Summary
The gut microbiota co-develops with the immune system beginning at birth. Mining the microbiota for bacterial strains responsible for shaping the structure and dynamic operations of the innate and adaptive arms of the immune system represents a formidable combinatorial problem but one that needs to be overcome to advance mechanistic understanding of microbial community-immune system co-regulation, and in order to develop new diagnostic and therapeutic approaches that promote health. Here, we discuss a scalable, less biased approach for identifying effector strains in complex microbial communities that impact immune function. The approach begins by identifying uncultured human fecal microbiota samples that transmit immune phenotypes to germ-free mice. Clonally-arrayed sequenced collections of bacterial strains are constructed from representative donor microbiota. If the collection transmits phenotypes, effector strains are identified by testing randomly generated subsets with overlapping membership in individually-housed germ-free animals. Detailed mechanistic studies of effector strain-host interactions can then be performed.
Introduction
First the good news: myriad aspects of host physiology, metabolism and immunity are being linked to the human gut microbiota. Now the disconcerting news: one of the biggest challenges for current research in this area is how to move from observational studies where the configuration of a person’s microbiota is correlated with their health status, to mechanistic studies that dissect how organisms impact host biology, and how we might take advantage of the knowledge gained to develop new medical treatments.
Our adult intestines harbor a microbiota composed of tens of trillions of microbes representing all three domains of life (and their viruses); most belong to the domain Bacteria. Current evidence from studies of healthy adults living in the USA or Europe is that we harbor roughly 100 species of gut bacteria, various combinations of which could be responsible for various functions and effects (Qin et al., 2010; Faith et al., 2013). This means that we have an immense combinatorial problem namely, how to efficiently search the vast number of possible species combinations to find those “effector strains” that individually or in concert affect one or more of our biological features. The lack of unbiased methods to identify the contributions of various community members to different human phenotypes has hampered efforts to harness the diagnostic potential of the microbiota. These effector strains also represent the starting point for discovery and development programs designed to identify next-generation probiotics for new types of microbiota-directed therapeutics, including but not limited to those involving various aspects of immune function.
Previous Approaches For Identification Of Effector Bacterial Strains
A widely practiced approach for identifying effector strains has been to employ culture-independent, DNA sequence-based methods to compare the abundance of bacterial taxa in gut microbiota associated with a given host phenotype to their abundance in communities where this phenotype is quantitatively or qualitatively different. This search is less daunting in mouse models since host genetic and environmental factors can be constrained in ways that are not practically or ethically feasible in human populations. The hope is that what is found in the animal models will be translatable to humans.
A ‘classic’ illustration of this approach involves segmented filamentous bacteria (SFB) and its discovery as an effector of intestinal T helper 17 (Th17) cell responses. Comparing the gut microbiota of conventionally-raised C57BL/6 mice from Jackson Labs that lack Th17 cells in their small intestinal lamina propria to the gut microbiota of C57BL/6 mice from Taconic Farms that have abundant Th17 cells in this compartment (Ivanov et al, 2008), revealed two bacterial taxa that were significantly enriched (>25 fold) in mice originating from Taconic Farms. One was SFB, which was subsequently shown to be sufficient in of itself for Th17 cell induction (Ivanov et al, 2009; Gaboriau-Routhiau, 2009). Unfortunately, such clean-cut examples are rare. Add in another fact of life: differences in abundance alone may not be the best metric to use when searching for members of a gut microbial community that are causally related to an immune phenotype. For example, Bloom and coworkers used a mouse model of inflammatory bowel disease (IBD) where signaling of the anti-inflammatory cytokines TGF-β and IL-10 was abrogated to show that Bacteroides thetaiotaomicron was causally related to severe intestinal pathology even though its relative abundance in the microbiota was not significantly different in inflammatory versus non-inflammatory states, whereas E. coli, which was enriched under inflammatory conditions, lacked colitogenic activity (Bloom et al., 2011). In another model of IBD involving compound homozygous T-bet- and Rag-deficient mice, Klebsiella pneumoniae and Proteus mirabilis were associated with inflammation but insufficient to produce disease when transferred to germ-free recipients. However, they were pathogenic when added to specific pathogen-free (SPF) mice with already established gut communities (Garrett et al., 2010).
The Challenge of Looking Beyond Species-level Diversity to Strain-level Diversity
Another challenge is that we need to look beyond (bacterial) species-level diversity and consider that different strains of a given species-level phylogenetic type may have considerably different biological effects. This strain level diversity exists within individuals. Until recently, it has been difficult to reliably assign strain-level designations to bacterial members of the microbiota using high-throughput DNA sequencing methods that focus on amplicons generated from the phylogenetic marker gene encoding the principal ribosomal RNA molecule in the small ribosomal subunit (16S rRNA). However, this problem is being overcome with new low-error amplicon sequencing techniques (Faith et al., 2013).
Recent studies emphasize the importance of considering strain-level differences. For example, analysis of Il10−/− mice treated with the alkylating mutagen azoxymethane (AOM), revealed that E. coli strains possessing the polyketide synthase (pks) genotoxic island elicit DNA damage in cultured intestinal epithelial cells and promote tumorigenesis in the colon. Neither a naturally occurring pks-minus strain of E. coli, nor one with an engineered deletion of pks elicited DNA damage in vitro to an extent equivalent to that of the pks-positive strain. Moreover, AOM treated Il10−/− recipients of the deletion mutant displayed reduced tumor multiplicity and invasion relative to recipients of the pks-positive strain (Arthur et al., 2012). In multiple intestinal neoplasia (Min) mice heterozygous for a null allele of the Apc gene, a B. fragilis strain expressing the toxin BFT produced colitis while strains lacking the toxin did not (Wu et al., 2009). These findings suggest that surveying bacterial isolates from gut microbiota samples obtained from an individual with a given immune phenotype, rather than ‘domesticated’ strains of the same species that are available as ‘off the shelf’ reagents from existing culture repositories, may be a more informative way for identifying effector strains and characterizing the nature of their interactions with a host.
The Challenge of Capturing the Bacterial Diversity Present in the Gut in Culture
Other challenges need to be added to the list. A number of labs have worked to improve our ability to culture the range of bacterial diversity present in the human gut microbiota. This cultured diversity can be quantified either in terms of community membership (e.g., 50 out of 100 total strains) or proportionally weighted by the abundance of each microbe (e.g., 80% of the sequenced metagenome). Using a single rich medium and anaerobic growth conditions, approximately 50% of the bacterial species-level taxonomic diversity (97%ID OTUs) and 60% of the proportionally weighted genus-level diversity present in previously frozen fecal samples can be captured in clonally arrayed culture collections generated by limiting dilution in multi-well format or by direct plating (Goodman et al., 2011 Ridaura et al., 2013; Faith et al., 2013). This approach allows a collection of bacterial strains that have co-existed and co-evolved in an individual’s gut to be recovered, archived and characterized in vitro and/or in vivo. Lagier et al. (2012) tested 212 different culture conditions and identified 20 different types of media that were sufficient to isolate over 70% of the bacterial strain-level diversity present across three individuals. Nonetheless, much work remains to be done to further expand our ability to culture previously unculturable microbes that inhabit this ecosystem.
Applying high-fidelity, low error 16S rRNA amplicon sequencing methods (e.g., LEA-Seq) to microbiota samples (Faith et al., 2013) enables more precise quantification of the efficiency of isolation at the strain-level and the potential to quantify cultured diversity by community membership: this capacity enables media conditions to be optimized so that the largest proportion of a microbiota can recovered with a minimized number of culture conditions.
Using the 16S rRNA gene to confidently identify strain-level variants of a given species requires a high degree of sequencing accuracy. This can be achieved with redundant sequencing of the DNA template. The goal for redundant sequencing is to generate fewer molecules to be sequenced than the available sequencing depth. For example, if the aim is to sequence 1,000,000 molecules in a sample, by starting with only 10,000 molecules of template you have the potential to sequence each template, or at least a copy of each template, up to 100 times. This redundancy enables error-correction of the resultant reads as multiple sequences from each template DNA molecule allow removal of errors as long as the downstream sequencing technology is unbiased in its error profile and generates true nucleotides more often than erroneous ones. This reduction in the number of template molecules relative to the sequencing capacity is the opposite of what is done in most sequencing contexts where a vast excess of template molecules are present and a subset are sequenced. To create a finite DNA pool that is smaller than the amount of sequencing effort applied (that is, to create a bottleneck), a method for labeling the molecules in the pool is needed. One approach for redundantly sequencing PCR amplicons is to dilute the initial template DNA to create a bottleneck. However, this dilution would need to be determined for every input sample. In LEA-Seq, a bottleneck is created with a linear PCR extension of the template amplicon using a dilute barcoded oligonucleotide primer solution. Each oligonucleotide is labeled with a random barcode, 5′ to the universal 16S rRNA primer sequence: the complexity of barcodes is far greater than the number of unique amplicons to be sequenced, thus ensuring that having the same barcode attached to multiple amplicons is an extremely improbable event. Another feature of the primer design is placement of a short adapter sequence at its 5′ end that contains a fragment of the sequencing primer needed for the DNA sequencing platform of choice. Placing this primer (or any adapter region) on the 5′ end enables the unique amplification of the linear PCR only without the need to remove existing template DNA or purify the linear primer. This feature enables an “add-only” reaction with few steps and ease of automation.
The Challenge of Improving the ‘Throughput’ for Functional Analyses of Microbial Consortia in Gnotobiotic Mice
Another obstacle that stands squarely in the path of systematic dissection of the contributions of gut microbial community members to host phenotypes is the way gnotobiotic experiments are currently performed (Faith et al., 2010). The development of gnotobiotic animal models has enabled the effects of individual microbial species or strains, or consortia composed of defined collection of organisms recovered from a human gut microbiota to be assayed. Traditionally, germ-free animals are housed inside a gnotobiotic isolator; controlled entry of sterilized food, water, bedding and other consumables allows experimenters to maintain animals under germ-free conditions until such time as they are colonized (‘gnotobiotic’ is derived from the Greek ‘gnosis’, meaning known, and ‘bios’, meaning life). Although many cages can be housed inside one large gnotobiotic isolator, it is virtually impossible to maintain multiple cages, each containing mice harboring different microbial communities, in the isolator without cross-cage contamination. Therefore, testing large numbers of communities is impractical given the large number of gnotobiotic isolators that would be required.
Past Successes in Identifying Bacterial Strains that Modulate Regulatory T cells
Despite these challenges, researchers have succeeded in identifying bacterial species that modulate immune responses. One key area where great success has been achieved is in examining effects on regulatory T cells. Germ-free mice have a paucity of Tregs in their colonic lamina propria compared to mice harboring a gut microbiota (Atarashi et al., 2013; Atarashi et al., 2011; Geuking et al., 2011; Weiss et al., 2012). Thus, the expansion of the colonic Treg compartment and the induction of peripheral Tregs (pTregs) in the colon (Atarashi et al., 2013; Atarashi et al., 2011; Geuking et al., 2011; Lathrop et al, 2011; Round and Mazmanian, 2010; Weiss et al., 2012), represents an attractive model for assessing how gut bacteria invoke tolerance to foreign antigens; the results have therapeutic relevance given the role of pTreg in preventing inflammation (Bilate and Lafaille, 2011; Curotto de Lafaille and Lafaille, 2009; Haribhai et al., 2009; Haribhai et al., 2011; Mucida et al., 2005). One approach used to identify these effector strains has been to fractionate microbial communities to reduce the diversity of potential candidates that mediate an immune phenotype. In an elegant set of studies, Atarashi and colleagues treated fecal microbiota from mice (Atarashi et al., 2011) or a human donor (Atarashi et al., 2013) with chloroform to enrich for spore-forming bacteria that survive this harsh treatment. This allowed isolation of a collection of chloroform-resistant Clostridia strains that boosted colonic Treg cell generation and/or suppressive function, and promoted pTreg cell accumulation in gnotobiotic mice. Nonetheless, approaches that selectively kill particular subsets of bacteria, including those that use narrow spectrum antibiotics (Ivanov et al., 2008; Atarashi et al., 2011) are biased and impede characterization of immune phenotypes that require the concerted action of bacterial strains with very different phylogenetic, physical, physiologic, and/or metabolic properties.
A Path Forward for Overcoming the Combinatorial Challenge in Identifying Effector Strains
Although the picture we have painted appears intimidating, newer methods will help overcome these obstacles. Whereas no single approach is devoid of limitations, the one outlined below should help advance studies aimed at understanding the interplay between microbes and the immune system.
Exhaustively testing all combinations of microbes (bacteria) for their ability to modulate a given phenotype requires testing an inordinate number of combinations – requiring more than a thousand, million, and billion combinations of strains, even for smaller communities of 10, 20, and 30 strains respectively. Thus, a brute-force approach is not feasible. Heuristic approaches are needed to more efficiently sort through this complexity. Given that many microbe-driven immune phenotypes are likely to be partially additive and involve no more than a few higher-order interactions (i.e., situations where two or more specific microbes must be present to elicit a response), this complexity can be reduced dramatically (Box et al., 2005; Hastie et al., 2009).
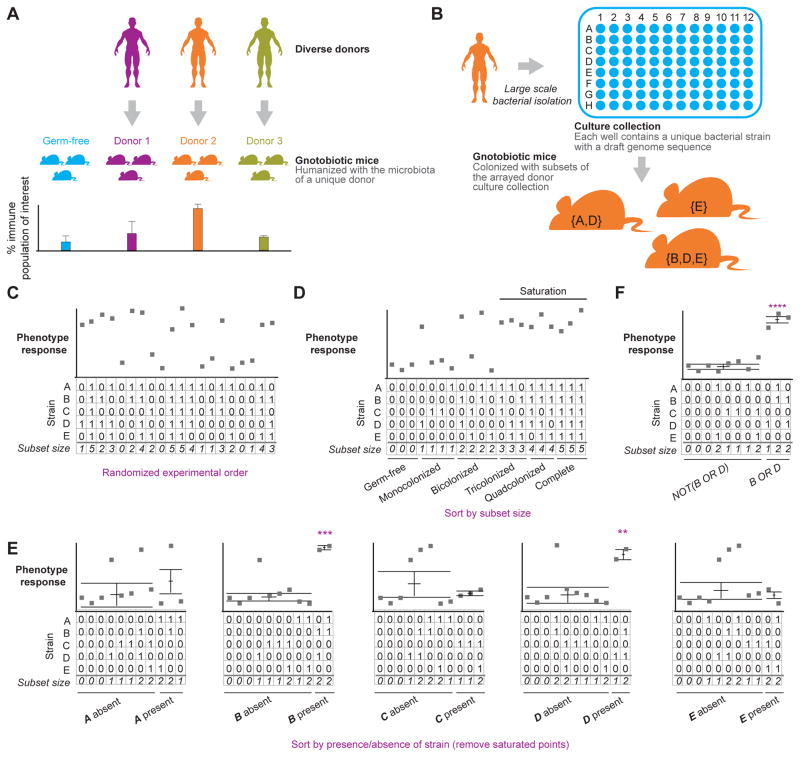
Fig. 1 describes an approach for tackling the combinatorial challenge faced by microbiota researchers. The initial step is to transplant intact uncultured human gut (fecal) microbiota samples representing different donor physiologic or disease states, or lifestyles, or geographies of interest, into germ-free mice. These screens test the ability of donor microbiota to transmit a whole range of phenotypes to recipient animals, from effects on metabolism to modulating immune function. By screening multiple donors it is possible to ascertain whether the responses of gnotobiotic transplant recipients occur with all or a majority of tested microbiota, or are highly donor-specific (Fig. 1A). The information gleaned is then used to identify microbiota samples that induce a response that is representative of the donor population or subpopulations.
Figure 1. An experimental pipeline for dissecting the interactions of effector strains present in complex microbial communities with the host immune system.
(A) The first step involves transplanting intact uncultured fecal microbiota from human donors to test the capacity of their gut microbial communities to impact immune phenotypes in recipient germ-free mice. In the case shown, the effects of different donor microbiota on the percent representation of an immune cell type of interest within a population of immune cells recovered from recipient gnotobiotic mice is plotted. Note that in principle, the approach can be applied to any phenotype of interest. (B) A representative human donor microbiota, nominated from the initial screen, is used to prepare a clonally arrayed culture collection in a multi-well plate, and the genome of each cultured member is sequenced. Subsets of members of this community are generated: subsets are either rationally designed, or cultured strains are randomly assigned to subsets. Subsets (defined consortia) are then administered to recipient gnotobiotic mice. (C) The order the subsets are introduced into different mice is randomized to minimize batch effects and biases, and the phenotype of interest is assayed. (D) Sorting the tested subsets by their size (i.e., the total number of strains in the subset) enables estimation of the phenotype’s saturation point, if there is one. In the situation depicted, one can begin to observe phenotype saturation with subset sizes of three, while higher variation is observed in community sizes smaller than this. Therefore, using mice that are mono- or bi-colonized would be an effective strategy for identifying which strains modulate this phenotype. (E) In cases where few members contribute to the phenotypic variation, effector strains can be identified by sorting the subsets based on the presence or absence of each strain individually, with p-values determined using a t-test. In the example shown, strains A, C and E have little effect on the phenotype being measured whether they are present or absent, while strains B and D are present in most cases where the phenotype is observed. (F) More complex interactions involving multiple effector strains can be more accurately inferred with model-based approaches that consider the combined influence of all community members on the phenotype. In the example shown, feature selection can be used to demonstrate that the phenotype is manifest in cases where strain B or D are present and absent in all cases where B and D are absent, thus identifying the strains B or D as key effector strains for this phenotype. In E and F, the central horizontal bars show the mean value and the error bars represent the SEM. Each point represents a hypothetical response measured in an individual recipient mouse.
The second step is to determine whether the culturable component of a representative microbiota sample can transmit the phenotype or phenotypes observed with the intact uncultured sample (Fig. 1B). A system for generating arrayed collections of cultured anaerobic bacterial members of a human gut microbiota sample in multi-well plates has been described (Goodman et al., 2011). Based on Poisson distribution, a dilution of the sample is selected so that there is 95% probability that if growth is observed in a given well of the multi-well plate, then that well will be monoclonal (i.e. growth derives from a single starting bacterial cell). In general, this means that the dilution will yield growth in one third of the wells. The resulting library of primary isolates is then ‘compressed’ robotically so that derivative multi-well plates are constructed where each well contains a cultured strain. The identity of each isolate in the clonally arrayed library is then determined by first sequencing its 16S rRNA gene and then its whole genome. In principle, altering the dilution used to generate the initial library to increase the number of starting cells in each well from one to two provides a way to increase the yield of culturable organisms, since those strains that have obligate nutrient sharing (syntrophic) relationships with other strains may survive together whereas in the absence of the partner, neither is culturable (the ‘cost’ of this approach is that simultaneous shotgun sequencing of the genomes of the two organisms that occupy a given well requires the ability to accurately assemble each strain’s genome). Whereas the majority of culturing efforts have focused on aerobic, microaerophilic or anaerobic bacteria, analogous strategies could be used to recover members of other domains of life represented in the microbiota to assess their contributions to host biology.
Having a clonally arrayed library of cultured bacterial isolates that have co-evolved together in a given donor’s gut sets the stage for selecting subsets of the collection for testing in gnotobiotic mice. An empirical approach is utilized that allows identification of the size of consortia derived from a culture collection that are capable of generating high variation in a host phenotype of interest. This process involves a number of steps. First, subsets of a nominal size (e.g. 3, 5, 10 members, etc.) are generated (Fig. 1B). Each subset of a given size represents a community of unique composition but with overlapping membership. Second, each of these communities is introduced into a germ-free recipient mouse, with those harboring each subset housed separately (see below). The phenotype of interest is then measured in recipient mice after a defined period of time has elapsed following oral gavage of the consortia (Fig. 1C). Both the composition of each subset of bacterial strains and the order in which the subsets are tested are randomized to minimize biases from strain selection, and batch effects. Biological repetition for each bacterial strain is obtained because subsets have overlapping membership. Strains whose presence in a community best explain the observed phenotypic variation are identified (i.e., those strains that are common to subsets capable of inducing the phenotypic response) and these are then advanced to a validation step where they alone, or in combination with other microbes whose presence are necessary for the phenotype to manifest, are introduced into germ-free recipients.
A key element in these approaches is the concept of phenotype saturation, which refers to the situation where multiple strains individually or in concert can induce a response such that the addition of more effector bacterial strains no longer alters the response (Fig. 1D). To identify potential effector strains there must be biological variation in the phenotype across the tested subsets. If all tested subsets of given size induce the phenotype relative to a germ-free control, and no subset induces a response significantly greater than any other, then the only information gained from each subset is that its evoked host response differs from what is observed in a germ-free mouse. However, if some subsets induce a response and others do not, orif some subsets induce a more intense response than others, then it is possible to use strain membership between subsets to identify modulators (effectors) of the phenotype (Fig. 1E). The number of subsets tested and subset size need to be determined empirically for each phenotype and will depend on the number of effector strains for the phenotype and the saturation profile. A good initial starting point is to first test the extremes, as large subsets and the complete cultured community will determine if the effector strains are in the culture collection while smaller communities (<4 strains) will identify if the phenotype can be saturated by a large proportion of the community members.
A Conceptual Illustration of the Approach
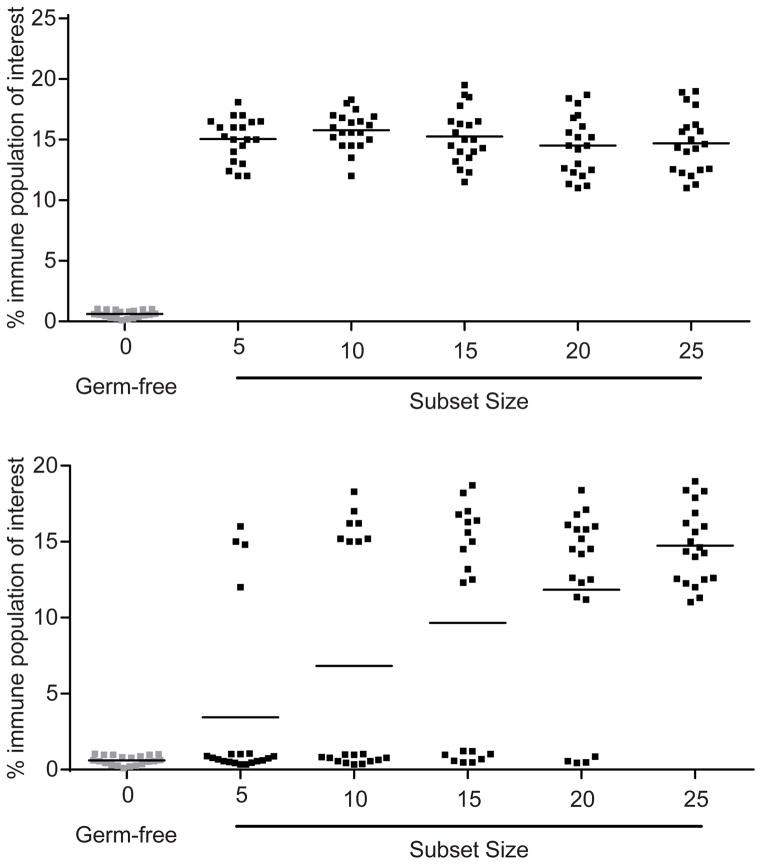
This approach can be illustrated by considering an imagined example where differentiation of an immune cell population of interest occurs upon colonization with a 25-member community. If we imagine a situation where all microbes present in this community could induce these cells, then regardless of subset size, no variation will be observed and increased numbers of cells will be seen in recipients of all communities (Fig. 2A). Thus, although community composition will be different across many subsets, this cannot be linked to variation in the response itself and no information can be gleaned from the study other than the fact that a large proportion of the community (perhaps all members) can elicit the response. If we now consider that the response is driven by a single strain, and that no other members of the culture collection possess this capacity, we will observe a different pattern. The only mice that should display an increase in these immune cells will be those recipients of a subset containing this unique effector strain; significant variation will be evident in the cellular response of recipient mice since the strain will not be present in every subset (recall that strain distribution among the subsets is random). From a probabilistic point of view, as community subset size is altered, variation in response among gnotobiotic recipients will change; e.g., the more members present in a given subset, the greater the likelihood that any one particular member, such as the hypothetical unique effector strain, will be present (Fig. 2B). Variation will be at its highest when 50% of the community is present, because half of the subsets will contain the effector strain while half will not. Smaller subset sizes will have lower variation because most gnotobiotic recipients will lack the effector strain and thus lack the immune cells being studied. Larger subset sizes will have lower variation because most recipients will contain the effector strain and thus induction of the immune cell type will be observed. One can then compare the membership of subsets across all recipients where induction has or has not been observed, and look for strains whose presence correlates significantly with the response. In cases where the number of effector strains is small relative to the total number of strains in the cultured community (i.e., effector strains ≪ total strains), the significance of such a result can be estimated with a t-test comparing immune cell number when the candidate effector strain is absent versus when it is present. Applying this absent versus present t-test-based approach to each strain in a culture collection provides an easy first-pass method to identify effector strains, although application of this method or any of the model-based approaches discussed below is more likely to be successful if data collected for subsets larger than the saturation point are removed (Fig. 1D,E). More complex situations can be envisioned where a small proportion of community members (greater than 1) can elicit a response, or where the combined activity of a few community members is required. The experimental approach described above can be used to identify such relationships but more complex model-based analyses need to be employed. One advantage of the model-based approach is that it can simultaneously consider the contributions of all strains in the culture collection, and will therefore better explain the observed variation when multiple strains contribute to the phenotypic response (Fig. 1F). In addition, there is a wealth of existing modeling tools designed to identify interactions between variables and other non-linearities (e.g., if the immune cell type is induced only with strain A AND strain B). A category of modeling termed feature selection provides a rigorous means to identify strains whose presence best explains the observed phenotypic variation. Feature selection tools include stepwise regression, lasso (Efron and Hastie, 2004) and random forests (Breiman, 2001). These techniques search the model variables for the subset that explains the most variation. One of the simplest and most common algorithms is stepwise regression with forward selection, where the variables are added to the model one-at-a-time, always adding the variable that improves the model the most until including additional variables does not improve the model above a predefined limit. Feature selection techniques can be run on thousands of response variables to identify those best explained by microbiota composition, as is the case for assays like RNA-Seq and metabolomics that measure numerous responses in parallel. Such approaches are also likely to be of benefit when multiple features of a given host response are being tested. For example, the immune cell population of interest may be heterogenous expressing a variety of cytokines, some of which have distinct functions (e.g., Th17 cells). Similarly, the cells may express a variety of immunosuppressive molecules (e.g., Tregs). The diverse effector strain-evoked phenotypes observed in these cells may be further modified, in a complex fashion, by numerous members of the microbiota, or in response to complex sets of interactions, even if the initial phenotypic response measured is induced by a smaller subset of the community. Such processes are difficult to identify using more traditional approaches. Based on the modeling results, more targeted follow up colonizations can be used to assess and confirm the validity of the results.
Fig. 2. Outline of how random subsetting can identify immunogenic effector strains in a hypothetical gut community composed of 25 members.
The figure shows hypothetical results of the induction of an immune cell type being studied, in order to illustrate the process by which random subsetting of clonally arrayed collections of cultured bacteria can be used to identify effector strains. (A) If all or many members of a community are capable of eliciting a phenotype, then recipients of all subset sizes will manifest the response. Despite each subset having different composition, no information is gained as to which strains are responsible, as there is low variation in the response and it already saturates at subset sizes as low as five bacteria (see text for further discussion of saturation). (B) If the ability to modify a phenotype is rare or restricted or just a single community member, then the value of the approach becomes evident. In the graph, there is a bimodal distribution in the response. By comparing the membership of consortia that promote induction a population of interest to those that completely lack such populations one can obtain a catalogue of all strains that are (i) always present when the response is observed and (ii) always absent when the response is not manifest. In the example shown in (B), although many communities elicit induction of the immune cell population of interest, there is only one effector strain common to all subsets. Follow-up colonizations with reduced size subsets can be used to confirm that the approach has indeed identified the correct effector strain, and whether additional membership explains the capacity of a strain to imprint the phenotype. Horizontal lines represent mean values; each point depicts the immune cell response in an individual recipient mouse.
Improving the Throughput for Functional Screening with ‘Out of the Isolator’ Gnotobiotics
A large number of individual gnotobiotic isolators would be required to perform these experiments given the need for a separate isolator for each group of recipient mice that receives a given subset of the culture collection. To overcome this challenge, we developed a simple scalable system termed “out-of-the-isolator” gnotobiotics. It involves sterilization of a rayon filter-topped cage together with enclosed bedding, food, water, syringe and a gavage needle. Germ-free mice, reared in a traditional gnotobiotic isolator are gavaged with bacterial subsets of interest in a laminar flow hood and then housed in these cages on a standard cage rack. Cross-cage contamination can be prevented by adhering to the following practices: (i) only a single cage should handled at any one time in the laminar flow hood, (ii) once an animal is introduced into a cage (with or without gavage of microbes), the lid needs to be securely attached (with taping) and all efforts made to minimize subsequent movement of the cage for the duration of the experiment, (iii) the laminar flow hood should be sterilized using 70% ethanol and the experimenter should change his/her gloves and gown after each cage is set up; and (iv) animals should be maintained in their cages in a room under positive pressure supplied with HEPA-filtered air. Using these procedures, over the course of the two-week period, >95% of animals remain free of unwanted environmental contamination when assayed by metagenomic sequencing of fecal DNA, essentially mimicking the situation in a traditional gnotobiotic isolator. This means that a single gnotobiotic isolator can be used to generate the germ-free mice required to test hundreds of subsets (Faith et al., 2014)
An Example of How these Methods have been used to Identify Effector Strains that Induce Treg Cells
We recently used the approach described above to identify human gut bacterial strains capable of inducing Treg cells (Faith et al., 2014). We generated an arrayed culture library comprised of 17 bacterial strains, isolated from the fecal microbiota of a lean healthy female living in the USA whose complete uncultured community promoted increases in colonic Treg cells when transplanted to gnotobiotic mice. To address the issue of saturation, we randomly selected subsets of different size from the initial 17 strains, as outlined above, and administered these subsets to germ-free recipients using the “out-of-the-isolator” system. By plotting subset-size versus phenotype response, we obtained an approximation as to whether the cultured community was capable of saturating a response of interest and if so, what proportion of community members were capable of inducing the saturation. This allowed us to perform targeted follow-up experiments using consortia sizes that produced high phenotypic variance and to avoid consortia that were larger than the phenotype saturation point (and thus uninformative for identifying effector strains). To our surprise, we found that colonic Treg cells saturated at small community sizes (subsets with as few as 1 strain), suggesting that a large proportion of the cultured component of the donor’s microbiota was capable of inducing the phenotype, rather than one or two specialized effector strains. In such cases, small communities (colonization of germ-free mice with one or two members in mono- and bi-colonization experiments) were necessary to identify the effector strains responsible, because no phenotypic variation was observed when larger community sizes were tested. The experimental approach expanded our knowledge of the range of bacterial taxa that could induce a response, prominent among them being (genetically manipulatable) Bacteroides caccae and Bacteroides thetaiotaomicron. We observed a favored accumulation of Nrp1lo/− pTregs (Faith et al., 2014). Given recent demonstrations that gut bacteria boost pTreg cell generation in response to non-bacterial orally administered antigens (Furusawa et al., 2013), identifying effector strains and the mechanisms by which they stimulate pTreg generation may provide strategies for promoting Treg cell responses to bacterial or food-derived antigens.
The modeling approaches we describe are valuable when a combination of different strains is required to induce a phenotype of interest. Strains identified from such models can then be added, as a consortium, to germ-free mice or to mice colonized with microbes incapable of eliciting the phenotype to confirm results from the model. Further proof of the requirement for each member of the consortium can be obtained by performing “leave one out” style experiments, where all members save one are administered to recipient animals.
Generalizing the Combinatorial Approach to Study How the Microbiota Affects Many Features of Immunity
Metabolism represents an important but still poorly characterized point of intersection between the microbiota and immune responses. For example, short chain fatty acids, bacterial end products of fermentation, support development of an anti-inflammatory environment in the intestine (Arpaia et al., 2013; Chang et al., 2014; Furusawa et al., 2013; Smith et al., 2013b), whereas generation of an arylhydrocaron receptor agonist by Lactobacilli reuteri in the intestine can activate host defense pathways (Zelante et al, 2013). There is likely a plethora of immuno-modulatory metabolites waiting to be discovered that reflect metabolism by individual microbes, co-metabolism by members of the microbiota, as well as host-microbial co-metabolism. A first step in dissecting these contributions is to identify effector strains whose presence or absence leads to accumulation or removal of specific metabolites. Whereas significant modulation of colonic Treg cells were documented in mono-colonization studies, higher order interactions were observed to determine the concentration of a number of metabolites in the distal gut (e.g., amounts were determined by the presence of two particular species, or the presence of one and the absence of another; Faith et al., 2014).
Documenting that the presence or absence of a metabolite correlates with the presence or absence of particular strains begs for follow-up mechanistic analyses that could involve genetic or biochemical approaches, or combinations of the two (Wieland Brown et al., 2013). Targeted depletion of metabolites of interest can be challenging. For example, knowledge of the pathways that regulate the amounts of the metabolite of interest is required for targeted genetic interventions. Alternatively, biochemical manipulation of the activities of enzymes that control production or further chemical modification of a metabolite of interest depends on the availability of inhibitors (or agonists) of the enzyme. Alternatively, the fate of metabolites can be followed using labeled compounds or their precursors, but this approach can be costly.
Interactions between members of the microbiota and other immune cell populations can be characterized using this combinatorial approach, including interactions involving CD8+ T cells, B cells, γδ T cells, or various components of innate immunity. How the gut microbiota is able to shape local and systemic T and B cell repertoires remains a major area of investigation. Experimental systems that use restricted T cell repertoires have produced varied results (Lathrop et al, 2011; Cebula et al, 2013) regarding the mechanisms that generate antigen-specific populations of Treg cells. Modeling-based approaches, akin to those employed to identify effector strains that modulate cecal metabolites, could be used in conjunction with sequenced T cell receptor and B cell receptor repertoires in recipient mice colonized with microbial consortia of varying size and composition to help establish which strains are linked to particular repertoires. Used in conjunction with more reductionist approaches, such strategies should help address these issues. Furthermore, germ-free recipients that have been subjected to various genetic manipulations can be used, or environmental conditions such as diet can be systematically varied. The latter analysis could involve a combinatorial approach with different diets, each containing different proportions of a set of ingredients, fed in ordered sequence to individual mice colonized with a given effector bacterial strain previously shown to modulate an immune cell population in the context of the ‘parental’ (unmanipulated) diet. Feature selection can be employed to identify which component of the diet is the principal driver of the (immune) phenotypic variation (Faith et al., 2011). These types of defined and manipulated gnotobiotic models can be analyzed at increasing depth and breadth, including for example, assaying the effects of the interaction between different diet ingredient combinations with the effector strain on microbial metabolism, or the patterns of gene expression and metabolism of the immune cell population whose representation is being affected by these manipulations.
Finally, it is reasonable to ask why there is a need to identify as many strains as possible that are capable of modulating a given immune phenotype? We believe that the reasons are two-fold. First, if we are to use gut microbes as a predictor of whether or not an individual will be susceptible to immunopathology due, for example, to a lack of Treg cell-inducing organisms, it will be essential to know all effector strains. If we only identify 10% of the strains then we may inaccurately predict that an individual is at risk for intestinal inflammation. Second, although studying an individual bacterial strain can yield important insights, characterizing the response to many different strains and species represents a starting point for identifying generalizable “rules” underlying the interaction rather than peculiarities of a particular system. This is analogous to the benefit derived from studying a single transgenic TCR and model antigen system, versus a collection of such model systems, each with different affinities for the antigen. The take home message is that by identifying effector strains, the complexity of the microbiota can be reduced and mechanisms deciphered, while at the same time the task of mining the microbiota for its diagnostic and therapeutic potential can be facilitated.
Acknowledgments
This work was supported in part by grants from the NIH (DK30292, DK70977) and the Crohns and Colitis Foundation of America to J.I.G, and a Henry Wellcome Postdoctoral Fellowship (096100) to P.P.A.
Footnotes
Author Contributions – P.P.A, J.J.F, and J.I.G. wrote the manuscript. The effort in reviewing the literature, developing the combinatorial approach for identifying effector strains described in this Perspectus and designing the Figures was shared equally by P.P.A and J.J.F.
Conflict of Interest – P. P.A and J.J.F declare no conflicts. J.I.G. is the co-founder of Matutu, Inc, a company that is characterizing the role of diet-by-microbiota interactions in defining health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilate AB, Lafaille JJ. It takes two to tango. Immunity. 2011;35:6–8. doi: 10.1016/j.immuni.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box GEP, Hunter JS, Hunter WG. Wiley series in probability and statistics. 2. Wiley-Interscience; Hoboken, N.J: 2005. Statistics for experimenters: design, innovation, and discovery; p. xvii.p. 633. [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Lease angle regression. The Annals of statistics. 2004;32:407–499. [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Science Translational Med. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Rey FE, O’Donnell D, Karlsson M, McNulty NP, Kallstrom G, Goodman AL, Gordon JI. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4:1094–1098. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, Li SH, Simpson PM, Chatila TA, Williams CB. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D, Williams JB, Jia S, Nickerson D, Schmitt EG, Edwards B, Ziegelbauer J, Yassai M, Li SH, Relland LM, et al. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 2011;35:109–122. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning: data mining, inference, and prediction. 2. Springer; New York, NY: 2009. p. xxii.p. 745. Springer series in statistics. [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, de Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, Curotto de Lafaille MA. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013a;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013b;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, Xiong H, Dolpady J, Frey AB, Ruocco MG, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–1742. S1721. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, Sonnenburg JL, Comstock LE, Bluestone JA, Fischbach MA. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS biology. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]