Abstract
Autophagy as a conserved degradation and recycling process in eukaryotic cells, occurs constitutively, but is induced by stress. A fine regulation of autophagy in space, time, and intensity is critical for maintaining normal energy homeostasis and metabolism, and to allow for its therapeutic modulation in various autophagy-related human diseases. Autophagy activity is regulated in both transcriptional and post-translational manners. In this review, we summarize the cytosolic regulation of autophagy via its molecular machinery, and nuclear regulation by transcription factors. Specifically, we consider Ume6-ATG8 and Pho23-ATG9 transcriptional regulation in detail, as examples of how nuclear transcription factors and cytosolic machinery cooperate to determine autophagosome size and number, which are the two main mechanistic factors through which autophagy activity is regulated.
Keywords: autophagy, lysosome, phagophore, stress, vacuole, yeast
Background
Macroautophagy, hereafter referred to as autophagy, is a conserved process among eukaryotic cells, through which cytoplasmic components are delivered to the vacuole (in yeasts or plants) or lysosomes (in mammals) for degradation [1]. The autophagy-lysosome system and the ubiquitin-proteasome system (UPS) are the two major subcellular degradation systems. However, in contrast to the UPS, which is primarily a degradative pathway, autophagy has a much greater range of functions, participating in cellular adaptation and remodeling, the recycling of macromolecular building blocks, and even biosynthetic trafficking [2].
The morphological hallmark of autophagy is a double-membrane vesicle, termed an autophagosome, which is assembled de novo. That is, formation of the autophagosome occurs by a process that is distinct from vesicle formation throughout the secretory pathway. In the latter, transient transport vesicles bud off from a pre-existing organelle already containing their cargo [3, 4]. In contrast, the autophagosome is formed in a step-wise manner, providing a tremendous flexibility and capacity with regard to cargo sequestration. In yeast cells, autophagy occurs at a low basal, constitutive level. In response to various types of stress, or changing nutrient conditions, autophagy is upregulated. The induction of autophagy involves the recruitment of various autophagy-related (Atg) proteins to a peri-vacuolar site, termed the phagophore assembly site (PAS), where they nucleate and assemble into the initial sequestering compartment, a double-membrane structure named the phagophore. The phagophore can surround cytoplasmic materials randomly, or in the case of selective autophagy targeted cargo molecules are sequestered through direct interaction between receptors, scaffold proteins, and protein components in the phagophore membrane. Upon completion of elongation the phagophore seals to generate an intact double-membrane vesicle; at this stage, formation of the autophagosome is completed. Subsequently, the outer membrane of the autophagosome fuses with the vacuole, exposing the inner vesicle to the hydrolytic environment of the vacuole lumen. Breakdown of the vesicle membrane, and degradation of the cargo ensues (Figure 1). The resulting macromolecules are released back into the cytosol through various permeases and recycled [1].
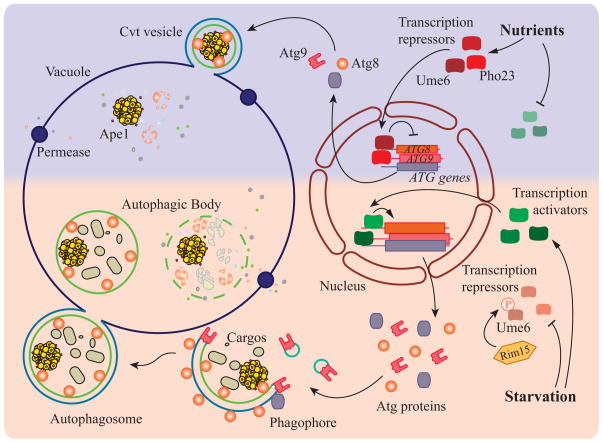
Figure 1.
A model of transcriptional regulation of autophagy in yeast. When nutrients are present, the transcription of most ATG genes is repressed due to the inhibition of autophagy transcription activators, and/or activation of transcription repressors. For example, Ume6 represses the transcription of ATG8, and Pho23 represses the expression of ATG9. Repression of transcription in growing conditions leads to a relatively low amount of Atg proteins that are sufficient for basal autophagy, and consequently the autophagosomes that are formed under these conditions are predicted to be smaller in size (due to limited Atg8) and generated at a slower rate (due to limited Atg9). The Cvt pathway, which is the main type of autophagy-like process in growing conditions, generates Cvt vesicles to sequester prApe1 dodecamers and deliver them to the vacuole to allow maturation of the zymogen. After starvation, transcription activators of autophagy become functional, and the repressors are inhibited. For example, Ume6 is phosphorylated by Rim15 kinase, releasing its repression of ATG8 transcription. Upregulation of ATG gene expression allows larger amounts of Atg components, such as Atg8 and Atg9, to participate in the formation of the autophagosome. As a result, more and larger autophagosomes are generated. During starvation-induced autophagy, Atg proteins are initially recruited to a peri-vacuolar site to generate the double-membrane structure named the phagophore. The phagophore randomly sequesters cytoplasmic material, and after the expansion phase is complete it seals to generate the double-membrane autophagosome. The outer membrane of the autophagosome fuses with vacuole, releasing the inner vesicle, now termed an autophagic body. This single-membrane vesicle along with its cargo is degraded in the vacuole lumen, and the resulting macromolecules are released back into the cytosol.
While the UPS is restricted to the degradation of ubiquitinated proteins that must be unfolded to gain access to the associated proteases, autophagy mediates the degradation of a wider range of targets, including soluble proteins, protein aggregates, damaged organelles, or even invasive pathogens. Nonselective autophagy is primarily a starvation response in which cytoplasm, potentially including organelles that are randomly sequestered, is delivered to the vacuole to provide macromolecules for catabolism (to provide energy), or for the synthesis of essential proteins. In contrast, during selective autophagy, the phagophore membrane is in close apposition to the cargo, preventing bulk cytoplasm from being sequestered [5]. Various types of cargo are selectively degraded through autophagy including protein aggregates (aggrephagy), mitochondria (mitophagy), peroxisomes (pexophagy), lipid droplets (lipophagy), and pathogens (xenophagy) [6]. Finally, although most types of autophagy are degradative in nature, selective autophagy can also serve a biosynthetic function. For example, during the cytoplasm-to-vacuole targeting (Cvt) pathway the precursor form of aminopeptidase I (prApe1) forms into a dodecamer in the cytosol. The dodecamer subsequently assembles into a larger self-interacting complex that is selectively sequestered by a phagophore. Upon completion, the double-membrane compartment is referred to as a Cvt vesicle. This vesicle targets to and fuses with the vacuole, again releasing the inner vesicle into the lumen. In this case, after lysis of the single-membrane vesicle, the contents are not degraded. Instead, a propeptide that keeps prApe1 inactive is proteolytically removed to generate the mature, active form of the hydrolase (Figure 1) [7].
Cytoplasmic regulation of autophagy: Atg proteins
Most of our understanding of the molecular machinery of autophagy has been achieved within the past two decades. In particular, studies relying on genetic analyses in yeast contributed to the identification of the majority of the known protein components that participate in autophagy. At present, more than 30 Atg proteins have been identified in yeast, and orthologs of most of these proteins are present in higher eukaryotes, indicating an evolutionarily conserved molecular machinery. A subset of the Atg proteins that is required for autophagosome formation (or for the generation of selective double-membrane compartments including the Cvt vesicle) is referred to as the molecular core machinery of autophagy. These proteins participate in different steps of autophagy, and, based on their roles in autophagosome formation and interaction with each other, they can be divided into several functional units.
Atg1 kinase complex and induction
The yeast Atg1 kinase complex contains the only kinase of the core machinery, Atg1, and its regulatory subunit, Atg13. The Atg1 complex senses the signals of autophagy induction, delivered from several upstream signaling inputs, such as the target of rapamycin (TOR) [8], protein kinase A (PKA), and Sch9 pathways [9–11]. Atg1 is a serine/threonine protein kinase [12,13], and the kinase activity of Atg1 requires its interaction with Atg13 [14]. During starvation-induced autophagy, Atg1 kinase activity is upregulated, which requires association of Atg1-Atg13 with a stable ternary complex composed of Atg17-Atg31-Atg29 [14–16]. Atg17 is proposed to be the first Atg protein that is recruited to the starvation-specific PAS upon autophagy induction (Atg11 serves this role in growing conditions [17]), and its correct localization is essential for the nucleation of other Atg proteins at this site [18]. Atg29 is a phosphoprotein that contains a C-terminal inhibitory domain; phosphorylation of the C terminus is necessary to relieve inhibition, although the mechanism and the relevant kinase are not known [19]. Similarly, the targets of Atg1 kinase that are involved in autophagy have not been well characterized. Atg1 can be autophosphorylated at Thr266 in the activation loop, and this modification is required for Atg1 kinase activity [20]. BECN1, the mammalian homolog of yeast Vps30/Atg6, is phosphorylated by ULK1 (a mammalian Atg1 homolog) [21], and Atg9 has been identified as one of the direct targets of Atg1 in yeast [22].
PtdIns3K complex and nucleation
Vps34 is the only phosphatidylinositol 3-kinase (PtdIns3K) in yeast, and the autophagy-specific PtdIns3K complex I, which is composed of Vps34, Vps15, Vps30, Atg14 and Atg38 is thought to act downstream of the Atg1 kinase complex (complex II, which contains Vps38 instead of Atg14, generates PtdIns3P at the endosome) [23–25]. The main function of the PtdIns3K complex is to generate PtdIns3P at the PAS, and recruit PtdIns3P binding proteins, such as Atg18, to the PAS [26, 27].
Ubl conjugation system and expansion
Atg8 and Atg12 are two ubiquitin-like (Ubl) proteins that belong to two distinct conjugation systems that are part of the core autophagy machinery. These proteins are not homologs of ubiquitin, but have structural similarity, containing ubiquitin folds. Both of the Ubl systems are essential for the expansion of the phagophore [28, 29]. A C-terminal glycine of Atg12 is stoichiometrically conjugated to an internal lysine of Atg5 in a process that is very reminiscent of ubiquitination; the first step involves activation of Atg12 by the E1-like enzyme Atg7 (a homolog of the ubiquitin-activating enzyme), which is followed by conjugation to Atg5 via an E2-like enzyme, Atg10 [29, 30]. An E3 ubiquitin ligase-like enzyme for Atg12–Atg5 conjugation has not been identified. Atg12–Atg5 further forms a complex with Atg16, a small coiled-coil protein [31], and the Atg12–Atg5-Atg16 complex forms larger oligomers via homo-oligomerization mediated by the Atg16 coil-coiled domain [32].
Atg8, the second Ubl protein of autophagy, undergoes a unique type of post-translational modification. Atg8 is initially synthesized with a C-terminal extension (a single arginine residue in yeast) that is proteolytically removed by the Atg4 cysteine protease. The same E1-like enzyme that is used in the Atg12–Atg5 conjugation system, Atg7, activates the modified Atg8; however, conjugation requires a separate E2-like enzyme, Atg3, which attaches the exposed C-terminal glycine to the lipid phosphatidylethanolamine (PE) [28]. The Atg12–Atg5-Atg16 complex may act as the E3 ligase for Atg8–PE conjugation [33], although it is not essential [34]. Besides its putative E3-like activity, Atg12–Atg5-Atg16 is also required for the PAS localization of Atg8 [18]. Unlike the Atg12–Atg5-Atg16 complex that localizes exclusively on the outer membrane of the phagophore [35], Atg8–PE is initially located on both sides of the phagophore during expansion; Atg8 on the outer surface of the completed autophagosome is removed by a second Atg4-dependent cleavage (referred to as deconjugation), whereas some of the Atg8 on the inner surface remains inside the completed autophagosome [36, 37]. The Atg8 that lines the concave surface of the phagophore plays an important role in cargo recognition during selective types of autophagy [38]. In contrast, the population located on the outer surface is involved in determining the size of the autophagosome and may play a role in the formation of a coat-like structure by interdigitating with the Atg12–Atg5-Atg16 complex [39, 40].
Atg9 and its cycling system
Atg9 is the only transmembrane protein in the core machinery [41]. Atg9 has a unique localization pattern different from the other core machinery proteins. Most Atg proteins are present in two populations, one being diffuse in the cytosol, and the second, that is thought to be the active pool, localized at the PAS; the latter appears as a single punctum in growing conditions that becomes much more intense (based on monitoring of fluorophore-tagged Atg proteins by microscopy) following autophagy induction. Interestingly, Atg9 localizes to multiple punctate structures. One of these punctate structures is the PAS, but the other sites are located proximal to the mitochondria; these latter populations are called Atg9 peripheral sites, Atg9 reservoirs or tubulovesicular clusters (TVCs) [41–43]. Based on studies with a temperature-sensitive Atg1 mutant, Atg9 is proposed to cycle between the peripheral sites/TVCs and the PAS. Anterograde movement to the PAS is dependent on Atg11, Atg23, Atg27 and the Arp2/3 complex, whereas retrograde trafficking depends on the Atg1-Atg13, and Atg2-Atg18 proteins [42, 44, 45]. The PtdIns3K complex is also required for Atg9 trafficking, which may reflect the fact that Atg18 is a PtdIns3P-binding protein. The exact function of Atg9 is not known, but it is proposed to be involved in delivering membrane to the PAS, or in directing this process. Membrane-bound Atg9 moves between the PAS and the peripheral sites/TVCs on single-membrane vesicles [46, 47]. Atg9 PAS localization is dependent on Atg17; the Atg17-Atg31-Atg29 complex forms a crescent-shaped structure that may recruit the Atg9 vesicles to the early phagophore [19, 48].
Physiology and pathology
Autophagy occurs at a constitutive basal level, but is upregulated when it is induced by different types of stress such as starvation or shifting to an alternative carbon source, hypoxia, or pathogen infection. In a nutrient-rich condition the basal level of autophagy is critical for normal homeostasis, performing housekeeping roles such as the removal of misfolded and damaged proteins, the biosynthetic delivery of vacuole-resident enzymes, or maintaining proper metabolism. Recent studies have mostly focused on stress-induced autophagy; however, basal autophagy plays a very important role as part of the quality control machinery, limiting the accumulation of damaged organelles and misfolded proteins that may contribute to many types of neurodegenerative diseases, such as Huntington, Alzheimer and Parkinson disease [49]. In the heart, basal autophagy is important for maintaining cardiomyocyte size and global cardiac structure and function [50], whereas in the liver it may play a role in preventing misfolding diseases such as α1-antitrypsin deficiency [51]. The degradation of depolarized mitochondria by basal mitophagy also prevents spurious inflammation caused by reactive oxygen species and DNA released from damaged mitochondria [52].
When a cell senses stress, autophagy is induced. Under starvation conditions, cells degrade proteins and lipids through autophagy to maintain energy homeostasis. Autophagy-defective cells display a tremendous reduction in viability during starvation, and autophagy-defective mice cannot survive after birth, when the maternal trans-placental nutrient supply is disrupted [53]. In addition autophagy triggered in oocytes by fertilization is essential for preimplantation development in mammals [54]. As mentioned above, autophagy is also induced by pathogen infection, and the selective autophagy of pathogens, xenophagy, can target and kill invasive pathogens, including bacteria and viruses; therefore, this process is essential for maintaining a healthy innate immune system. Conversely, some microbes have evolved to evade or even subvert autophagy for their own purposes, establishing a replicative niche within autophagosomes, or relying on autophagy to provide nutrients [55]. Besides its functions in preventing infectious diseases, autophagy is involved in immunity by delivering antigens for major histocompatibility (MHC) presentation. The intracellular substrates of autophagy can be loaded on MHC class II, which is usually considered to present extracellular antigens, explaining how the MHC class II pathway contributes to intracellular antigen presentation [56].
Too little autophagy is harmful, but too much autophagy can also be deleterious—uncontrolled autophagy may cause cell death. Autophagy functions as a mechanism of tumor suppression, but cancer cells can also use autophagy to help them to survive in a hypoxic and low-nutrient environment, or to carry out its cytoprotective function following anticancer treatments that may damage organelles. Furthermore, autophagy plays different roles in different tissues. Inhibition of autophagy in liver cells causes accumulation of hepatic lipids, while inhibition of autophagy in adipose tissue causes decreased white adipose mass. Therefore, a tight regulation of autophagy in terms of its magnitude and in a tissue-specific manner will be crucial for the therapy of autophagy-related diseases.
Nuclear regulation: transcriptional regulation of autophagy
Like many other cellular processes, the induction of autophagy involves not only the post-translational modification and action of the Atg proteins, but also a coordinated transcriptional activation of the ATG genes. The abnormal expression of many ATG genes has been observed in various human diseases [57–60] suggesting the physiological significance of transcriptional regulation of autophagy. Although the transcriptional induction of ATG genes was first reported more than ten years ago, at present our knowledge of the transcription factors that are involved in autophagy regulation is very limited, even in the yeast system, where the molecular mechanism of autophagy has been most intensively studied. Some progress has been made with regard to transcriptional regulation in the mammalian system in recent years, and we briefly highlight these findings along with data from yeast below.
FOXO transcription factors
The conserved forkhead box-containing protein O subfamily (FOXO) transcription factors regulate autophagy in different cell types and organisms. The FOXO proteins shuttle between the cytoplasm and the nucleus, which is regulated by phosphorylation, and they have both transcription-dependent and -independent roles in autophagy regulation. Here we consider the transcription-dependent roles of FOXO family members. There is only one FOXO protein in worms and flies, whereas there are four members, FOXO1, FOXO3, FOXO4, and FOXO6, in mammals [61]. In general, FOXO transcription factors function as activators of autophagy. Under stress stimuli such as nutrient deprivation, phosphorylation of FOXO proteins by AKT1 is blocked, and FOXO transcription factors translocate into the nucleus. In mouse muscle cells, some of the Atg genes including Lc3b, and Gabarapl1 are directly targeted and activated by FOXO3 as indicated by chromatin immunoprecipitation (ChIP) assays [62, 63]. In neurons, transcription of Atg genes such as Atg3, Atg5, and Atg12 is activated by FOXO1 [64]. Transcriptional regulation of autophagy by FOXO transcription factors has also been reported in various other types of cells, including cardiomyocytes, hepatocytes, and primary renal proximal tubular cells [61]. The positive regulation of autophagy by FOXO transcription factors in muscle is conserved in Drosophila [65]. In budding yeast, the role of Fhl1, the homolog of FOXO transcription factors, in transcriptional regulation of autophagy has not been directly tested. However, CHIP-chip data from several independent studies revealed that this protein directly targets to the promoters of several ATG genes including ATG13, ATG27 and ATG31 [8, 66], indicating that a conserved regulation may exist in the yeast system.
TFEB vs ZKSCAN3
The autophagosome itself is not a degradative compartment. Accordingly, the autophagosome cooperates in the degradation of its cargos with the lysosome in order to fulfill its roles in the degradation and recycling of cytoplasmic materials. Under starvation conditions, both autophagosome and lysosome biogenesis are upregulated. For the transcriptional regulation, two master switches, transcription factor EB (TFEB) as the activator, and ZKSCAN3 as the repressor, have been reported to regulate both autophagosome and lysosome biogenesis. Overexpression of TFEB induces autophagy, while TFEB RNA interference decreases autophagy. Starvation induces TFEB translocation to the nucleus and activates the transcription of several lysosomal and autophagy genes by directly binding to their promoters [67]. This organized pattern of control is referred to as the coordinated lysosomal expression and regulation (CLEAR) network [68]. Conversely, silencing ZKSCAN3, a zinc family DNA-binding repressor protein, upregulates the mRNA level of several autophagy and lysosomal genes, and ChIP experiments suggest that ZKSCAN3 represses transcription of these target genes by direct binding at the respective promoters [69]. Interestingly, in contrast to TFEB, ZKSCAN3 accumulates in the cytosol during starvation.
E2F1 vs NFKB
BNIP3 is an activator for hypoxia-induced autophagy [70], and its transcription is regulated in an opposite manner by two transcription factors, E2F1 and NFKB. Bnip3 is a direct target of E2F1, which functions as a transcription activator. NFKB competes with E2F1 for binding at the Bnip3 promoter, and therefore represses its expression [71]. Besides BNIP3, E2F1 also activates additional genes associated with autophagy including LC3, ULK1, and ATG5, and induces autophagy activity [72]. NFKB represses BNIP3, however, NFKB activates other ATG genes such as BECN1 [73].
GATA family factors
GATA family zinc finger transcription factors regulate nitrogen catabolite repression (NCR)-sensitive genes, which produce enzymes and permeases required for using an alternative (nonpreferred) nitrogen source. Gln3, a yeast GATA family transcription activator, is regulated by Tor, and Gln3 is reported to positively regulate ATG14 transcription [74]. This regulatory event is one of the first examples of transcriptional control of autophagy, and this regulation is conserved in Drosophila [75]. In yeast, GATA family factors include the relatively well-studied activators Gat1 and Gln3, and the repressors Gzf3 and Dal80. In addition to these factors, there are other DNA-binding proteins containing the conserved GATA-type zinc finger in yeast, such as Gat2, Gat3 and Gat4, whose functions are not clear. Whether other GATA family factors besides Gln3, have roles in the regulation of autophagy is not yet known.
In mammals, GATA1 activates the transcription of ATG4, ATG12, BNIP3, and mammalian homologs of ATG8 (MAP1LC3B, GABARAP, GABARAPL1, and GABARAPL2/GATE-16). GATA1 directly binds at the promoters of BNIP3, MAP1LC3B, GABARAP, and GABARAPL2 [76]. Interestingly, GATA1 activates the transcription of FOXO3, and transcriptional regulation of MAP1LC3B requires FOXO3. In contrast, knockdown of TFEB does not affect GATA1-mediated autophagy gene induction [76]. Another GATA transcription factor, GATA4, inhibits autophagy by inducing the transcription of BCL2, and repressing the transcription of ATG5, ATG7, ATG12, and BECN1 [77], which is surprising because GATA4 is generally known as a transcriptional activator. Whether GATA4 represses transcription of ATG genes through direct binding at their promoters is unclear at this time.
Other transcription factors
Other transcription factors such as TP53/p53 and STAT are involved in autophagy regulation, and their roles have been reviewed recently [78, 79]. The function of the yeast transcriptional repressor Ume6 in autophagy regulation is discussed in detail below.
How autophagy activity is regulated: Size and number of autophagosomes
Autophagy activity, which corresponds to the amount of cytoplasmic material, whether random portions of the cytoplasm or selective cargos, sequestered by autophagosomes per unit time, followed by degradation within the vacuole and permease efflux, is modulated primarily by two features of autophagosomes: Size and number.
Regulation of the size of autophagosomes
A Cvt vesicle, which is the predominant type of sequestering compartment formed in growing conditions, is approximately 140–160 nm in diameter. Upon autophagy induction, much larger autophagosomes are formed; nitrogen-starvation induced autophagosomes range from 300–900 nm in diameter [80]. Therefore, the mechanism involved in determining the size of the autophagosome, that is, which Atg (or other) proteins play a central role in the formation of the larger autophagosome is an interesting question. The flexibility in the size of autophagosomes allows the sequestration of different sizes and amounts of cargo by autophagy. During nonselective autophagy, premature closure of the phagophore would result in smaller autophagosomes, which would in turn attenuate the efficiency of autophagic sequestration and degradation. During selective autophagy, the size of the cargo may determine the size of the sequestering compartment directly through protein-protein interactions between the cargo/ligand-receptor and Atg8 family proteins that are located on the concave face of the phagophore. However, there is a limit to the size of the autophagosomes that form during selective autophagy, indicating that this simple model is not the complete explanation, and there likely exists other machinery (which may be the same as that used in nonselective autophagy) to control the expansion of the phagophore. Under physiological conditions the diameter of the Cvt vesicle is essentially the size of the cargo, the Cvt complex composed primarily of prApe1 and its receptor Atg19. When the prApe1 cargo protein is overexpressed, a larger Cvt complex is formed that cannot be efficiently sequestered by a Cvt vesicle [80]. However, after starvation, when autophagy is induced and larger autophagosomes are generated, the larger Cvt complex can be sequestered and delivered to the vacuole. For both mitophagy and pexophagy, a fission complex is needed for the efficient sequestration of the targeted organelles [81, 82], suggesting that matching the size of the cargo with the phagophore may be important. Therefore, for selective autophagy, the size of the autophagosome not only regulates efficiency, but may also be critical for the cargo selectivity.
A series of studies in our lab on ATG8 and its regulation provides some clues for the regulation of autophagosome size. The amounts of both ATG8 mRNA and its protein product are elevated dramatically within a short time following nitrogen starvation or rapamycin (an inhibitor of TOR) treatment. To reveal the roles of the increased level of Atg8 during autophagy induction, several strains with various, but reduced, expression levels of ATG8 were generated through the use of heterologous promoters. Based on transmission electron microscopy, in conjunction with other autophagy assays, the correlation between Atg8 protein amount and autophagosome size/autophagy activity was determined [39]. It is still unclear how Atg8 regulates the size of the autophagosome. The observation that the Atg12–Atg5-Atg16 complex together with Atg8 can form highly oligomerized network structures on giant unilamellar vesicles in vitro, indicates the possibility that Atg8 along with the Atg12–Atg5-Atg16 complex forms a protein coat on the outer membrane of the phagophore to regulate its curvature, as discussed above [40]. As PE, a cone-shaped lipid that is the target of Atg8 conjugation, can modulate membrane curvature [83], Atg8 may also regulate the size of the autophagosome through its binding to PE, possibly due to differential distribution on the two faces of the phagophore.
Considering that the amount of Atg8 plays an important role in determining the size of the autophagosome, we became interested in understanding how the transcription of ATG8 is regulated. We identified a transcriptional repressor, Ume6, which can directly bind at the promoter of ATG8 and suppress its transcription in nutrient-rich conditions. In agreement with our previous study, ume6, cells, which express an elevated amount of Atg8 even in nutrient-rich conditions, generate larger autophagosomes than the wild-type cells. Phosphorylation of Ume6 by Rim15 after starvation releases its suppression, and these findings suggest a model for a regulatory signaling axis of autophagy: In nutrient-rich conditions upstream sensors such as PKA inhibit the effector kinase Rim15, resulting in active Ume6 that suppresses ATG8 transcription; the resulting low level of Atg8 is sufficient to generate the small Cvt vesicles. During starvation, inhibition of PKA allows the now active Rim15 to inhibit Ume6; the resulting increase in Atg8 allows the formation of larger autophagosomes (Figure 1) [84]. Therefore, the Ume6-dependent control of ATG8 transcription is an example of size regulation during autophagosome formation.
Regulation of the number of autophagosomes
The second feature, which is as important as the size of autophagosomes, is their number, which can be viewed as the frequency of autophagosome formation. In budding yeast cells, usually only one PAS is observed, but the frequency of autophagosome formation can be increased by autophagy induction to approach a condition of continuous and accelerated autophagy. Data suggest that the half-life of prApe1 processing through the Cvt pathway is approximately 45 min [85], whereas after starvation-induction the half-life of an autophagosome is approximately 10 min [86]. There are multiple PAS sites in fission yeast, and in mammalian cells phagophore nucleation is not limited to a single location. In these systems, as a consequence of increased autophagosome formation frequency, the number of PAS is increased by autophagy induction. It had been not clear, however, which Atg proteins play a role in determining the number of autophagosomes. From our recent studies of transcriptional regulators of autophagy, we found that cells deleted for the PHO23 gene, encoding a transcription repressor, formed more (but not larger) autophagosomes relative to the wild-type cells. Pho23 regulates transcription of more than one ATG gene, but ATG9 expression is changed the most among these genes in the pho23Δ mutant (Figure 1) [87]. To determine the contribution of Atg9 to the pho23Δ phenotype we used yeast strains with different ATG9 gene expression levels. We found that the amount of Atg9 protein correlated with the numbers of autophagic bodies, supporting our hypothesis that Atg9 levels determine the number of autophagosomes [87]. Considering the role of Atg9 in autophagy as a potential membrane carrier or in directing the delivery of membrane to the PAS, we conclude that Atg9 may regulate the rate of phagophore expansion, and consequently the time required for generating each autophagosome through its cycling between the PAS and the peripheral sites/TVCs.
Mathematical modeling of the energy status of double-membrane structures suggests that a minimal change of the rim curvature on the edge of the double-membrane sheet may drive its bending and closure to form a vesicle [88]. Interestingly, Atg9 appears to localize only at the edge of the phagophore [89], suggesting another possibility that Atg9 regulates closure of the phagophore by affecting the rim curvature on the edge of this structure. If this is the case, the Atg17-Atg31-Atg29 scaffold, which forms a crescent structure with a similar curvature as that of the phagophore rim and also only localizes on the edge of the phagophore, may cooperate with Atg9 in this aspect of autophagy regulation [19, 48, 89]. Our studies with Atg8 and Atg9 indicate that the size and number of autophagosomes may be separately regulated by different autophagy components. This makes sense considering that modulating size may affect primarily cargo selectivity, while regulating the number of autophagosomes could be carried out to regulate mostly autophagy flux. Interestingly, Pho23 and Ume6 both play roles in the Rpd3 large complex, but Pho23 regulates ATG9 transcription in a Ume6-independent manner, suggesting a dual function of the Rpd3 complex in regulating autophagy activity.
Future directions
How the size and number of autophagosomes are regulated by different subgroups of Atg proteins is an interesting question to pursue, as it will reveal the different modulations of autophagy activity and selectivity by different autophagy proteins. Furthermore, identifying new transcriptional regulators of autophagy and the corresponding gene targets will help to dissect their functions in modulating autophagy activity. Considering that most regulatory pathways of autophagy are conserved among different organisms, a future genomic study in yeast to reveal the autophagy transcriptome would provide valuable data for these studies, and may provide information that can be used for the therapeutic treatment of autophagy-related diseases.
Abbreviations
- Ape1
aminopeptidase I
- Atg
autophagy-related
- ChIP
chromatin immunoprecipitation
- Cvt
cytoplasm-to-vacuole targeting
- TFEB
transcription factor EB
- UPS
ubiquitin-proteasome system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–61. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–5. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- 4.Kovács AL, Palfia Z, Rez G, Vellai T, Kovacs J. Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy. 2007;3:655–62. doi: 10.4161/auto.4590. [DOI] [PubMed] [Google Scholar]
- 5.Sawa-Makarska J, Abert C, Romanov J, Zens B, Ibiricu I, Martens S. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat Cell Biol. 2014;16:425–33. doi: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mijaljica D, Nazarko TY, Brumell JH, Huang WP, Komatsu M, Prescott M, Simonsen A, Yamamoto A, Zhang H, Klionsky DJ, Devenish RJ. Receptor protein complexes are in control of autophagy. Autophagy. 2012;8:1701–5. doi: 10.4161/auto.21332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–66. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–9. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–50. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 13.Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3875–83. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–53. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. A multiple ATG gene knockout strain for yeast two-hybrid analysis. Autophagy. 2009;5:699–705. doi: 10.4161/auto.5.5.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–81. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 19.Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A. 2013;110:E2875–84. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185:871–82. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan K-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, Lee SS, Brezovich A, Lou JH, Turk BE, Aebersold R, Ammerer G, Peter M, Kraft C. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–83. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203:299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jao CC, Ragusa MJ, Stanley RE, Hurley JH. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci U S A. 2013;110:5486–91. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–66. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stromhaug PE, Reggiori F, Guan J, Wang C-W, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–66. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 29.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 30.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–41. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–96. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–25. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 33.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 34.Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182:703–13. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–51. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 37.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–81. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 41.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–80. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 43.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–22. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–38. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 45.Yen W-L, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007;18:581–93. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–9. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–12. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–97. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 50.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 51.Perlmutter DH. Autophagic disposal of the aggregation-prone protein that causes liver inflammation and carcinogenesis in α-1-antitrypsin deficiency. Cell Death Differ. 2009;16:39–45. doi: 10.1038/cdd.2008.103. [DOI] [PubMed] [Google Scholar]
- 52.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol. 2012;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 54.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–20. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 55.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 56.Nedjic J, Aichinger M, Mizushima N, Klein L. Macroautophagy, endogenous MHC II loading and T cell selection: the benefits of breaking the rules. Curr Opin Immunol. 2009;21:92–7. doi: 10.1016/j.coi.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 57.Chen D, Pang S, Feng X, Huang W, Hawley RG, Yan B. Genetic analysis of the ATG7 gene promoter in sporadic Parkinson’s disease. Neurosci Lett. 2013;534:193–8. doi: 10.1016/j.neulet.2012.12.039. [DOI] [PubMed] [Google Scholar]
- 58.Liu H, He Z, von Rutte T, Yousefi S, Hunger RE, Simon HU. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med. 2013;5:202ra123. doi: 10.1126/scitranslmed.3005864. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Pan XL, Ding LJ, Liu DY, Da-Peng L, Jin T. Aberrant expression of Beclin-1 and LC3 correlates with poor prognosis of human hypopharyngeal squamous cell carcinoma. PLoS One. 2013;8:e69038. doi: 10.1371/journal.pone.0069038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jo YK, Kim SC, Park IJ, Park SJ, Jin DH, Hong SW, Cho DH, Kim JC. Increased expression of ATG10 in colorectal cancer is associated with lymphovascular invasion and lymph node metastasis. PLoS One. 2012;7:e52705. doi: 10.1371/journal.pone.0052705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 63.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 64.Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–22. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–25. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, Pugh BF. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell. 2011;41:480–92. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–66. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 69.Chauhan S, Goodwin JG, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaw J, Yurkova N, Zhang T, Gang H, Aguilar F, Weidman D, Scramstad C, Weisman H, Kirshenbaum LA. Antagonism of E2F-1 regulated Bnip3 transcription by NF-κB is essential for basal cell survival. Proc Natl Acad Sci U S A. 2008;105:20734–9. doi: 10.1073/pnas.0807735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 73.Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan TF, Bertram PG, Ai W, Zheng XF. Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J Biol Chem. 2001;276:6463–7. doi: 10.1074/jbc.M008162200. [DOI] [PubMed] [Google Scholar]
- 75.Bánréti A, Lukácsovich T, Csikós G, Erdélyi M, Sass M. PP2A regulates autophagy in two alternative ways in Drosophila. Autophagy. 2012;8:623–36. doi: 10.4161/auto.19081. [DOI] [PubMed] [Google Scholar]
- 76.Kang YA, Sanalkumar R, O’Geen H, Linnemann AK, Chang CJ, Bouhassira EE, Farnham PJ, Keles S, Bresnick EH. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012;32:226–39. doi: 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi S, Volden P, Timm D, Mao K, Xu X, Liang Q. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem. 2010;285:793–804. doi: 10.1074/jbc.M109.070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol. 2013;23:310–22. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 79.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 80.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–95. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mao K, Liu X, Feng Y, Klionsky DJ. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy. 2014;10:652–61. doi: 10.4161/auto.27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013;1831:543–54. doi: 10.1016/j.bbalip.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 84.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, Inoki K, Klionsky DJ. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A. 2012;109:11206–10. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–99. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008;182:129–40. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014 doi: 10.1016/j.cub.2014.04.048. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Knorr RL, Dimova R, Lipowsky R. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS One. 2012;7:e32753. doi: 10.1371/journal.pone.0032753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–44. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]