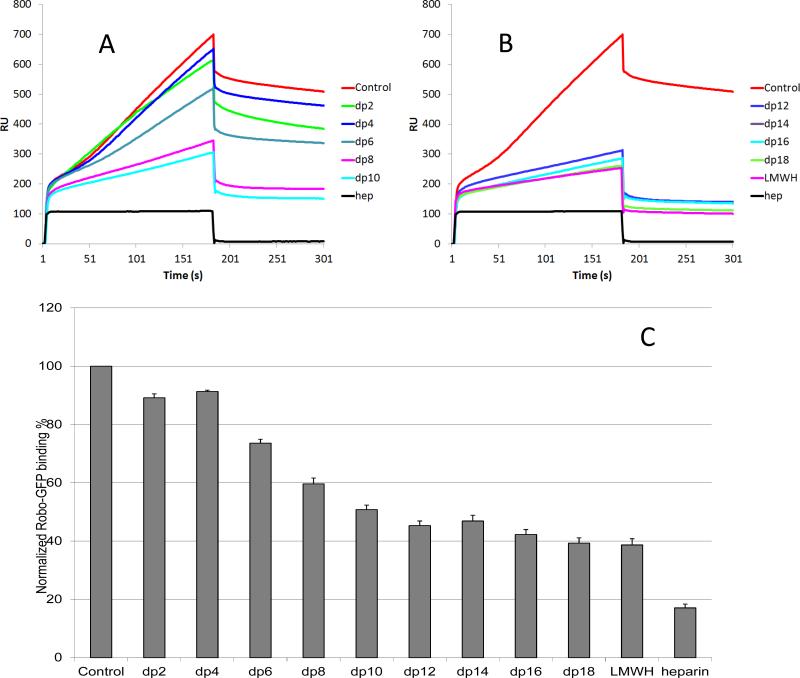
In the case of Robo1-GFP, the impact of saccharide chain size on the heparin-protein interaction seemed greatly enhanced compared to that observed for Robo1. Slight competition effect was observed using 1000 nM of disaccharide and tetrasaccharide in the protein solution. Larger oligosaccharides (dp 6 to dp18) markedly decreased the binding of Robo1-GFP to the surface immobilized heparin (Fig. 4). The decrease of the Robo1-GFP binding observed suggests that the interaction between Robo1-GFP and heparin is chain-length dependent and minimum size of heparin is larger than a hexasaccharide in length. Meanwhile, this data is in consistent with the observation that the binding affinity of Robo1-GFP to heparin is lower than that of Robo1, explaining the same concentrations of heparin oligosaccharides that competitively inhibit Robo1-GFP binding to immobilized heparin but not obvious for the Robo1 binding at the same experimental setting.
Figure 4.
Oligosaccharide competition experiments for Robo1-GFP-heparin binding. A and B: Sensorgrams of solution heparin oligosaccharides /surface heparin competition. Robo1-GFP concentration was 500 nM, and concentrations of heparin oligosaccharides in solution were 1000 nM. C: Normalized Robo1-GFP binding to surface immobilized heparin in the presence of different size heparin oligosaccharides in solution in competitive SPR analysis. The data was summarized from triplicate experiments and are presented as mean with standard deviation.
Discussion
Green fluorescent proteins (GFPs) and their derivatives are widely used as markers in many research areas, such as the localization and trafficking of proteins, protein–protein interactions, which can be directly detected in a living cell in real time. There is no doubt that GFP has revolutionized the perception in molecular biology and cell biology. In the heparin-based interactome studies, proteins are often prepared as GFP fusion proteins in order to allow a direct visualization of the binding processes [3, 37]. One issue is the size of GFP (27 kDa), which appears rarely to affect the subcellular localization of the tagged protein, controls are still required to ensure that the tagged protein behaves as the native counterpart [38], however. There are a number of reports indicating that the use of GFP may impact the biological activity of the fusion proteins and that these tags may not be as innocuous in all systems as previously believed [9-16]. Based on literature, apparently there is no study whether the GFP tagging impacts protein-heparin interaction. In the present study we have examined how GFP tagging on the heparin-binding interactions with Robo1 using SPR spectroscopy. To identify the molecular mechanisms involved, we analyzed (i) binding kinetics; (ii) possible heparin binding-induced conformation change; (iii) heparin chain size requirement for the binding.
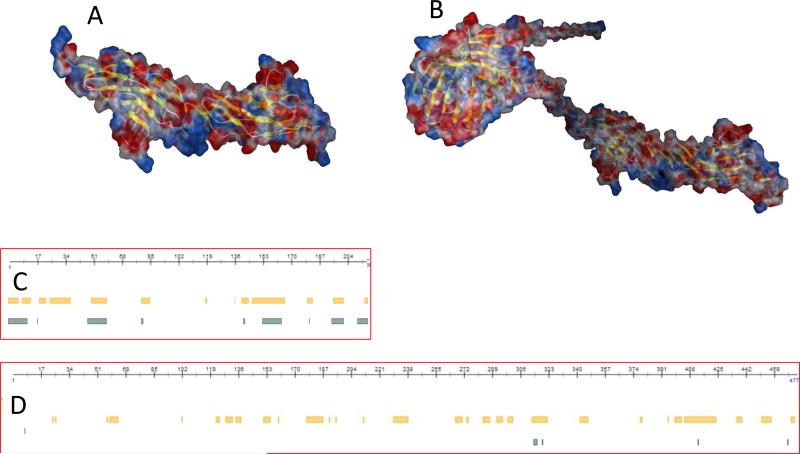
The heparin-binding proteins (HBPs) interact with sulfated domains of HS/heparin chains by ionic attraction between negatively charged groups in HS/heparin chains and basic amino acid residues in the protein. Visual inspection of the electrostatic potential on the Robo surface using MOE modeling (Fig.5A and B) revealed a very basic region at the domain interface [39]. The basic region on Robo is responsible for heparin binding with fair high affinity (KD for Robo1-heparin interaction: 0.65 μM; GFP-tagged Robo1-heparin interaction: 5.5 μM). Apparently the affinity of Robo1 binding to heparin is reduced with GFP tagging. This might due to the overall electrostatic potential change after GFP tagging. Using the PROTEIN CALCULATOR v3.3 (http://www.scripps.edu/~cdputnam/protcalc.html), it is calculated that the values of surface charges at pH 7.4: Robo1 = -1.9; Robo1-GFP = -10.0. The increased negative charges on the GFP-tagged Robo1 may have more repulsive potency to the negatively charged groups on heparin chains than the tag-free Robo1.
Figure 5.
A and B: Electrostatic surfaces of Robo1 and Robo1-GFP modeling using MOE 13.08 and the AMBER99 forcefield with 0.15 salt at 25°C. The values of surface charges at pH 7.4 were estimated: Robo1 = -1.9; Robo1-GFP = -10.0. C and D: Intrinsic protein disorder and flexibility prediction based on protein sequences of Robo1 and Robo1-GFP using the protein prediction program from PredictProtein (www.predictprotein.org). Yellow bars: predicted disordered/flexible regions using the protocol from [33]; Green bars: predicted disordered/flexible regions using the protocol from [34].
When a protein binds to heparin, it can induce a conformation change in the protein [40, 41]. For example, the interaction between antithrombin and heparin, the initial interaction induces a conformational change in antithrombin that enables additional interactions between antithrombin and heparin, resulting in stronger binding [40]. Our SPR data (Fig.2) indicate that the binding of Robo1 to heparin might be a two-state process involving binding and/or a conformational change [35], no conformational change takes place on Robo1-GFP binding to heparin. To explain this observation, we did protein flexibility prediction from sequence using the programs from The PredictProtein server (www.predictprotein.org) [42]. The flexibility prediction data (Fig.5C and D) show that flexible residuals/regions (yellow bars and green bars) in Robo1 are greatly reduced when Robo1 tagged with GFP.
Conclusion
In this study, our results clearly show that GFP tagging reduces the binding affinity of Robo1 to heparin and hinders heparin binding-induced Robo1 conformation change. Our study suggests that examination of heparin-ligand interaction using GFP-tagged protein needs to be cautious and the potential “GFP effect” needs to be considered.
Acknowledgments
This work was supported by grants from the National Institutes of Health in the form of GM-38060 to R.J.L. and NIH R01HL093339 (L.W.), RR005351/GM103390 (L.W. and K.M.)
Abbreviations
- GFPs
green fluorescent proteins
- SPR
surface plasmon resonance
- GAG
glycosaminoglycan
- Robo1
Roundabout homolog 1
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HS
heparan sulfate
- RU
resonance unit
- dp
degree of polymerization
Contributor Information
Fuming Zhang, Department of Chemical and Biological Engineering, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
Heather A. Moniz, Department of Biochemistry and Molecular Biology, Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, USA
Benjamin Walcott, Department of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
Kelley W. Moremen, Department of Biochemistry and Molecular Biology, Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, USA
Lianchun Wang, Department of Biochemistry and Molecular Biology, Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, USA.
Robert J. Linhardt, Department of Chemical and Biological Engineering, Department of Chemistry and Chemical Biology, Departments of Biology and Biomedical Engineering, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, NY 12180, USA.
References
- 1.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962;59:223–39. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Kremers GJ, Gilbert SG, Cranfill PJ, Davidson MW, Piston DW. Fluorescent proteins at a glance. J. Cell. Sci. 2011;124:157–60. doi: 10.1242/jcs.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 4.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr. Opin. Struct. Biol. 2006;16:714–21. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc. Natl. Acad. Sci. U. S.A. 2001;98:462–7. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. The 2.0-A crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J. Biol. Chem. 2003;278:44626–31. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Davidson MW, Campbell RE. Engineered fluorescent proteins: innovations and applications. Nat. Methods. 2009;6:713–17. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- 9.Agbulut O, Coirault C, Niederlander N, Huet A, Vicart P, Hagege A, Puceat M, Menasche P. GFP expression in muscle cells impairs actin-myosin interactions: implications for cell therapy. Nat.Methods. 2006;3:331. doi: 10.1038/nmeth0506-331. [DOI] [PubMed] [Google Scholar]
- 10.Agbulut O, Huet A, Niederlander N, Puceat M, Menasche P, Coirault C. Green fluorescent protein impairs actin-myosin interactions by binding to the actin-binding site of myosin. J. Biol. Chem. 2007;282:10465–71. doi: 10.1074/jbc.M610418200. [DOI] [PubMed] [Google Scholar]
- 11.Baens M, Noels H, Broeckx V, Hagens S, Fevery S, Billiau AD, Vankelecom H, Marynen P. The dark side of EGFP: defective polyubiquitination. PLoS One. 2006;1:e54. doi: 10.1371/journal.pone.0000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is green fluorescent protein toxic to the living cells? Biochem. Biophys. Res. Commun. 1999;260:712–7. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 13.Mak GW, Wong CH, Tsui SK. Green fluorescent protein induces the secretion of inflammatory cytokine interleukin-6 in muscle cells. Anal. Biochem. 2007;362:296–8. doi: 10.1016/j.ab.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 14.Koelsch KA, Wang Y, Maier-Moore JS, Sawalha AH, Wren JD. GFP affects human T cell activation and cytokine production following in vitro stimulation. PLoS One. 2013;8:e50068. doi: 10.1371/journal.pone.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang WY, Aramburu J, Douglas PS, Izumo S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat. Med. 2000;6:482–3. doi: 10.1038/74914. [DOI] [PubMed] [Google Scholar]
- 16.Krestel HE, Mihaljevic AL, Hoffman DA, Schneider A. Neuronal co-expression of EGFP and beta-galactosidase in mice causes neuropathology and premature death. Neurobiol. Dis. 2004;17:310–8. doi: 10.1016/j.nbd.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Capila I, Linhardt RJ. Heparin-protein interactions. Angew. Chem. Int. Ed. Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 18.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell .Biol. 2005;6:530–41. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 19.Parish CR. The role of heparan sulphate in inflammation. Nat. Rev. Immunol. 2006;6:633–43. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 20.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 21.Sasisekharan R, Raman R, Prabhakar V. Glycomics approach to structure-function relationships of glycosaminoglycans. Annu. Rev. Biomed. Eng. 2006;8:181–231. doi: 10.1146/annurev.bioeng.8.061505.095745. [DOI] [PubMed] [Google Scholar]
- 22.Qiu H, Jiang JL, Liu M, Huang X, Ding SJ, Wang L. Quantitative phosphoproteomics analysis reveals broad regulatory role of heparan sulfate on endothelial signaling. Mol. Cell. Proteomics. 2013;12:2160–73. doi: 10.1074/mcp.M112.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat. Immunol. 2005;6:902–10. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 24.Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 25.Jones CA, et al. Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat. Med. 2008;14:448–53. doi: 10.1038/nm1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Dietrich UM, Geng JG, Bicknell R, Esko JD, Wang L. Repulsive axon guidance molecule Slit3 is a novel angiogenic factor. Blood. 2009;114:4300–9. doi: 10.1182/blood-2008-12-193326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Condac E, et al. The C-terminal fragment of axon guidance molecule Slit3 binds heparin and neutralizes heparin's anticoagulant activity. Glycobiology. 2012;22:1183–92. doi: 10.1093/glycob/cws087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, Prestegard JH. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry. 2012;51:4618–26. doi: 10.1021/bi300319q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandersall-Nairn AS, Merkle RK, O'Brien K, Oeltmann TN, Moremen KW. Cloning, expression, purification, and characterization of the acid alpha-mannosidase from Trypanosoma cruzi. Glycobiology. 1998;8:1183–94. doi: 10.1093/glycob/8.12.1183. [DOI] [PubMed] [Google Scholar]
- 30.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–9. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 32.Hernaiz M, Liu J, Rosenberg RD, Linhardt RJ. Enzymatic modification of heparan sulfate on a biochip promotes its interaction with antithrombin III. Biochem. Biophys. Res. Commun. 2000;276:292–297. doi: 10.1006/bbrc.2000.3453. [DOI] [PubMed] [Google Scholar]
- 33.Schlessinger A, Punta M, Rost B. Natively unstructured regions in proteins identified from contact predictions. Bioinformatics. 2007;23:2376–2384. doi: 10.1093/bioinformatics/btm349. [DOI] [PubMed] [Google Scholar]
- 34.Schlessinger A, Punta M, Yachdav G, Kajan L, Rost B. Improved Disorder Prediction by Combination of Orthogonal Approaches. PLoS ONE. 2009;4:e4433–e4433. doi: 10.1371/journal.pone.0004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang F, Moniz HA, Walcott B, Moremen KW, Linhardt RJ, Wang L. Characterization of the interaction between Robo1 and heparin/glycosaminoglycans. Biochemie. 2013;95:2345–2353. doi: 10.1016/j.biochi.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prince RN, Schreiter ER, Zou P, Wiley HS, Ting AY, Lee RT, Lauffenburger DA. The heparin-binding domain of HB-EGF mediates localization to sites of cell-cell contact and prevents HB-EGF proteolytic release. J. Cell. Sci. 2010;123:2308–18. doi: 10.1242/jcs.058321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt M, Govindasamy L, Afione S, Kaludov N, Agbandje-McKenna M, Chiorini JA. Molecular characterization of the heparin-dependent transduction domain on the capsid of a novel adeno-associated virus isolate, AAV(VR-942). J. Virol. 2008;82:8911–6. doi: 10.1128/JVI.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavaré JM, Fletcher LM, Welsh GI. Using green fluorescent protein to study intracellular signalling. J. Endocrinol. 2001;170:297–306. doi: 10.1677/joe.0.1700297. [DOI] [PubMed] [Google Scholar]
- 39.Fukuhara N, Howitt JA, Hussain SA, Hohenester E. Structural and functional analysis of slit and heparin binding to immunoglobulin-like domains 1 and 2 of Drosophila Robo. J. Biol. Chem. 2008;283:16226–16234. doi: 10.1074/jbc.M800688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson ST, Bjork I, Sheffer R, Craig PA, Shore JD, Choay J. Role of the antithrombin-binding pentasaccharide in heparin acceleration of antithrombin-proteinase reactions. Resolution of the antithrombin conformational change contribution to heparin rate enhancement. J. Biol. Chem. 1992;267:12528–12538. [PubMed] [Google Scholar]
- 41.Futamura M, Dhanasekaran P, Handa T, Phillips MC, Lund-Katz S, Saito H. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J.Biol. Chem. 2005;280:5414–22. doi: 10.1074/jbc.M411719200. [DOI] [PubMed] [Google Scholar]
- 42.Schlessinger A, Rost B. Protein flexibility and rigidity predicted from sequence. Proteins. 2005;61:115–26. doi: 10.1002/prot.20587. [DOI] [PubMed] [Google Scholar]