Abstract
Background
Owing to the number of genetic mutations that contribute to malformations of cortical development, identification of causative mutations in candidate genes is challenging. To overcome these challenges, we performed whole-exome sequencing in this study.
Case presentation
A Japanese patient presented with microcephaly and severe developmental delay. Brain magnetic resonance imaging showed the presence of colpocephaly associated with lateral ventricle dilatation and the presence of a simplified gyral pattern. Hypoplasia of the corpus callosum and cerebellar vermis were also noted. Because Sanger sequencing is expensive, laborious, and time-consuming, whole-exome sequencing was performed and a de novo missense mutation in TUBA1A (E27Q) was identified.
Conclusion
The novel mutation identified in this study was located in the genetic region that encodes the N-terminal domain of TUBA1A, a region of TUBA1A with few reported mutations. Retrospective assessment of the clinical and radiological features of this patient―i.e., microcephaly, lissencephaly (pachygyria) with cerebellar hypoplasia, and corpus callosum hypoplasia―indicated that the TUBA1A mutation did not lead to any contradictions. Because rapid and comprehensive mutation analysis by whole-exome sequencing is time- and cost-effective, it might be useful for genetic counseling of patients with sporadic malformations of cortical development.
Keywords: Whole-exome sequencing, TUBA1A, Malformations of cortical development, Lissencephaly, Microcephaly
Background
Malformations of cortical development (MCD) are a cause of motor and intellectual disabilities and severe epilepsy [1]. Although various types of MCD have been described, the most typical type is lissencephaly, a group of malformations caused by abnormal neuronal migration [2]. The gene encoding platelet-activating factor acetylhydrolase isoform 1b regulatory subunit 1 (PAFAH1B1, formerly LIS1) was initially identified as the causative gene of lissencephaly. Advances in neuroradiological imaging and genetic diagnosis have since improved our understanding of the underlying pathogenesis of such diseases. Mutations in tubulin genes, encoding tubulin alpha 1a (TUBA1A), beta 2B class IIb (TUBB2B), and beta 3 class III (TUBB3), are now recognized as major causes of MCD [3,4]. Moreover, the rapid development of new revolutionary molecular technologies has enabled us to identify rare genetic variants in sporadic MCD cases [5].
In this study, we examined a Japanese patient with sporadic MCD. As mentioned above, causative genes are difficult to identify, owing to the large number of MCD candidate genes. Sanger sequencing of potential causative genes is expensive, laborious, and time-consuming; thus, we performed whole-exome sequencing. Using this method, we identified a novel TUBA1A mutation. Here, we discuss the advantages of using whole-exome sequencing for the rapid detection of causative mutations and genetic counseling of patients with sporadic MCD.
Case presentation
Patient description
A Japanese baby girl was born at 41 weeks of gestation with a birth weight of 3,400 g (+1.0 standard deviation [SD]), length of 49.5 cm (+0.5 SD), and an occipitofrontal circumference (OFC) of 31.5 cm (-1.0 SD). Her Apgar score was 7/8 (1 min/5 min). Her 30-year-old mother was gravida 0, para 0. At 22 weeks of gestation, fetal microcephaly was detected by echosonography. Soon after delivery, the sucking ability of the infant was good despite mild muscle hypotonia. She showed marked posterior sloping of the forehead, a high-arched palate, and microretrognathia. At 6 days after birth, brain magnetic resonance imaging (MRI) revealed colpocephalic configulation of the lateral ventricle dilatation associated with a simplified gyral pattern (Figure 1).The sucking ability of the infant gradually decreased; at 2 months of age, tube feeding was initiated. At this age, she weighed 4.7 kg (-0.8 SD) and measured 59.4 cm (+1.2 SD) in length, with an OFC of 34.0 cm (-3.3 SD), indicating severe microcephaly. She could not pursue objects. Although her muscle tone was within normal limits, she often showed an opisthotonic posture. At the age of 3 months, brain MRI examination was performed again, and brain hypoplasia associated with diffuse pachygyria and lateral ventricle dilatation was found, which is atypical for lissencephaly patients (Figure 1).
Figure 1.
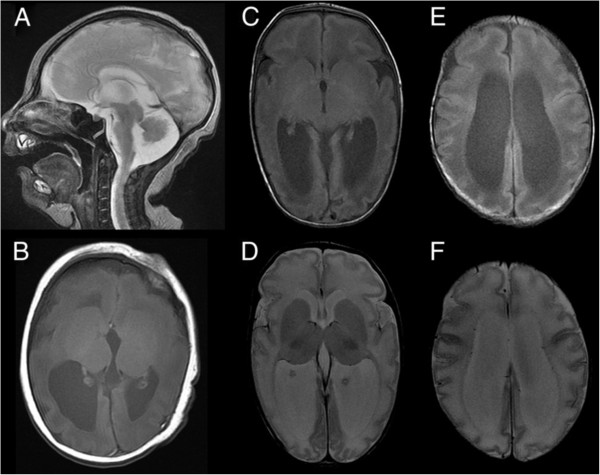
Brain magnetic resonance imaging of the patient. T2-weighted sagittal (A) and T1-weighted axial (B) images at 6 days after birth indicate hypoplastic vermis of the cerebellum, cortical dysgenesis with a simplified gyral pattern, and dilatation of the lateral ventricles. T1-weighted (C and E) and T2-weighted (D and F) axial images at 3 months of age indicate colpocephalic appearance associated with dilatation of the lateral ventricles. Cortical gyration is poor and reduced volume of the white matter is apparent.
Conventional karyotyping revealed a normal female karyotype with 46,XX. Because this child was the first born, genetic counseling was provided and both parents expressed a desire for their daughter to be genotyped.
We searched for the causative mutation(s) by whole-exome sequencing by using a next generation sequencer. After obtaining written informed consent, we obtained blood samples from the patient and her biological parents. This study was approved by the ethical committee at Tokyo Women’s Medical University.
Whole-exome sequencing
Genomic DNA collected from the patient and her parents were used as trio samples for whole-exome sequencing. DNA was extracted using a QIAamp DNA extraction kit (Qiagen, Hilden, Germany). Using a SureSelect whole exon kit (Agilent Technologies, Santa Clara, CA), 3 μg of DNA was processed according to the manufacturer’s instructions. Captured DNA molecules were analyzed using a SOLiD3® system by paired-end analysis (Life Technologies, Foster City, CA).
Color space reads were mapped to the UCSC hg19 reference genome using SOLiD® BioScope™ software (version 1.3; Life Technologies). Single-nucleotide variants (SNVs) were subsequently called by the DiBayes algorithm using medium call stringency. Small insertions and deletions (indels) were detected using the SOLiD Small InDel Tool (Life Technologies). Called SNVs and indels were filtered based on depth and quality, combined, and the annotated using ANNOVAR and a custom analysis pipeline [6]. Data for annotations was downloaded from the UCSC database (http://genome.ucsc.edu/).
As a priority, we focused only on non-synonymous coding variants, splice acceptor and donor site mutations, and frameshift indels. We extracted the candidate variants that were not observed in the 1000 Genomes Project (http://www.1000genomes.org/) and the dbSNP 132 database (http://www.ncbi.nlm.nih.gov/projects/SNP/). Because we assumed that an autosomal dominant trait existed in this family, variants identified only in the proband, and not in the unaffected parents (de novo variants), were selected. Missense mutations were tested for mutational effects by using amino acid substitution prediction tools such as PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT (http://sift.jcvi.org/) [7,8]. The flow chart used to select candidate genes is shown in Figure 2.
Figure 2.
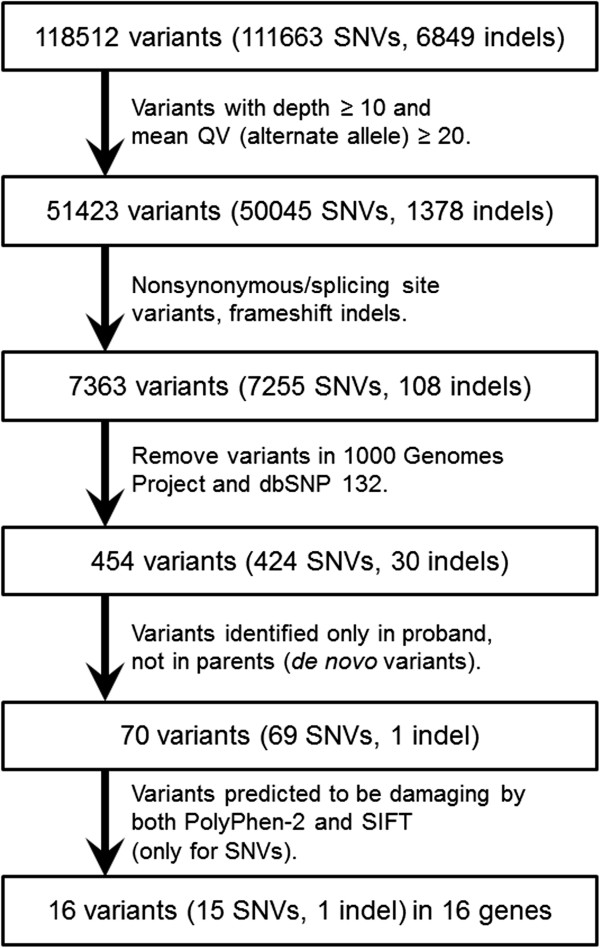
Flow chart indicating the validation process for variants. After 5 steps of validation, 16 genetic variants are finally selected.
The potential genetic variants identified by whole-exome sequencing were reconfirmed using standard polymerase chain reaction (PCR)-direct sequencing analysis (Sanger sequencing), as described previously [9,10]. The effects of the identified amino acid changes were analyzed using web-based software such as Pôle Bioinformatique Lyonnais Network Protein Sequence Analaysis (http://doua.prabi.fr/) and PSIPRED Protein Sequence Analysis Workbench (http://bioinf.cs.ucl.ac.uk/psipred/).
Whole-exome sequencing generated an average of 223 × 106 mapped reads that could be mapped to the reference genome—98% could be mapped accurately—with a mean coverage of 93 (Table 1). After filtering, 1 indel and 15 SNVs remained for the de novo mutation model (Table 2). For this model, the status of both parents was required to define the wild-type state. No genetic variation remained for the X-linked recessive and autosomal recessive models.
Table 1.
Whole-exome sequencing output
| Sample | Proband | Father | Mother |
|---|---|---|---|
| Total reads |
406,845,894 |
437,902,198 |
396,963,736 |
| Mapped reads |
252,829,848 |
214,184,732 |
201,932,294 |
| Properly mapped reads |
248,030,760 |
209,714,312 |
197,748,762 |
| Total bases mapped |
9,844,652,145 |
9,331,887,457 |
8,502,528,820 |
| Total bases uniquely mapped |
7,330,875,865 |
6,294,992,574 |
5,842,891,532 |
| Total bases mapping to target |
5,449,744,981 |
4,422,564,459 |
4,033,208,506 |
| Mean target coverage |
109.336672 |
88.706873 |
81.004891 |
| % target bases with ≥10x coverage | 85.51% | 85.04% | 83.56% |
Table 2.
Filtered variations
| Chromosome | Position | Region | Gene | Function | dbSNP132 | 1000G_2010Nov_ALL | PolyPhen2 | SIFT | ref_allele | alter_allele | Proband | Father | Mother |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1 |
223,175,758 |
exonic |
DISP1 |
nonsynonymous SNV |
- |
- |
1 |
0 |
G |
A |
△ |
× |
× |
| chr1 |
240,371,141 |
exonic |
FMN2 |
nonsynonymous SNV |
- |
- |
0.76515 |
0 |
C |
T |
△ |
× |
× |
| chr1 |
247,615,260 |
exonic |
OR2B11 |
Deletion and frameshift > premature terminal codon |
- |
- |
|
|
A |
- |
△ |
× |
× |
| chr2 |
189,860,860 |
exonic |
COL3A1 |
nonsynonymous SNV |
- |
- |
0.784653 |
0 |
G |
A |
△ |
× |
× |
| chr2 |
190,718,672 |
exonic |
PMS1 |
nonsynonymous SNV |
- |
- |
0.996 |
0 |
G |
A |
△ |
× |
× |
| chr6 |
30,861,156 |
exonic |
DDR1 |
nonsynonymous SNV |
- |
- |
0.16 |
0.03 |
G |
A |
△ |
× |
× |
| chr6 |
105,824,051 |
exonic |
PREP |
nonsynonymous SNV |
- |
- |
0.241 |
0 |
C |
T |
△ |
× |
× |
| chr8 |
113,277,807 |
exonic |
CSMD3 |
nonsynonymous SNV |
- |
- |
0.985 |
0 |
C |
T |
△ |
× |
× |
| chr11 |
124,267,132 |
exonic |
OR8B3 |
nonsynonymous SNV |
- |
- |
0.275 |
0.02 |
A |
G |
△ |
× |
× |
| chr12 |
49,580,541 |
exonic |
TUBA1A |
nonsynonymous SNV |
- |
- |
0.777136 |
0 |
C |
G |
△ |
× |
× |
| chr16 |
75,682,281 |
exonic |
TERF2IP |
nonsynonymous SNV |
- |
- |
0.214 |
0.02 |
G |
A |
△ |
× |
× |
| chr17 |
74,288,545 |
exonic |
QRICH2 |
nonsynonymous SNV |
- |
- |
0.995 |
0.01 |
C |
G |
△ |
× |
× |
| chr19 |
1,055,329 |
exonic |
ABCA7 |
nonsynonymous SNV |
- |
- |
0.996 |
0 |
A |
G |
△ |
× |
× |
| chr21 |
10,942,950 |
exonic |
TPTE |
nonsynonymous SNV |
- |
- |
0.999 |
0.01 |
T |
C |
△ |
× |
× |
| chr22 |
25,011,077 |
exonic |
GGT1 |
nonsynonymous SNV |
- |
- |
0.318 |
0.01 |
C |
T |
△ |
× |
× |
| chrX | 153,008,788 | exonic | ABCD1 | nonsynonymous SNV | - | - | 1 | 0 | G | A | △ | × | × |
ref, reference; alter, alteration; exonic, exonic region; SNV, single nucleotide variation; △, heterozygous; ×, no SNV.
Among the 16 candidate variants in the de novo mutation model, a TUBA1A variant, E27Q, was included. The damaging effects of the variations were scored by PolyPhen-2 and SIFT; the TUBA1A E27Q variant showed a possible damaging effect. We considered the TUBA1A variant as the strongest candidate mutation. Subsequent Sanger sequencing confirmed the presence of this variant in the proband and absence in the genomes of both parents (Figure 3A). No variants in the other candidate genes related to MCD were observed. Therefore, we concluded that MCD in this patient was the consequence of a de novo mutation in TUBA1A. The risk of recurrence in this family was estimated to be the same as that for the general population.
Figure 3.
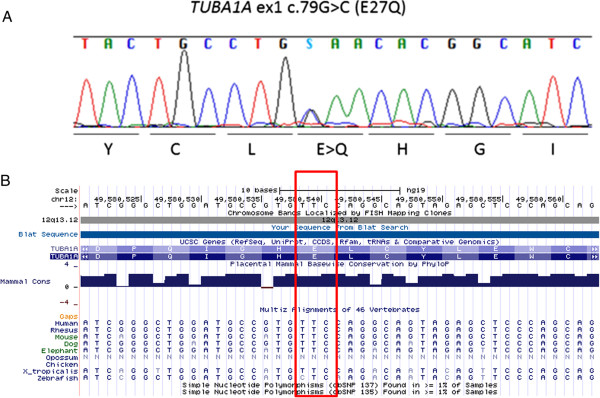
Results of Sanger sequencing and a map of the identified mutation locus. (A) A Sanger sequencing electropherogram identifying the missense mutation, c.79G > C (E27Q), is shown. (B) A genome map of the locus surrounding the mutation captured from the UCSC genome browser is depicted. Because TUBA1A is encoded in an antisense manner, the reverse-complement nucleotide sequence is shown. The affected amino acid is indicated by a red rectangle. The c.79C residue is conserved among many species as shown.
Discussion
MCD is the consequence of abnormal neuronal migration, which results in intellectual impairment and epilepsy [11]. Many of the genes responsible for MCD are related to autosomal dominant traits caused by de novo mutations—e.g., mutations in PAFAH1B1, TUBA1A, TUBB2B, and TUBB3. However, some genes are related to autosomal recessive MCD phenotypes, including the genes encoding reelin (RELN), very-low-density lipoprotein receptor (VLDLR), tubulin alpha-8 (TUBA8), as well as X-linked genes encoding doublecortin (DCX), aristaless related homeobox (ARX), and filamin A alpha (FLNA) [1,12,13]. Therefore, molecular diagnostic testing in patients with MCD is an important prerequisite for genetic counseling, because the risk of recurrence cannot be estimated in cases that lack a definitive molecular diagnosis.
In this study, we successfully identified a de novo TUBA1A mutation in a patient with sporadic MCD using whole-exome sequencing of trio samples. Among the genes in the tubulin family, TUBA1A was the first to be identified as a causative factor of MCD. The TUBA1A protein is thought to function as a scaffold protein [14].
Although there are at least 34 known TUBA1A mutations, and many of these mutations are located in the C-terminal half of the amino acid sequence [3,11,14-27], this report is the first to describe the E27Q mutation (Additional file 1: Figure S1). In contrast to most of the previously reported mutations, this mutation is located in the N-terminal region (Additional file 1: Figure S1). Because the mutation is located in a conserved region of tubulin (Additional file 1: Figure S1), and the mutated residue is conserved among many species (Figure 3B), we predicted that this mutation might be pathogenic. We analyzed the effects of this mutation by using web-based tools to predict the secondary structure of the mutated protein. Of the two different computational simulations, one predicted a significant change in the structure of TUBA1A (Additional file 2: Figures S2 and Additional file 3: Figure S3), suggesting that this mutation is likely pathogenic.
Amino acid substitutions may affect heterodimer stability and/or microtubule dynamics by altering the structure of tubulin during nucleotide exchange and hydrolysis. Microtubule stability may be diminished during the critical process of cell migration [28]. All mutations reported thus far in TUBA1A, TUBB2B, and TUBB3 are heterozygous missense mutations [3,4,29-31]. Missense mutations in the absence of nonsense mutations, frameshifts, or genomic deletions support altered protein function, rather than haploinsufficiency, as the primary genetic etiology of these tubulin-related disorders [28].
Retrospectively, we evaluated the MRI findings of this patient. Brain MRI showed a colpocephalic appearance of the lateral ventricle dilatation associated with a simplified gyral pattern. Hypoplastic corpus callosum and cerebellar vermis hypoplasia were also noted. According to the classification proposed by Kumar et al. [11], lissencephaly with cerebellar hypoplasia (LCH) in group 3 would be most compatible with this patient, because of the involvement of the corpus callosum and cerebellum. Ross et al. proposed genotype-phenotype correlations in LCH cases encompassing heterogeneous disorders such as those classified as LCH types a—d [32]. LCHa cases are related to mutations in PAFAH1B1 or DCX, whereas LCHb cases are linked to a mutation in RELN. Based on the LCH pattern of the patient presenting microcephaly (≤ - 3 SD), this case may be classified as LCHd, which could be related to a mutation in TUBA1A. Therefore, no contradiction in the etiology of the TUBA1A mutation was observed.
Generally, young couples whose first child has been afflicted with a sporadic case of severe disease are eager to have a healthy child in their next pregnancy [33]. Therefore, a precise genetic diagnosis is necessary. However, many MCD candidate genes have been identified, and Sanger sequencing for predicted causative genes is rather expensive, laborious, and time-consuming [34]. In comparison, in this study, we show that whole-exome sequencing is rapid and comprehensive [35]. We further emphasize that the results of this method provided us not only with confirmation of the causative genes but also simultaneously rules out mutations in other candidate genes. Therefore, we could completely negate the contribution of other candidate genes in this patient. In the future, targeted panel sequencing rather than whole-exome sequencing may become the standard method used to evaluate human disorders derived from multiple candidate genes.
Conclusion
We successfully identified a causative TUBA1A mutation in a patient with sporadic MCD associated with a simplified gyral pattern by using whole-exome sequencing. The identified novel mutation (E27Q) was located in the N-terminal region of the amino acid sequence. Rapid and comprehensive mutation analysis by using whole-exome sequencing may be useful for genetic counseling in sporadic cases of human disorders derived from multiple candidate genes.
Consent
Written informed consent was obtained from the patient’s parents for publication of this Case Report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal. Blood samples from the patient and her biological parents were obtained with parental consent. This study was approved by the ethical committee at our institution.
Abbreviations
MCD: Malformations of cortical development; LCH: Lissencephaly with cerebellar hypoplasia; PAFAH1B1: Platelet-activating factor acetylhydrolase isoform 1b regulatory subunit 1; TUBA1A: Tubulin alpha 1a; TUBB2B: Tubulin beta 2B class IIb; TUBB3: Tubulin beta 3 class III; SD: Standard deviation; MRI: Magnetic resonance imaging; SNV: Single-nucleotide variant; indel: Insertions and deletion; PCR: Polymerase chain reaction; RELN: Reelin; VLDLR: Very-low-density lipoprotein receptor; TUBA8: Tubulin alpha-8; DCX: Doublecortin; ARX: Aristaless related homeobox; FLNA: Filamin A alpha; LCH: Lissencephaly with cerebellar hypoplasia.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
KS analyzed and interpreted the data, evaluated the genotype-phenotype correlation, and drafted the paper. TF performed whole-exome sequencing. AS analyzed the sequencing data. AN and YM correlated clinical information. TY designed the study and reviewed this article. All authors read and approved the final manuscript.
Supplementary Material
Location of the identified TUBA1A mutation and a comparison of the TUBA1A amino acid sequence with tubulin family members. (Top) Although many mutations are located in the C-terminal region, the mutation identified in this patient (E27Q; indicated by red characters), is located in the N-terminal region. (Bottom) E27 is conserved among tubulin family member (red rectangle).
Predicted conformational changes in the protein structure caused by the identified mutation. The Pôle Bioinformatique Lyonnais Network Protein Sequence Analysis predicted a conformational change caused by the E27Q mutation. The location of the mutation (E27Q) is indicated by a red arrow.
The predicted secondary structure of TUBA1A and a simulation of the conformational change. The PSIPRED Protein Sequence Analysis Workbench predicted no definite change between the wild-type and mutant protein.
Contributor Information
Keiko Shimojima, Email: shimojima.keiko@twmu.ac.jp.
Aya Narita, Email: aya.luce@nifty.com.
Yoshihiro Maegaki, Email: maegaki@med.tottori-u.ac.jp.
Akira Saito, Email: saito@stagen.co.jp.
Toru Furukawa, Email: furukawa.toru@twmu.ac.jp.
Toshiyuki Yamamoto, Email: yamamoto.toshiyuki@twmu.ac.jp.
Acknowledgements
We thank the patient and their parents for their cooperation. This work was partially supported by the JST PRESTO program (KS) and Grant-in-Aid for Scientific Research on Innovated Areas “Foundation of Synapse and Neurocircuit Pathology” and Grant-in-Aid for scientific research from the Health Labor Sciences Research Grants from the Ministry of Health, Labor, and Welfare, Japan (TY).
References
- Liu JS. Molecular genetics of neuronal migration disorders. Curr Neurol Neurosci Rep. 2011;11(2):171–178. doi: 10.1007/s11910-010-0176-5. [DOI] [PubMed] [Google Scholar]
- Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. A developmental and genetic classification for malformations of cortical development: update 2012. Brain. 2012;135(Pt 5):1348–1369. doi: 10.1093/brain/aws019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushion TD, Dobyns WB, Mullins JG, Stoodley N, Chung SK, Fry AE, Hehr U, Gunny R, Aylsworth AS, Prabhakar P, Uyanik G, Rankin J, Rees MI, Pilz DT. Overlapping cortical malformations and mutations in TUBB2B and TUBA1A. Brain. 2013;136(Pt 2):536–548. doi: 10.1093/brain/aws338. [DOI] [PubMed] [Google Scholar]
- Poirier K, Saillour Y, Bahi-Buisson N, Jaglin XH, Fallet-Bianco C, Nabbout R, Castelnau-Ptakhine L, Roubertie A, Attie-Bitach T, Desguerre I, Genevieve D, Barnerias C, Keren B, Lebrun N, Boddaert N, Encha-Razavi F, Chelly J. Mutations in the neuronal ss-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum Mol Genet. 2010;19(22):4462–4473. doi: 10.1093/hmg/ddq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, Parrini E, Valence S, Pierre BS, Oger M, Lacombe D, Genevieve D, Fontana E, Darra F, Cances C, Barth M, Bonneau D, Bernadina BD, N’Guyen S, Gitiaux C, Parent P, des Portes V, Pedespan JM, Legrez V, Castelnau-Ptakine L, Nitschke P, Hieu T, Masson C, Zelenika D, Andrieux A, Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013. [DOI] [PMC free article] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Okumura A, Hayashi M, Shimojima K, Ikeno M, Uchida T, Takanashi JI, Okamoto N, Hisata K, Shoji H, Saito A, Furukawa T, Kishida T, Shimizu T, Yamamoto T. Whole-exome sequencing of a unique brain malformation with periventricular heterotopia, cingulate polymicrogyria and midbrain tectal hyperplasia. Neuropathology. 2012. [DOI] [PubMed]
- Shimojima K. Novel compound heterozygous mutations of POLR3A revealed by whole-exome sequencing in a patient with hypomyelination. Brain Dev. in press. [DOI] [PubMed]
- Kumar RA, Pilz DT, Babatz TD, Cushion TD, Harvey K, Topf M, Yates L, Robb S, Uyanik G, Mancini GM, Rees MI, Harvey RJ, Dobyns WB. TUBA1A mutations cause wide spectrum lissencephaly (smooth brain) and suggest that multiple neuronal migration pathways converge on alpha tubulins. Hum Mol Genet. 2010;19(14):2817–2827. doi: 10.1093/hmg/ddq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HM, Wynshaw-Boris A. Cytoskeleton in action: Lissencephaly, a neuronal migration disorder. Wiley Interdiscip Rev Dev Biol. 2013;2(2):229–245. doi: 10.1002/wdev.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G, Marcorelles P, Saugier-Veber P, Quille ML, Marret S, Laquerriere A. Role of cytoskeletal abnormalities in the neuropathology and pathophysiology of type I lissencephaly. Acta Neuropathol. 2011;121(2):149–170. doi: 10.1007/s00401-010-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, Pinon MC, Dear N, Valdar W, Brown SD, Davies KE, Rawlins JN, Cowan NJ, Nolan P, Chelly J, Flint J. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128(1):45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Keays DA, Francis F, Saillour Y, Bahi N, Manouvrier S, Fallet-Bianco C, Pasquier L, Toutain A, Tuy FP, Bienvenu T, Joriot S, Odent S, Ville D, Desguerre I, Goldenberg A, Moutard ML, Fryns JP, van Esch H, Harvey RJ, Siebold C, Flint J, Beldjord C, Chelly J. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A) Hum Mutat. 2007;28(11):1055–1064. doi: 10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- Bahi-Buisson N, Poirier K, Boddaert N, Saillour Y, Castelnau L, Philip N, Buyse G, Villard L, Joriot S, Marret S, Bourgeois M, Van Esch H, Lagae L, Amiel J, Hertz-Pannier L, Roubertie A, Rivier F, Pinard JM, Beldjord C, Chelly J. Refinement of cortical dysgeneses spectrum associated with TUBA1A mutations. J Med Genet. 2008;45(10):647–653. doi: 10.1136/jmg.2008.058073. [DOI] [PubMed] [Google Scholar]
- Fallet-Bianco C, Loeuillet L, Poirier K, Loget P, Chapon F, Pasquier L, Saillour Y, Beldjord C, Chelly J, Francis F. Neuropathological phenotype of a distinct form of lissencephaly associated with mutations in TUBA1A. Brain. 2008;131(Pt 9):2304–2320. doi: 10.1093/brain/awn155. [DOI] [PubMed] [Google Scholar]
- Morris-Rosendahl DJ, Najm J, Lachmeijer AM, Sztriha L, Martins M, Kuechler A, Haug V, Zeschnigk C, Martin P, Santos M, Vasconcelos C, Omran H, Kraus U, Van der Knaap MS, Schuierer G, Kutsche K, Uyanik G. Refining the phenotype of alpha-1a Tubulin (TUBA1A) mutation in patients with classical lissencephaly. Clin Genet. 2008;74(5):425–433. doi: 10.1111/j.1399-0004.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Poirier K, Friocourt G, Jaglin X, Goldenberg A, Saugier-Veber P, Chelly J, Laquerriere A. Human lissencephaly with cerebellar hypoplasia due to mutations in TUBA1A: expansion of the foetal neuropathological phenotype. Acta Neuropathol. 2010;119(6):779–789. doi: 10.1007/s00401-010-0684-z. [DOI] [PubMed] [Google Scholar]
- Tian G, Jaglin XH, Keays DA, Francis F, Chelly J, Cowan NJ. Disease-associated mutations in TUBA1A result in a spectrum of defects in the tubulin folding and heterodimer assembly pathway. Hum Mol Genet. 2010;19(18):3599–3613. doi: 10.1093/hmg/ddq276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AC, Oostra A, Desprechins B, De Vlaeminck Y, Verhelst H, Regal L, Verloo P, Bockaert N, Keymolen K, Seneca S, De Meirleir L, Lissens W. TUBA1A mutations: from isolated lissencephaly to familial polymicrogyria. Neurology. 2011;76(11):988–992. doi: 10.1212/WNL.0b013e31821043f5. [DOI] [PubMed] [Google Scholar]
- Sohal AP, Montgomery T, Mitra D, Ramesh V. TUBA1A mutation-associated lissencephaly: case report and review of the literature. Pediatr Neurol. 2012;46(2):127–131. doi: 10.1016/j.pediatrneurol.2011.11.017. [DOI] [PubMed] [Google Scholar]
- Okumura A, Hayashi M, Tsurui H, Yamakawa Y, Abe S, Kudo T, Suzuki R, Shimizu T, Shimojima K, Yamamoto T. Lissencephaly with marked ventricular dilation, agenesis of corpus callosum, and cerebellar hypoplasia caused by TUBA1A mutation. Brain Dev. 2013;35(3):274–279. doi: 10.1016/j.braindev.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Poirier K, Saillour Y, Fourniol F, Francis F, Souville I, Valence S, Desguerre I, Marie Lepage J, Boddaert N, Line Jacquemont M, Beldjord C, Chelly J, Bahi-Buisson N. Expanding the spectrum of TUBA1A-related cortical dysgenesis to Polymicrogyria. Eur J Hum Genet. 2013;21(4):381–385. doi: 10.1038/ejhg.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanni G, Colafati GS, Barresi S, Randisi F, Talamanca LF, Genovese E, Bellacchio E, Bartuli A, Bernardi B, Bertini E. Description of a novel TUBA1A mutation in Arg-390 associated with asymmetrical polymicrogyria and mid-hindbrain dysgenesis. Eur J Paediatr Neurol. 2013;17(4):361–365. doi: 10.1016/j.ejpn.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Hikita N, Hattori H, Kato M, Sakuma S, Morotomi Y, Ishida H, Seto T, Tanaka K, Shimono T, Shintaku H, Tokuhara D. A case of TUBA1A mutation presenting with lissencephaly and Hirschsprung disease. Brain Dev. 2014;36(2):159–162. doi: 10.1016/j.braindev.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Romaniello R, Arrigoni F, Cavallini A, Tenderini E, Baschirotto C, Triulzi F, Bassi MT, Borgatti R. Brain malformations and mutations in alpha- and beta-tubulin genes: a review of the literature and description of two new cases. Dev Med Child Neurol. 2014;56(4):354–360. doi: 10.1111/dmcn.12370. [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Cederquist GY, Gupta ML Jr, Engle EC. Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr Opin Genet Dev. 2011;21(3):286–294. doi: 10.1016/j.gde.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, Castelnau-Ptakhine L, Odent S, Loget P, Kossorotoff M, Snoeck I, Plessis G, Parent P, Beldjord C, Cardoso C, Represa A, Flint J, Keays DA, Cowan NJ, Chelly J. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41(6):746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Luchniak A, Tischfield MA, Peeva M, Song Y, Menezes MP, Chan WM, Andrews C, Chew S, Jamieson RV, Gomes L, Flaherty M, Grant PE, Gupta ML Jr, Engle EC. An inherited TUBB2B mutation alters a kinesin-binding site and causes polymicrogyria, CFEOM and axon dysinnervation. Hum Mol Genet. 2012;21(26):5484–5499. doi: 10.1093/hmg/dds393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniello R, Tonelli A, Arrigoni F, Baschirotto C, Triulzi F, Bresolin N, Bassi MT, Borgatti R. A novel mutation in the beta-tubulin gene TUBB2B associated with complex malformation of cortical development and deficits in axonal guidance. Dev Med Child Neurol. 2012;54(8):765–769. doi: 10.1111/j.1469-8749.2012.04316.x. [DOI] [PubMed] [Google Scholar]
- Ross ME, Swanson K, Dobyns WB. Lissencephaly with cerebellar hypoplasia (LCH): a heterogeneous group of cortical malformations. Neuropediatrics. 2001;32(5):256–263. doi: 10.1055/s-2001-19120. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nishikawa T, Yamanaka M, Takada F, Kurosawa K. Couple’s decision-making after birth of a child with an unbalanced chromosomal translocation. J Pediatr Neurol. 2008;6:203–208. [Google Scholar]
- Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15(12):1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470(7333):198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of the identified TUBA1A mutation and a comparison of the TUBA1A amino acid sequence with tubulin family members. (Top) Although many mutations are located in the C-terminal region, the mutation identified in this patient (E27Q; indicated by red characters), is located in the N-terminal region. (Bottom) E27 is conserved among tubulin family member (red rectangle).
Predicted conformational changes in the protein structure caused by the identified mutation. The Pôle Bioinformatique Lyonnais Network Protein Sequence Analysis predicted a conformational change caused by the E27Q mutation. The location of the mutation (E27Q) is indicated by a red arrow.
The predicted secondary structure of TUBA1A and a simulation of the conformational change. The PSIPRED Protein Sequence Analysis Workbench predicted no definite change between the wild-type and mutant protein.


