A ribokinase from V. cholerae O395 has been cloned, overexpressed, purified and crystallized both in a native form and in a ribose- and ADP-bound form. Both crystals belonged to space group P1. Apo crystals of ribokinase diffracted to 3.4 Å resolution and the co-crystals diffracted to 1.75 Å resolution.
Keywords: Vibrio cholerae, ribokinase, ribose, ATP
Abstract
Ribokinase (RK) is one of the principal enzymes in carbohydrate metabolism, catalyzing the reaction of d-ribose and adenosine triphosphate to produce ribose-5-phosphate and adenosine diphosphate (ADP). To provide further insight into the catalytic mechanism, the rbsK gene from Vibrio cholerae O395 encoding ribokinase was cloned and the protein was overexpressed in Escherichia coli BL21 (DE3) and purified using Ni2+–NTA affinity chromatography. Crystals of V. cholerae RK (Vc-RK) and of its complex with ribose and ADP were grown in the presence of polyethylene glycol 6000 and diffracted to 3.4 and 1.75 Å resolution, respectively. Analysis of the diffraction data showed that both crystals possess symmetry consistent with space group P1. In the Vc-RK crystals, 16 molecules in the asymmetric unit were arranged in a spiral fashion, leaving a large empty space inside the crystal, which is consistent with its high Matthews coefficient (3.9 Å3 Da−1) and solvent content (68%). In the Vc-RK co-crystals four molecules were located in the asymmetric unit with a Matthews coefficient of 2.4 Å3 Da−1, corresponding to a solvent content of 50%.
1. Introduction
d-Ribose is one of the vital pentose sugars that are distributed in all three kingdoms of life, serving not only as an energy source but also as a component of RNA, DNA and many cofactors. Phosphorylation by ribokinase (RK; EC 2.7.1.15) enables ribose to enter into the pentose phosphate pathway and also into the synthesis of nucleotides and amino acids such as tryptophan and histidine (Anderson & Cooper, 1969 ▶; Lopilato et al., 1984 ▶). During phosphorylation, RK specifically transfers the γ-phosphate of adenosine triphosphate (ATP) to O5′ of d-ribose to form d-ribose-5-phosphate. Besides d-ribose and 2-deoxy-d-ribose, RK has also been found to catalyze the 5-O-phosphorylation of d-arabinose, d-xylose and d-fructose in the presence of ATP and potassium and magnesium ions (Chuvikovsky et al., 2006 ▶).
Although RK activity was first reported in calf liver in 1956 (Agranoff & Brady, 1956 ▶), detailed biochemical and structural results on RK were first reported from Escherichia coli (Sigrell et al., 1998 ▶, 1999 ▶; Andersson & Mowbray, 2002 ▶). The structure of human adenosine kinase (Mathews et al., 1998 ▶), another member of the RK family, exhibits a similar fold and mechanism, but the enzyme acts as a monomer. The structure and catalytic mechanism of human RK (Park et al., 2007 ▶) are very similar to those of E. coli RK. Recently, crystal structures of Sa239, an RK from Staphylococcus aureus (Li et al., 2012 ▶), and of an RK homologue from Thermococcus kodakarensis (Sato et al., 2013 ▶) have been reported. Structural studies on E. coli RK revealed that each monomer consists of two domains. A larger α/β domain that provides the binding site for sugar and ATP is comprised of a twisted nine-stranded β-sheet flanked by α-helices: four on one side and five on the other. A smaller β-sheet domain makes crucial contacts for dimerization and also acts as a lid over the catalytic domain to completely bury the sugar upon complex formation. An Asp residue in the α/β domain serves as a base that is responsible for activating O5′ of ribose, making it a better nucleophile to attack the γ-phosphate of ATP. The phosphorylation reaction is catalyzed by divalent cations such as Mg2+, Mn2+, Co2+, Ca2+, Ni2+ and Cu2+ (Chuvikovsky et al., 2006 ▶; Park & Gupta, 2008 ▶), whereas monovalent cations such as Na+, K+, Cs+ and NH4 + serve as an activator of RK (Andersson & Mowbray, 2002 ▶; Chuvikovsky et al., 2006 ▶). Large relative motion between the domains in the same subunit upon substrate binding is thought to be responsible for the entry and release of ribose.
Vibrio cholerae shows a biphasic life style with a periodic transition between environmental and host-associated niches. During this process, the metabolic reactions and gene expression of V. cholerae are modified depending on the physiological conditions. However, the detailed mechanism of ribose metabolism has not yet been reported. RK from V. cholerae O395 (Vc-RK; UniProtKB accession code A5F1B7; gene name rbsK) possesses 306 amino acids with a molecular weight of 32 kDa. In order to gain further insight into the catalytic function at the atomic level, we report the cloning, overexpression, purification, crystallization and preliminary structural analysis of Vc-RK in the apo form, in a sugar- and adenosine diphosphate (ADP)-bound form and in a sugar-, ADP- and Cs+-bound form at resolutions of 3.4, 1.75 and 2.37 Å, respectively.
2. Materials and methods
2.1. Cloning, expression and purification of Vc-RK
The rbsK gene encoding Vc-RK was amplified using polymerase chain reaction (PCR) from the chromosomal DNA of V. cholerae (strain O395) using specific primers (forward, 5′-GGAATTC CATATGAATAAGTTGGTGGTTCTGGGTAGCGTC-3′; reverse, 5′-CG GGATCCCTATGAGTGTTCTGCTAAAAAGGCCTCAAC-3′). The primers were synthesized (NeuPro-Cell) with adaptors (bold) for the restriction enzymes NdeI and BamHI (shown in italics). The purified PCR product was cloned into the NdeI and BamHI sites of the expression vector pET-28a(+) (Novagen), which adds six consecutive histidines to the N-terminus of the desired protein followed by a thrombin cleavage site. The clones were selected appropriately using E. coli XL1-Blue cells with kanamycin resistance. The recombinant protein was overexpressed in E. coli strain BL21 (DE3) in the presence of kanamycin (30 µg ml−1). For overexpression, a single colony was picked, transferred into 100 ml Luria–Bertani (LB) medium and grown overnight. 1 l of LB broth was inoculated with 10 ml overnight culture and the culture was grown at 310 K until the OD600 reached 0.6; the cells were then transferred to 293 K. The cells were induced with 0.2 mM IPTG and incubated for 16 h at 293 K following growth to mid-log phase. The cells were harvested at 4500g for 20 min and the pellet was resuspended in 25 ml ice-cold lysis buffer [50 mM Tris–HCl pH 8.0, 300 mM NaCl, 2 mM phenylmethanesulfonyl fluoride (PMSF), 5 mM β-mercaptoethanol]. The cells were lysed by the addition of lysozyme followed by sonication on ice. The cell lysate was then centrifuged at 12 000g for 45 min. The supernatant was applied onto a nickel–nitrilotriacetic acid (Ni2+–NTA) affinity chromatography column (Qiagen) which was previously equilibrated with buffer A (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol). The protein was eluted with a linear gradient of 5–150 mM imidazole in buffer A.
The 6×His tag was cleaved using restriction-grade thrombin (Novagen) and final purification of the protein from contaminating proteins, thrombin and cleaved 6×His tag was achieved by gel filtration using a Sephacryl S-100 (GE Healthcare) column pre-equilibrated with buffer B (50 mM Tris–HCl pH 8.0, 100 mM NaCl). RK was concentrated to 20 mg ml−1 (Amicon ultracentrifugation unit; molecular-weight cutoff 10 kDa) and used for crystallization.
2.2. Crystallization of Vc-RK
Initial crystallization trials for both unliganded Vc-RK and ADP- and ribose-bound Vc-RK were performed by the hanging-drop vapour-diffusion method at 293 and 277 K with Grid Screen Ammonium Sulfate, Grid Screen PEG 6000, Crystal Screen and Crystal Screen 2 from Hampton Research (Jancarik & Kim, 1991 ▶). A Hampton Research 24-well crystallization tray (Hampton Research, Laguna Niguel, California, USA) was used to explore the initial crystallization conditions with 600 µl reservoir solution and 4–5 µl drop size (protein and precipitant varied around a 1:1 volume ratio). Initial trials using PEG 6000 as precipitant always produced very tiny crystals covered with protein skin, but the size and quality of the crystals could not be improved further by using different protein:precipitant ratios or reservoir strength. Although fresh protein samples appeared to be homogeneous, a nonreducing 10%(w/v) SDS–PAGE of few-days-old samples indicated a heterogeneous population with the appearance of higher oligomeric species. However, these higher oligomers disappeared on a reducing gel (Fig. 1 ▶). The amino-acid sequence of Vc-RK contains five cysteine residues and there is a strong probability that unpaired cysteines form disulfide-linked oligomers. Therefore, we maintained a β-mercaptoethanol concentration of 5 mM in all crystallization experiments. For the co-crystals, Vc-RK was incubated with ribose (5 mM) and ADP (5 mM) for 1 h before crystallization setup. Both apo Vc-RK and ribose- and ADP-bound Vc-RK crystallized using Grid Screen PEG 6000 condition A3 consisting of 5% PEG 6000 in 0.1 M MES pH 6.0 as the precipitant and 35%(w/v) PEG 6000 as the reservoir within 7 d at 293 K (Fig. 2 ▶).
Figure 1.
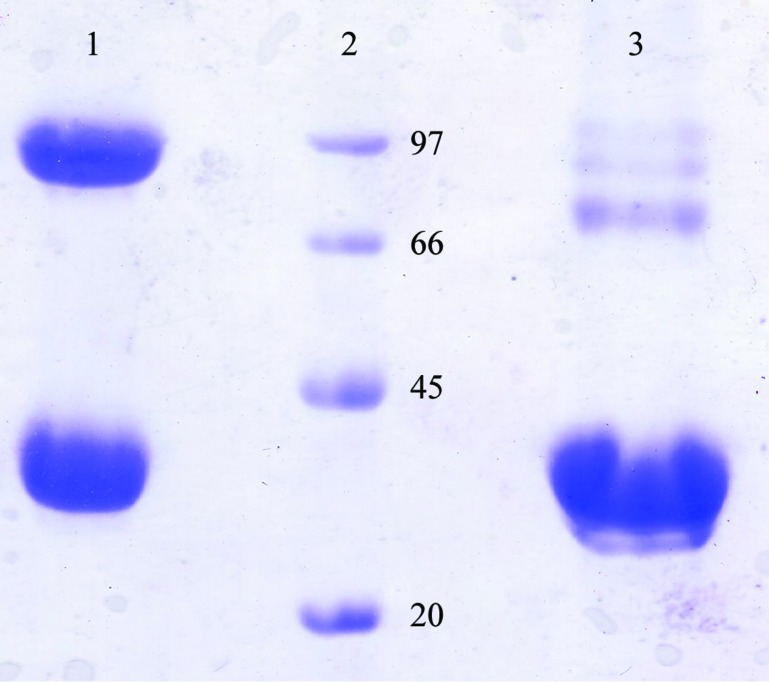
The homogeneity of the purified Vc-RK protein as checked by 10% SDS–PAGE. Lane 1, Vc-RK protein sample without β-mercaptoethanol; lane 2, molecular-weight markers (labelled in kDa); lane 3, Vc-RK protein sample treated with β-mercaptoethanol.
Figure 2.
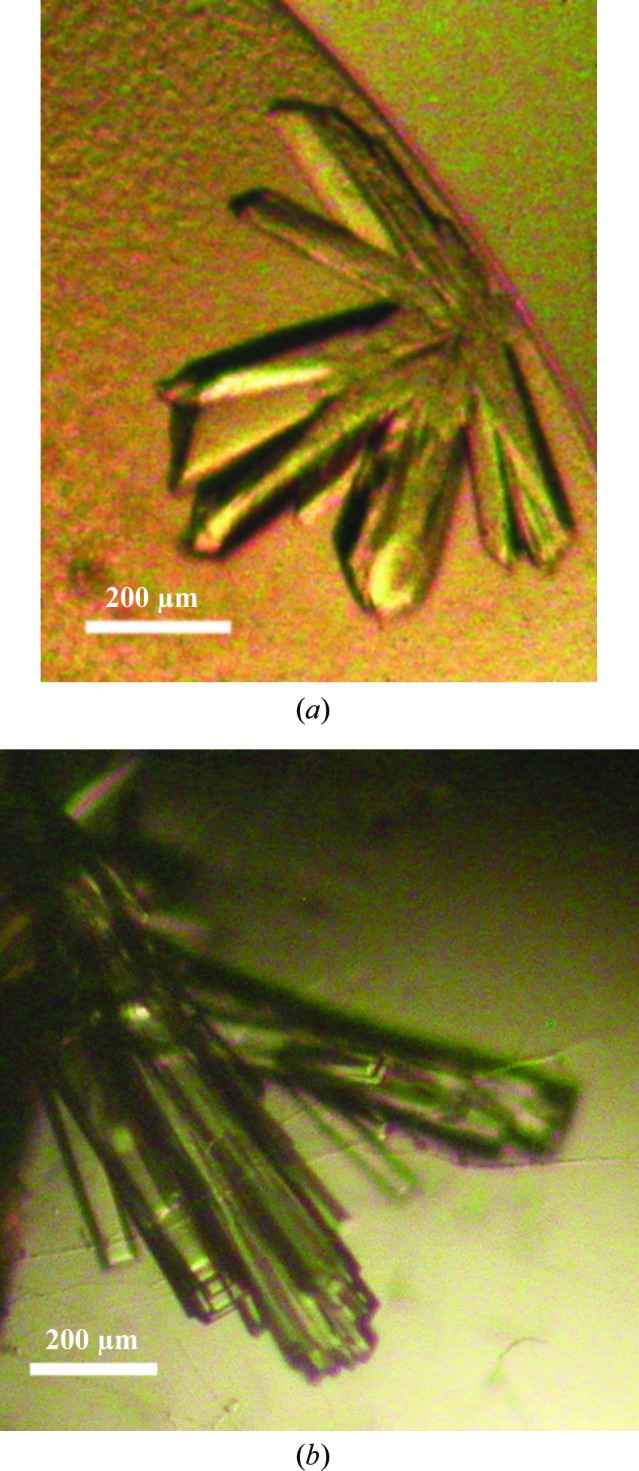
Crystals of native Vc-RK and its co-crystals with ribose and ADP grown using 5% PEG 6000 at pH 6 as precipitant. (a) Crystals of native Vc-RK; the maximum dimensions of the crystals were 0.2 × 0.2 × 0.3 mm. (b) Co-crystals of Vc-RK; the maximum dimensions of the crystals were 0.4 × 0.3 × 0.2 mm.
2.3. Data collection and preliminary X-ray diffraction of Vc-RK
Crystals of apo Vc-RK and its co-crystal with ribose and ADP were looped out from the crystallization drops using a 20 mm nylon loop and flash-cooled in a stream of nitrogen (Oxford Cryosystems) at 100 K. Diffraction data were collected on a MAR 345 CCD on BM14 at the European Synchrotron Radiation Facility (ESRF), Grenoble. The incident X-ray beam had a wavelength of 0.97 Å and 360 frames were recorded with non-overlapping 1° oscillations. For apo Vc-RK crystals 12 s exposure per image was used, whereas for the co-crystals the exposure time was only 1 s. Data were processed and scaled using iMosflm (Battye et al., 2011 ▶), POINTLESS and SCALA (Evans, 2006 ▶). Data for the Cs+-bound crystals were collected by soaking ribose- and ADP-bound Vc-RK crystals with a cryoprotectant containing 20 mM CsCl. Data-collection and processing statistics are given in Table 1 ▶.
Table 1. Data-collection and processing parameters for Vc-RK crystals.
Values in parentheses are for the outermost resolution shell.
| Apo Vc-RK | Vc-RK + ribose + ADP + Na | Vc-RK + ribose + ADP + Cs | |
|---|---|---|---|
| Space group | P1 | P1 | P1 |
| Unit-cell parameters (Å, °) | a = 129.22, b = 130.85, c = 145.69, α = 110.52, β = 90.00, γ = 119.59 | a = 59.42, b = 70.70, c = 79.91, α = 106.31, β = 97.66, γ = 98.69 | a = 59.30, b = 69.55, c = 78.31, α = 106.99, β = 98.76, γ = 98.46 |
| Resolution (Å) | 3.40 | 1.75 | 2.37 |
| Oscillation range (°) | 1 | 1 | 1 |
| Molecules in asymmetric unit | 16 | 4 | 4 |
| Matthews coefficient V M (Å3 Da−1) | 3.9 | 2.4 | 2.3 |
| Solvent content (%) | 68 | 50 | 48 |
| No. of unique reflections | 97045 | 117357 | 42241 |
| Multiplicity | 2.1 (2.5) | 4.0 (3.9) | 2.07 (2.11) |
| Mosaicity (°) | 1.5 | 0.8 | 0.7 |
| Completeness (%) | 92.9 (85.6) | 95.1 (95.1) | 93.8 (93.4) |
| R merge (%) | 15.1 (45.9) | 9.3 (56.7) | 6.4 (28.37) |
| Average I/σ(I) | 4.7 (2.0) | 8.5 (2.4) | 5.1 (1.5) |
3. Results and discussion
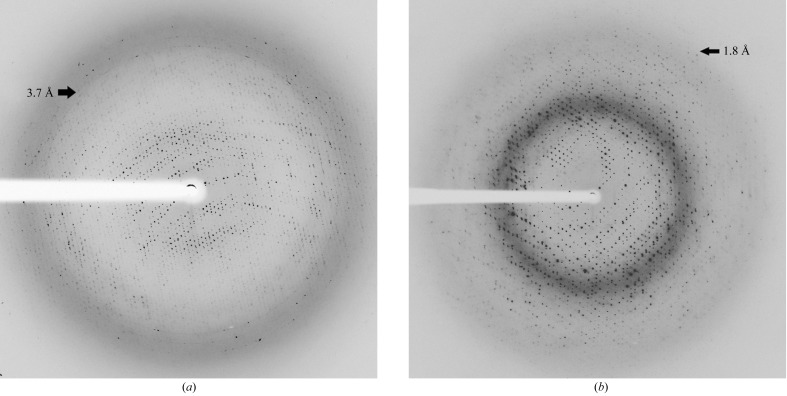
Apo Vc-RK crystals diffracted poorly and we could process the data to a resolution of 3.4 Å (Fig. 3 ▶ a, Table 1 ▶). Crystals of apo Vc-RK have symmetry consistent with the triclinic space group P1 with unit-cell parameters a = 129.22, b = 130.85, c = 145.69 Å, α = 110.52, β = 90.0, γ = 119.59°. However, the co-crystals of Vc-RK produced excellent quality diffraction data to a resolution of 1.75 Å with symmetry consistent with the triclinic space group P1 with unit-cell parameters a = 59.42, b = 70.70, c = 79.91 Å, α = 106.31, β = 97.66, γ = 98.69° (Fig. 3 ▶ b, Table 1 ▶).
Figure 3.
(a) X-ray diffraction image of an unliganded Vc-RK crystal that diffracted to a resolution of 3.4 Å (black arrow). (b) X-ray diffraction image of a Vc-RK, ADP and ribose co-crystal that diffracted to a resolution of 1.75 Å (black arrow).
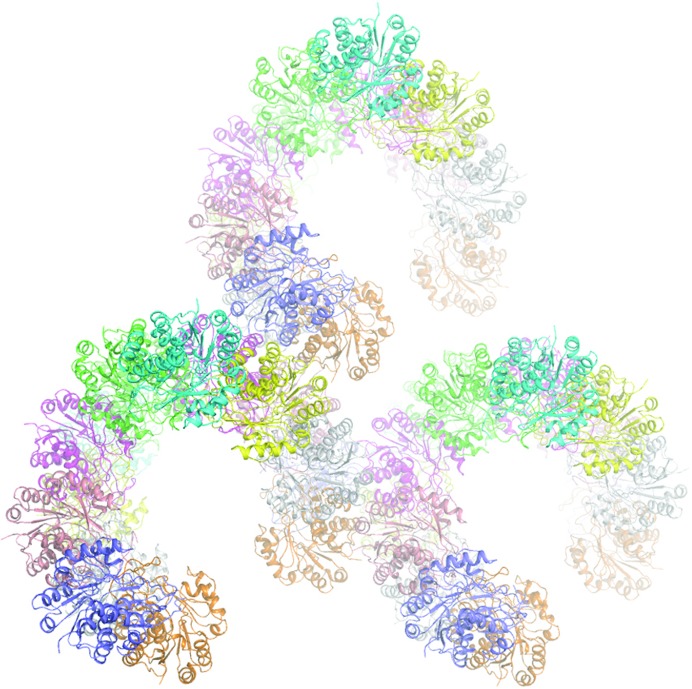
A BLAST (Altschul et al., 1990 ▶) search for homologous structures showed that Vc-RK has the highest identity (54%) to RK from E. coli (PDB entry 1rk2; Sigrell et al., 1999 ▶). A model was prepared using the coordinates of 1rk2, where the mismatched residues between Vc-RK and 1rk2 were truncated to Ala. This model was subsequently used as a search model during molecular-replacement (MR) calculations. We first solved the co-crystal structure of Vc-RK using the high-resolution diffraction data. Molecular-replacement calculations using the program Phaser (McCoy et al., 2007 ▶) located four molecules in the asymmetric unit with an LLG value of 12686 and an R factor of 43.2%. Considering the molecular weight of 32 kDa for Vc-RK, four molecules in the asymmetric unit corresponds to a Matthews coefficient V M (Matthews, 1968 ▶) of 2.4 Å3 Da−1 and a solvent content of 50%. A few cycles of refinement of this structure using REFMAC5 (Murshudov et al., 2011 ▶) and PHENIX (Adams et al., 2010 ▶) produced an R factor of 34.7% (R free = 36.2%) and a 2F obs − F calc map calculated using this model showed continuous unambiguous electron density for each Vc-RK molecule. This refined Vc-RK molecule was used to solve the apo Vc-RK structure. 16 molecules of this modified model in the asymmetric unit produced an LLG value of 7531 with an R factor of 45.5% with data between 30.0 and 3.4 Å resolution. Packing considerations with 16 molecules in the asymmetric unit correspond to a Matthews coefficient V M (Matthews, 1968 ▶) of 3.9 Å3 Da−1 and a solvent content of 68% (Table 1 ▶). Packing of the Vc-RK molecules inside the crystal shows that they form a spiral type of arrangement, leaving a huge empty space at the centre accounting for the high solvent content (68%; Table 1 ▶; Fig. 4 ▶). Analysis of the packing ruled out the presence of further molecules in the asymmetric unit.
Figure 4.
Arrangement of ribokinase molecules within the crystal; the large empty space within the crystal is evident. The figure was generated using PyMOL v.1.5 (DeLano, 2002 ▶).
Acknowledgments
The laboratory of US is supported by the MSACR project, DAE, Government of India and SINP. This work is supported by the Department of Biotechnology (DBT), Government of India (the BM14 beamline project for synchrotron data collection at the ESRF, Grenoble). MDP thanks the DBT for a DBT-RA fellowship.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Agranoff, B. W. & Brady, R. O. (1956). J. Biol. Chem. 219, 221–229. [PubMed]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol. 215, 403–410. [DOI] [PubMed]
- Anderson, A. & Cooper, R. A. (1969). Biochim. Biophys. Acta, 177, 163–165. [DOI] [PubMed]
- Andersson, C. E. & Mowbray, S. L. (2002). J. Mol. Biol. 315, 409–419. [DOI] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Chuvikovsky, D. V., Esipov, R. S., Skoblov, Y. S., Chupova, L. A., Muravyova, T. I., Miroshnikov, A. I., Lapinjoki, S. & Mikhailopulo, I. A. (2006). Bioorg. Med. Chem. 14, 6327–6332. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Li, J., Wang, C., Wu, Y., Wu, M., Wang, L., Wang, Y. & Zang, J. (2012). J. Struct. Biol. 177, 578–582. [DOI] [PubMed]
- Lopilato, J. E., Garwin, J. L., Emr, S. D., Silhavy, T. J. & Beckwith, J. R. (1984). J. Bacteriol. 158, 665–673. [DOI] [PMC free article] [PubMed]
- Mathews, I. I., Erion, M. D. & Ealick, S. E. (1998). Biochemistry, 37, 15607–15620. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Park, J. & Gupta, R. S. (2008). Cell. Mol. Life Sci. 65, 2875–2896. [DOI] [PMC free article] [PubMed]
- Park, J., van Koeverden, P., Singh, B. & Gupta, R. S. (2007). FEBS Lett. 581, 3211–3216. [DOI] [PubMed]
- Sato, T., Fujihashi, M., Miyamoto, Y., Kuwata, K., Kusaka, E., Fujita, H., Miki, K. & Atomi, H. (2013). J. Biol. Chem. 288, 20856–20867. [DOI] [PMC free article] [PubMed]
- Sigrell, J. A., Cameron, A. D., Jones, T. A. & Mowbray, S. L. (1998). Structure, 6, 183–193. [DOI] [PubMed]
- Sigrell, J. A., Cameron, A. D. & Mowbray, S. L. (1999). J. Mol. Biol. 290, 1009–1018. [DOI] [PubMed]