Abstract
Antibiotics and antibiotic resistant bacteria enter wastewater treatment plants (WWTPs), an environment where resistance genes can potentially spread and exchange between microbes. Several antibiotic resistance genes (ARGs) were quantified using qPCR in three WWTPs of decreasing capacity located in Helsinki, Tallinn, and Tartu, respectively: sulphonamide resistance genes (sul1 and sul2), tetracycline resistance genes (tetM and tetC), and resistance genes for extended spectrum beta-lactams (blaoxa-58, blashv-34, and blactx-m-32). To avoid inconsistencies among qPCR assays we normalised the ARG abundances with 16S rRNA gene abundances while assessing if the respective genes increased or decreased during treatment. ARGs were detected in most samples; sul1, sul2, and tetM were detected in all samples. Statistically significant differences (adjusted p<0.01) between the inflow and effluent were detected in only four cases. Effluent values for blaoxa-58 and tetC decreased in the two larger plants while tetM decreased in the medium-sized plant. Only blashv-34 increased in the effluent from the medium-sized plant. In all other cases the purification process caused no significant change in the relative abundance of resistance genes, while the raw abundances fell by several orders of magnitude. Standard water quality variables (biological oxygen demand, total phosphorus and nitrogen, etc.) were weakly related or unrelated to the relative abundance of resistance genes. Based on our results we conclude that there is neither considerable enrichment nor purification of antibiotic resistance genes in studied conventional WWTPs.
Introduction
Antibiotic resistance (AR) has become a worldwide problem, making infectious diseases more resilient thus making treatment more difficult and costly [1]. AR is not confined to hospital environments, and is able to spread between both human dominated and natural environments. Increased amounts of antibiotic residues [2]–[4] and antibiotic resistant bacteria (ARB) are found in human-related environments such as agricultural settings (e.g. farms, soil etc.) [5], [6] and surface-, drinking- and wastewaters [7]–[11]. Wastewater treatment plants (WWTPs) receive sewage from various sources, including hospitals and households which are both important sources of antibiotics and their residues [12]–[14]. and antibiotic resistant bacteria (ARB) [15]–[17]. The presence of antibiotics and antibiotic residues [18]. ARB, and antibiotic resistance genes (ARG) have been confirmed in many WWTPs [19]–[23]. Bacteria from various environments, including human, soil, and activated sludge, are mixed in WWTPs and therefore these facilities are considered to be important “hot-spots” for AR and spread of resistance genes [18], [24]–[26]. The presence of antibiotics, ARB, and ARG in the same setting creates an environment that selects for AR and provides an opportunity for genetic material housing ARGs to transfer between bacterial species via horizontal gene transfer [13], [21], [24], [25], [27], [28]. In a metagenomic study of plasmids it was shown that numerous medically relevant ARG can be found in WWTPs (140 ARGs in a single WWTP and 123 in the effluent water) [29]. Therefore, there is a concern that resistance genes will spread in the bacterial population and further into more natural environments less impacted by human activity [30]–[33].
Many studies of antibiotic resistant bacteria and resistance genes have used culture-based assays, which are biased towards specific cultivable pathogenic or environmental species [17], [19], [21], [23], [34]. Culture-dependent data does not reflect the real variability or the actual amount of resistance genes present in a given WWTP, so these studies normally characterize only a small subset of the total population [35], [36]. Surprisingly, only a limited number of studies have used quantitative methods to investigate resistance genes in the total communities in effluent waters [21], [33], [37]–[40]. The small number of quantitative studies could be one reason why the overall impact of WWTPs in spreading resistance to the environment has not yet been properly evaluated.
The current study was designed to investigate the role of three conventional WWTPs in the distribution of ARGs using culture-independent quantitative methods. We thus focus on quantitative whole community level measurements on the water-phase of the inflow and effluent of WWTPs which has not been studied using quantitative methods in sufficient detail to assess the impact of WWTPs on the distribution of ARGs. To quantify the number of ARGs we analysed the total DNA from wastewater and effluent samples using quantitative real-time PCR (qPCR). Our original hypothesis was that conventional WWTPs increase the relative abundance of ARGs during processing because they do not employ technologies that target the removal of genetic elements.
Materials and Methods
1.1. Sample collection
Both raw wastewater and final effluent water samples were collected from three city WWTPs of decreasing capacity located in the Baltic Sea catchment area: large (Helsinki, Finland), medium (Tallinn, Estonia), and small (Tartu, Estonia). No specific permissions were required for sampling these locations, and the sampling was carried out in collaboration with each WWTP staff. The wastewater treatment technology employed in these three WWTPs is similar and typical of other facilities located in both Nordic (Finland, Sweden, Norway) and Eastern European (Estonia, Latvia, Lithuania, Poland) countries (EEA 2013). The WWTP in Helsinki is both the largest wastewater treatment plant in Finland and all Nordic countries; with about 0.8 million residents in the Helsinki metropolitan area. The majority of wastewater in Estonia is produced in Tallinn (∼350 000 residents) and Tartu (∼100 000). To abide by European law (91/271/EEC), the main steps of treatment are: primary treatment - mechanical treatment steps (sand, grit, fat and grease removal, pre-sedimentation); secondary treatment - biological treatment (activated sludge); followed by tertiary treatment - final deep purification using a combination of methods (chemical, mechanical and biological as in secondary sedimentation, bio-filters etc.). The WWTP effluent in both Helsinki and Tallinn is directed into the Baltic Sea while the WWTP effluent in Tartu flows into the Emajõgi River which forms part of the Baltic Sea catchment area. The main steps in the WWTP process and water-phase treatment are presented in Figure S1 in File S1.
Samples were collected over a one year period from December 2010 to December 2011 at five different time points, each representing a different season (four seasons; winter was sampled twice, Table S1A in File S1). In each sampling period three consecutive samples were taken on separate days at 1–3 day intervals; the exact sampling dates and monitored variables are given in Table S1 in File S1. Composite samples were taken, collected over 24 h periods, except in Tartu in the 2011 winter season when a grab sample was taken owing to technical problems (the automatic sampler was frozen because of extremely low temperatures).
1.2. Collection of total microbial community
The water samples were stored at 4°C pending filtration (within a couple of hours). Ten ml of influent water and 100 ml of effluent water were filtered through polycarbonate filters (pore size 0.22 µm, diameter 47 mm, GE Water & Process Technologies). For the last two time points at the Tallinn WWTP 200 ml of effluent water was filtered because a new treatment step (bio-filter) was added in September 2011.
1.3. DNA extraction
For the first three time points (Dec 2010; March 2011; June 2011) from the Helsinki WWTP, DNA was extracted from the samples using a MoBio PowerWater DNA isolation kit (MoBio Laboratories, Inc., CA, USA). For all other samples, the DNA was extracted using the modified bead beating and silica-membrane method (nucleic acid binding on to silica particles [41]. Method in brief: for lysis: 400 µl TE+50 µl lysozyme (50 mg/ml from egg yolk) was added to the filter and incubated at 37°C for 15 min. Fifty µl of 10% SDS and 500 µl lysis buffer BQ1 (NucleoSpin, Macherey-Nagel)+zirconium beads (0.1 mm diameter; burned at 500°C) were added and the samples were processed by a 5 min beating on a bead beater (Biospec products, Minibead beater). Thereafter, proteinase K was added followed by 15 min at 65°C with constant shaking. Five hundred µl of 96% ethanol was added and the whole volume was applied to commercial silica-membrane columns (NucleoSpin Macherey-Nagel). Finally, the total DNA was recovered in 50 µl of elution buffer BE (NucleoSpin Macherey-Nagel, 5 mM Tris/HCl pH 8.5). The concentration of extracted DNA was measured using a NanoDrop Spectrophotometer ND-1000 (absorption readings at 260 nm). The extracted DNA was stored at −20°C pending further analysis.
1.4. Detection and quantification of ARG copy number by qPCR
Seven resistance genes were surveyed: sul1, sul2, tetM, tetC, blashv-34, blactx-m-32, and blaoxa-58. For the first three time point samples (Dec 2010; March 2011; June 2011) from the Helsinki WWTP (Assay 1), qPCR was performed using a Dynamo Flash SYBR Green qPCR kit (Thermo Scientific, Lithuania) and a 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The thermal cycling conditions were as follows: 95°C for 7 min, 40 cycles at 95°C for 10 s and Tm for 30 s. A melting curve was obtained to confirm specificity of amplification. Reactions were conducted in 10 µl volumes on 96-well plates containing 1×Dynamo Flash SYBR Green master mix, 0.3 µM of each primer and 1×ROX passive reference dye. Template DNA was used in qPCR reactions in the range 2–12 ng DNA per reaction; a fixed dilution of raw DNA exctract was used. In parallel with the ARGs, the16S rRNA gene copy numbers were quantified.
The qPCR for detecting 16S rRNA gene and ARGs for all the other samples (Assay 2) used the 7900HT Fast Real-Time PCR System (Applied Biosystems). Reactions were conducted in 10 µl volumes on 384-well plates containing 1×Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific), 0.4 µM of each primer. The two-step thermal cycling conditions for detecting 16S rRNA gene were as follows: 95°C for 10 min, 40 cycles of 95°C 15 s, 60°C 1 min. For ARGs the thermal cycling conditions were as follows: 95°C for 10 min, 40 cycles at 95°C for 15 s, Tm °C for 30 s and 72°C for 30 s. Melting curves were obtained to confirm specificity of amplification. Template DNA was used in the qPCR reaction in the range 3×10−4 to 22.4 ng DNA per reaction. This was obtained by a wider range of dilutions (to avoid inhibition) of the raw DNA extraction product, three levels of 10 fold dilutions were used. Primers and annealing temperature, Tm (°C), are given in Table 1. Number of technical replicates in both qPCR assays was 3.
Table 1. Primers used for detecting the target genes and the melting temperatures (Tm) used for primers.
| Gene name | Primers | |||||
| Forward | Reverse | Tm °C | Ref. | Amplification efficiency | ||
| Assay 1 | Assay 2 | |||||
| 16S rRNA gene | 5′-AGA GTT TGA TCC TGG CTC AG-3′ | [60] | ||||
| 5′-CTG CTG CSY CCC GTA GGA-3′ | 60 | [61] modif. | 82–100 | |||
| 5′-CTG CTG CCT CCC GTA GG-3′ | 60 | [61] | 87 | |||
| tetC | 5′-TGC GTT GAT GCA ATT TCT ATG C-3′ | 5′-GGC GCC TAC AAT CCA TG-3′ | 64 | [62] | 80 | 93–101 |
| tetM | 5′-GCA ATT CTA CTG ATT TCT GC-3′ | 5′-CTG TTT GAT TAC AAT TTC CGC-3′ | 60 | [61] | 90–93 | 89–108 |
| sul1 | 5′-CGG CGT GGG CTA CCT GAA CG-3′ | 5′-GCC GAT CGC GTG AAG TTC CG-3′ | 64 | [63] | 90–93 | 81–88 |
| sul2 | 5′-GCG CTC AAG GCA GAT GGC ATT-3′ | 5′-GCG TTT GAT ACC GGC ACC CGT-3′ | 64 | [62] | 102 | 91–103 |
| blactx-m-32 | 5′-CGT CAC GCT GTT GTT AGG AA-3′ | 5′-CGC TCA TCA GCA CGA TAA AG-3′ | 64 | [59] | 87 | 89–101 |
| blashv-34 | 5′-GCG TTA TTT TCG CCT GTG TA-3′ | 5′-AGG TGC TCA TCA TGG GAA AG-3′ | 60 | [63] | 92–94 | 97–108 |
| blaoxa-58 | 5′-GCA ATT GCC TTT TAA ACC TGA-3′ | 5′-CTG CCT TTT CAA CAA AAC CC-3′ | 60 | [63] | 90 | 97–111 |
qPCR amplification efficiency is given for 16S RNA gene and for ARGs, R2 of the linear range of standards was always >0.99.
1.5. Standards used for quantification
A plasmid vector and fragments of ARGs were constructed and used as standards for quantifying the raw qPCR results. The standard plasmids were checked for the correct inserts by sequencing. In Assay 1 for the three Helsinki time points (Dec 2010; March 2011 and June 2011) the 16S rRNA gene quantification standard was genomic DNA from E. coli K12 (genome size 4.6 Mbp with seven copies of the rRNA operon). In Assay 2, used for all other samples, the standard for quantifying 16S rRNA gene was constructed from a 16S rRNA gene fragment from the natural aquatic Chryseobacterium strain isolated from Emajõgi River, which receives WWTP effluent water, and was cloned into a plasmid and validated by sequencing. Information about the plasmids used and Genbank accession numbers are given in Table 2.
Table 2. Plasmids and PCR fragments used as standards.
| Gene name | Standard constructs | Accession number | Reference |
| 16S rRNA gene | Assay 1 | ||
| Genomic DNA from E. coli K12 genome size 4.6 Mbp with 7 copies of rRNA operon | |||
| Assay 2 | |||
| PCR product cloned in plasmid PGEM-T Easy Vector System (Promega) | KF737394 | present study | |
| tetC | pDrive (Qiagen) | [62] | |
| tetM | pDrive (Qiagen) | [62] | |
| sul1 | R388 | [64] | |
| sul2 | RSF1010 | [64] | |
| blactx-m-32 | pUC19 | KF737395 | present study |
| blashv-34 | PGEM-T Easy Vector System (Promega) | KF737397 | present study |
| blaoxa-58 | pUC19 | KF737396 | present study |
1.6. Quantification and normalisation of ARGs
Standard curves (Ct per log copy number) for 16S rRNA gene and ARG quantification were obtained for each run using the plasmid constructs (or genomic E. coli DNA for 16S rRNA gene in Assay 1) (Table 2) in ten-fold serial dilutions. The gene copy number of a standard was determined from the plasmid/genomic DNA concentration (measured using a NanoDrop in Assay 1 or by fluorescent staining with PicoGreen (Invitrogen) and using VICTOR X3 Multilabel Plate Readers (Perkin-Elmer) in Assay 2). The ARG levels in the sample were calculated using the standard curve equation and measured Ct value, the quality control of raw Ct values for standard curve and unknown samples was done before further analysis. The limit of quantification (LOQ) was defined as the lowest point of the linear part of standard curve: Assay 1, ARGs 100 and 16S rRNA gene 1000 gene copy number per reaction; Assay 2, ARGs and 16S rRNA gene 100, except sul1 with 10 copy numbers per reaction. The negative control was the reaction mix with nuclease-free water instead of the template DNA. The negative control had always a Ct value at least 3.3 cycles lower than the smallest standard used for calculation of LOQ. Technical replicates were incorporated in the statistical analysis to reflect differences in quantification (see below for details).
1.7. Water quality variables
WWTPs in Europe analyse water quality parameters according to the EU directive for urban wastewater treatment 91/271/ECC. These parameters are monitored regularly in accredited laboratories according to standard methods. The parameters used in this study were: biochemical oxygen demand (BOD7) - standard method EN 1899-2; total suspended solids (SS) - EN 872; total phosphorus (Ptot) – EN ISO 6878; total nitrogen (Ntot) – EN ISO 11905. In addition, the automated measurements of flow rate and water temperature in the process were recorded. We received the data for water variables from the staff of each WWTP.
1.8. Statistics
Linear mixed models were fitted to the data using the functionality of the package lme4 [42]. the statistical software used was R version 3.0.1 [43]. Average levels of gene copy numbers (16S rRNA gene or ARGs) were modelled using the location of the WWTP (Helsinki, Tallinn, Tartu), sample source (IF/EF) and method protocol (Assay1/Assay2) as fixed effects and technical and biological replicates as random effects. Significance of fixed effects was assessed by an F-test using a (pre-specified) significance level of 1%. In addition, the combined effect of each of the environmental variables BOD7, SS, Ptot, Ntot, flow rate (i.e water discharge - WD) and temperature in inflowing versus effluent waste on gene copy numbers was described by a linear mixed model version of an analysis of covariance model. Model checking was based on residual plots and normal probability plots using the raw residuals. Models were reduced using the likelihood ratio test. A 1% or 5% significance level was used. Pairwise comparisons were evaluated based on adjusted p-values obtained using the single-step method [44]. Values below LOQ were not included in the analyses.
Principal component analysis (PCA) and its extensions to between groups (BGA) and within groups (WGA) analyses (ade4 package in R) was used to analyse the grouping of inflowing/effluent samples by monitored water quality variables (nutrients, suspended solids, biological oxygen demand, water discharge and temperature).
Results and Discussion
2.1. Selection of ARGs for the study
Initially, we made a screening for 25 selected genes that could spread with higher probability from any WWTPs into environment. Initial selection based on four major criteria: (i) clinically relevant genes (risk to human health) previously detected in WWTPs (e.g. [29]); (ii) genes found in various mobile elements, demonstrating their potential for transfer between bacteria [45]–[48]. (iii) high consumption antibiotics–sulphonamides, tetracyclines, and beta-lactams; (iv) incorporation of long-used antibiotics (tetracycline, sulfonamides) and the newer extended spectrum beta-lactams (carbapenems and 3rd and 4th generation cephalosporins) and their resistance genes. Altogether 12 bla, 6 tet, 3 sul, 3 qnr (floroquinolones) and 1 vancomycin resistant gene were tested using traditional PCR. Finally, two resistance genes for sulphonamide (sul1 and sul2), two for tetracycline (tetM and tetC), and three for extended spectrum beta-lactams (blaoxa-58, blashv-34, and blactx-m-32) remained in the study based on their abundance and frequency in screening study. It was necessary to choose a small number of relevant target genes because the variety of different ARGs is large. In the Comprehensive Antibiotic Resistance Database [49] (http://arpcard.mcmaster.ca/accessed Sept. 23, 2013), the number of ARGs is 2153 and testing all these genes quantitatively in one study would be prohibitively difficult.
2.2. Abundance of genes (16S rRNA gene and ARGs)
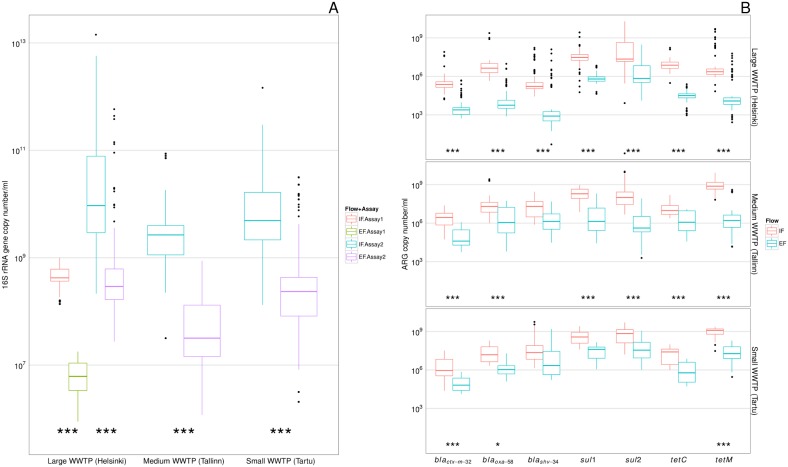
To evaluate the abundance of the total bacterial community, we quantified 16S rRNA gene in the samples using qPCR. The amplification efficiency is given in Table 1. Initially, the raw gene copy numbers were used to estimate the general changes of bacterial levels during wastewater purification. The copy number of 16S rRNA gene was several orders of magnitude lower in the effluent (EF) than the inflow (IF) (Figure 1A, Table S2 in File S1). The differences between the IF and EF samples were statistically significant in all cities (adjusted p<0.01) (Figure 1A).
Figure 1. Raw gene copy numbers detected in a WWTP sample (copy number/ml).
A - 16S rRNA gene in inflow (IF) and effluent (EF). Assay1 was used only for samples from large WWTP (Helsinki) from Winter 2010 to Autumn 2011; B–Antibiotic resistance genes (ARGs). Statistical significance between inflow wastewater and effluent samples: *** - p<0.01; *0.03>p>0.01. For the pairs not marked the statistical difference between inflow and outflow was statistically insignificant. The line in each box marks the median and boxes: 25th and 75th percentiles; whiskers: 5th and 95th percentiles and outliers ±1.5 * IQR. See Figure S2 in File S1 for abundances of same genes presented by each sampling event.
The raw gene copy numbers of ARGs/ml decreased during processing in the WWTP water phase. The levels of ARGs detected in the EF were lower than IF in all three plants (Figure 1B, Table S2 in File S1). The decrease of abundance from IF to EF was statistically significant (p<0.01) for all ARGs in the large (Helsinki) WWTP. In the medium (Tallinn) WWTP, for all ARGs except blashv-34 (p>0.05), the decrease was statistically significant (p<0.01). In the small WWTP (Tartu), the raw abundance of ARGs decreased after purification but the decrease was statistically significant only for blactx-m-32 and tetM (p<0.01), and weakly significant for blaoxa-58 (p = 0.03). Earlier studies have demonstrated the large variation of treatment plant efficiency in removing microorganisms [50] and micropollutants [51]. which also depends on the capacity of the WWTP plant. Low capacity treatment facilities are more vulnerable to changes in inflowing wastewater composition and flow rates. In addition, the WWTP in Tartu did not have biological post-filtration at the time of sampling.
ARGs are surprisingly rarely quantified directly using community DNA in both IF and EF water samples from WWTPs. As in our study (Figure 1B), a few other studies have found from 100 to 1000 fold reductions of raw ARG copy numbers during the purification process; e.g. sul1, tetW; [52]; tetC, tetA; [53]. tetG, tetQ [54]. In one study, the relative abundance of sul1 increased, while sul2 decreased slightly in WWTP effluent [31]. In our study, resistance genes for “older” antibiotics (with exception of tetC) were more commonly detected. sul1, sul2, and tetM were present above LOQ in all sites and samples (Table 3 and Figure S2 in File S1). High abundances of various tetracycline resistance genes and sulphonamide resistance genes were also demonstrated in other studies [21], [23]. Quantitative studies that target resistance to the newer beta-lactams in community DNA in WWTPs are almost completely absent–only blaTEM by Lachmayr et al. [38]. in addition, bla genes were quantified in the river water under the influences of wastewater but not directly in the WWTP effluent [55].
Table 3. Detection of ARGs in different WWTPs (total of all analyses per gene, n = 15).
| ARG | City | IF %(number) detected | EF %(number) detected |
| tetC | Helsinki | 93 (14) | 80 (12) |
| Tallinn | 67 (10) | 27 (4) | |
| Tartu | 67 (10) | 73 (11) | |
| blaoxa-58 | Helsinki | 100 (15) | 87 (13) |
| Tallinn | 100 (15) | 47 (7) | |
| Tartu | 87 (13) | 80 (12) | |
| blashv-34 | Helsinki | 100 (15) | 100 (15) |
| Tallinn | 87 (13) | 87 (13) | |
| Tartu | 100 (15) | 100 (15) | |
| blactx-m-32 | Helsinki | 100 (15) | 100 (15) |
| Tallinn | 87 (13) | 40 (6) | |
| Tartu | 80 (12) | 47 (7) |
Only genes that were sometimes not detected are given. sul1, sul2 and tetM were detected 100% in all IF and EF samples from the WWTPs.
2.3. Normalised/Relative abundances of ARGs
To avoid inconsistencies among qPCR assays, including sub-optimal efficiency in some cases, we used 16S rRNA gene-normalised values, and the different protocol (Assay1/Assay2) was added as an additional fixed effect into the statistical models. This type of data analysis allows one to quantify the relative changes in ARG abundances, whether more or fewer ARGs appear per microbial genome. When relative abundances were compared, statistically significant (p<0.01) differences between IF and EF were detected in only four cases (Figure 2). For blaoxa-58 and tetC in the Helsinki plant we observed a relative decrease after purification. In addition, tetM decreased in the Tallinn WWTP EF samples. The only increase in EF was observed for blashv-34 in the Tallinn WWTP. In all other cases the purification process had no significant effect on the relative abundances of resistance genes.
Figure 2. Normalised ARG abundances.
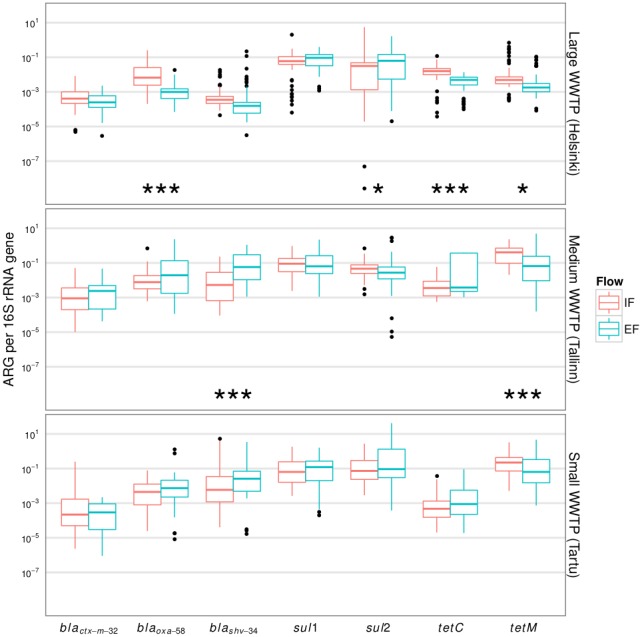
Antibiotic resistance gene copy numbers normalised to 16S rRNA gene copy numbers. The results are given for all samples for one gene for a WWTP, no seasonal comparison. Statistically significant comparison results are marked with *** at p<0.01, *0.03>p>0.01. The line in each box marks the median and boxes: 25th and 75th percentiles; whiskers: 5th and 95th percentiles and outliers ±1.5 * IQR.
We conclude from our study that there is neither considerable enrichment (selection) nor purification of ARGs during processes in WWTPs (Figure 2) at the whole community level. Effective selection would be assumed when there are appropriate conditions, i.e. increased concentration of ABs occur. In two WWTPs studied, the measured levels of AB concentrations of several compounds were very low but measurable compared to highly labile beta-lactams [56]. This suggests that conditions could favour enrichment of at least tet and sul genes within studied WWTPs. Although, quantitative enrichment of ARGs (sul) responsible for resistance against refractory ABs with longer history of usage has been demonstrated in some studies [31]. reduction or no change has been observed in most studies [21], [23], [40]. To date, only one study demonstrated similar case of positive selection for newer ARG possessing organisms in a WWTP, which suggests that bacteria harbouring blaTEM are released more from effluent water compared to wastewater [38].
2.4. Abundance of ARGs and treatment efficiency of wastewater
In EU countries, treatment efficiency of WWTPs is estimated by monitoring a few water quality measures, according to European directive 271/1991/EC. The compulsory parameters monitored are total nitrogen (Ntot) and phosphorus (Ptot), Biological Oxygen Demand (BOD7), and suspended solids (SS). In addition, a few generic background parameters are measured in all WWTPs e.g. Water Discharge (WD) and temperature. Such water quality parameters are good for evaluating wastewater purification in the traditional sense–removal of excess nutrients, labile organic compounds etc. Obviously, the wastewater was purified of excess nutrients and organic compounds in the WWTPs studied because all parameters were up to an order of magnitude lower in EF than IF. We observed a change of between 6 to 45 fold depending on the parameter. Averages in IF: BOD7 - 231; Ptot 8; Ntot 51 and SS 306 mg/l and in EF: BOD7 5; Ptot 0.6; Ntot 8 and SS 8 mg/l (Table S1B in File S1). Volumetric concentrations of nutrients, SS and labile organic compounds were higher in the IF of the small (Tartu) than in medium (Tallinn) and large (Helsinki) WWTP (Figure S4 in File S1; IF samples are strongly associated with these variables, permutation test, 1000 replicates, p<0.01). This could be caused by shorter solid retention time in smaller plants [57]. At the same time, the efficiency of purification in the traditional sense did not differ dramatically among plants (Figure S3 in FileS1; residual differences among WWTPs disappear after decomposing the IF/EF level differences, permutation test, 1000 replicates, p>0.05). A relationship between the change of ARG abundances and the efficiency of nutrient removal and temperature has been reported previously (e.g. [58], [59]). These studies suggest that changes in ARG abundance could depend on processes and conditions in the WWTP. However, none of these parameters are designed to estimate threats associated with the spread of either ARB or ARGs. Indeed, in our study, temperature, water discharge and concentrations of nutrients did not help in estimating the efficiency of ARG removal. This was demonstrated by the absence of the combined effect of monitored environmental variables and abundances of ARGs (Figure S4 in File S1). Moreover, a new treatment step (installation of biological post-filtration for final effluent treatment mainly for nitrogen removal) was added during the study period in the Tallinn WWTP (July 2011). However, no statistically significant effect was observed on ARG removal after this event. The Helsinki WWTP had the biological post-filtration installed throughout the study period.
In conclusion, these results force us to reject our original hypothesis. All ARGs were detected in most wastewater and effluent samples, however, the conventional WWTPs under study seem not to be important sites for changes in the relative abundance of ARGs at the whole community level: no enrichment in relative abundance was observed. Furthermore, no additional reduction of ARGs occurred; raw abundance changed in proportion to the decrease of bacterial abundance. We conclude that many unknown factors may influence the biological purification processes in conventional WWTPs and the evaluation of their relationship to ARG removal or selection requires more complex case studies.
Supporting Information
Includes Tables S1 and S2; Figures S1, S2, S3 and S4.
(PDF)
Acknowledgments
We acknowledge the staff from HSY at Viikinmäki, Tartu Veevärk AS and Tallinna Vesi AS for providing samples, and Dr. David Schryer for language revision.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. In addition, all sequences are available from GenBank (KF737394–KF737396).
Funding Statement
The research was funded by the Academy of Finland and EnSTe graduate school, and the European Regional Development Fund through the Centre of Excellence in Chemical Biology in Estonia. Additional funding was provided by COST Action TD0803. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10: S122–9. [DOI] [PubMed] [Google Scholar]
- 2. Lindberg RH, Wennberg P, Johansson MI, Tysklind M, Andersson BA V (2005) Screening of Human Antibiotic Substances and Determination of Weekly Mass Flows in Five Sewage Treatment Plants in Sweden. Environ Sci Technol 39: 3421–3429. [DOI] [PubMed] [Google Scholar]
- 3. Le-Minh N, Khan SJ, Drewes JE, Stuetz RM (2010) Fate of antibiotics during municipal water recycling treatment processes. Water Res 44: 4295–4323. [DOI] [PubMed] [Google Scholar]
- 4. Novo A, André S, Viana P, Nunes OC, Manaia CM (2013) Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater. Water Res 47: 1875–1887 Available: http://www.sciencedirect.com/science/article/pii/S0043135413000274 Accessed 23 January 2013 [DOI] [PubMed] [Google Scholar]
- 5. Khachatourians GG (1998) Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria. Can Med Assoc J 159: 1129–1136. [PMC free article] [PubMed] [Google Scholar]
- 6. Durso LM, Miller DN, Wienhold BJ (2012) Distribution and Quantification of Antibiotic Resistant Genes and Bacteria across Agricultural and Non-Agricultural Metagenomes. PLoS One 7: e48325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwartz T, Kohnen W, Jansen B, Obst U (2003) Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol 43: 325–335. [DOI] [PubMed] [Google Scholar]
- 8. Kümmerer K (2004) Resistance in the environment. J Antimicrob Chemother 54: 311–320. [DOI] [PubMed] [Google Scholar]
- 9. Baquero F, Martínez J-L, Cantón R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19: 260–265. [DOI] [PubMed] [Google Scholar]
- 10. Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, et al. (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8: 251–259 Available: 10.1038/nrmicro2312 Accessed 21 March 2014. [DOI] [PubMed] [Google Scholar]
- 11. Stoll C, Sidhu JPS, Tiehm A, Toze S (2012) Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ Sci Technol 46: 9716–9726. [DOI] [PubMed] [Google Scholar]
- 12. Brown KD, Kulis J, Thomson B, Chapman TH, Mawhinney DB (2006) Occurrence of antibiotics in hospital, residential, and dairy effluent, municipal wastewater, and the Rio Grande in New Mexico. Sci Total Environ 366: 772–783. [DOI] [PubMed] [Google Scholar]
- 13. Duong HA, Pham NH, Nguyen HT, Hoang TT, Pham HV, et al. (2008) Occurrence, fate and antibiotic resistance of fluoroquinolone antibacterials in hospital wastewaters in Hanoi, Vietnam. Chemosphere 72: 968–973. [DOI] [PubMed] [Google Scholar]
- 14. Chang X, Meyer MT, Liu X, Zhao Q, Chen H, et al. (2010) Determination of antibiotics in sewage from hospitals, nursery and slaughter house, wastewater treatment plant and source water in Chongqing region of Three Gorge Reservoir in China. Environ Pollut 158: 1444–1450. [DOI] [PubMed] [Google Scholar]
- 15. Reinthaler F, Posch J, Feierl G, Wüst G, Haas D, et al. (2003) Antibiotic resistance of E. coli in sewage and sludge. Water Res 37: 1685–1690. [DOI] [PubMed] [Google Scholar]
- 16. Kim S, Aga DS (2007) Potential Ecological and Human Health Impacts of Antibiotics and Antibiotic-Resistant Bacteria from Wastewater Treatment Plants. J Toxicol Environ Heal Part B 10: 559–573. [DOI] [PubMed] [Google Scholar]
- 17. Figueira V, Vaz-Moreira I, Silva M, Manaia CM (2011) Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res 45: 5599–5611. [DOI] [PubMed] [Google Scholar]
- 18. Michael I, Rizzo L, McArdell CS, Manaia CM, Merlin C, et al. (2013) Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res 47: 957–995. [DOI] [PubMed] [Google Scholar]
- 19. Łuczkiewicz A, Jankowska K, Fudala-Książek S, Olańczuk-Neyman K, Łuczkiewicz A, et al. (2010) Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res 44: 5089–5097 Available: http://www.sciencedirect.com/science/article/pii/S0043135410005713 Accessed 23 January 2013. [DOI] [PubMed] [Google Scholar]
- 20. Li D, Qi R, Yang M, Zhang Y, Yu T (2011) Bacterial community characteristics under long-term antibiotic selection pressures. Water Res 45: 6063–6073. [DOI] [PubMed] [Google Scholar]
- 21. Munir M, Wong K, Xagoraraki I (2011) Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res 45: 681–693. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X-X, Zhang T (2011) Occurrence, Abundance, and Diversity of Tetracycline Resistance Genes in 15 Sewage Treatment Plants across China and Other Global Locations. Environ Sci Technol 45: 2598–2604. [DOI] [PubMed] [Google Scholar]
- 23. Gao P, Munir M, Xagoraraki I (2012) Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci Total Environ 421–422: 173–183. [DOI] [PubMed] [Google Scholar]
- 24. Guardabassi L, Lo Fo Wong DM, Dalsgaard A (2002) The effects of tertiary wastewater treatment on the prevalence of antimicrobial resistant bacteria. Water Res 36: 1955–1964. [DOI] [PubMed] [Google Scholar]
- 25. Moura A, Henriques I, Smalla K, Correia A (2010) Wastewater bacterial communities bring together broad-host range plasmids, integrons and a wide diversity of uncharacterized gene cassettes. Res Microbiol 161: 58–66. [DOI] [PubMed] [Google Scholar]
- 26. Jury KL, Khan SJ, Vancov T, Stuetz RM, Ashbolt NJ (2011) Are Sewage Treatment Plants Promoting Antibiotic Resistance? Crit Rev Environ Sci Technol 41: 243–270. [Google Scholar]
- 27. Courvalin P (1994) Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob Agents Chemother 38: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schlüter A, Szczepanowski R, Pühler A, Top EM, Schlüter A, et al. (2007) Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev 31: 449–477. [DOI] [PubMed] [Google Scholar]
- 29. Szczepanowski R, Linke B, Krahn I, Gartemann K, Guetzkow T, et al. (2009) Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology-Sgm 155: 2306–2319. [DOI] [PubMed] [Google Scholar]
- 30. West BM, Liggit P, Clemans DL, Francoeur SN (2010) Antibiotic Resistance, Gene Transfer, and Water Quality Patterns Observed in Waterways near CAFO Farms and Wastewater Treatment Facilities. Water, Air, Soil Pollut 217: 473–489. [Google Scholar]
- 31.Czekalski N, Berthold T, Caucci S, Egli A, Bürgmann H (2012) Increased Levels of Multiresistant Bacteria and Resistance Genes after Wastewater Treatment and Their Dissemination into Lake Geneva, Switzerland. Front Microbiol 3. [DOI] [PMC free article] [PubMed]
- 32. Pruden A, Arabi M, Storteboom HN (2012) Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Technol 46: 11541–11549. [DOI] [PubMed] [Google Scholar]
- 33. Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, et al. (2013) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci Total Environ 447: 345–360. [DOI] [PubMed] [Google Scholar]
- 34. Da Silva MF, Tiago I, Verssimo A, Boaventura RAR, Nunes OC, et al. (2006) Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol Ecol 55: 322–329. [DOI] [PubMed] [Google Scholar]
- 35. Levy SB (2002) Factors impacting on the problem of antibiotic resistance. J Antimicrob Chemother 49: 25–30 Available: http://jac.oxfordjournals.org/content/49/1/25. [DOI] [PubMed] [Google Scholar]
- 36. Jury KL, Khan SJ, Vancov T, Stuetz RM, Ashbolt NJ (2011) Are Sewage Treatment Plants Promoting Antibiotic Resistance? Crit Rev Environ Sci Technol 41: 243–270. [Google Scholar]
- 37. Auerbach EA, Seyfried EE, McMahon KD (2007) Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res 41: 1143–1151. [DOI] [PubMed] [Google Scholar]
- 38. Lachmayr KL, Kerkhof LJ, DiRienzo AG, Cavanaugh CM, Ford TE (2009) Quantifying Nonspecific TEM β-Lactamase (blaTEM) Genes in a Wastewater Stream. Appl Environ Microbiol 75: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang T, Zhang M, Zhang X, Fang HH (2009) Tetracycline Resistance Genes and Tetracycline Resistant Lactose-Fermenting Enterobacteriaceae in Activated Sludge of Sewage Treatment Plants. Environ Sci Technol 43: 3455–3460 Available: 10.1021/es803309m Accessed 27 March 2013. [DOI] [PubMed] [Google Scholar]
- 40. Börjesson S, Mattsson A, Lindgren P-E (2010) Genes encoding tetracycline resistance in a full-scale municipal wastewater treatment plant investigated during one year. J Water Health 08: 247. [DOI] [PubMed] [Google Scholar]
- 41. Boom R, Sol CJ, Salimans MM, Jansen CL, Dillen PMW, et al. (1990) Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bates D, Bolker MM and Ben (2013) lme4: Linear mixed-effects models using S4 classes.
- 43.R Development Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
- 44. Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50: 346–363. [DOI] [PubMed] [Google Scholar]
- 45. Toleman MA, Bennett PM, Walsh TR (2006) Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J Antimicrob Chemother 58: 1–6. [DOI] [PubMed] [Google Scholar]
- 46. Bennett PM (2008) Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol 153: S347–S357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roberts AP, Mullany P (2009) A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol 17: 251–258. [DOI] [PubMed] [Google Scholar]
- 48. Liu M, Zhang Y, Yang M, Tian Z, Ren L, et al. (2012) Abundance and Distribution of Tetracycline Resistance Genes and Mobile Elements in an Oxytetracycline Production Wastewater Treatment System. Environ Sci Technol 46: 7551–7557. [DOI] [PubMed] [Google Scholar]
- 49. McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. (2013) The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57: 3348–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koivunen J, Siitonen A, Heinonen-Tanski H (2003) Elimination of enteric bacteria in biological/chemical wastewater treatment and tertiary filtration units. Water Res 37: 690–698 10.1016/S0043-1354(02)00305-6 [DOI] [PubMed] [Google Scholar]
- 51.Luo Y, Guo W, Ngo HH, Nghiem LD, Hai FI, et al. (2014) A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci Total Environ 473<unicod: 619–641. doi:10.1016/j.scitotenv.2013.12.065. [DOI] [PubMed]
- 52. Gao P, Munir M, Xagoraraki I (2012) Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci Total Environ 421–422: 173–183. [DOI] [PubMed] [Google Scholar]
- 53. Zhang T, Zhang M, Zhang X, Fang HH (2009) Tetracycline Resistance Genes and Tetracycline Resistant Lactose-Fermenting Enterobacteriaceae in Activated Sludge of Sewage Treatment Plants. Environ Sci Technol 43: 3455–3460. [DOI] [PubMed] [Google Scholar]
- 54. Auerbach EA, Seyfried EE, McMahon KD (2007) Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res 41: 1143–1151. [DOI] [PubMed] [Google Scholar]
- 55. Graham DW, Olivares-Rieumont S, Knapp CW, Lima L, Werner D, et al. (2011) Antibiotic Resistance Gene Abundances Associated with Waste Discharges to the Almendares River near Havana, Cuba. Environ Sci Technol 45: 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lillenberg M (2011) Residues of some pharmaceuticals in sewage sludge in estonia, their stability in the environment and accumulation into food plants via fertilizing residues of some pharmaceuticals in sewage sludge in estonia, their stability in the environment and accumul Estonian University of Life Sciences. Available: http://dspace.emu.ee/xmlui/bitstream/handle/10492/146/Thesis_Lillenberg_2011.pdf.
- 57. Clara M, Kreuzinger N, Strenn B, Gans O, Kroiss H (2005) The solids retention time-a suitable design parameter to evaluate the capacity of wastewater treatment plants to remove micropollutants. Water Res 39: 97–106. [DOI] [PubMed] [Google Scholar]
- 58. Novo A, Manaia CM (2010) Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl Microbiol Biotechnol 87: 1157–1166 Available: http://www.ncbi.nlm.nih.gov/pubmed/20396880. [DOI] [PubMed] [Google Scholar]
- 59. Nõlvak H, Truu M, Tiirik K, Oopkaup K, Sildvee T, et al. (2013) Dynamics of antibiotic resistance genes and their relationships with system treatment efficiency in a horizontal subsurface flow constructed wetland. Sci Total Environ 461–462: 636–644. [DOI] [PubMed] [Google Scholar]
- 60. Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17: 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. López-Gutiérrez JC, Henry S, Hallet S, Martin-Laurent F, Catroux G, et al. (2004) Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J Microbiol Methods 57: 399–407. [DOI] [PubMed] [Google Scholar]
- 62. Tamminen M, Karkman A, Lohmus A, Muziasari WI, Takasu H, et al. (2010) Tetracycline Resistance Genes Persist at Aquaculture Farms in the Absence of Selection Pressure. Environ Sci Technol 45: 386–391. [DOI] [PubMed] [Google Scholar]
- 63. Pei R, Kim S-C, Carlson KH, Pruden A (2006) Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40: 2427–2435. [DOI] [PubMed] [Google Scholar]
- 64. Heuer H, Smalla K (2007) Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ Microbiol 9: 657–666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Includes Tables S1 and S2; Figures S1, S2, S3 and S4.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. In addition, all sequences are available from GenBank (KF737394–KF737396).