Abstract
The family Cudoniaceae (Rhytismatales, Ascomycota) was erected to accommodate the “earth tongue fungi” in the genera Cudonia and Spathularia. There have been no recent taxonomic studies of these genera, and the evolutionary relationships within and among these fungi are largely unknown. Here we explore the molecular phylogenetic relationships within Cudonia and Spathularia using maximum likelihood and Bayesian inference analyses based on 111 collections from across the Northern Hemisphere. Phylogenies based on the combined data from ITS, nrLSU, rpb2 and tef-1α sequences support the monophyly of three main clades, the /flavida, /velutipes, and /cudonia clades. The genus Cudonia and the family Cudoniaceae are supported as monophyletic groups, while the genus Spathularia is not monophyletic. Although Cudoniaceae is monophyletic, our analyses agree with previous studies that this family is nested within the Rhytismataceae. Our phylogenetic analyses circumscribes 32 species-level clades, including the putative recognition of 23 undescribed phylogenetic species. Our molecular phylogeny also revealed an unexpectedly high species diversity of Cudonia and Spathularia in eastern Asia, with 16 (out of 21) species-level clades of Cudonia and 8 (out of 11) species-level clades of Spathularia. We estimate that the divergence time of the Cudoniaceae was in the Paleogene approximately 28 Million years ago (Mya) and that the ancestral area for this group of fungi was in Eastern Asia based on the current data. We hypothesize that the large-scale geological and climatic events in Oligocene (e.g. the global cooling and the uplift of the Tibetan plateau) may have triggered evolutionary radiations in this group of fungi in East Asia. This work provides a foundation for future studies on the phylogeny, diversity, and evolution of Cudonia and Spathularia and highlights the need for more molecular studies on collections from Europe and North America.
Introduction
Fungi are the principal degraders of biomass in terrestrial ecosystems [1], [2]. As decomposers of organic matter, they form a significant component of forest ecosystems. However, compared to the 5.1 million estimated fungal species [3], our current understanding of fungal evolutionary diversity is limited [4], [5] with an estimated 100,000 species currently recognized [3]. This lack of basic information on diversity has significantly hampered our interpretations of biogeographic patterns in fungi [6], [7]. This is particularly true for saprotrophic fungi, such as the Earth Tongue mushrooms, which are ephemeral, easily overlooked on the forest floor, and have no commercial value as food.
The family Cudoniaceae (Rhytismatales, Ascomycotina) was erected by P. F. Cannon to accommodate the genera Cudonia Fr. and Spathularia Pers. [8]. Members of this family are usually referred to in fungi guidebooks as “earth tongues” or “fairy fans” because of their shapes (Fig. 1). These fungi form small fruiting bodies that have a flattened to club-shaped apex. Cudonia and Spathularia are restricted to the Northern Hemisphere with highest species diversity in temperate habitats and only a few species reported from subtropical areas. However, species of Cudonia and Spathularia have not been recovered in molecular surveys as root endophytes or mycorrhizas and are thus presumed to be soil and leaf litter saprotrophs.
Figure 1. Macroscopic and microscopic morphological features of Cudonia and Spathularia species.
A. Cudonia circinans (M. Carbone 313); B. Cudonia confusa (M. Carbone 314); C. Cudonia sp1 (Z.-W. Ge 729); D. Cudonia lutea (Z.-W. Ge 1634); E. Cudonia sp10 (Z. L. Yang 4297); F. Cudonia sp9 (X. H. Wang 2324); G. Cudonia sp12 (Z.-W. Ge 829); H. Spathularia sp5 (Z. L. Yang 5385); I. Spathularia sp4 (L. P. Tang 252); J. Spathularia velutipes (Z.-W. Ge 2232); K. Spathularia rufa (M. Carbone 309); L. Spathularia flavida (M. Carbone 313); M. Hymenium of Spathularia flavida (Z.-W. Ge 3348); N. Asci and paraphyses of Spathularia flavida (Z.-W. Ge 3348); O. Ascospores of Spathularia flavida (Z.-W. Ge 3348).
Cudonia differs from Spathularia in that the Spathularia species have flattened fruiting bodies (e.g. like a spatula) whereas Cudonia species have rounded and club-shaped fruiting bodies (Fig. 1). However, the micromorphology of these two genera is similar: both genera have curved paraphyses, club-shaped asci, and long, thin, hyaline ascospores that are enveloped in gelatinous sheaths [8], [9]. These characters are also shared by many species of Rhytismataceae Chevall.
Members of Cudonia and Spathularia differ morphologically from other species in the family Rhytismataceae, such as Rhytisma Fr. species that cause “tar spot” on leaves [10], Coccomyces De Not. species that cause “leaf blights” [11], and some Lophodermium Chevall. species that cause “needle cast” diseases on conifers [12]. These members of Rhytismatales M. E. Barr. ex Minter have inconspicuous fruiting bodies where the spore-bearing hymenium is covered by tissue (at least during early development). Although Cudonia and Spathularia have distinct fruit bodies, with a stipe and spore bearing hymenium, the hymenium is covered in early development by a thin veil [13]-[17]. This veil can be seen in mature specimens of Cudonia and Spathularia as a skirt-like structure at the margin of the hymenium and this feature may be homologous to the covering layer that is present in some Rhytismataceae species (e.g. Rhytisma spp. [10]).
Because of the unique morphology of Cudonia and Spathularia species, there have been a large number of taxonomic studies of this group in Europe [18]-[20], North America [14]-[16], [21], and Asia [9], [22], [23]. Cudonia and Spathularia were traditionally placed in the order Helotiales Nannf. ex Korf & Lizon, with Cudonia in the Leotiaceae Corda and Spathularia in Geoglossaceae Corda [24]. However, the Geoglossaceae has recently been redefined and placed it in its own order (Geoglossales) and class (Geoglossomycetes) [25]. When P. F. Cannon erected Cudoniaceae with Cudonia as the type genus, the group was considered to belong in Helotiales [8] but was later transferred to Rhytismatales [26], [27].
Although the taxonomy of Cudonia and Spathularia has received ample attention in the past, this group has rarely been examined using molecular tools. Most studies that have included sequences of Cudonia and Spathularia have been focused on family-level, order-level, or class-level phylogenetic relationships, and thus have incorporated only a few species from these genera. Phylogenetic analyses based on the 18S nuc-SSU nrDNA [28]-[31] showed that C. circinans and S. flavida formed a monophyletic clade. This relationship was later confirmed based on the ITS and 28S nrLSU [9], [17], [32] and other genes [33]-[35]. Although only a few species of Cudonia and Spathularia included, the most recent findings based on nrLSU and mtSSU indicate an affinity between Cudonia, Spathularia, and members of Coccomyces, Colpoma Wallr., and Tryblidiopsis P. Karst. (Rhytismataceae, Rhytismatales) [27], suggesting that the Cudoniaceae arose from within Rhytismataceae.
Until now, only one phylogenetic analysis [9] has examined relationships within Cudonia and Spathularia. The combined ITS-nrLSU analysis recovered both genera as monophyletic but with low bootstrap support. However, this study was based on a small number of specimens from a few species and the ribosomal genes were insufficient to adequately resolve the relationships within the group. Thus, there are still many open questions about the relationships both within and between Cudonia and Spathularia. For instance, no studies have examined this group based on a geographically and phylogenetically broad sampling of specimens, no studies have used a multi-gene phylogeny to study this group, and no studies have inferred biogeographic or morphological evolutionary patterns within Cudonia and Spathularia. In recent years, multi-gene datasets have proved successful for species delimitation and for inferring robust phylogenies of major fungal clades [36]. Well-resolved phylogenies have subsequently facilitated biogeographic studies of several widespread and ecologically important fungal groups [37]-[42].
This study investigates the species diversity, biogeography, and phylogenetic relationships within and between Cudonia and Spathularia. To accomplish this goal we have performed maximum likelihood (ML) and Bayesian inference (BI) phylogenetic analyses based on four DNA regions: the nuclear internal transcribed spacers (ITS), the nuclear large subunit ribosomal DNA (nrLSU), the second largest subunit of RNA polymerase II (rpb2), and elongation factor 1-α (tef-1α). We also performed Bayesian molecular clock analysis to estimate evolutionary divergence times for Cudonia and Spathularia and we used Likelihood analysis of geographic range evolution to hypothesize the putative center of origin for this group. The specific aims of this study were: (i) to examine genetic differentiation and species diversity among a global sampling of Cudonia and Spathularia specimens; (ii) to infer the phylogenetic relationships within this group using ribosomal DNA sequences (ITS and nrLSU) and two single-copy nuclear genes (rpb2 and tef-1α); (iii) to infer the biogeography of Cudonia and Spathularia and to elucidate both its center of diversity and the main biogeographic events; and (iv) to discuss the morphological evolution that occurred during their radiation in the Northern Hemisphere.
Materials and Methods
Taxon sampling
We obtained 111 collections of Cudonia and Spathularia from throughout the Northern Hemisphere (Table S1). Most collections (105) were obtained over the last decade where Cudonia and Spathularia are known to occur; Asia (China), Europe (Finland, Switzerland) and North America (Canada, United States). The remaining six specimens were obtained from the fungal herbaria at Jilin Agricultural University (HMJAU) (2 specimens) and the University of Tennessee (TENN) (4 specimens). Since species within both Cudonia and Spathularia have no protected status, no specific permits were required for collecting these fungi. Voucher specimens are housed in the Cryptogamic Herbarium of the Kunming Institute of Botany, Chinese Academy of Sciences (HKAS) and the Farlow Herbarium of Harvard University (FH).
DNA extraction, PCR and Sequencing
DNA was extracted from approximately 20 mg of dried fruiting body tissue using the DNeasy Plant Mini Kit (Qiagen, Maryland, US) according to the manufacturer's recommendations or using a modified CTAB method [43].
PCR reactions were performed on an ABI 2720 Thermal Cycler (Applied Biosystems, Foster City, CA, USA) or an Eppendorf Master Cycler (Eppendorf, Netheler-Hinz, Hamburg, Germany), using Biomed taq and their suggested protocols (Biomed, Beijing 100097, China).
We used published primers to PCR-amplify fragments from the ITS, nrLSU, rpb2 and tef-1α DNA regions: ITS1F/ITS4 for ITS [44], [45], LR0R/LR5 for nrLSU [46], 983F/1567R for tef-1α [47], and RPB2-6F and RPB2-7R for rpb2 [48]. The rpb2 region was difficult to amplify with published primers, so for difficult samples we designed a new primer pair (RPB2_6Cudf: 5′- TCAGGCTTGTGGTCTGGT -3′; RPB2-7Cudr: 5′- GGGAAGGGAATGATGGAT -3′) using the online software package Primer3 [49].
For PCR amplification, we used published thermocycler conditions [50] except that annealing temperatures were optimized for each gene region: 53°C for ITS, 50°C for nrLSU, 60°C for rpb2, or 55°C for tef-1α. Amplicons were electrophoresed in 1% agarose gels stained with SYBR Green I (Molecular Probes, Eugene, OR, USA) or Gel View (BioTeke Corporation, Beijing, China) and then visualized under UV light. PCR products were purified using a QIAquick PCR purification kit (Qiagen Science, USA) or EXO and SAP enzymes [51]. When multiple amplicons were present, we excised amplicons of the appropriate size using a Gel Extraction & PCR Purification Combo Kit (Spin-column) (Bioteke, Beijing, China; Qiagen Science, USA).
Sequencing was performed with the same PCR primers using the Big Dye Sequencing Kit v.3.1 on an ABI-3730-XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA) or sent to Shanghai Sangon Biological Engineering Technology and Service Co. for sequencing. For samples that failed in direct sequencing, PCR products were gel purified and then cloned in the TakaRa pMD18-T Vector. Two to three clones per amplicon were sequenced. Sequence chromatograms were compiled with Sequencher v4.1.4 (Gene Codes Corp., Ann Arbor, MI, USA) or Seqman (DNA STAR Package; DNAStar, Madison, WI, USA). All sequences have been deposited in GenBank (Table S1).
Data matrices and phylogenetic analyses
Coccomyces dentatus (J.C. Schmidt &Kunze) Sacc. and Tryblidiopsis pinastri (Pers.) P. Karst. (Rhytismatales) were selected as outgroups for phylogenetic analyses except for the ITS dataset. These genera are considered close relatives of Cudonia and Spathularia by previous studies [27], [31], [52], and our preliminary analysis, including diverse Rhytismatales and Helotiales, verified that these taxa are appropriate outgroups.
After adding Cudonia and Spathularia sequences from GenBank (16 ITS, 17 nrLSU, 2 rpb2, and 1 tef-1α – see Tables S2 and S3) with sequences from our 111 collections, phylogenetic reconstructions were conducted to identify well-supported clades of Cudonia and Spathularia. Alignments were performed for each gene (ITS, nrLSU, rpb2 and tef-1α) using Mafft v6.8 [53] with manual improvements including trim the ends and adjust the obviously misaligned base pairs in Bioedit v7.0.9 [54]. The alignments (sites excluded from the analyses listed after the alignment file) have been submitted to TreeBase with submission number S15957 (http://purl.org/phylo/treebase/phylows/study/TB2:S15957). In the 127 specimens ITS alignment, 75 out of 587 sites have been excluded for analyses, most of these sites are gaps due to sequences downloaded from GenBank (particularly Cudonia monticola and Spathularia sp.). These gaps are obvious introns that are clearly visible and easy to interpret (Data S1). The sites excluded from analyses are: 11, 35, 61, 69-70, 78, 82-85, 91, 106, 117, 123-125, 144-158, 167, 180-181, 193-194, 211-215, 224-225, 229, 392-393, 410-414, 440-447, 482-483, 501-503, 517, 524, 544, 554-560, and 567.
A reciprocal 70% bootstrap support approach [55] was used to compare the tree topologies from individual genes. The result revealed there is no significant incongruence between the data sets, so the ITS, nrLSU, rpb2, tef-1α were concatenated for phylogenetic reconstruction.
Maximum Likelihood analyses were performed to infer phylogenetic relationships for each gene and for the combined ITS-nrLSU-rpb2-tef-1α dataset using RAxML version 7.2.3 [56]. Optimal substitution models for each dataset were determined using Akaike Information Criterion (AIC) implemented in MrModeltest v2.3 [57]: GTR+G+I for ITS, HKY+I+G for nrLSU, GTR+G for rpb2, SYM+I+G for tef-1α, and HKY+G+I for 5.8S. The GTR+G+I model of evolution was selected for the multi-gene dataset. The ML analyses were implemented using default settings with gaps treated as missing data and branch support was assessed through 1000 bootstrap partitions (BP) with the rapid bootstrap option.
Bayesian Inference (BI) analysis was conducted for the combined dataset using MrBayes 3.1.2 [58]. Bayesian posterior probabilities were determined twice by running one cold and three heated chains for fifty million generations using selected models, saving trees every 1000th generation. Burn-ins were determined by checking the likelihood trace plots in Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/) and subsequently discarded. The partitioned mixed model, which allows for model parameters estimated separately for each gene, was used in the combined analysis. Chain convergence was determined using Tracer v1.5 to ensure most of the effective sample size (ESS) values were above 200.
Phylogenetic species determination
To circumscribe species within Cudonia and Spathularia that have multiple collections and sequences from multiple loci available, we applied the multilocus genealogical phylogenetic species recognition approach [59], [60]. A phylogenetic species is recognized when it satisfies either of two criteria: (1) a genealogical concordant group that is present in the majority of the single-locus genealogies or (2) a clade that is strongly supported by at least one single-locus genealogy and is not contradicted by any other locus. For putatively distinct taxa with only a single ITS nrDNA sequence, we calculated the genetic distance compared to the ITS nrDNA for all other recognized species to assess the likelihood that the organism with the unique sequence should be recognized as a distinct species [37], [61]. Intra- and inter-specific variation within species of Cudonia and Spathularia were separately calculated using ITS alignments. If the sequence variation between the putative new species and all other recognized species was higher than the variation among the known species, then the putative species was recognized as a separate taxon. To assist in species delimitation, barcode gaps between the infraspecific and interspecific divergence were also calculated using the Automatic Barcode Gap Discovery (ABGD) method [62].
Diversification time estimates
To infer the “time to most recent common ancestor” (tMRCA), we used the secondary calibration strategy implemented by Renner [63]. Initially, to infer the stem node age of Cudoniaceae, we built a 5.8S-nrLSU-rpb2-tef-1α dataset that included 33 taxa from across major clades of Pezizomycotina [34], [64]. We then used this stem node age to calibrate the same node in a multi-gene (ITS-nrLSU-rpb2-tef-1α) analysis that included 2 taxa for calibration and 25 species of Cudonia and Spathularia.
The initial, phylogenetically broad analysis included representatives from most major clades within Pezizomycotina as well as representatives from Saccharomycotina, Taphrinomycotina, and Basidiomycota. Rhizopus oryzae, a member of Zygomycota, was chosen as the outgroup taxon (Table S2). Three calibration points were used for the initial analysis: the stem of Ascomycota (575 ± 37.5 Mya), the stem of Pezizomycotina (460 ± 30 Mya), and the crown of Pezizomycotina (360 ± 20 Mya). These values are the mean ranges of dates (500–650 Mya, 400-520 Mya, and 320-400 Mya respectively), with a standard deviation that produces a central 95% range of dates corresponding to those reported by Lücking et al. [65] for these nodes.
Both the initial and secondary tMRCA analyses were performed using BEAST v. 1.5.3 [66]. We used a Yule tree prior that assumes a constant lineage birth rate for each branch in the tree and we also used a relaxed lognormal molecular clock prior so that substitution rates were allowed to vary across branches. Priors of the calibration points were constrained to be normally distributed with a standard deviation as mentioned above.
For each analysis, two independent MCMC analyses were performed with parameter values sampled every 1,000 cycles over 50,000,000 MCMC steps. Convergence and acceptable mixing of the sampled parameters was verified using the program Tracer v. 1.5 [66]. After discarding the burn-in, the mean ± standard error and the 95% highest posterior density (HPD) interval for the divergence dates of the ancestral nodes were calculated from combined BEAST log files.
Ancestral area reconstruction analysis
To reconstruct the possible ancestral areas for Cudonia and Spathularia, ML-based dispersal-extinction-cladogenesis module was performed using LAGRANGE [67]. We used our multi-gene ML phylogeny and the diversification time of Cudonia and Spathularia estimated by BEAST to reconstruct the possible historical distribution of these genera. For the analyses of range evolution, a species was considered endemic if it was restricted to Eurasia or North America. We determined the presence or absence of each species of Cudonia or Spathularia in each of these world regions: Asia (A), Europe (E), and North America (N).
Results
Phylogenetic analyses of Cudonia and Spathularia based on individual genes
We generated 372 new sequences from 111 collections of Cudonia and Spathularia, including 111 sequences of ITS, 94 of nrLSU, 74 of rpb2, and 93 of tef-1α.
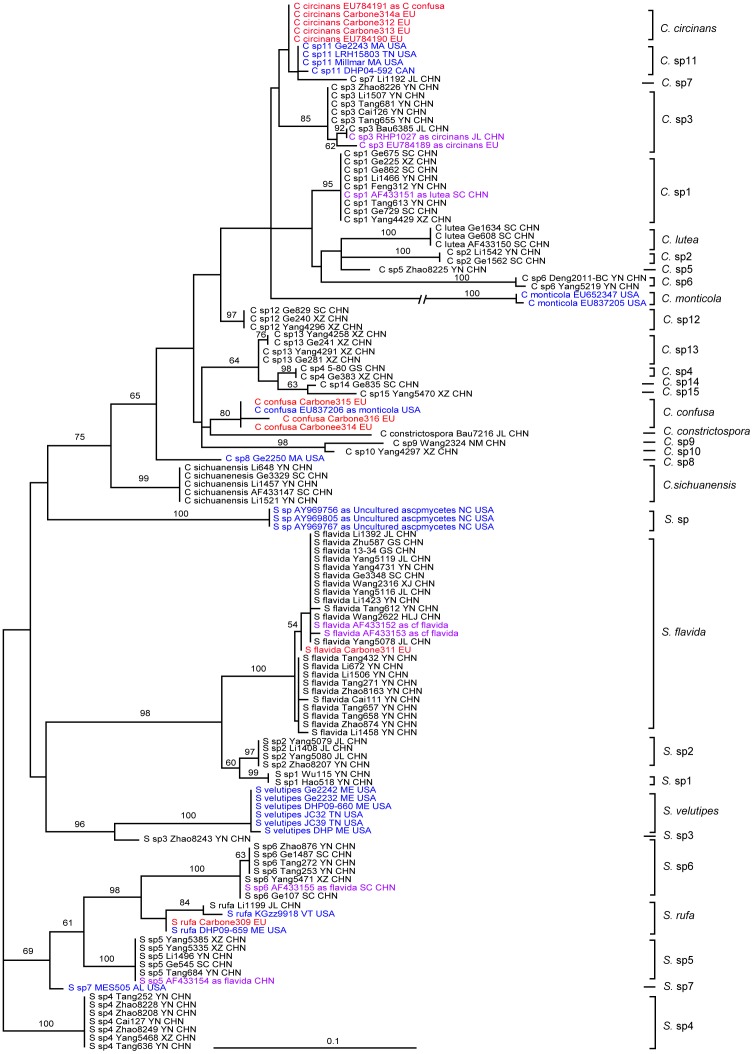
The aligned ITS dataset consisted of 127 sequences and was 587 nucleotides in length. We excluded 75 ambiguously aligned sites that consisted of introns found only in the divergent GenBank sequences of Cudonia monticola and three sequences labeled as “Uncultured Ascomycete”. The final analysis of the ITS region consisted of 512 clearly aligned sites. Maximum likelihood analysis resulted in one ML tree with final ML optimization Likelihood: -3018.425987. Using a phylogenetic species criterion in combination with the Automatic Barcode Gap Discovery (ABGD) method, a total of 32 species-level clades were circumscribed in the ITS gene tree, with 21 species-level clades in Cudonia and 11 in Spathularia.
Maximum likelihood analysis of the nrLSU data yielded an optimal ML tree with a log likelihood score -4099.381378 (Figure S1). The nrLSU phylogeny recovered a strongly supported Cudoniaceae clade derived from within the Rhytismataceae. Within Cudoniaceae clade, several subclades within Cudonia and Spathularia were recovered but bootstrap support for the nrLSU dataset was generally weak compared with the ITS dataset.
The optimal ML tree based on the rpb2 dataset had a log likelihood score of -3549.890566 (Figure S2). The rpb2 analysis recovered a monophyletic Cudoniaceae clade and a monophyletic Cudonia with strong bootstrap support. Spathularia was paraphyletic and divided into two major clades. One Spathularia clade, represented by S. velutipes Cooke & Farl. and allies, is resolved as sister to Cudonia. The other Spathularia clade, represented by S. flavida and two undescribed species, is sister to the rest of the group.
The tef-1α ML analysis produced a tree with the optimization likelihood value -2044.088513 (Figure S3). The tef-1α analysis recovered a strongly supported, monophyletic Cudoniaceae and genus Spathularia, but Cudonia was paraphyletic with Spathularia nested inside of Cudonia.
Phylogenetic species delimitation
In total, 32 putative species of Cudonia and Spathularia were recognized based on ITS data using the ABGD method (Figure 2), with 21 species-level clades in Cudonia and 11 in Spathularia. Among the 32 putative species, 20 species (8 in Spathularia and 12 in Cudonia) were further confirmed by the multilocus genealogical phylogenetic species recognition approach since there are multiple collections and sequences from multiple loci available for these species; two additional species (Cudonia monticola and Spathularia sp.) were recognized as distinct species because they form monophyletic clades with strong statistical support in the ITS analysis (both got 100 bootstrap support).
Figure 2. Maximum likelihood phylogeny of Cudonia and Spathularia species based on 127 ITS ribosomal DNA sequences.
ML bootstrap support values are shown above the internal branches. Sequences from Asia are in black text, sequences from North America are in blue text, and sequences from Europe are in red text. Mislabeled sequences from GenBank and sequences from misidentified herbarium collections are shown in purple text.
Ten putative species represented by only a single specimen were identified based on the uniqueness of their ITS sequences. The ITS variation within species ranged from 0 to 0.005 in the known Cudonia and Spathularia species (Table S4). Cudonia monticola displayed the highest ITS rDNA hetereogeneity (0.005), and was chosen as the cutoff value for the species identification using ITS sequences. Taking this value as a threshold, Cudonia ‘sp5’, Cudonia ‘sp7’, Cudonia ‘sp8’, Cudonia ‘sp9’, Cudonia ‘sp10’, Cudonia ‘sp14’, Cudonia ‘sp15’, Spathularia ‘sp3’ and Spathularia ‘sp7’ were recognized as putative species based on the ABGD analyses (Table S4).
Phylogenetic analysis of Cudonia and Spathularia based on a multi-gene dataset
Using the reciprocal 70% bootstrap support approach, there is very little evidence for incongruence among the individual genes. Accordingly, all four genes were concatenated for ML and BI analyses to assess the phylogeny of Cudonia and Spathularia.
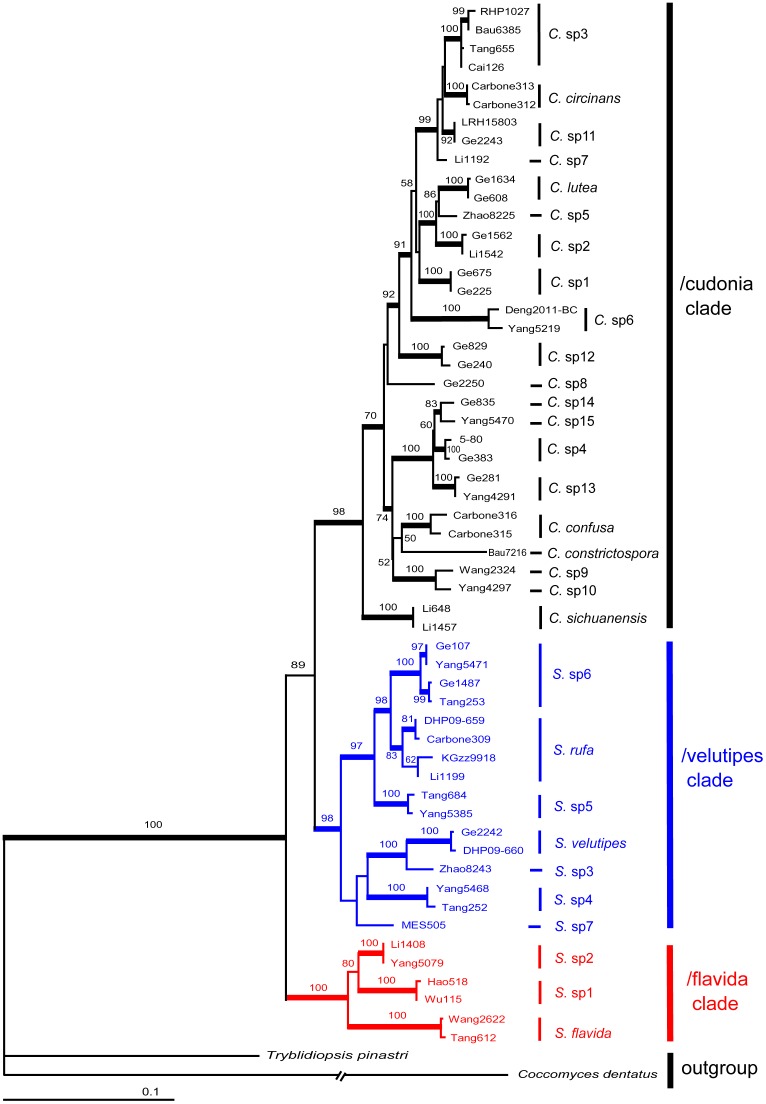
Cudonia monticola and the novel “uncultured ascomycete” from North Carolina forest soil were not included in the multilocus phylogenetic analyses because only ITS sequences were available for these taxa and these sequences were challenging to align with other members of Cudonia and Spathularia. Because the sampling is unbalanced for certain taxa in the 96 specimen phylogeny (for instance, Spathularia flavida has 15 collections in Figure S5), and to display the phylogeny effectively, we randomly chose two collections with the most complete sequences for each species and built a 58 specimen phylogeny for Cudonia and Spathularia (Figure 3). This analysis utilized 2,583 aligned nucleotides from 58 specimens, including representatives of all major clades. After excluding ambiguously aligned positions (mainly gaps introduced by the outgroup taxa), the final alignment was 2,458 nucleotides in length. The likelihood value of the final ML tree was lnL = -10065.428556 (Figure 3), and the likelihood value of the consensus Bayesian tree were lnL = -9973.42 and -9979.74 for the cold chain runs.
Figure 3. Four-locus (ITS, nrLSU, rpb2 and tef-1α) maximum likelihood (ML) phylogeny depicting phylogenetic relationships among 58 sequences of Cudonia and Spathularia.
Bootstrap support values from ML (1000 bootstrap replicates) analyses are shown above the internal branches. Bayesian posterior probabilities (BPP) greater than 0.95 are indicated by thickened branches. Branch lengths are proportional to the average number of substitutions per site except for dashed lines, which indicate longer branches that are not shown to scale.
The topology of the tree identified by ML analysis (see Figure 3) was almost identical to that found by the Bayesian analyses. Regardless of the analytical method, a highly resolved phylogeny of Cudonia and Spathularia was recovered from the multi-gene dataset. The family Cudoniaceae and the genus Cudonia were both strongly supported as monophyletic groups (Figure 3) whereas Spathularia was divided into two major clades: the /flavida clade and /velutipes clade. These three major clades (e.g. /cudonia, /flavida, and /velutipes) were recovered with strong support by both methods.
The /flavida clade forms a sister clade to a clade jointly formed by /velutipes clade and /cudonia clade. The /velutipes and /cudonia clades are supported as sister to one another, but this relationship received moderate to weak support (ML = 88, BI = 0.90). Overall, the multi-gene analysis identified 30 species level clades and different methods generally recovered similar relationships between the groups. The only difference is that S. sp7 is sister to a clade formed by S. sp5, S. sp6 and S. rufa in the Bayesian analyses, instead of sister to a clade formed by S. Sp3, S. Sp4, and S. velutipes in the ML analysis. Within Cudonia we identified 20 putative species, with C. sichuanensis resolved as the earliest diverging clade that is sister to the remaining taxa with strong statistical support (Figure 3). Among these 20 putative species, scientific names can only be confidently applied to five species whereas the other 15 clades appear to represent novel taxa due to their unique morphology and/or consistent and identifiable habitat preferences. Similarly, of the 10 Spathularia species-level clades that were recovered from the phylogeny, names can only confidently be applied to three clades. The /flavida clade is comprised of three taxa; the widely distributed Spathularia flavida Pers. and two undescribed species, Spathularia sp1 and sp2. Spathularia sp1 from Southwestern China and Spathularia sp2 from Northeastern China are sister taxa that form a joint clade that is sister to S. flavida. The /velutipes clade includes Spathularia velutipes Cooke & Farl., Spathularia rufa Schmidel and five unidentified species from southwestern China and/or North America.
Divergence time estimation and ancestral area reconstruction for Cudonia and Spathularia
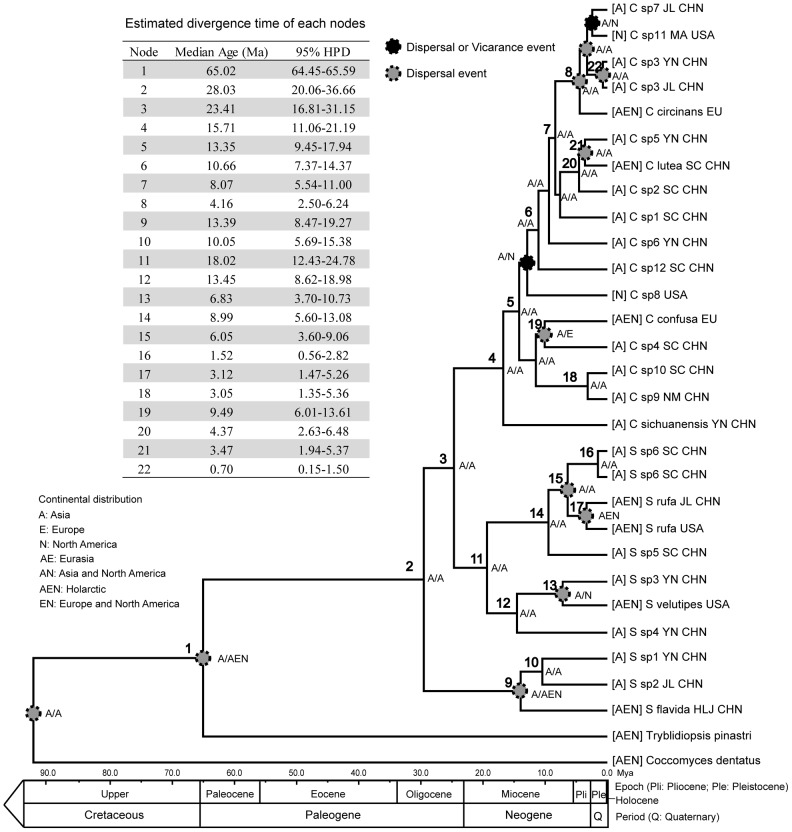
The estimated mean divergence time of Cudoniaceae from Tyblidiopsis pinastri (Rhytismatales) was during the beginning of the Paleogene in the Early Tertiary at 65.03 ± 0.29 Mya (95% highest posterior density [HPD] interval: 41.97-90.21). Our analysis suggests that diversification within Cudonia and Spathularia began during the Middle Oligocene 28.65 ± 0.14 Mya (95% HPD: 15.25-42.87) in late Paleogene (Figure S4). Divergence time of the Cudoniaceae members estimated using the above values (Cudoniaceae stem and crown) as calibration points are summarized in Figure 4.
Figure 4. Ancestral area reconstruction and chronogram of clade of Cudonia and Spathularia based on BEAST analyses using Cudonia stem (65.03 ± 0.29 MYA, node 1) as calibration.
Estimated mean divergence times and 95% highest posterior density of resolved nodes in phylogenetic analyses are summarized in the upper left panel. Geological time scale according to Gradstein et al. (2004) is shown along the bottom for scale.
From the chronogram in Figure 4, /flavida clade is estimated to have diverged from /velutipes clade and /cudonia clade at 28.03 Mya (95% HPD: 20.06-36.66) in the Oligocene. The divergence between /velutipes clade and /cudonia clade is estimated to have occurred between late Oligocene and early Miocene (23.41 mya, with 95% HPD 16.81-31.15). The /velutipes clade is estimated to have diverged at 18.02 Mya (12.43-24.78) in the early Miocene, and the /cudonia clade is estimated to have diverged at 15.71 mya (95% HPD: 11.06-21.19) in the Middle Miocene. Subsequent divergence times within the /velutipes clade were estimated for the /velutipes subclade at 13.45 Mya (95% HPD: 8.62-18.98) and for the /rufa subclade at 8.99 Mya (95% HPD: 5.60-13.08). Speciation events within the /rufa subclade were estimated to have occurred during the Pliocene and Pleistocene. Spathularia sp3 diverged from /velutipes in the late Miocene.
Speciation events of most /cudonia clades were estimated to be diverged in Miocene and Pliocene, with a few diverged within Pleistocene. The morphologically unique species Cudonia sp9 and Cudonia sp10 were estimated to have diverged from C. confusa and Cudonia sp4 at approximately 9.49 Mya (95% HPD: 6.01-13.61), and they split from each other at roughly 3.05 Mya (95% HPD: 1.35-5.36).
Based on the dispersal-extinction-cladogenesis analysis generated with the LAGRANGE software package, our ML-based ancestral area reconstruction analysis determined that the most recent common ancestor of Cudoniaceae most likely lived in Asia (Figure 4), followed by dispersal of the /flavida clade to Europe and North America. Similarly, we also inferred that the most recent common ancestor of both the /velutipes clade and /cudonia clade is likely to have arisen in Asia.
Discussion
Molecular phylogeny and species diversity of Cudonia and Spathularia
A recent study estimated that there are 5.1 million species of fungi on Earth but only about 100,000 fungi are currently described in the literature [3]. Two important reasons for this disconnect between estimated and described fungal species are: 1) there has been inadequate sampling of various fungal groups in many world regions, and 2) many reproductively isolated, cryptic fungal species are “hidden” within morphological species complexes and can only be elucidated by in-depth molecular studies.
Our results suggest that Cudonia and Spathularia are two such understudied genera that have been poorly sampled and contain cryptic species diversity within complexes of morphologically similar taxa. Cudonia and Spathularia have been under-collected, overlooked, and rarely studied using molecular tools. In this study, we use sequences of ITS, nrLSU, rpb2 and tef-1α to elucidate diversity within Cudonia and Spathularia and also to clarify broader biogeographical and evolutionary patterns within the group. Our results support the hypothesis of a monophyletic Cudoniaceae, as suggested by previous molecular studies that included only a few Cudonia and Spathularia species [27]-[32].
Our molecular analysis of Cudonia and Spathularia has several implications for the taxonomy and systematics of this group. First, our individual (Figure S1) and multi-gene analyses (Figure S4) agree with the phylogenetic results of Lantz et al. [27] that the Cudoniaceae is nested within the Rhytismataceae. Since this situation renders Rhytismataceae a paraphyletic group, we suggest that Cudoniaceae be abandoned in favor of a broadly circumscribed Rhytismataceae.
A second major issue is that Cudonia is a monophyletic group whereas Spathularia is not. At first glance it might seem appealing to split the genus Spathularia in two since there are already two generic names available: the /flavida clade corresponds to Spathularia, and the /velutipes clade to Spathulariopsis Maas Geest. Geesteranus erected Spathulariopsis to accommodate Spathularia velutipes based on hemiangiocarpous development (the hymenium becomes exposed before maturity) and the hyphal structure of the medulla layer in the stipe [68]. However, similar structures can also be found in Cudonia sichuanensis [9], C. lutea [14], Cudonia ‘sp12’ (Z-W Ge and ZL Yang, personal observation), and early developmental stages of other Cudonia and Spathularia species [13], [15], [16]. Imai also erected a separate genus, Pachycudonia S. Imai, for Cudonia constrictospora S. Ito et S. Imai [69]. He suggested that the long-tailed asci, elongated spores with constricted middle sections, and paraphyses with coiled ends were diagnostic for this genus [69]. However, our phylogenetic placement of C. constrictospora and C. monticola in the /cudonia clade suggests that Pachycudonia can be considered a synonym of Cudonia.
Instead of recognizing three genera we suggest a more straightforward approach of recognizing the single genus Spathularia. This genus name has priority because it was described by Persoon in 1794 whereas Cudonia was not described until 1849 [70], [71]. The main rationale for recognizing the single genus Spathularia is that there are very few diagnostic morphological characters to readily separate the major clades in this group of fungi. Traditionally, the main character that has been used to distinguish Spathularia from Cudonia was that Spathularia species have flattened, spatula-shaped ascoma (e.g. S. flavida and S. velutipes in Figure 1) whereas Cudonia species have rounded ascoma (e.g. C. circinans, C. lutea, C. confusa in Figure 1). However, we have documented two apparently undescribed species from China that are morphologically unique and are not easily pigeonholed into Cudonia or Spathularia. Cudonia ‘sp10’ has ascoma that are intermediate between the spatulate and rounded forms whereas Cudonia ‘sp9’ forms smooth, clavate ascoma that macroscopically resemble the fruiting bodies of the basidiomycete genus Clavariadelphus (Figure 1). Since major nomenclatural amendments are beyond the scope of this paper, we retained the genus names Cudonia and Spathularia to describe the overall morphology of our undescribed species in this publication.
In addition to clarifying the major groups within Spathularia and Cudonia, and assessing their relationships to one another, our molecular analyses also identified 32 distinct species-level clades based on our herbarium collections and GenBank sequences (30 species were identified based on both a multi-locus phylogenetic species recognition approach and the Automatic Barcode Gap Discovery (ABGD) method whereas the remaining two species were identified based only on ITS sequences using the ABGD approach[62]). We detected high species diversity in East Asia, including 16 species-level clades in Cudonia, and 8 in Spathularia. The large number of species in East Asia suggests that this region is a center of diversity for Spathularia and Cudonia. However, since our field sampling was most intense in East Asia, it is also possible that other world regions host unrecognized cryptic diversity of Spathularia and Cudonia. Our analysis includes 6 out of 13 named species of Cudonia and 3 out of 6 named Spathularia species. There are currently five described Cudonia species (C. circinans, C. lutea, C. grisea, C. monticola, and C. ochroleuca) and three described Spathularia species (S. clavata S. rugosa and S. velutipes) recognized in North America [14], [16], [21], [72]. We identified two additional North American species based on herbarium specimens (Cudonia ‘sp8’ and Spathularia ‘sp7’) as well as a clade that is only known from soil DNA sequences (Spathularia ‘sp’). In Europe, two Cudonia species (C. circinans, C. confusa), and two Spathularia species (S. flavida and S. rufa) are commonly recognized [19], [20]. In addition, several infraspecific varieties of S. flavida have been described in Europe [73], [74], but were subsequently ignored as synonyms by most mycologists. Based on the unexpectedly high diversity and the large number of “hidden” taxa detected in this study, we expect that these varieties probably represent distinct species and will need to be reevaluated with molecular techniques.
Our molecular phylogeny points to the need for a major taxonomic revision within Cudonia and Spathularia. In addition to the major taxonomic changes highlighted above, it will be important to identify and describe the putative new species that we detected in East Asia and also to reevaluate the previously described species in Europe and North America based on molecular data. Results from the ITS phylogeny suggest that specimens in Cudonia and Spathularia are routinely mislabeled. For example, sequence EU837206 was deposited as Cudonia monticola but is resolved in the C. confusa clade, sequence EU784191 was deposited as C. confusa but falls in the C. circinans clade, and sequence EU784189 was deposited as C. circinans but is in fact Cudonia ‘sp3’. In Spathularia, two sequences deposited as S. flavida (AF433154, AF433155) are resolved in the Spathularia ‘sp6’ clade and Spathularia ‘sp5’ clade, respectively. Three soil clone sequences from North Carolina (USA) pine forests (as “Uncultured Ascomycete” – AY969756, AY969805, AY969767) apparently represent a novel Spathularia species. Further taxonomic work will also help to clarify the species delimitations of described taxa and may help to alleviate the large number of misidentified sequences deposited in GenBank (Figure 2).
Based on our extensive field collections (Table S1), we have observed that some species have consistent and identifiable habitat preferences that have been mostly ignored in previous studies. For example, Spathularia velutipes is usually found on decayed logs, Spathularia sp4 usually fruits directly on soil in angiosperm-dominated forests, and Cudonia sichuanensis, Cudonia ‘sp10’ (Figure 1E), and Cudonia ‘sp12’ (Figure 1G) all fruit among mosses in dense, mature coniferous forests. These observations suggest that ecological factors may be useful in clarifying the taxonomy and ecology of species in this group.
Morphological evolution and adaptations to a temperate environment
Most species within Rhytismatales produce their spores within a dark stroma that is immersed in host tissues and is sometimes covered by a layer of fungal hyphae [17]. The vast majority of species in the order also produce fruiting bodies that are small, thin, and inconspicuous whereas those of Spathularia and Cudonia are relatively noticeable and large (stipes up to 7 cm long and pilei up to 2 cm broad). Like most other Rhytismatales, Spathularia and Cudonia species also have a veil that shields the hymenium during development (Figure 1G. Cudonia ‘sp12’). This type of development (called “hemiangiocarpous development”) may help to protect the hymenium from adverse environmental conditions such as freezing or desiccation. We also detected the production of conidia (asexual spores) directly on or within the fruiting bodies of Cudonia ‘sp10’ and ‘sp12’, and this phenomenon was previously reported for C. lutea, C. circinans, S. flavida and S. velutipes [15], [16]. We think this strategy of asexual spore production may be an additional adaptation to environmental stress, but more studies are needed to fully understand the role of conidia in the biology of Spathularia and Cudonia.
Members of the order Rhytismatales are considered plant endophytes or pathogens [10]-[12], [17], whereas Spathularia and Cudonia have been considered by some authors to be white-rot saprobes [75]. In an evolutionary context, the transition from an endophytic or pathogenic to a saprobic lifestyle may have been an important evolutionary event during the history of Spathularia and Cudonia. It is possible that the large ascoma are an adaptation to saprotrophism; it has been hypothesized that large fruiting bodies can generate more spores and their fleshy stalks can more easily rise above the surface of the leaf litter. The idea that fleshy sporocarps are more likely to occur in saprobic taxa has also been suggested as a wider pattern within Ascomycota [17]. Given that the ecology of these fungi is so poorly known, it also remains possible that Spathularia and Cudonia species have cryptic plant endophytic, mycoparasitic, or plant pathogenic stages. We think this is unlikely, however, because most environmental sequences of Spathularia and Cudonia are from soil. For example, we detected three sequences from organic soil in a North Carolina pine forest (AY969756, AY969805 and AY969767), one sequence from soil under Pinus sylvestris in Sweden (FJ475669), and one from soil in a mixed forest in New Hampshire (HQ022046). In contrast, we detected only one environmental DNA sequence from roots; sequence HQ260170 was recovered from Ericaceae roots in Alaska. These environmental sequence data suggest that Spathularia and Cudonia are probably most common in soil but also reinforce that taxa in this group tend to be rare and their ecology is not well understood.
Biogeographic history of Cudonia and Spathularia and their diversification in East Asia
The divergence patterns of fungi are still poorly understood because there are only a handful of fossils that can be used to definitively date the emergence of particular fungal clades [76], [77]. However, molecular dating and phylogenetics tools have proved useful for estimating the divergence times and centers of origin for some fungal groups. For example, recent studies have examined patterns and timing of evolution in ascomycetes such as Hypocreales [78], Morchella [41], [79], Tuber [80], [81], Golovinomyces [82], and some lichen-forming clades [83]-[85].
Our relaxed clock analyses, which allow the molecular rate to vary among clades, suggest the diversification of Cudoniaceae started around 28.65 Mya (95% HPD: 42.87-15.25) (Node 2 in Figure 4). Compared to some other Pezizomycetes that are thought to have diverged during the Cretaceous (e.g. Morchella [41]; Tuber [80], our analysis suggests that Cudoniaceae is a relatively young clade whose divergence times are more similar to lichen-forming fungi in Parmeliaceae (Lecanoromycetes) [83].
Our ancestral area reconstruction analyses suggest that Cudoniaceae may have evolved in Asia and then later dispersed to Europe and North America (Figure 4). We detected 24 species of Spathularia (8 species) and Cudonia (16 species) in Asia whereas only 9 species were found in North America and only 5 species were found in Europe. Some of this high species diversity in Asia versus other continents may reflect our intensive field sampling in Asia and the fact that some uncommon species originally described from North America (e.g. Cudonia ochroleuca [Cooke & Harkn.] E.J. Durand, Cudonia grisea Mains) or Europe (Spathularia alpestris [Rehm] Rahm, Spathularia crispata Fuckel) were not available for molecular analysis. However, Asian Spathularia and Cudonia species exhibit higher morphological diversity than species from other regions (including ascocarp shape and color, spore morphology, stipe anatomy, and the structure of the hymenial veil). These fungi are also found in a wide variety of habitats in Asia, including alpine, subalpine, and subtropical habitats. Furthermore, Asia has been implicated as a center of diversity for several major groups of fungi such as Amanita, Boletus, Flammulina, and Morchella [37], [79], [86], [87], [106].
The dates for major radiations in Spathularia and Cudonia are estimated to coincide with one of the two pulses of rapid uplifting of the Tibetan plateau which occurred during 25-30 Mya [88]. This time period was characterized by a global long-term glaciation and massive geological uplifting in the Himalaya region in Asia that contributed to both regional and global cooling trends [89]-[94]. During this time, the mountains of northwestern China became arid and subtropical evergreen forests were restricted to southern and coastal Asia [95]. These large-scale geological and climatic events in Oligocene (e.g. the period of global cooling from 34–23 Mya and the uplift of the Tibetan plateau) are strongly correlated with diversification in Spathularia and Cudonia. This unique geological history of the Tibetan Plateau is thought to have generated heterogeneity in the soils, climate, and elevation across the region [87]. These changes are implicated as potential drivers of evolutionary radiations in many organisms, including key clades of forest trees [96], [97]. The rapid drop in temperature during this period may have stimulated the radiation of other cold-adapted groups in fungi such as Amanita and Boletus [37], [38], in plants such as Quercus [98], [99], and in insects such as Bombus and Myrmica [100], [101]. We hypothesize that the global cooling and expansion of temperate forests across south Asia may have triggered evolutionary radiations in this group of fungi, either directly through the separation of fungal populations into refugia or indirectly through changes in the composition of forest communities.
Long-distance spore dispersal and ancient vicariance are the main mechanisms used to explain the intercontinental distributions for fungi, although more recent movements have sometimes been due to human-mediated introductions [102]-[104]. In combination with the divergence time estimates, our LAGRANGE analyses suggest that dispersal is the dominant mechanism behind the continental distribution of Spathularia and Cudonia species. Due to the large gelatinous spore sheaths that cover ascospores at maturity (Figure 1O), we suspect that Spathularia and Cudonia ascospores are not easily wind dispersed. Rain-splash dispersal and/or insect vectors may be involved in ascospore dispersal and we hypothesize that the asexual conidia we observed might also be important for occasional, long-distance dispersal. However, more detailed ecological studies are needed. In addition to the inferred instances of dispersal, two vicariance events may also have occurred in the evolutionary history of the /cudonia clade (inferred at around 12 Mya and 3 Mya respectively – Figure 4). However, these events can also be explained by dispersal because the Bering Land Bridge has been a viable route for terrestrial plants (and fungi) since the Paleocene [105]. One species within the /cudonia clade, Cudonia monticola, demonstrated an extremely long branch within the ITS phylogeny (Figure 2). This may be a signature of rapid evolution and/or a selection bottleneck during dispersal to western North America but more work is needed to resolve the placement and history of this species.
This study clearly shows that the species-level diversity of Spathularia and Cudonia is higher than was previously thought and we have increased the total number of putative taxa to 32 phylogenetic species based on a combination of multi-gene phylogenetic analyses and the Automatic Barcode Gap Discovery (ABGD) method. Although the diversity of Spathularia and Cudonia was particularly high in East Asia and our analyses suggest that this region is likely an important center of diversity for the group, the finding of many apparently new cryptic species based on molecular data suggests that other global regions may also hold unsampled or cryptic diversity. Accordingly, the biogeographic hypotheses outlined here must remain preliminary until more complete sampling is completed in key parts of Europe and North America. We suggest that additional sampling of Spathularia and Cudonia will further elucidate the systematics, ecology, and evolution of this group of fungi.
Supporting Information
Maximum likelihood phylogeny of Cudonia and Spathularia species based on 113 nrLSU sequences with bootstrap values for 1000 replicates shown above the internal branches. Species of Cudonia and Spathularia are shaded in yellow, and Rhytismataceae species are shaded in green.
(TIF)
Maximum likelihood tree of the Cudonia and Spathularia taxa based on 78 rpb2 sequences with bootstrap values for 1000 replicates shown above the internal branches.
(TIF)
Maximum likelihood tree of the Cudonia and Spathularia taxa based on 96 tef-1α sequences with bootstrap values for 1000 replicates shown above the internal branches.
(TIF)
Chronogram of clades of Cudonia and Spathularia based on BEAST analyses. Three calibration points were used for the initial analysis based on results from Lücking et al. (2009): the stem of Ascomycota (575 ± 37.5 Mya, node A), the stem of Pezizomycotina (460 ± 30 Mya, node B), and the crown of Pezizomycotina (360 ± 20 Mya, node C). Estimated mean divergence times and 95% highest posterior densities of major nodes are summarized the upper left panel.
(TIF)
Maximum likelihood (ML) cladogram for 96 specimens depicting the phylogenetic relationships among Cudonia and Spathularia based on ITS-LSU-rpb2-tef-1α. Bootstrap support values above 50 (from 1000 bootstrap replicates) are shown above the branches.
(TIF)
GenBank accession numbers and voucher information for specimens used in molecular studies.
(DOC)
GenBank accession numbers of GenBank sequences used in this study.
(DOCX)
GenBank accession numbers of sequences used in the divergence time estimation.
(DOCX)
Evolutionary divergence of ITS sequence pairs within and between putative species of Cudonia and Spathularia.
(DOC)
The 127 specimens ITS alignment with the exact nucleotides to be excluded.
(NEX)
Acknowledgments
The authors are very grateful to Mr. Qi Zhao, Drs. Yan-Chun Li, Li-Ping Tang, Qing Cai, Xue-Tai Zhu, Yan-Jia Hao, Xiang-Hua Wang, Gang Wu, Bang Feng, Xiao-Juan Deng (Kunming Institute of Botany, Chinese Academy of Sciences) for providing specimens for this study. We thank Dr. Ronald H. Petersen (The University of Tennessee, USA) and Dr. Kentaro Hosaka (National Museum of Nature and Science, Japan) for arranging and loaning herbarium specimens, and the anonymous reviewers for their valuable comments. We also thank the librarians in Farlow Library for assistances with providing literature.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Science Foundation of China (No. 31270074), the Joint Funds of the National Natural Science Foundation of China and Yunnan Provincial Government (No. U1302263), the Knowledge Innovation Program of the Chinese Academy of Sciences (No. KSCX2-EW-J-24), and the Farlow Fellowships from the Friends of Farlow to Zai-Wei Ge. Field work was jointly supported by the CAS/SAFEA International Partnership Program for Creative Research Teams and the U. S. National Science Foundation (No. DEB-0321846 to D. E. Boufford), and the University of Florida's Institute for Food and Agricultural Sciences (IFAS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Swift MJ, Heal OW, Anderson JM (1979) Decomposition in terrestrial ecosystems: Univ of California Press.
- 2. Jones MDM, Forn I, Gadelha C, Egan MJ, Bass D, et al. (2011) Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474: 200–203. [DOI] [PubMed] [Google Scholar]
- 3. Blackwell M (2011) The Fungi: 1, 2, 3… 5.1 million species? American Journal of Botany 98: 426–438. [DOI] [PubMed] [Google Scholar]
- 4. Mueller GM, Schmit JP (2007) Fungal biodiversity: what do we know? What can we predict? Biodiversity and Conservation 16: 1–5. [Google Scholar]
- 5. Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycological Research 105: 1422–1432. [Google Scholar]
- 6.Hawksworth DL, Mueller GM (2005) Fungal communities: their diversity and distribution. In: Dighton J, Oudemans P, White J (eds) The fungal community: its organization and role in the ecosystem, 3rd edn Marcel Dekker, New York, pp 27–37.
- 7.Mueller GM, Foster MS, Bills GF (2004) Biodiversity of fungi: inventory and monitoring methods: Academic Press.
- 8.Kirk P, Cannon P, David J, Staplers JA (2001) Bisby's Dictionary of the Fungi. 672 p. [Google Scholar]
- 9. Wang Z, Binder M, Hibbett DS (2002) A new species of Cudonia based on morphological and molecular data. Mycologia 94: 641–650. [DOI] [PubMed] [Google Scholar]
- 10. Hou C-L, Piepenbring M (2005) Known and two new species of Rhytisma (Rhytismatales, Ascomycota) from China. Mycopathologia 159: 299–306. [DOI] [PubMed] [Google Scholar]
- 11. Lin Y, Liu H, Tang Y, Hu B (1994) Two new species of the genus Coccomyces from China. Acta Mycologica Sinica 13: 8–12. [Google Scholar]
- 12. Hanso M, Drenkhan R (2012) Lophodermium needle cast, insect defoliation and growth responses of young Scots pines in Estonia. Forest Pathology 42: 124–135. [Google Scholar]
- 13.Duff GH (1922) Development of the Geoglossaceae. Botanical gazette: 264–291.
- 14.Durand EJ (1908) The Geoglossaceae of North America. Annales Mycologici 6.
- 15. Mains EB (1955) North American Hyaline-Spored Species of the Geoglosseae. Mycologia 47: 846–877. [Google Scholar]
- 16. Mains EB (1956) North American Species of the Geoglossaceae. Tribe Cudonieae. Mycologia 48: 694–710. [Google Scholar]
- 17.Wang Z, Johnston PR, Yang ZL, Townsend JP (2009) Evolution of Reproductive Morphology in Leaf Endophytes. PLoS One 4. [DOI] [PMC free article] [PubMed]
- 18.Lloyd CG (1916) The Geoglossaceae: (viz., The Genus Geoglossum and Related Genera).
- 19. Nannfeldt JA (1942) The Geoglossaceae of Sweden (with regard also to the surrounding countries). Arkiv för Botanik 30A: 1–67. [Google Scholar]
- 20. Rahm E (1966) Geoglossaceae im Hochtal von Arosa. Schweiz Z Pilzk 44: l65–179. [Google Scholar]
- 21. Mains EB (1940) New and unusual species of the Geoglossaceae. American Journal of Botany 27: 322–326. [Google Scholar]
- 22. Imai S (1941) Geoglossaceae Japoniae. Journal of the Faculty of Agriculture, Hokkaido Imperial University 45: 155–264. [Google Scholar]
- 23. Sharma M, Rawla G (1983) The genus Cudonia Fr. in India. India J Crammer's Bibliotheca Mycologica 91: 203–211. [Google Scholar]
- 24.Korf RP (1973) Discomycetes and Tuberales. In: Ainsworth G, Sparrow F, Sussman A, editors. The fungi: an advanced treatise. London & New York: Academic Press. pp. 249–319.
- 25. Schoch CL, Wang Z, Townsend JP, Spatafora JW (2009) Geoglossomycetes cl. nov., Geoglossales ord. nov and taxa above class rank in the Ascomycota Tree of Life. Persoonia 22: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirk PM, Cannon PF, Minter D, Stalpers JA (2008) Dictionary of the Fungi: CABI.
- 27. Lantz H, Johnston PR, Park D, Minter DW (2011) Molecular phylogeny reveals a core clade of Rhytismatales. Mycologia 103: 57–74. [DOI] [PubMed] [Google Scholar]
- 28. Landvik S, Eriksson OE, Gargas A, Gustafsson P (1993) Relationships of the genus Neolecta (Neolectales ordo nov., Ascomycotina) inferred from 18S rDNA sequences. Systema Ascomycetum 11: 107–118. [Google Scholar]
- 29. Gargas A, DePriest PT, Grube M, Tehler A (1995) Multiple origins of lichen symbioses in fungi suggested by SSU rDNA phylogeny. Science 268: 1492–1495. [DOI] [PubMed] [Google Scholar]
- 30. Döring H, Triebel D (1998) Phylogenetic relationships of Bulgaria inferred by 18S rDNA sequence analysis. Cryptogamie Bryologie Lichenologie 19: 123–136. [Google Scholar]
- 31. Gernandt DS, Platt JL, Stone JK, Spatafora JW, Holst-Jensen A, et al. (2001) Phylogenetics of Helotiales and Rhytismatales based on partial small subunit nuclear ribosomal DNA sequences. Mycologia 93: 915–933. [Google Scholar]
- 32. Wang Z, Binder M, Schoch CL, Johnston PR, Spatafora JW, et al. (2006) Evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): A nuclear rDNA phylogeny. Molecular Phylogenetics and Evolution 41: 295–312. [DOI] [PubMed] [Google Scholar]
- 33. Platt JL, Spatafora JW (2000) Evolutionary Relationships of Nonsexual Lichenized Fungi: Molecular Phylogenetic Hypotheses for the Genera Siphula and Thamnolia from SSU and LSU rDNA. Mycologia 92: 475–487. [Google Scholar]
- 34. Spatafora JW, Sung G-H, Johnson D, Hesse C, O'Rourke B, et al. (2006) A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. [DOI] [PubMed] [Google Scholar]
- 35. Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G, et al. (2004) Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. American Journal of Botany 91: 1446–1480. [DOI] [PubMed] [Google Scholar]
- 36. Yang ZL (2011) Molecular techniques revolutionize knowledge of basidiomycete evolution. Fungal Diversity 50: 47–58. [Google Scholar]
- 37. Feng B, Xu J, Wu G, Zeng N-K, Li Y-C, et al. (2012) DNA Sequence Analyses Reveal Abundant Diversity, Endemism and Evidence for Asian Origin of the Porcini Mushrooms. PLoS One 7: e37567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geml J, Laursen G, O'NEILL K, Nusbaum HC, Taylor DL (2006) Beringian origins and cryptic speciation events in the fly agaric (Amanita muscaria). Molecular Ecology 15: 225–239. [DOI] [PubMed] [Google Scholar]
- 39. Hosaka K, Castellano MA, Spatafora JW (2008) Biogeography of Hysterangiales (Phallomycetidae, Basidiomycota). Mycological Research 112: 448–462. [DOI] [PubMed] [Google Scholar]
- 40. Matheny PB, Aime MC, Bougher NL, Buyck B, Desjardin DE, et al. (2009) Out of the Palaeotropics? Historical biogeography and diversification of the cosmopolitan ectomycorrhizal mushroom family Inocybaceae. Journal of Biogeography 36: 577–592. [Google Scholar]
- 41. O'Donnell K, Rooney AP, Mills GL, Kuo M, Weber NS, et al. (2011) Phylogeny and historical biogeography of true morels (Morchella) reveals an early Cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genetics and Biology 48: 252–265. [DOI] [PubMed] [Google Scholar]
- 42.Skrede I, Engh IB, Binder M, Carlsen T, Kauserud H, et al. (2011) Evolutionary history of Serpulaceae (Basidiomycota): molecular phylogeny, historical biogeography and evidence for a single transition of nutritional mode. BMC Evolutionary Biology 11. [DOI] [PMC free article] [PubMed]
- 43. Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- 44.White TJ, Bruns TD, Lee S, Taylor J (1990) Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genes. San Diego: Academic. 315–322.
- 45. Gardes M, Bruns T (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- 46. Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- 48. Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- 49. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 50. Ge ZW, Yang ZL, Vellinga EC (2010) The genus Macrolepiota (Agaricaceae, Basidiomycota) in China. Fungal Diversity 45: 81–98. [Google Scholar]
- 51. Dugan KA, Lawrence HS, Hares DR, Fisher CL, Budowle B (2002) An improved method for post-PCR purification for mtDNA sequence analysis. Journal of forensic sciences 47: 811–818. [PubMed] [Google Scholar]
- 52. Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS (2006) Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia 98: 1065–1075. [DOI] [PubMed] [Google Scholar]
- 53. Katoh K, Asimenos G, Toh H (2009) Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537: 39–64. [DOI] [PubMed] [Google Scholar]
- 54.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; 1999. pp. 95–98.
- 55. Mason-Gamer RJ, Kellogg EA (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Systematic Biology 45: 524–545. [Google Scholar]
- 56. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- 57.Nylander J (2004) MrModeltest v2. 3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University Available: http://wwwabcse/~nylander/mrmodeltest2/mrmodeltest2html.
- 58. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 59. Dettman JR, Jacobson DJ, Taylor JW (2003) A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- 60. Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- 61. Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, et al. (2012) Estimating divergence times in large molecular phylogenies. Proceedings of the National Academy of Sciences 109: 19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21: 1864–1877. [DOI] [PubMed] [Google Scholar]
- 63. Renner SS (2005) Relaxed molecular clocks for dating historical plant dispersal events. Trends in plant science 10: 550–558. [DOI] [PubMed] [Google Scholar]
- 64. Schoch CL, Sung G-H, López-Giráldez F, Townsend JP, Miadlikowska J, et al. (2009) The Ascomycota Tree of Life: A Phylum-wide Phylogeny Clarifies the Origin and Evolution of Fundamental Reproductive and Ecological Traits. Systematic Biology 58: 224–239. [DOI] [PubMed] [Google Scholar]
- 65. Lücking R, Huhndorf S, Pfister DH, Plata ER, Lumbsch HT (2009) Fungi evolved right on track. Mycologia 101: 810–822. [DOI] [PubMed] [Google Scholar]
- 66. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology 57: 4–14. [DOI] [PubMed] [Google Scholar]
- 68. Geesteranus RA (1972) Spathularia and Spathulariopsis . Proceedings of the Koninklijke Nederlandse Akademie van Wetenschappen Series C Biological and medical sciences 45: 243–255. [PubMed] [Google Scholar]
- 69.Imai S (1955) Contributiones ad studia monographica Geoglossacearum. II. Science Reports of the Yokohama National University.
- 70.Fries E (1849) Summa vegetabilum Scandinaviae. Discomycetes. 343–374.
- 71. Persoon CH (1794) Neuer Versuch einer systematischen Eintheilung der Schwämme. Roemer's Neues Magazin für die Botanik 1: 116. [Google Scholar]
- 72. Durand EJ (1921) New or noteworthy Geoglossaceae. Mycologia 13: 184–187. [Google Scholar]
- 73.Schumacher HCF (1803) Enumeratio Plantarum in Partibus Sællandiae Septentrionalis et Orientalis Crescentium 2: 1-489. Denmark, Copenhagen, Hafniae; Fridericum Brummer, typis E.M. Cohen.
- 74. Rehm H (1904) Beiträge zur Pilzflora von Südamerika XIV. Hedwigia 44: 1–13. [Google Scholar]
- 75.Hintikka V (1970) Studies on white-rot humus formed by higher fungi in forest soils. Metsatieteellisen tutkimuslaitoksen julkaisuja 69.
- 76. Berbee ML, Taylor JW (2010) Dating the molecular clock in fungi – how close are we? Fungal Biology Reviews 24: 1–16. [Google Scholar]
- 77. Taylor JW, Berbee ML (2006) Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia 98: 838–849. [DOI] [PubMed] [Google Scholar]
- 78. Sung G-H, Poinar GO, Spatafora JW (2008) The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal–arthropod symbioses. Molecular Phylogenetics and Evolution 49: 495–502. [DOI] [PubMed] [Google Scholar]
- 79. Du X-H, Zhao Q, O'Donnell K, Rooney AP, Yang ZL (2012) Multigene molecular phylogenetics reveals true morels (Morchella) are especially species-rich in China. Fungal Genetics and Biology 49: 455–469. [DOI] [PubMed] [Google Scholar]
- 80. Bonito G, Smith ME, Nowak M, Healy RA, Guevara G, et al. (2013) Historical Biogeography and Diversification of Truffles in the Tuberaceae and Their Newly Identified Southern Hemisphere Sister Lineage. PLoS One 8: e52765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jeandroz S, Murat C, Wang Y, Bonfante P, Le Tacon F (2008) Molecular phylogeny and historical biogeography of the genus Tuber, the 'true truffles'. Journal of Biogeography 35: 815–829. [Google Scholar]
- 82. Takamatsu S, Matsuda S, Niinomi S, Havrylenko M (2006) Molecular phylogeny supports a Northern Hemisphere origin of Golovinomyces (Ascomycota: Erysiphales). Mycological Research 110: 1093–1101. [DOI] [PubMed] [Google Scholar]
- 83. Amo de Paz G, Cubas P, Divakar PK, Lumbsch HT, Crespo A (2011) Origin and Diversification of Major Clades in Parmelioid Lichens (Parmeliaceae, Ascomycota) during the Paleogene Inferred by Bayesian Analysis. PLoS One 6: e28161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Divakar PK, Del-Prado R, Lumbsch HT, Wedin M, Esslinger TL, et al. (2012) Diversification of the newly recognized lichen-forming fungal lineage Montanelia (Parmeliaceae, Ascomycota) and its relation to key geological and climatic events. American Journal of Botany 99: 2014–2026. [DOI] [PubMed] [Google Scholar]
- 85. Leavitt SD, Esslinger TL, Divakar PK, Lumbsch HT (2012) Miocene divergence, phenotypically cryptic lineages, and contrasting distribution patterns in common lichen-forming fungi (Ascomycota: Parmeliaceae). Biological Journal of the Linnean Society 107: 920–937. [Google Scholar]
- 86. Ge Z-W, Yang Z, Zhang P, Matheny P, Hibbett D (2008) Flammulina species from China inferred by morphological and molecular data. Fungal Diversity 32: 59–68. [Google Scholar]
- 87.Yang ZL (2005) Diversity and biogeography of higher fungi in China. In: Xu J, editor. Evolutionary genetics of fungi Horizon Bioscience, Norfolk. pp. 35–62.
- 88. Wang E, Kirby E, Furlong KP, van Soest M, Xu G, et al. (2012) Two-phase growth of high topography in eastern Tibet during the Cenozoic. Nature Geosci 5: 640–645. [Google Scholar]
- 89. Chung SL, Lo CH, Lee TY, Zhang Y, Xie Y, et al. (1998) Diachronous uplift of the Tibetan plateau starting 40 Myr ago. Nature 394: 769–773. [Google Scholar]
- 90. Dupont-Nivet G, Krijgsman W, Langereis CG, Abels HA, Dai S, et al. (2007) Tibetan plateau aridification linked to global cooling at the Eocene–Oligocene transition. Nature 445: 635–638. [DOI] [PubMed] [Google Scholar]
- 91.Gradstein FM, Ogg JG, Smith AG (2004) A geologic time scale 2004. Cambridge: Cambridge University Press.
- 92. Kraatz BP, Geisler JH (2010) Eocene–Oligocene transition in Central Asia and its effects on mammalian evolution. Geology 38: 111–114. [Google Scholar]
- 93. Raymo M, Ruddiman WF (1992) Tectonic forcing of late Cenozoic climate. Nature 359: 117–122. [Google Scholar]
- 94. Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- 95. Sun X, Wang P (2005) How old is the Asian monsoon system?—Palaeobotanical records from China. Palaeogeography, Palaeoclimatology, Palaeoecology 222: 181–222. [Google Scholar]
- 96. Fang M, Fang R, He M, Hu L, Yang H, et al. (2005) Rhododendron . Flora of China 14: 260–455. [Google Scholar]
- 97. Ran JH, Wei XX, Wang XQ (2006) Molecular phylogeny and biogeography of Picea (Pinaceae): Implications for phylogeographical studies using cytoplasmic haplotypes. Molecular Phylogenetics and Evolution 41: 405–419. [DOI] [PubMed] [Google Scholar]
- 98. Axelrod DI (1983) Biogeography of oaks in the Arcto-Tertiary province. Annals of the Missouri Botanical Garden 70: 629–657. [Google Scholar]
- 99. Manos PS, Stanford AM (2001) The historical biogeography of Fagaceae: tracking the tertiary history of temperate and subtropical forests of the Northern Hemisphere. International Journal of Plant Sciences 162: S77–S93. [Google Scholar]
- 100. Hines HM (2008) Historical biogeography, divergence times, and diversification patterns of bumble bees (Hymenoptera: Apidae: Bombus). Systematic Biology 57: 58–75. [DOI] [PubMed] [Google Scholar]
- 101. Jansen G, Savolainen R, Vepsäläinen K (2010) Phylogeny, divergence-time estimation, biogeography and social parasite–host relationships of the Holarctic ant genus Myrmica (Hymenoptera: Formicidae). Molecular Phylogenetics and Evolution 56: 294–304. [DOI] [PubMed] [Google Scholar]
- 102. Hibbett DS, Pine EM, Langer E, Langer G, Donoghue MJ (1997) Evolution of gilled mushrooms and puffballs inferred from ribosomal DNA sequences. Proceedings of the National Academy of Sciences of the United States of America 94: 12002–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Moncalvo JM, Buchanan PK (2008) Molecular evidence for long distance dispersal across the Southern Hemisphere in the Ganoderma applanatum-australe species complex (Basidiomycota). Mycological Research 112: 425–436. [DOI] [PubMed] [Google Scholar]
- 104. Pringle A, Adams RI, Cross HB, Bruns TD (2009) The ectomycorrhizal fungus Amanita phalloides was introduced and is expanding its range on the west coast of North America. Molecular Ecology 18: 817–833. [DOI] [PubMed] [Google Scholar]
- 105. Tiffney BH, Manchester SR (2001) The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. International Journal of Plant Sciences 162: S3–S17. [Google Scholar]
- 106. Cai Q, Tulloss RE, Tang LP, Tolgor B, Zhang P, et al. (2014) Multi-locus phylogeny of lethal amanitas: Implications for species diversity and historical biogeography. BMC Evolutionary Biology 14: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood phylogeny of Cudonia and Spathularia species based on 113 nrLSU sequences with bootstrap values for 1000 replicates shown above the internal branches. Species of Cudonia and Spathularia are shaded in yellow, and Rhytismataceae species are shaded in green.
(TIF)
Maximum likelihood tree of the Cudonia and Spathularia taxa based on 78 rpb2 sequences with bootstrap values for 1000 replicates shown above the internal branches.
(TIF)
Maximum likelihood tree of the Cudonia and Spathularia taxa based on 96 tef-1α sequences with bootstrap values for 1000 replicates shown above the internal branches.
(TIF)
Chronogram of clades of Cudonia and Spathularia based on BEAST analyses. Three calibration points were used for the initial analysis based on results from Lücking et al. (2009): the stem of Ascomycota (575 ± 37.5 Mya, node A), the stem of Pezizomycotina (460 ± 30 Mya, node B), and the crown of Pezizomycotina (360 ± 20 Mya, node C). Estimated mean divergence times and 95% highest posterior densities of major nodes are summarized the upper left panel.
(TIF)
Maximum likelihood (ML) cladogram for 96 specimens depicting the phylogenetic relationships among Cudonia and Spathularia based on ITS-LSU-rpb2-tef-1α. Bootstrap support values above 50 (from 1000 bootstrap replicates) are shown above the branches.
(TIF)
GenBank accession numbers and voucher information for specimens used in molecular studies.
(DOC)
GenBank accession numbers of GenBank sequences used in this study.
(DOCX)
GenBank accession numbers of sequences used in the divergence time estimation.
(DOCX)
Evolutionary divergence of ITS sequence pairs within and between putative species of Cudonia and Spathularia.
(DOC)
The 127 specimens ITS alignment with the exact nucleotides to be excluded.
(NEX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.