The structure of AA(Gly)B, a dual-specificity blood group synthase, in complex with UDP-galactose, the O6′′ atom of which is protected by an 2-nitrobenzyl group, has been determined in two space groups. The so-called caged substrate exhibits various distinct conformations in the active site.
Keywords: glycosyltransferases, photo-activatable substrate, UDP-Gal derivative
Abstract
Glycosyltransferases (GTs) catalyse the sequential addition of monosaccharides to specific acceptor molecules and play major roles in key biological processes. GTs are classified into two main families depending on the inverted or retained stereochemistry of the glycosidic bond formed during the reaction. While the mechanism of inverting enzymes is well characterized, the precise nature of retaining GTs is still a matter of much debate. In an attempt to clarify this issue, studies were initiated to identify reaction-intermediate states by using a crystallographic approach based on caged substrates. In this paper, two distinct structures of AA(Gly)B, a dual-specificity blood group synthase, are described in complex with a UDP-galactose derivative in which the O6′′ atom is protected by a 2-nitrobenzyl group. The distinct conformations of the caged substrate in both structures of the enzyme illustrate the highly dynamic nature of its active site. An attempt was also made to photolyse the caged compound at low temperature, which unfortunately is not possible without damaging the uracil group as well. These results pave the way for kinetic crystallography experiments aiming at trapping and characterizing reaction-intermediate states in the mechanism of enzymatic glycosyl transfer.
1. Introduction
In all domains of life, the biosynthesis of complex glycans requires the concerted action of a multitude of glycosyltransferases (GTs) which catalyse the sequential addition of monosaccharides from glycosyl donors (usually nucleotide-sugars) to acceptor molecules (Lairson et al., 2008 ▶; Weadge & Palcic, 2008 ▶; Breton et al., 2006 ▶; Schuman et al., 2007 ▶). GTs play a key role in many fundamental biological processes underpinning human health and disease, such as cell signalling, cellular adhesion, carcinogenesis and cell-wall biosynthesis in human pathogens (Berg et al., 2007 ▶; Dube & Bertozzi, 2005 ▶; Marth & Grewal, 2008 ▶; Rexach et al., 2008 ▶).
GTs are classified as either inverting or retaining, depending on whether the anomeric configuration of the transferred monosaccharide in the product is inverted or remains the same as that in the donor substrate. With one exception (Lira-Navarrete et al., 2011 ▶), inverting reactions are suggested to occur in a single displacement SN2 mechanism via an oxocarbenium transition state assisted by a catalytic base. X-ray structures of several inverting GT enzymes show amino acids (usually an Asp, Glu or His) appropriately positioned for proton abstraction of the nucleophile hydroxyl group of the acceptor (Breton et al., 2012 ▶; Lairson et al., 2008 ▶). The mechanism of retaining GTs remains controversial. A double-displacement mechanism with the formation of a covalent glycosyl-enzyme intermediate has been proposed for some GTs (Monegal & Planas, 2006 ▶; Soya et al., 2011 ▶; Gastinel et al., 2001 ▶; André et al., 2003 ▶). An alternative ‘internal return-like’ mechanism, or ‘SNi-like’ mechanism, has also been suggested, involving a short-lived oxocarbenium-ion intermediate where the nucleophilic hydroxyl group of the acceptor attacks the anomeric C atom from the same side from which the leaving group departs (Ardèvol & Rovira, 2011 ▶; Errey et al., 2010 ▶; Goedl & Nidetzky, 2009 ▶; Lairson et al., 2008 ▶; Lee et al., 2011 ▶).
We have been using the blood group A and B synthesizing enzymes, GTA and GTB, respectively, as models for mechanistic and structure–function studies of retaining GTs (Alfaro et al., 2008 ▶; Marcus et al., 2003 ▶; Patenaude et al., 2002 ▶; Pesnot et al., 2010 ▶; Angulo et al., 2006 ▶; Soya et al., 2011 ▶; Jørgensen et al., 2012 ▶). GTA catalyses the transfer of N-acetylgalactosamine (GalNAc) from UDP-GalNAc to the H-antigen acceptor [α-l-Fuc-(1→2)-β-d-Gal-OR, where R is glycolipid or glycoprotein] to form the A antigen. GTB catalyses the transfer of galactose (Gal) from UDP-Gal to the same acceptor to form the B antigen. GTA and GTB are highly homologous enzymes differing in only four of 354 amino-acid residues (Arg/Gly176, Gly/Ser235, Leu/Met266, Gly/Ala268; Patenaude et al., 2002 ▶). The dual-specificity enzyme AA(Gly)B (Arg176, Gly235, Gly266 and Ala268) used in this study can use either UDP-Gal or UDP-GalNAc as a donor substrate and can transfer both Gal or GalNAc to the H-antigen with equal efficiency, producing both blood group A and blood group B structures (Yamamoto et al., 2001 ▶).
GTA and GTB are members of the GT-A fold type family, with a Rossmann-like domain and a DXD motif responsible for nucleotide-donor binding. These enzymes undergo extensive structural rearrangement during a catalytic cycle and high-resolution structures reveal ‘open’, ‘semi-closed’ and ‘closed’ conformations as the enzyme goes from the unliganded to the liganded state (Alfaro et al., 2008 ▶; Jørgensen et al., 2012 ▶; Pesnot et al., 2010 ▶). The observed conformational changes are the results of movements of two flexible polypeptide regions, one internal loop (residues 176–199) and the C-terminal tail (the last nine residues), that are disordered in the apoenzyme and which become ordered upon substrate binding. The transition between the open and closed forms of the enzyme demonstrates the necessity of ordered substrate binding with nucleotide-donor binding first, followed by acceptor binding. GTA and GTB are suggested to use a double-displacement mechanism where Glu303 serves as the catalytic nucleophile (Patenaude et al., 2002 ▶). Although the formation of covalent glycosyl-enzyme intermediates has been observed for mutant GTA and GTB enzymes (Soya et al., 2011 ▶), attempts to trap such covalent intermediates (or any other types of intermediates) in wild-type enzymes have been unsuccessful.
Since many proteins are active in the crystalline state, it is possible to obtain the structure of reaction-intermediate states by X-ray crystallography. Kinetic crystallography comprises the ensemble of techniques that are used in conjunction with classical crystallography in order to trap and characterize the structural changes associated with the buildup of unstable species (Bourgeois & Royant, 2005 ▶). In order to capture one, or several, putative covalent intermediates during the reaction of retaining GTs, we decided to perform a kinetic crystallography study of GT crystals containing a ‘caged’ substrate. Such an approach has been successful in the study of the GTPase activity of H-Ras p21 (Klink et al., 2006 ▶; Klink & Scheidig, 2010 ▶; Schlichting et al., 1990 ▶) and the transient opening of the so-called ‘back-door’ of acetylcholinesterase (Colletier et al., 2007 ▶). These experiments rely on the presence of substrate/product/cofactor that has been chemically modified by the addition of a photolabile group, the ‘cage’, the presence of which prevents the reaction from proceeding. Upon photocleavage, and depending on the available free energy of the system (commonly limited by the temperature), the cage is released and the reaction can progress in a controlled manner.
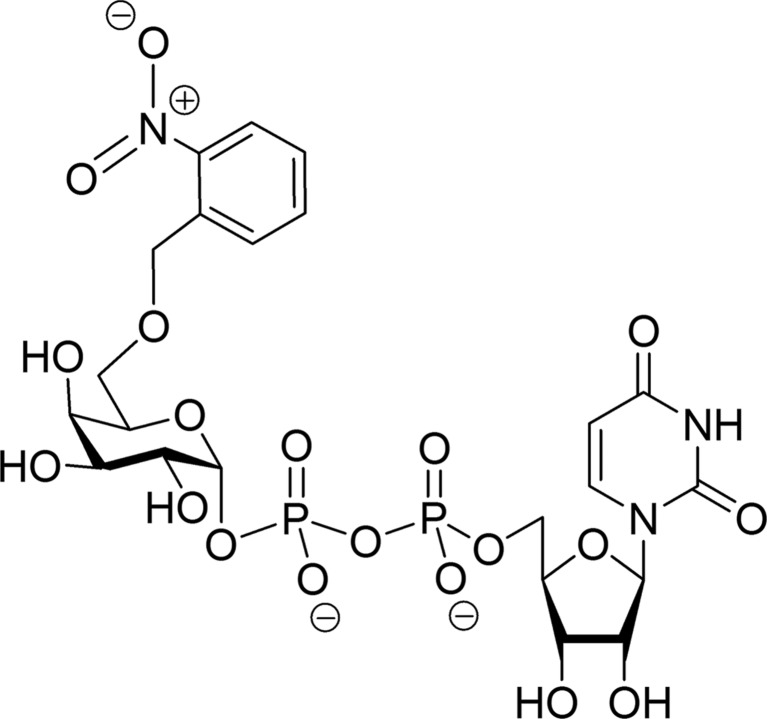
Here, we show that the recently described ‘caged’ 6′′-O-2-nitrobenzyl-UDP-Gal derivative (Fig. 1 ▶; CC01) (Mannerstedt & Hindsgaul, 2008 ▶) binds to the donor-binding site of a blood group GT in two different conformations. We have determined two high-resolution crystal structures of AA(Gly)B in complex with CC01 (cr1 and cr2) and, remarkably, the structures reveal different binding modes. In the first structure the galactose and 2-nitrobenzyl caging group interferes with the H-antigen acceptor binding site and in the other conformation the sugar has rotated such that the galactose and 2-nitrobenzyl group are in two different conformations, both interfering with the internal active-site loop in its closed conformation. We have also attempted to photolyse the CC01 compound at low temperature, which, unfortunately, does not occur without concomitant damage to the uracil group.
Figure 1.
Schematic illustration of the caged UDP-Gal derivative CC01 harbouring a 2-nitrobenzyl group at the O6 position of galactose (Mannerstedt & Hindsgaul, 2008 ▶).
2. Materials and methods
2.1. Expression and purification of AA(Gly)B
AA(Gly)B was cloned using standard mutagenesis techniques (Jørgensen et al., 2013 ▶), expressed in Escherichia coli and purified by ion-exchange and affinity chromatography with a final dialysis to remove UDP, as described previously (Seto et al., 1995 ▶, 1997 ▶, 2000 ▶), yielding ∼10–15 mg pure protein per litre of cell culture. At the end of purification of the enzyme batch used for crystallizing the cr2 complex, any remaining UDP bound to the active site of the protein after dialysis was removed from the protein solution by removing excess Mn2+. This was performed by adding a fivefold (v/v) excess of a buffer consisting of 50 mM EDTA, 50 mM MOPS pH 7.0, 0.1 M NaCl, 1 mM DTT and incubating on ice for 30 min. After EDTA treatment, the protein solution was dialysed overnight in the same buffer without EDTA and with 5 mM MnCl2 and was then concentrated to 15–17 mg ml−1. Complete removal of co-purified UDP from the binding site was confirmed by visual inspection of the X-ray crystal structure of an apo AA(Gly)B crystal (data not shown).
2.2. Synthesis of caged substrate (CC01)
The ‘caged’ 6′′-O-2-nitrobenzyl-UDP-Gal derivative (Fig. 1 ▶) was synthesized as described previously (Mannerstedt & Hindsgaul, 2008 ▶). Briefly, synthetic 6-O-2-nitrobenzyl-d-galactose was converted to the α-1-phosphate, which was coupled to uridine-5′-monophosphate using the morpholidate procedure.
2.3. Crystallization of AA(Gly)B and soaking procedures
AA(Gly)B was crystallized by hanging-drop vapour diffusion against a reservoir solution consisting of 7–12% PEG 3350, 0.1–0.3 M ammonium sulfate, 0.05 M MnCl2, 0.05 M MOPS pH 7.0 as described previously (Pesnot et al., 2010 ▶). The soaking and cryocooling of the crystals were performed by two different approaches. Crystal 1 (cr1) was first soaked in a buffer consisting of 0.025 M caged compound, 10% PEG 3350, 0.2 M ammonium sulfate, 0.05 M MnCl2, 0.05 M MOPS pH 7.0 for 45 min before being transferred to Paratone-N (Hampton Research) for cryoprotection and flash-cooled in liquid N2. Crystal 2 (cr2) was cryoprotected by adding 15% PEG 3350, 0.2 M ammonium sulfate, 0.05 M MOPS pH 7, 0.05 M MnCl2, 0.025 M caged compound, 22.3% glycerol directly to the drop containing crystals. Again, soaking was carried out for 45 min before flash-cooling in liquid nitrogen. All soaking experiments were carried out in a laboratory with very limited ambient light and crystal drops were covered by tinfoil during soaking.
2.4. X-ray data collection
X-ray diffraction data for AA(Gly)B in complex with CC01 were collected from two crystals prepared using the different soaking conditions described above. The first data set (cr1) was collected to 1.7 Å resolution on MAX-lab beamline I911-5 (λ = 0.908 Å, 100 K). The second data set (cr2) was collected to 1.9 Å on beamline ID29 at the ESRF, Grenoble, France (λ = 0.97625 Å, 100 K).
2.5. Structure determination and refinement
Both data sets were indexed, integrated and scaled using the XDS package (Kabsch, 2010 ▶). After integration and scaling, the structures were solved by molecular replacement using the CCP4 module Phaser (McCoy et al., 2005 ▶) with the previously solved structure of wild-type GTB (PDB entry 2rit; Alfaro et al., 2008 ▶) as the search model. For both structures, initial rigid-body and restrained refinement was carried out using REFMAC5 (Murshudov et al., 2011 ▶) before iteratively rebuilding the structure using Coot (Emsley & Cowtan, 2004 ▶); finally, cr1 was refined in PHENIX (Afonine et al., 2012 ▶) including TLS in an attempt to obtain the best possible model. Structures were deposited in the Protein Data Bank as entries 3zgf for cr1 and 3zgg for cr2.
2.6. Spectroscopic photolysis experiments
Photolysis experiments were performed at 100 K at the Cryobench in crystallo spectroscopy facility (Royant et al., 2007 ▶) of the ESRF, Grenoble, France. Samples consisted of solutions of mother liquor containing 25 mM CC01 and supplemented with 22.3%(w/w) glycerol or of 125 mM UDP-Gal supplemented with 25%(w/w) glycerol. Sample irradiation was carried out using a 266 nm laser (Changchun New Industries) providing 14 mW power after laser-to-fibre coupling and 3.5 mW at the sample position. Absorption spectra were recorded using a QE65 Pro spectrometer (Ocean Optics) providing high sensitivity.
3. Results and discussion
3.1. Structure determination
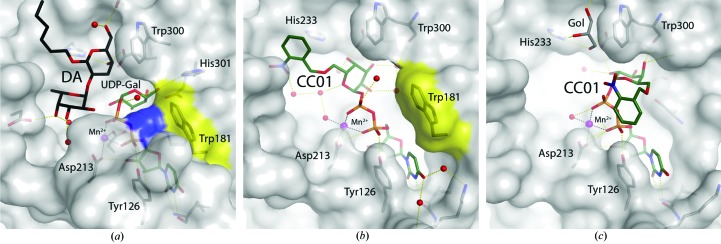
To understand the molecular basis of the binding mode of the caged UDP-Gal derivative (Fig. 1 ▶), we solved two high-resolution crystal structures of AA(Gly)B with compound CC01 bound to the active site (see Tables 1 ▶ and 2 ▶ for data-collection and refinement statistics). Previously, it was shown that upon binding of both UDP-Gal and the acceptor analogue [αFuc-(1–2)-β-3-deoxy-Gal-O-(CH2)7CH3; deoxy analogue; DA] to the active site of the AABB chimeric enzyme, the internal active-site loop (residues 176–199) and the final nine C-terminal residues undergo a conformational change from an open to a closed form where the internal loop and the C-terminus come together (Alfaro et al., 2008 ▶). In this closed form, Trp181 in the internal loop and Arg352 in the C-terminus are π–π stacking directly above the uracil of the donor as the only direct contact between the internal loop and the C-terminus (Fig. 2 ▶ a).
Table 1. Data collection and processing.
Values in parentheses are for the outer shell.
| Unphotolysed AA(Gly)B–CC01 complex (cr1) | Unphotolysed AA(Gly)B–CC01 complex (cr2) | |
|---|---|---|
| Diffraction source | I911-5, MAX-lab | ID29, ESRF |
| Wavelength (Å) | 0.9077 | 0.9763 |
| Temperature (K) | 100 | 100 |
| Detector | MAR CCD | Pilatus 6M |
| Crystal-to-detector distance (mm) | 120 | 414 |
| Rotation range per image (°) | 0.5 | 0.1 |
| Total rotation range (°) | 180 | 150 |
| Exposure time per image (s) | 10 | 0.08 |
| Space group | P212121 | C2221 |
| a, b, c (Å) | 52.8, 78.5, 155.1 | 52.7, 150.7, 79.4 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Mosaicity (°) | 0.27 | 0.47 |
| Resolution range (Å) | 20.0–1.70 (1.85–1.70) | 49.74–1.90 (2.00–1.90) |
| Total No. of reflections | 497893 | 96364 |
| No. of unique reflections | 71207 (11865) | 24091 (2905) |
| Completeness (%) | 99.0 (96.2) | 95.2 (80.6) |
| Multiplicity | 7.0 (6.3) | 4.0 (2.4) |
| 〈I/σ(I)〉 | 14.9 (3.0) | 9.4 (3.0) |
| R r.i.m. † (%) | 8.4 (68.8) | 12.1 (50.5) |
| Overall B factor from Wilson plot (Å2) | 20.2 | 21.7 |
The redundancy-independent merging R factor R r.i.m. is estimated by multiplying the R merge value by the factor [N/(N − 1)]1/2, where N is the data multiplicity.
Table 2. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Unphotolysed AA(Gly)B–CC01 complex (cr1) | Unphotolysed AA(Gly)B–CC01 complex (cr2) | |
|---|---|---|
| Resolution range (Å) | 19.98–1.70 (1.74–1.70) | 42.18–1.90 (1.95–1.90) |
| Completeness (%) | 99.3 (90.0) | 95.0 (75.6) |
| σ Cutoff | 0.0 | 0.0 |
| No. of reflections, working set | 71130 (4110) | 22844 (1326) |
| No. of reflections, test set | 2134 (127) | 1232 (67) |
| Final R cryst (%) | 18.16 (33.78) | 18.36 (23.40) |
| Final R free (%) | 21.73 (41.86) | 22.62 (30.60) |
| Cruickshank DPI | 0.10 | 0.18 |
| No. of molecules in the asymmetric unit | 2 | 1 |
| No. of non-H atoms | ||
| Protein | 4718 | 2228 |
| Ion | 2 | 1 |
| Ligand | 81 | 52 |
| Water | 537 | 134 |
| Total | 5338 | 2415 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.009 | 0.009 |
| Angles (°) | 1.11 | 1.34 |
| Average B factors (Å2) | ||
| Overall | 26.7 | 26.4 |
| Protein | 26.7 | 26.7 |
| Ion | 26.3 | 28.5 |
| Ligand | 40.8 | 42.8 |
| Water | 32.5 | 31.9 |
| Ramachandran plot | ||
| Most favoured (%) | 97.0 | 96.2 |
| Allowed (%) | 3.0 | 3.8 |
| Disallowed (%) | 0.0 | 0.0 |
Figure 2.
Surface representation of the binding pocket of human blood group GTs. (a) Structure of the previously determined AABB chimeric mutant in complex with UDP-Gal and a deoxy acceptor (DA) in the closed conformation (PDB entry 2rj7; Alfaro et al., 2008 ▶). The position of Trp181 in the internal loop and Arg352 in the C-terminus are shown in yellow and blue, respectively. (b) AA(Gly)B–CC01 complex (cr1). Trp181 is shown in yellow. (c) AA(Gly)B–CC01 complex (cr2) in the presence of glycerol (Gol).
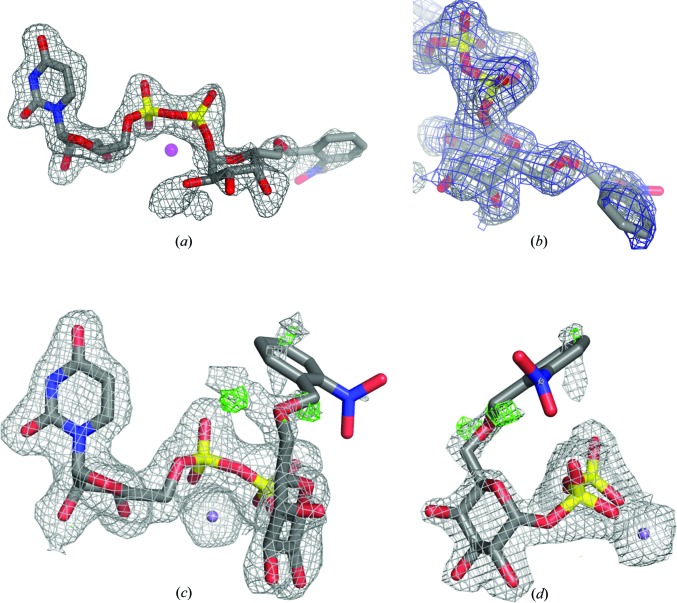
The first AA(Gly)B structure in complex with CC01 (cr1) was solved to 1.7 Å resolution and belonged to space group P212121. Of the two molecules in the asymmetric unit, only chain B shows an intact CC01 molecule together with a Mn2+ ion bound to the active site (Fig. 2 ▶ b). The electron density is well defined for the entire UDP-Gal moiety, whereas the density for the 2-nitrobenzyl protecting group is less ordered (Figs. 3 ▶ a and 3 ▶ b). The internal active-site loop from residues 176–199 in chain B is folded over the active site, bringing Trp181 into the closed position directly above the 5′ position of the uridine (Fig. 2 ▶ b). The C-terminus, however, is disordered and not visible in the electron density. This ‘semi-closed’ conformation of chain B is usually generated by binding of the donor substrate to the binding site (Alfaro et al., 2008 ▶). In chain A only the UDP moiety and a Mn2+ ion can be seen (data not shown). The presence of UDP and Mn2+ in chain A is presumably because this protein batch was not EDTA-treated before crystallization. Although no acceptor is present, the active site is in a fully closed conformation, with both the internal loop and the C-terminus fully ordered and folded over the active site. Such a conformation has only been observed previously when both donor and acceptor are bound in the active site and there seems to be no interaction with symmetry-related molecules in the crystal, which could help to explain the ordering of the last nine residues (Alfaro et al., 2008 ▶).
Figure 3.
(a) Electron-density F o − F c OMIT map around CC01 in the binding site of structure cr1 of the AA(Gly)B–CC01 complex contoured at 2.5σ. (b) 90° rotation of the view shown in (a), including a refined 2F o − F c map contoured at 0.85σ (blue). (c) 2F o − F c and F o − F c electron-density maps around CC01 in the binding site of structure cr2 before modelling of the cage contoured at 0.85 and 3.0σ, respectively. For clarity, the 2F o − F c electron density around Mn2+ has been contoured at 3.0σ. Residual density next to the O6′ atom of the galactose indicates the presence of at least one additional conformation of the cage. (d) 90° rotation of the view shown in (c).
The second AA(Gly)B structure in complex with CC01 (cr2) was solved to 1.9 Å resolution and belonged to space group C2221 with only one molecule in the asymmetric unit. This structure also shows an almost intact CC01 molecule together with a Mn2+ ion bound to the active site (Fig. 2 ▶ c). The electron density for the UDP-Gal part is again clearly defined but with the 2-nitrobenzyl group highly disordered in at least two distinct conformations (Figs. 3 ▶ c and 3 ▶ d). In this structure, in addition to a disordered C-terminus with the last nine residues not visible in the electron-density map, the internal loop is also disordered from residue 177 to residue 182. Apart from the internal loop and C-terminus, the overall conformations of all three molecules, the two molecules in the asymmetric unit of the cr1 structure and the single molecule in the cr2 structure, are highly isomorphous to each other, with r.m.s.d. values ranging from 0.28 to 0.36 Å using between 271 and 280 Cα atoms.
The partial disorder of the caging group in both structures raised the question of whether specific X-ray damage had occurred upon data collection. It is difficult to imagine that a ten-atom group could go from ordered to disordered in a crystal cooled to 100 K. In addition, the photo-cleavable bond links an O atom of the galactose and a C atom of the cage. Because X-ray-induced and light-induced photochemistry are similar processes (see, for instance, the X-ray- and light-induced decarboxylation of green fluorescent protein; Royant & Noirclerc-Savoye, 2011 ▶; van Thor et al., 2002 ▶), we would expect X-ray damage to break this specific bond. In the F o − F c electron density for cr2 (Fig. 3 ▶ c), two positive peaks can be unambiguously identified as two possible locations for the first C atom of the cage, strongly suggesting the presence of two instances of that cleavable bond. Hence, we are inclined to believe that the lack of density for the cage in cr2 is due to static and dynamic disorder rather than radiation damage, especially since it is oriented towards the bulk solvent and a disordered protein loop. Furthermore, in the 2F o − F c map of the caged donor in chain B of the cr1 structure the density of the link between the galactose and the caging group is continuous at a contour level of 0.9σ. This further indicates that the photo-cleavable bond is intact, although the position of the caging group is somewhat flexible.
3.2. Binding of CC01
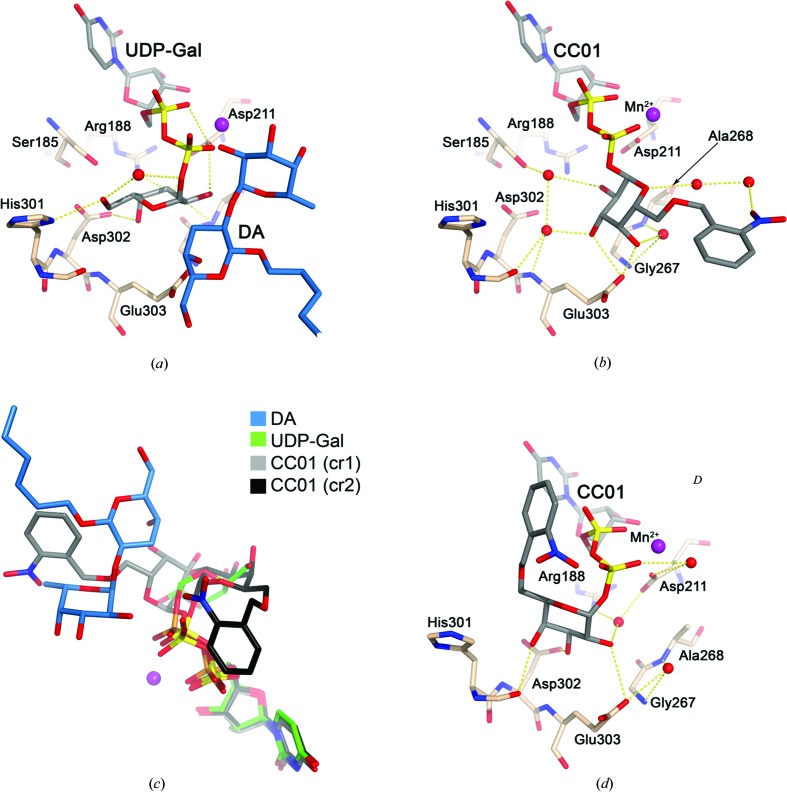
In the previously reported ternary complex AABB–UDP-Gal–DA (Alfaro et al., 2008 ▶), the sugar moiety of UDP-Gal is tucked underneath the pyrophosphoryl group, exposing the glycosidic bond to the solvent in what is often referred to as a ‘tucked-under’ conformation. The galactose residue is stabilized by hydrogen bonds to Arg188, Asp211, His301 and Asp302 (Fig. 4 ▶ a). In the cr1 structure, CC01 binds to the active site in an extended conformation where the galactose is rotated approximately 90° compared with the ‘tucked-under’ galactose of UDP-Gal (Fig. 4 ▶ b) and is only hydrogen bonded to Glu303. In this position, the O6 atom of the galactosyl is pointing towards the acceptor-binding site, resulting in an overlap of the 2-nitrobenzyl group to the place where the acceptor fucose binds in the active site (Fig. 4 ▶ c).
Figure 4.
Hydrogen bonds involving the galactose residue. (a) Structure of the ternary complex AABB–DA–UDP-Gal (PDB entry 2rj7). Amino-acid residues surrounding the active site are shown as light brown sticks. UDP-Gal and DA are shown as sticks with grey and blue C atoms, respectively. (b) The AA(Gly)B–CC01 (cr1) structure coloured as in (a). (c) Deviation from the ‘tucked-under’ conformation. Superposition of the AA(Gly)B–CC01 (cr1) structure (grey), the AA(Gly)B–CC01 (cr2) structure (black) and the AABB–DA–UDP-Gal structure (green; PDB entry 2rj7). (d) The AA(Gly)B–CC01 (cr2) structure coloured as in (a).
In the cr2 structure the conformation of CC01 is dramatically different. The galactose in cr2 is rotated approximately 120° compared with cr1, with hydrogen bonds to Trp300 (Fig. 2 ▶ c), Asp302 and Glu303 (Fig. 4 ▶ d) with the O6 of the galactosyl group pointing in the direction of the internal loop. Therefore, instead of overlapping with the acceptor-binding site the 2-nitrobenzyl group overlaps with the position where Trp181 and Arg352 are normally stacking in the closed conformation (Figs. 2 ▶ a and 2 ▶ c). This is most likely to be the reason why the internal loop is destabilized in this structure.
It is therefore clearly shown that CC01 is capable of adopting several conformations (Fig. 4 ▶ c), which can have different effects on the active-site dynamics and binding properties for the enzyme. The crystal which was used to solve the cr2 structure was soaked in a solution containing 22.3% glycerol for cryoprotection. As a result, a glycerol molecule is found in the acceptor-binding site, where it occupies the classical galactose-binding site and forms hydrogen bonds to His233, Thr245 and Glu303 (Fig. 2 ▶ c). The glycerol is partly overlapping with the position of CC01 in cr1 and therefore could be the reason why the conformation of CC01 in cr2 is rotated.
3.3. Spectroscopic photolysis experiments
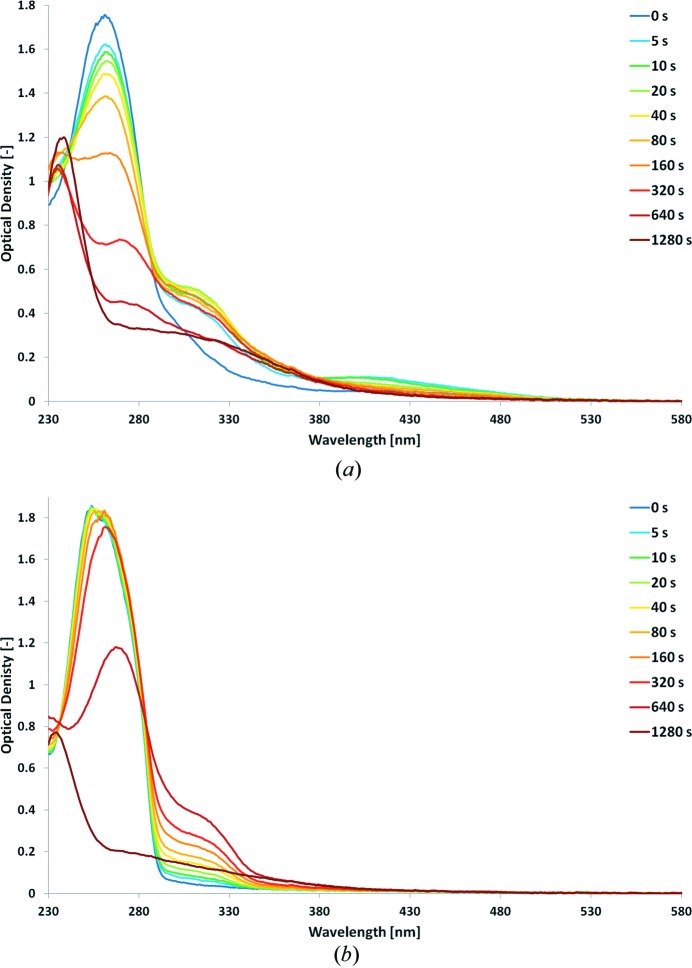
Photolysis of CC01 was tested in the crystallization buffer at cryogenic temperature. As photolysis was originally performed by 365 nm light at room temperature (Mannerstedt & Hindsgaul, 2008 ▶), we initially used light from a 355 nm laser but failed to observe significant changes in the absorption spectrum of the sample at 100 K, which can be attributed to the usual narrowing of absorption bands when going from room to low temperature. We then used light from a 266 nm laser and monitored the progressive conversion of the initial absorption spectrum on a 20 min time scale (Fig. 5 ▶ a). The process appears to be complex. The 260 nm peak decreases in two steps: in the first one (between 0 and 40 s) it decreases slowly; in the second one (after 40 s) the decay is faster and completely disappears after 20 min. During this second step, the peak progressively red-shifts until 280 nm. Two peaks quickly build up at 315 and 410 nm, which reach maxima at 5 and 20 s, respectively, and then decrease until only a broad and shallow peak at 330 nm remains. In order to interpret these data, we exposed solutions of noncaged UDP-Gal to 266 nm irradiation (Fig. 5 ▶ b). During the first 20 s of exposure, the 253 nm peak (corresponding to the uracil group) appears to be hardly affected, while a broad absorption band builds up between 300 and 330 nm. Starting at 40 s, the peak starts to shift towards 260 nm, while the peak below 330 nm continues to rise. Between 5 and 10 min, the 260 nm peak collapses by ∼40% and is completely gone after 20 min. The peak below 330 nm reaches a maximum at 10 min and is also gone after 20 min.
Figure 5.
Photolysis at 100 K of (a) CC01 and (b) UDP-Gal. Absorption spectra are represented at increasing times of irradiation by a 266 nm laser.
Overall, these data suggest that the compound is quickly de-caged (as shown by the buildup of the 410 nm peak), but that the uracil group is also affected, yet on a slower time scale. An optimal duration of irradiation must exist between 0 and 10 s (the maximum of the 410 nm peak). Because in crystallo photolysis experiments have to be performed on material strongly absorbing below 280 nm, the precise monitoring of cage photocleavage is difficult. Our initial attempts to photolyse CC01 in AA(Gly)B crystals at 100 K showed a broad increase of optical density between 300 and 540 nm which hampers the precise identification of the 410 nm peak, with the risk that significant damage is done to the uracil group of the compound. As a consequence, it appears that designing a caged compound absorbing away from the 250–300 nm region of the UV–visible spectrum is highly desirable to (i) keep the integrity of the substrate and (ii) allow the precise monitoring of optimal photocleavage.
4. Conclusion
We have obtained two distinct structures of the glycosyltransferase AA(Gly)B in complex with a caged version of UDP-Gal, the donor substrate of the glycosylation reaction. The two structures exhibit different orientations of the galactose and distinct locations of the cage. These differences must stem from subtle rearrangements of the enzyme structure between the two space groups and illustrate the highly dynamic nature of its active site. We have shown that the caged substrate could be photocleaved at 100 K, but that concomitant damage to the uracil was difficult to avoid. These two preliminary results are prerequisites for experiments that aim at characterizing putative covalent intermediate states. We have shown that these are within reach, and we will now attempt to design a caged compound that exhibits absorption at higher wavelengths which will prevent damage to the substrate and facilitate determination of the optimal time of irradiation. A logical follow-up to our experiments would consist of obtaining the structure of the enzyme with caged UDP-Gal and an acceptor molecule. This may prove difficult since the acceptor site is occupied in both structures, either by the cage of UDP-Gal or a glycerol molecule originating from the cryoprotection condition. However, kinetic crystallography experiments can be attempted without the acceptor molecule in the active site, since the galactose can be transferred from UDP-Gal to a solvent water molecule with the same retention of configuration as seen for the transfer of galactose to the H-antigen acceptor by GTB (Sindhuwinata et al., 2010 ▶).
Supplementary Material
PDB reference: AA(Gly)B, 3zgf
PDB reference: 3zgg
Acknowledgments
We thank T. G. Larsen and G. Gotthard for technical assistance. We are grateful for help with X-ray data collection provided by the beamline staff at I911-5 at MAX-lab, Lund, Sweden. The ESRF is acknowledged for financial support through a PhD fellowship to GB and for providing in-house research access to beamlines. The Cryobench is a platform of the Grenoble Instruct centre (ISBG UMS 3518 CNRS–CEA–UJF–EMBL) with support from ESRF, FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). We thank the French Embassy in Denmark for financial support through a French–Danish cooperation grant (to AR and MMP). This work was supported by DANSCATT and the Danish Council for Independent Research/Natural Sciences (FNU to MMP) and by the French National Research Agency (grant ANR-13-BSV8-0011-02 to CB).
References
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Alfaro, J. A., Zheng, R. B., Persson, M., Letts, J. A., Polakowski, R., Bai, Y., Borisova, S. N., Seto, N. O. L., Lowary, T. L., Palcic, M. M. & Evans, S. V. (2008). J. Biol. Chem. 283, 10097–10108. [DOI] [PubMed]
- André, I., Tvaroska, I. & Carver, J. P. (2003). Carbohydr. Res. 338, 865–877. [DOI] [PubMed]
- Angulo, J., Langpap, B., Blume, A., Biet, T., Meyer, B., Krishna, N. R., Peters, H., Palcic, M. M. & Peters, T. (2006). J. Am. Chem. Soc. 128, 13529–13538. [DOI] [PubMed]
- Ardèvol, A. & Rovira, C. (2011). Angew. Chem. Int. Ed. 50, 10897–10901. [DOI] [PubMed]
- Berg, S., Kaur, D., Jackson, M. & Brennan, P. J. (2007). Glycobiology, 17, 35R–56R. [DOI] [PubMed]
- Bourgeois, D. & Royant, A. (2005). Curr. Opin. Struct. Biol. 15, 538–547. [DOI] [PubMed]
- Breton, C., Fournel-Gigleux, S. & Palcic, M. M. (2012). Curr. Opin. Struct. Biol. 22, 540–549. [DOI] [PubMed]
- Breton, C., Šnajdrová, L., Jeanneau, C., Koča, J. & Imberty, A. (2006). Glycobiology, 16, 29R–37R. [DOI] [PubMed]
- Colletier, J.-P., Royant, A., Specht, A., Sanson, B., Nachon, F., Masson, P., Zaccai, G., Sussman, J. L., Goeldner, M., Silman, I., Bourgeois, D. & Weik, M. (2007). Acta Cryst. D63, 1115–1128. [DOI] [PubMed]
- Dube, D. H. & Bertozzi, C. R. (2005). Nature Rev. Drug Discov. 4, 477–488. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Errey, J. C., Lee, S. S., Gibson, R. P., Martinez Fleites, C., Barry, C. S., Jung, P. M., O’Sullivan, A. C., Davis, B. G. & Davies, G. J. (2010). Angew. Chem. Int. Ed. 49, 1234–1237. [DOI] [PubMed]
- Gastinel, L. N., Bignon, C., Misra, A. K., Hindsgaul, O., Shaper, J. H. & Joziasse, D. H. (2001). EMBO J. 20, 638–649. [DOI] [PMC free article] [PubMed]
- Goedl, C. & Nidetzky, B. (2009). Chembiochem, 10, 2333–2337. [DOI] [PubMed]
- Jørgensen, R., Grimm, L. L., Sindhuwinata, N., Peters, T. & Palcic, M. M. (2012). Angew. Chem. Int. Ed. 51, 4171–4175. [DOI] [PubMed]
- Jørgensen, R., Pesnot, T., Lee, H. J., Palcic, M. M. & Wagner, G. K. (2013). J. Biol. Chem. 288, 26201–26208. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Klink, B. U., Goody, R. S. & Scheidig, A. J. (2006). Biophys. J. 91, 981–992. [DOI] [PMC free article] [PubMed]
- Klink, B. U. & Scheidig, A. J. (2010). BMC Struct. Biol. 10, 38. [DOI] [PMC free article] [PubMed]
- Lairson, L. L., Henrissat, B., Davies, G. J. & Withers, S. G. (2008). Annu. Rev. Biochem. 77, 521–555. [DOI] [PubMed]
- Lee, S. S., Hong, S. Y., Errey, J. C., Izumi, A., Davies, G. J. & Davis, B. G. (2011). Nature Chem. Biol. 7, 631–638. [DOI] [PubMed]
- Lira-Navarrete, E., Valero-González, J., Villanueva, R., Martínez-Júlvez, M., Tejero, T., Merino, P., Panjikar, S. & Hurtado-Guerrero, R. (2011). PLoS One, 6, e25365. [DOI] [PMC free article] [PubMed]
- Mannerstedt, K. & Hindsgaul, O. (2008). Carbohydr. Res. 343, 875–881. [DOI] [PubMed]
- Marcus, S. L., Polakowski, R., Seto, N. O., Leinal, E., Borisova, S., Blancher, A., Roubinet, F., Evans, S. V. & Palcic, M. M. (2003). J. Biol. Chem. 278, 12403–12405. [DOI] [PubMed]
- Marth, J. D. & Grewal, P. K. (2008). Nature Rev. Immunol. 8, 874–887. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Monegal, A. & Planas, A. (2006). J. Am. Chem. Soc. 128, 16030–16031. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Patenaude, S. I., Seto, N. O. L., Borisova, S. N., Szpacenko, A., Marcus, S. L., Palcic, M. M. & Evans, S. V. (2002). Nature Struct. Biol. 9, 685–690. [DOI] [PubMed]
- Pesnot, T., Jørgensen, R., Palcic, M. M. & Wagner, G. K. (2010). Nature Chem. Biol. 6, 321–323. [DOI] [PMC free article] [PubMed]
- Rexach, J. E., Clark, P. M. & Hsieh-Wilson, L. C. (2008). Nature Chem. Biol. 4, 97–106. [DOI] [PMC free article] [PubMed]
- Royant, A., Carpentier, P., Ohana, J., McGeehan, J., Paetzold, B., Noirclerc-Savoye, M., Vernède, X., Adam, V. & Bourgeois, D. (2007). J. Appl. Cryst. 40, 1105–1112.
- Royant, A. & Noirclerc-Savoye, M. (2011). J. Struct. Biol. 174, 385–390. [DOI] [PubMed]
- Schlichting, I., Almo, S. C., Rapp, G., Wilson, K., Petratos, K., Lentfer, A., Wittinghofer, A., Kabsch, W., Pai, E. F., Petsko, G. A. & Goody, R. S. (1990). Nature (London), 345, 309–315. [DOI] [PubMed]
- Schuman, B., Alfaro, J. A. & Evans, S. V. (2007). Top. Curr. Chem. 272, 217–257.
- Seto, N. O. L., Compston, C. A., Szpacenko, A. & Palcic, M. M. (2000). Carbohydr. Res. 324, 161–169. [DOI] [PubMed]
- Seto, N. O. L., Palcic, M. M., Compston, C. A., Li, H., Bundle, D. R. & Narang, S. A. (1997). J. Biol. Chem. 272, 14133–14138. [DOI] [PubMed]
- Seto, N. O. L., Palcic, M. M., Hindsgaul, O., Bundle, D. R. & Narang, S. A. (1995). Eur. J. Biochem. 234, 323–328. [DOI] [PubMed]
- Sindhuwinata, N., Munoz, E., Munoz, F. J., Palcic, M. M., Peters, H. & Peters, T. (2010). Glycobiology, 20, 718–723. [DOI] [PubMed]
- Soya, N., Fang, Y., Palcic, M. M. & Klassen, J. S. (2011). Glycobiology, 21, 547–552. [DOI] [PubMed]
- Thor, J. J. van, Gensch, T., Hellingwerf, K. J. & Johnson, L. N. (2002). Nature Struct. Biol. 9, 37–41. [DOI] [PubMed]
- Weadge, J. T. & Palcic, M. M. (2008). Wiley Encyclopedia of Chemical Biology, Vol. 2, edited by T. P. Begley, pp. 198–211. New York: Wiley.
- Yamamoto, M., Lin, X.-H., Kominato, Y., Hata, Y., Noda, R., Saitou, N. & Yamamoto, F. (2001). J. Biol. Chem. 276, 13701–13708. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AA(Gly)B, 3zgf
PDB reference: 3zgg