SUMMARY
R-loops exist at the murine IgH switch regions and possibly other locations, but their functional importance is unclear. In biochemical systems, R-loop initiation requires DNA sequence regions containing clusters of G nucleotides, but cellular studies have not been done. Here, we vary the G-clustering, total switch region length, and the number of target sites (WGCW sites for the activation-induced deaminase) at synthetic switch regions in a murine B cell line to determine the effect on class switch recombination (CSR). G-clusters increase CSR, regardless of their immediate proximity to the WGCW sites. This increase is accompanied by an increase in R-loop formation. CSR efficiency correlates better with the absolute number of WGCW sites in the switch region rather than the total switch region length or density of WGCW sites. Thus, the overall strength of the switch region depends on G-clusters, which initiate R-loop formation, and on the number of WGCW sites.
Keywords: recombination, activation-induced deaminase, AID, RNA:DNA hybrid, immunoglobulin, isotype switch, B cell, antibody, secondary response, genetic instability, chromosomal rearrangement, gene rearrangement, S9.6 antibody
INTRODUCTION
Immunoglobulin heavy chains (IgH) have a variable exon assembled by V(D)J recombination and constant domain exons (Chaudhuri et al., 2007; Hackney et al., 2009) (Figure S1A). The IgH constant domains can be changed at the DNA level by Ig class switch recombination (CSR), and this allows the same antigen binding pocket to be directed to different locations in the organism and to elicit different immune responses. The initial immunoglobulin, IgM, encoded by the Cμ constant domain exons, that are immediately downstream of the V(D)J exon. But other IgH isotypes, including IgG, IgA, and IgE, are located over a region of several hundred kb downstream. Each set of IgH isotype exons for the constant region is preceded by a sterile transcript promoter and a switch region that is repetitive and rich in WGCW sites.
CSR is particularly efficient and well developed in mammals, where a majority of the antibody in the blood is generated from an IgH locus that has undergone CSR (Murphy, 2012). Efficient CSR may be essential for organisms with well-developed lungs to transport IgA into the airways in order to minimize pulmonary infections from viruses, bacteria and fungi. The rapid IgH switch from IgM to IgA requires DNA recombination at the IgH locus. Rapid IgH isotype switch is also important in the gastrointestinal and urogenital tracts in mammals (Murphy, 2012).
AID is a cytidine deaminase which initiates both somatic hypermutation (SHM) and CSR (Muramatsu et al., 2007; Muramatsu et al., 1999). AID requires single-stranded DNA (ssDNA) in order to deaminate C to U (Bransteitter et al., 2003; Pham et al., 2011; Yu et al., 2004). How ssDNA arises for SHM is not clear but it may be a simple consequence of transcription. Such a mechanism of generating ssDNA would be inefficient, and this may explain why SHM occurs relatively slowly, over a period of weeks in a typical immune response (Murphy, 2012). In contrast, CSR occurs much faster over a period of days, typically 48 to 72 hrs, using the same AID enzyme. A preferred site of action by AID is the WGCW motif, shown both biochemically (Bransteitter et al., 2003; Yu et al., 2004) and within the switch regions in the genome (Han et al., 2011).
The basis for the difference in efficiency between mammalian CSR and SHM has not been clear. One difference may be that R-loops form at mammalian IgH switch region DNA when B cells are activated for switch region transcription via cytokine stimulation (Yu et al., 2003; Yu and Lieber, 2003). The R-loops form when transcription extends through the repetitive switch regions, and the nascent RNA threads back (anneals) onto the template DNA strand before the nontemplate DNA and template DNA strands can reanneal (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). Thus, an R-loop is a three-stranded nucleic acid structure consisting of the two DNA strands of the normal duplex, but with the newly transcribed RNA base paired as a duplex with the template DNA strand. Therefore, this leaves the nontemplate DNA strand entirely single-stranded (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010; Yu et al., 2003; Yu and Lieber, 2003). As mentioned, a promoter is located upstream of each Ig class switch region, and yet no protein is produced from this transcript; hence, this is called a sterile transcript (Chaudhuri et al., 2007; Stavnezer et al., 2008; Wabl and Steinberg, 1996). The function of sterile transcripts has been unclear. We have proposed that one major function is to generate R-loops because these would provide a stable site of ssDNA for AID action in a very efficient manner, especially for organisms which require rapid IgH switching (Yu et al., 2003; Yu and Lieber, 2003). Independent genome-wide methods to document single-stranded DNA confirm that Ig switch regions are among the most single-stranded regions in the genomes of mammalian B cells (our analysis of (Kouzine et al., 2013), Z. Lu and MRL, unpubl.). It is readily apparent that an R-loop makes the nontemplate DNA strand available in ssDNA form for AID (Yu et al., 2005). Regarding the template DNA strand, we have described elsewhere how RNase H action can create zones of ssDNA on this remaining template strand (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010; Yu et al., 2003; Yu and Lieber, 2003).
In biochemical studies with purified DNA and prokaryotic RNA polymerases, we showed that G-clusters (consecutive G’s) are important for the newly transcribed RNA to thread back onto the template DNA strand to form an R-loop (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). This is almost indistinguishable from the formation of R-loops at DNA replication origins in prokaryotes, where a G-cluster of a newly transcribed RNA must anneal to the template DNA strand to initiate DNA replication (Masukata and Tomizawa, 1990). Factors that favor annealing of the RNA to the DNA template, such as negative superhelicity, nontemplate DNA strand nicks, and proximity of the G-cluster to the 5′ end of the RNA all favor more efficient R-loop formation (Baaklini et al., 2008; Broccoli et al., 2004; Drolet et al., 2003; Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). For switch sequences, we term the G-cluster rich region responsible for initiating the R-loops the “R-loop initiation zone” or RIZ. Once initiated in the RIZ, we find that the R-loop can extend downstream of the RIZ into a region with no G-clusters but in which the G-density is relatively high (35 to 50% G). We call this region the “R-loop elongation zone” or REZ.
Despite having shown that R-loops form in the genomes of mammalian B cells at Ig switch regions, the relationship between G-clusters and R-loop formation within cells has not been tested. This link is a critical one, especially since recent reports have proposed R-loops in key biological processes far outside of CSR (Balk et al., 2013; El Hage et al., 2010; Mischo et al., 2011; Nakama et al., 2012; Pfeiffer et al., 2013; Skourti-Stathaki et al., 2011; Stirling et al., 2012; Sun et al., 2013; Wahba et al., 2011; Yang et al., 2014). Genome biologists have highlighted Ig class switch-like sequences at other locations in the genome for the possibility that they have a propensity for DNA breakage; the hypothesis is that such breaks would direct chromosomal changes during evolutionary speciation (Bailey and Eichler, 2006). Other studies claim that half the genes in the genome are predicted to form R-loops (Wongsurawat et al., 2012). Some studies claim that R-loops exist at most CpG island promoters (Ginno et al., 2012). R-loops have been associated with genome instability (Li et al., 2005; Stirling et al., 2012; Wahba et al., 2011). Despite these additional roles, none of these studies have defined what are the essential DNA sequence elements or features needed to cause R-loop formation in the genome.
Here we examine the effects of the following on CSR: (a) G-clusters interspersed among WGCW sites; (b) G-clusters physically separated from the WGCW sites; (c) WGCW site number and density; and (d) total switch region length (determined by the number of switch repeats). We find that CSR is dependent on the presence of G-clusters, which are the key determinant of R-loop initiation, regardless of whether the G-clusters are interspersed within the switch repeats (as they are naturally in mammalian switch regions) or restricted to the upstream boundary in an RIZ. Once an R-loop is initiated, the R-loop final length determines how much ssDNA is exposed for AID action. AID preferentially acts at WGCW sites in ssDNA. Within the total switch region lengths studied here, we find that the total number of WGCW sites is more important than their density or the DNA length over which a given number of WGCW sites is distributed. These findings are important because they define what DNA sequence features are needed for efficient Ig CSR.
RESULTS
Experimental Strategy and Rationale
To study the factors important for R-loop formation and for Ig CSR, we used a system that allows us to exchange out the genomic switch region and replace it with any desired sequence (Han et al., 2011). The mouse B-cell line, CH12F3.2a, is able to specifically and efficiently switch to IgA upon cytokine stimulation (hereafter called the cellular system). Using this cellular assay system, the endogenous Sα locus in CH12F3.2a was replaced with a positive-negative selection cassette (PuroΔTK, provides puromycin resistance and ganciclovir sensitivity) flanked by two different loxP sites. The resulting cell line is called 1F7 (Han et al., 2011)(Figure S1B). The sequence of interest (Table S1) is cloned into an exchange vector with the same loxP sites as 1F7 cells. This exchange vector is cotransfected into 1F7 cells along with a Cre-expressing vector. Cre mediates recombination between the exchange vector and the 1F7 chromosome at the corresponding loxP sites. The successful replacement of the selection cassette by the sequence of interest gives rise to ganciclovir resistant clones, which are screened with a puromycin sensitivity test and PCR analysis for all clones (Table S2). Selected clones are confirmed by Southern blot.
Here we tested parameters that may be important for IgH CSR using the cellular assay system. CSR as a function of G-cluster number was a focus of primary interest. Given that efficient CSR requires WGCW sites (Han et al., 2011), we tested synthetic switch regions that had two fundamentally different configurations: (a) G-clusters interspersed with WGCW sites (designated the interspersed configuration) and (b) G-clusters in an upstream RIZ, followed by WGCW sites in a downstream REZ, which has no G-clusters, but has a 45% G-density (designated the RIZ/REZ configuration).
Naturally-occurring mammalian switch regions consist of repetitive units of 25 to 80 bp, and the natural mammalian Sα switch region has an 80 bp repeat (Dunnick et al., 1993; Snapper and Finkelman, 1999). The naturally-occurring switch regions have an interspersed configuration (i.e., many G-clusters and WGCW sites are mixed together within each repeat). For the synthetic switch regions studied here, we varied the number of 80 bp repeats between 4 and 28 (for a total switch region length of 320 to 2240 bp). For the interspersed configurations, we varied the G-cluster number in the repeat units (but not the WGCW number in each repeat unit). For the RIZ/REZ configurations, we varied the G-cluster number in the RIZ and the WGCW number in each repeat of the REZ.
Clusters of G Nucleotides Increase CSR
Biochemical studies showed that G-clusters are able to promote R-loop formation (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). In the genome of activated B cells in culture, we have determined that the R-loops typically begin near G-clusters and end when the G-density falls below 30% (Huang et al., 2007; Huang et al., 2006). However, a formal study of CSR as a function of G-cluster number has not been done in a cellular assay system. This aspect of the R-loop hypothesis is critical for the Ig class switch field to evaluate the extent to which R-loop formation is central to the high efficiency of mammalian CSR.
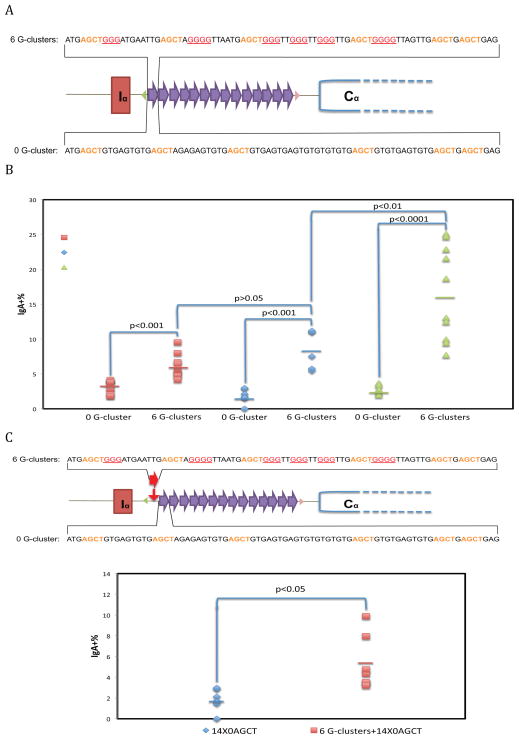
We created an 80 bp repeat unit with 6 G-clusters, and constructed exchange vectors with increasing numbers of these repeat units. The cellular clones with these sequences of interest were selected as described (Han et al., 2011)(Figure S1B). At least five clones were picked for CSR efficiency tests. As a control, a different 80 bp repeat unit with exactly the same nucleotide composition, but no G-clusters, was constructed (Figure 1A).
Figure 1. Effect of G-Clusters Interspersed within Switch Repeat Unit.
A. Construction of synthetic switch regions and the genomic configuration of Sα locus (Han et al., 2011). Each 80 bp switch repeat unit (with 6 G-clusters in the DNA sequence shown for the duplex above the diagram or with no G-clusters for the duplex below the diagram) was self-ligated and the products with the desired number of switch repeats were inserted into the exchange vector. An exchange vector and a Cre-expression vector were cotransfected into 1F7 cells. Triangles represent different loxP sties, and each oval represents one 80 bp repeat unit. Six to ten independent cellular clones were made for each switch construct.
B. FACS analysis of CSR is shown. Each data symbol represents an independent clone. The clones are organized in groups of two, based on the number of repeat units shown in the key on the right. The number on the bottom indicates the number of G-clusters within each repeat. The bars represent the mean of each group. The Student’s t test was used to analyze the effect of G-clusters.
C. One 80 bp repeat with 6 G-clusters (the red oval in the top sequence) was cloned immediately upstream of the fourteen 80 bp repeats with no G-clusters (purple ovals that have the DNA sequence shown under the diagram at the top of this panel). Seven independent cellular clones of each switch construct were analyzed by FACS. The horizontal colored bars represent the mean of each group.
We found that with 9 switch repeats having an interspersed configuration, G-clusters promote CSR by almost two-fold (Figure 1B). G-clusters appear to slightly inhibit germline transcription (GLT)(Figures S3 and S4A). When the repeat number rises from 9 to 14, CSR increases another 1.5-fold (borderline significance, p=0.07), whereas the CSR efficiency in cells without G-clusters remains at a low level.
We wanted to see whether CSR increases further with more G-clusters, so we constructed cellular clones with 28 repeats containing either 0 or 6 G-clusters interspersed within each switch repeat. CSR in cells with G-clusters increases significantly by another 2-fold over the cells without G-clusters for a switch region of 14 repeats, and the increase is even more significant in cells with 9 repeats (Figure 1B). Importantly, CSR is inefficient in cells without G-clusters even with 28 switch repeat units.
In all three pairs of genomic configurations, the presence of G-clusters in the synthetic switch repeats significantly increases the CSR ability. Based on this, we conclude that G-clusters are important for efficient CSR. Without G-clusters, CSR is inefficient, regardless of the number of switch repeats and, hence, regardless of the total switch region length. This clearly indicates that the presence of G-clusters is critical to promote CSR, and the total switch region length alone is insufficient to promote CSR. As we examine in more detail below, but is apparent here, if G-clusters are present, CSR also increases as the number of switch repeats (total switch region length) increases (Figure 1B).
G-Clusters Can Convert Less Efficient Class Switch Regions to More Efficient Ones
We wondered if we could convert a low efficiency synthetic switch region to a higher one by adding a short region of G-clustered DNA immediately upstream of a synthetic switch region without G-clusters. We inserted the same 80 bp unit with 6 G-clusters immediately upstream of the 14 switch repeats without any G-clusters (Figure 1C). We constructed and analyzed the cell clones as described above.
We found that with just one 80bp unit containing 6 G-clusters upstream of the repetitive sequence of 14 repeats lacking G-clusters, the CSR efficiency increases by almost three-fold (Figure 1C). Therefore, the increase in CSR is not simply due to the longer DNA sequence (insertion of the 80 bp sequence) since the cells with 28 repeats without G-clusters have relatively low CSR (Figure 1B). This suggests that the G-clusters play an important stimulatory role for switch region strength for initiating R-loop formation.
G-Clusters Near the Beginning of the Class Switch Region Have a Large Effect on CSR
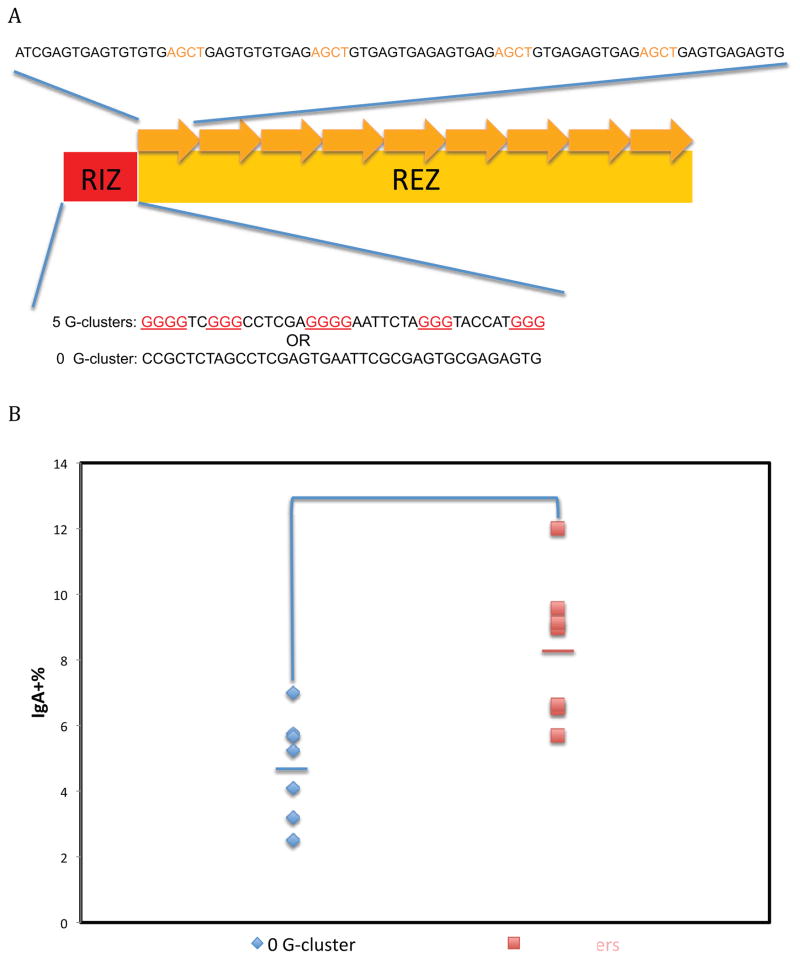
To investigate how G-clusters affect CSR further, we fully separated G-clusters from the WGCW sites by using an RIZ/REZ configuration (see Introduction). The 5 G-clusters are only located at the beginning of our synthetic switch regions (the RIZ), whereas the repetitive region (REZ) has 9 repeats with 4 AGCT sites, but no G-clusters. The REZ is also a region with 45% G-density to support R-loop elongation, once an R-loop is initiated within the RIZ. As a negative RIZ control, a piece of DNA with the same length but no G-clusters was used (Figure 2A). With G-clusters within the RIZ, CSR increases significantly by almost 2-fold (Figure 2B). This further indicates that G-clusters increase CSR even when G-clusters are only present at the beginning of a switch region.
Figure 2. Effect of G-Clusters at the Upstream Boundary of the Class Switch Region.
A. RIZ/REZ configuration of a synthetic switch region. Nine repeats of 80 bp oligonucleotides (top strand of duplex shown above the RIZ/REZ diagram) with 4 AGCT sites (orange) and alternating G nucleotides were used for the REZ in this synthetic switch region. The DNA sequence of the RIZ are shown below the RIZ/REZ diagram, with G-clusters in red. For synthetic switch regions in Figures 2, 4, 5, and 6, the RIZ and REZ are separated, in contrast to Figure 1 where the G-clusters are interspersed with the WGCW sites. The RIZ with zero G-clusters is a negative control for the RIZ with G-clusters.
B. Effect of CSR of G-clusters separate from WGCW motifs. Seven independent cellular clones were shown for each RIZ. The bars represent the mean of each group. The Student’s t test was used to analyze the effect of G-clusters.
G-Clusters Increase R-loop Formation within Cells
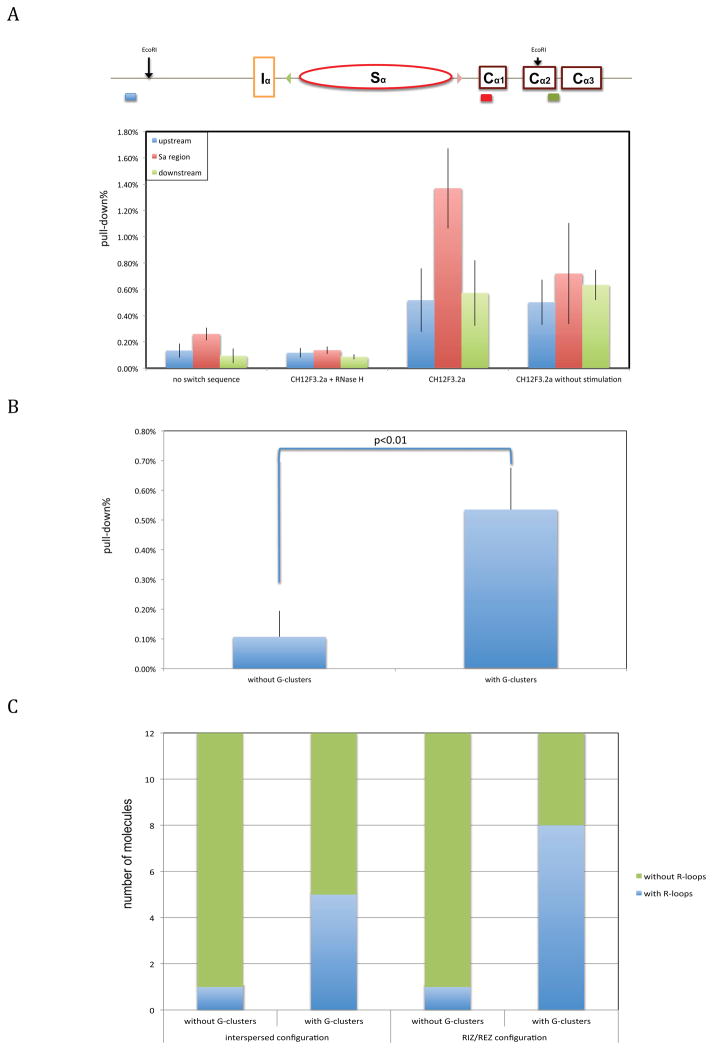
An antibody has been described which appears to bind RNA:DNA hybrids preferentially over DNA:DNA or RNA:RNA of the same sequence (Hu et al., 2006; Phillips et al., 2013), and we employed this antibody to detect R-loops (Ginno et al., 2012; Skourti-Stathaki et al., 2011; Stirling et al., 2012; Sun et al., 2013; Wahba et al., 2011). We stimulated cells of interest with cytokines for 24 hrs before the genomic DNA was harvested and digested with EcoRI for immunoprecipitation.
We initially compared wild type CH12F3.2a cells under different stimulation conditions and cellular clones that lack any switch sequences at the endogenous Sα locus. Due to the highly repetitive nature of switch regions, the most proximal region to Sα that is in common among all of the constructs was chosen for the qPCR detection (Table S3) of the Sα locus in the IP. In the CH12F3.2a group, the IP signal peaks in the fragment containing the Sα region, which indicates that R-loops are enriched in switch regions relative to neighboring DNA. Without stimulation, this signal drops approximately 50%, which agrees with the drop in their GLT (data not shown). All signals decrease significantly if genomic DNA is pretreated with RNase H, which specifically degrades RNA:DNA hybrids. All of the IP signals in the no-switch sequence group are relatively low, suggesting lower levels of R-loop formation (Figure 3A).
Figure 3. Effect of G-Clusters on R-Loop Formation within Cells.
A. Immunoprecipitation with the S9.6 antibody was performed on wild type CH12F3.2a cells and one mutant with no switch sequences at the endogenous Sα locus. The amplicons of qPCR are shown in different colors, corresponding to the bars on the bottom. Background signals from mock samples with no antibody were subtracted. Values were normalized to the total input DNA to calculate the pull-down percentage. Three independent IP experiments were performed for each cell line.
B. IP was performed on cellular constructs containing 14 repeats with and without G-clusters (the middle group in Figure 1B) using the S9.6 antibody. Four independent IP experiments were performed for each cellular construct. The pull-down percentage of the mutant with no switch sequence was subtracted as background. The Student’s t test was used to analyze the effect of G-clusters.
C. Bisulfite sequencing analysis was done on the cellular constructs containing 14 repeats with and without G-clusters, or the cellular constructs in Figure 2B with and without G-clusters. The y-axis gives the number of IgH alpha locus alleles, out of 12 examined (duplicates were eliminated), that have long (>60 bp) regions of C to T conversion due to bisulfite. Actual patterns of conversion along each allele are shown in Figures S5 and S6.
Having shown that G-clusters increase R-loop formation in a biochemical system (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010), we wondered whether this is true in the cellular system. We tested cells with 14 repeats of 80 bp synthetic switch sequence at the endogenous Sα locus. These repeats have the interspersed configuration (Figure 1B). We found that cells with G-clusters give us a consistently higher signal at the Sα locus than those without G-clusters, which indicates more R-loop formation when G-clusters are present in the switch sequence (Figure 3B).
To further examine whether G-clusters affect R-loop status within the switch regions, we did bisulfite sequencing for the same cellular constructs at the synthetic Igα switch regions (Table S4). When G-clusters are present, five out of twelve molecules have long stretches of C conversion on the non-template DNA strand (Figures 3C and S5). However, without G-clusters, this ratio drops to one out twelve. In addition to the interspersed configuration, we also wanted to evaluate the effect of G-clusters on R-loop formation in the RIZ/REZ configuration (shown in Figure 2). We found that eight out of twelve of the molecules have long stretches of conversion when G-clusters are present in the RIZ, and only one out of twelve when G-clusters are not present in the RIZ (Figure S6). Therefore, G-clusters lead to increased R-loop formation within the cells at this locus.
Strategy for Testing WGCW and CSR Efficiency
One of the central features of Ig switch regions we sought to understand is why they are so long. We define the total switch region length as the summation of the length of all of the repeat units (plus any RIZ). We plotted CSR as a function of the switch repeat number to investigate if the total class switch region length plays a role in CSR. We find that with identical repeat units, CSR correlates with total switch region length (Figure S2), as others have found using a murine knock-in system (Zarrin et al., 2005).
A longer switch region could affect CSR in either of two ways. Greater length could (A) provide more WGCW sites or (B) permit longer R-loops to form, thereby providing a greater length of ssDNA. In the RIZ/REZ configuration, R-loops merely initiate within the RIZ, whereas the REZ, with its high G-density, permits elongation of the R-loop to the end of the switch region (Effect B). In considering the contribution of WGCW sites to switch region strength (Effect A), there are three related factors: the number of WGCW sites, the density of WGCW sites, and the total switch region length. The total number of WGCW sites equals the number of WGCW sites within each repeat unit (the density of WGCW sites) times the repeat number (total switch region length divided by 80 bp).
Our assay system allows us to determine which of these factors contribute to CSR efficiency. Only one of the three factors can remain constant while the other two vary; therefore, three different scenarios were devised, and switching efficiency was measured.
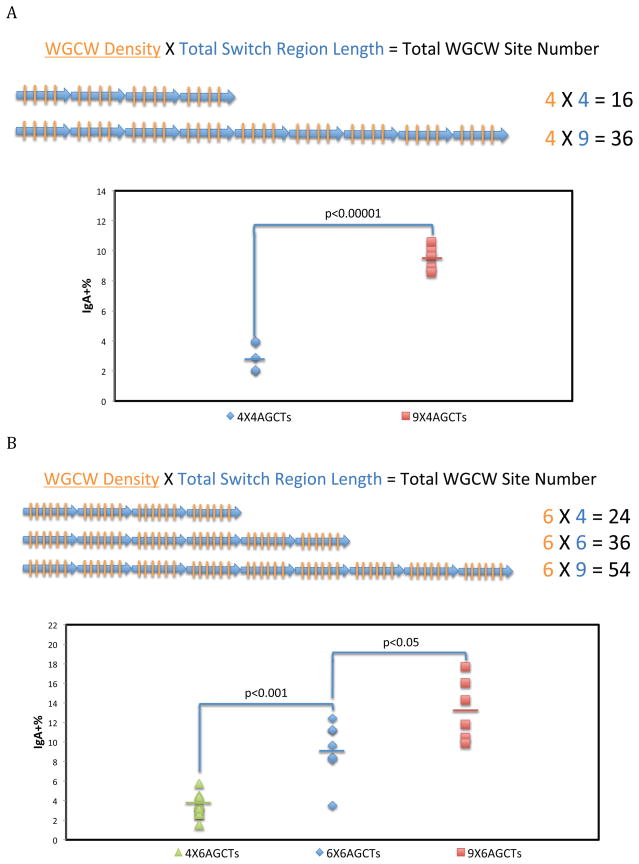
Constant WGCW Density But Varied Total Switch Region Length and Total WGCW Site Number
We first examined the effect of switch region length and total WGCW site number when the G-clusters are only located at the beginning of the switch region (RIZ/REZ configuration). In the earlier biochemical studies, we showed that a high G-density in the REZ could maintain R-loops (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). If this applies within cells, then with a few G-clusters at the beginning of a switch region in an RIZ and a high overall G-density in the REZ, longer switch regions would be expected to generate higher CSR. We constructed two cell lines with a single 23 bp RIZ containing 3 G-clusters and different total REZ lengths (four versus nine 80 bp synthetic Sα repeats containing 4 AGCT sites and alternating G nucleotides in each repeat, but no G-clusters). We found that 9 repeats result in significantly higher CSR compared to only 4 repeats (almost 3-fold)(Figure 4A).
Figure 4. Constant WGCW Density But Varied Total Switch Region Length and Total WGCW Site Number.
Each blue arrow and orange bar represents one repeat unit and one AGCT site, respectively.
A. An REZ consisting of four or nine 80 bp repeats with 4 AGCT sites each was constructed immediately downstream of an RIZ with three G-clusters. Each dot represents an independent clone. Five independent cellular clones of each cellular construct were analyzed by FACS. The bars represent the mean of each group. The Student’s t test was used to analyze the effect of switch region length. In the equation at the top of the panel, the underlined parameter is kept constant, and the remaining two parameters vary (this applies to Figures 5 and 6 also).
B. An REZ of four, six or nine 80 bp repeats with 6 AGCT sites was constructed immediately downstream of an RIZ containing five G-clusters. Six to ten independent cellular clones of each cellular construct were analyzed by FACS.
To further evaluate this, three constructs with the same REZ repeat unit were created, but with progressively greater numbers of switch repeats: 4, 6 and 9 repeats, each repeat containing 6 AGCT sites (Figure 4B). As in the case above (Figure 4A), we observe an increase in CSR with increased total switch region length and total WGCW number (Figure 4B).
Therefore, we have shown that with the same repeat unit (i.e., density of WGCW sites), CSR can increase if the total switch region length and total number of WGCW sites increase, suggesting that either total switch region length or the total numbers of WGCW sites within the region are important for switching.
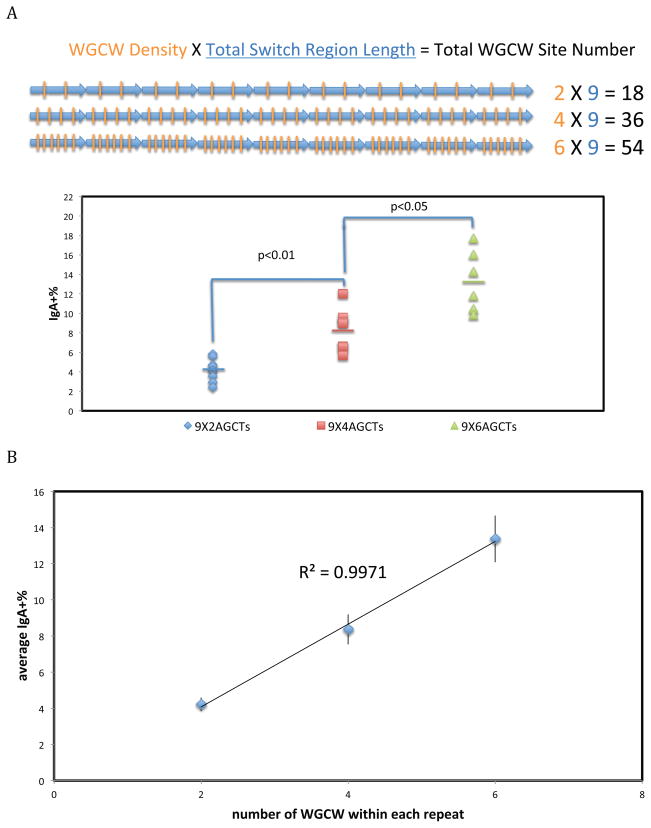
Constant Total Switch Region Length But Varied WGCW Density and Total WGCW Site Number
Next, we wanted to determine if WGCW number was the most important aspect of Effect A. We kept the RIZ constant and varied the WGCW number within each 80 bp REZ repeat. Each switch region in this comparison has nine REZ repeats (Figure 5A). Cells were screened and analyzed as previously described. When there are only two WGCW sites within each 80 bp repeat, we find that the average CSR is 4.2% (Figure 5A). If we increase the number of WGCW sites within each repeat to 4, then the average CSR doubles to 8.4%. When we increase the number of WGCW sites to 6, the average CSR increases to 13.4% (Figure 5A). Hence, we found that with more WGCW sites within each repeat unit, CSR increases significantly. A plot of CSR against the total number of WGCW sites within each 80 bp repeat (or across the entire switch region) shows a strong positive correlation (Figure 5B). Thus, when total switch region length is held constant, CSR increases as a function of the number of WGCW sites, strongly suggesting that the number of WGCW sites is important but total switch region length is not.
Figure 5. Constant Total Switch Region Length But Varied WGCW Density and Total WGCW Site Number.
Each blue arrow and orange bar represents one repeat unit and one AGCT site, respectively.
A. REZ with the same number of 80 bp repeat units with different numbers of AGCT sites in the repeats were constructed immediately downstream of an RIZ with five G-clusters. Each data symbol represents an independent clone. Six to nine independent cellular clones of each cellular construct were analyzed by FACS. The bars represent the mean of each group. The Student’s t test was used to analyze the effect of WGCW density.
B. Average CSR values of each group were plotted against their WGCW density. Error bars represent SEM.
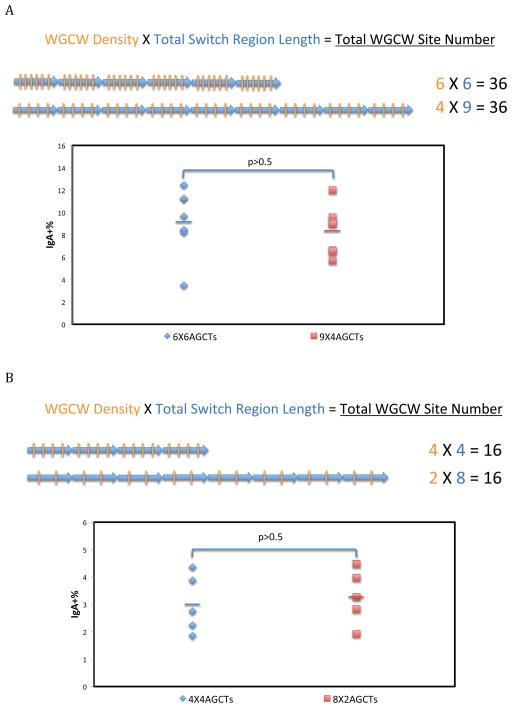
Constant Total Number of WGCW Sites But Variable WGCW Density and Switch Region Length
Having ruled out WGCW density and total switch region length, we wanted to further evaluate whether CSR is primarily dependent on total WGCW number (which, for identical repeats, as in our case, is a multiple of the WGCW site number in each repeat unit). We constructed a pair of cell lines with the same RIZ and the same total number of WGCW sites (Figure 6A). Within each synthetic switch region, we designed 6 repeats containing 6 AGCT sites each, for a total of 36 WGCW sites. We compared this to 9 repeats containing 4 AGCT sites each, for a total of 36 WGCW sites. Even though the 9 repeat switch region has a longer total switch region length (9 × 80 = 720 bp versus 6 × 80 = 420 bp), we found no significant difference in CSR (9.2% versus 8.4%).
Figure 6. Constant Total Number of WGCW Sites But Variable WGCW Density and Switch Region Length.
Each blue arrow and orange bar represents one repeat unit and one AGCT site, respectively. FACS analysis of CSR is shown. The same total number of WGCW sites were constructed immediately downstream of an RIZ containing 5 G-clusters, as shown on the bottom left. Both switch region length and WGCW density are different.
A. Six repeats of 80 bp oligonucleotides with 6 AGCT sites or nine repeats of 80 bp oligonucleotides with 4 AGCT sites were constructed downstream of an RIZ containing 5 G-clusters.
B. Four repeats of 80 bp oligonucleotides with 4 AGCT sites or eight repeats of 80 bp oligonucleotides with 2 AGCT sites were constructed downstream of an RIZ containing 6 G-clusters. Each data symbol represents an independent clone. Five to eight independent cellular clones were shown in each group. The color bars represent the mean of each group. The Student’s t test was used to analyze the effect of WGCW number.
In order to further evaluate this point, we made two additional cell lines with the same total number of WGCW sites as one another, but with different WGCW densities and total switch region lengths. Either 4 repeats with 4 WGCW sites (16 total WGCW site number) or 8 repeats with 2 WGCW sites (16 total WGCW site number) were integrated at the Sα locus. We found no difference in CSR (Figure 6B). Therefore, the total number of WGCW sites, rather than their density or the total switch region length determines CSR (Effect A).
Regarding R-loop length (Effect B), while R-loop initiation (RIZ) is important, the total length of the REZ (Figures 5 and 6), and hence, the total switch region length, are not important. We found that with the same RIZ, the R-loop distribution is similar among REZs that we have tested (Table S5). Hence, within the total switch region lengths studied here, as long as numerous WGCW sites are present within an R-loop zone, the length over which that R-loop is distributed is not critical (Figure 7).
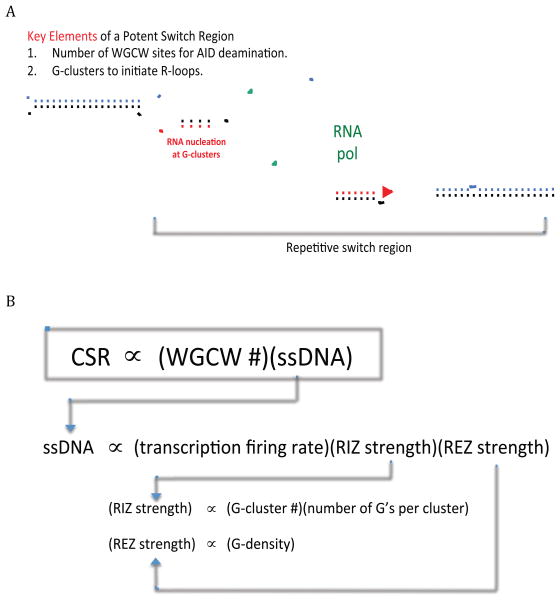
Figure 7. Model of CSR.
A. Features of typical Ig class switch recombination regions. AID is the key enzyme that initiates CSR. WGCW sites are the preferred DNA sequence motif targets of AID, but AID only recognizes cytosine deamination sites within ssDNA. The G-clusters initiate R-loops in mammalian switch regions and are proposed to provide the ssDNA for AID action. We have shown that G-clusters are important for efficient R-loop initiation within cells.
B. Proposed relationships in CSR. CSR efficiency is in direct proportion to the total number of WGCW sites and the amount of ssDNA. The amount of ssDNA is proportional to the transcription firing rate, RIZ strength and REZ strength. RIZ strength is a function of the total G-cluster number and the G nucleotide number within each G-cluster. REZ strength is dependent on overall G-density, which determines the length of the R-loop. Naturally-occurring switch regions have interspersed G-clusters, WGCW sites, and a high G-density. Hence, for naturally-occurring R-loops in cells, the ssDNA length (R-loop length) varies depending at which G-cluster the R-loop initiates (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010).
DISCUSSION
Here we define the major factors that determine switch region strength. AID requires WGCW sites to efficiently create the C deamination that begins the DNA breakage process. But these WGCW sites must be within ssDNA for AID to recognize them as substrates (Bransteitter et al., 2003; Pham et al., 2011; Yu et al., 2004). For SHM, transcription might inefficiently provide ssDNA to permit the relatively slow process of SHM to occur over weeks during an immune response. But CSR occurs typically within a few days in mammals. We have proposed that some of the more rapid AID action in CSR in mammals is due to the high G-clustering and G-density that is the distinctive feature of all mammalian IgH switch regions (Yu et al., 2003; Yu and Lieber, 2003) and not present in organisms that evolved CSR earlier, such as avians and amphibians (Hackney et al., 2009; Zhu et al., 2012). These considerations lead to the following questions. First, are the G-clusters that we define in our biochemical studies as R-loop initiation zones (RIZ) also important for cellular CSR? Second, can the G-clusters of the RIZ be separated from the G-dense R-loop elongation zone (REZ), or must these be interspersed, as they have evolved in mammals? Third, can the R-loop initiation zones (RIZ) be separated from the WGCW sites, which are located in the REZ in the RIZ/REZ configuration? Fourth, is total length of the switch region more important to provide a longer R-loop (i.e., ssDNA) or to provide a larger number of WGCW sites? Fifth, is it the total number of WGCW sites that is important; or is it the density of these sites or the length over which the WGCW sites are distributed that is the key determinant? Sixth, how do the answers to these questions relate to the function of IgH switch regions, to their evolution, and to earlier vertebrate switch regions that do not have G-clusters or do not have a high G-density?
G-Clusters and Switch Region Strength
Biochemical data had shown that G-clusters are extremely important to initiate the thread-back of the RNA onto the DNA template, which thereby initiates R-loop formation (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). Here, we have demonstrated that G-clusters are a very important determinant of whether a switch region leads to CSR, predominantly by causing R-loop formation. The G-clusters can be located at the leading edge of the switch region, thereby leading to R-loop formation that can allow downstream WGCW sites to be within a zone of ssDNA (and therefore be substrates for AID). Or the G-clusters can be interspersed among the WGCW sites, as they are with all mammalian switch regions. We find that both of these configurations can support CSR.
We have not extensively tested for variation in the number of G’s in each G-cluster. But most of our G-clusters contain three G’s. We have noted previously that any clustering of G’s appears to be sufficient, including GGG and GGGG. GGG clusters are not likely to form stable G-quartets (Sinden, 1994). We have previously noted that G-quartet formation is not necessary for R-loop formation in biochemical systems, where we have formally ruled them out in R-loop formation (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). G-quartet formation at switch regions also is unlikely within B cells for additional reasons that we have described elsewhere (Huang et al., 2007). We acknowledge that the G-clusters may contribute to Ig SCR in additional ways other than R-loop formation.
Number of WGCW Sites as a Determinant of Switch Region Strength
Previous work using this system showed that WGCW is the key DNA sequence required for CSR (Han et al., 2011). Building on this observation, we have varied the number and density of WGCW sites and have found that the number of WGCW sites is more critical than the density of these sites, at least for total switch region lengths of less than 1 kb.
Mammalian switch regions have evolved to be long. The longer switch regions typically provide more WGCW sites and more G-clusters, but the total switch region length itself (independent of WGCW sites and G-clusters) does not appear to be critical for CSR efficiency.
For SHM, WGCW sites must be in a ssDNA state, at least transiently, to be a substrate for AID. Transcription through random DNA (containing few or no G-clusters) appears to be able to induce such transient ssDNA states, though inefficiently. Once AID causes a U:G mismatch at a WGCW site, this is converted to a nick via UNG and APE (DiNoia and Neuberger, 2007; Masani et al., 2013). In biochemical systems, R-loops can form when a nick is present in the non-template strand of the DNA, even when there are no G-clusters and the G-density is low (Roy et al., 2010). This may be the basis for how AID can act during SHM, despite a lack of G-clusters.
AID acts more rapidly in CSR than in SHM. For CSR studied here, the total number of WGCW sites is critical, regardless of the length over which the R-loop extends. Hence, a given number of WGCW sites yields a certain level of CSR, regardless of whether the WGCW sites are spread over twice the length of DNA in the switch region. This indicates that the number of WGCW lesion sites is the rate-limiting factor for CSR, as long as they are located within the R-loop zone. The distance between WGCW sites (WGCW density) does not seem to be critical, and increased total length of the switch region is important only to the extent that it provides a greater number of WGCW sites.
Evolution of G-Clusters for CSR Efficiency
Given that both interspersed G-clusters and a separated zone of G-clusters (RIZ) are sufficient, why did the interspersed configuration evolve for mammalian switch regions? This may simply be a consequence of the mechanism by which switch regions became repetitive. DNA expands over evolutionary time via unequal sister chromatid exchange (Smith, 1974, 1976; Strachen and Read, 1999). Such an expansion requires that each repeat unit be nearly identical. This means that each 25 to 80 bp repeat unit in a mammalian switch region would harbor all of the key elements for CSR: WGCW sites, G-clusters, and an overall high G-density to permit R-loop elongation since the first repeat unit had to contain all of these elements. If each repeat unit carries its own G-clusters, then an interspersed configuration would be the result, once all of the repeats are assembled end to end. After sufficient evolutionary expansion of the switch regions, sequence divergence of the repeats from one another may then be selected for in order to minimize deletion of the repeats by homologous recombination within a switch region. Therefore, the repeats became not quite so identical over evolutionary time, since they only need to retain the three key mechanistic features: a high number of WGCW sites, G-clusters, and a high G-density.
Transcription Through R-loop Regions
We find that G-clusters increase CSR but typically reduce transcription. This is precisely what we see when studying R-loop formation in a biochemical system using T7 RNA polymerase and naked DNA templates (Daniels and Lieber, 1995). With each round of transcription, a new RNA polymerase must displace the RNA from the existing R-loop in order to progress through that region. Movement of the RNA polymerase through an R-loop would be expected to be slower due to the increased difficulty of separating three nucleic acid strands rather than only two.
We also observe that with the same repeat unit, a greater number of such repeats, namely, a longer switch region, causes a decrease in transcription (Figures S3 and S4). This is likely because the RNA polymerase movement through R-loops is slower than through duplex DNA, and longer R-loops are more difficult than shorter ones for the RNA polymerase.
When we plotted the transcription values against the CSR efficiency of individual cellular clones, the distribution is random (Figure S4). This suggests that above a certain level of transcription, there is no direct correlation between CSR and GLT, supporting previous observations (Dunnick et al., 2004).
Concluding Comments
Our studies here show that Ig switch region strength is dependent on R-loop formation and on the number of WGCW sites within the R-loop region. The R-loop formation requires transcription and a switch region with a nontemplate DNA strand that is rich in clusters of G nucleotides and an overall high G-density. The G-clusters can be interspersed with the WGCW sites, just as they are in naturally-occurring switch regions; or the G-clusters can be limited to the upstream edge of the WGCW-rich zone, as long as this WGCW-rich zone also has a high G-density to support elongation of the R-loop. Therefore, while the total number of WGCW sites is important in the context of our experiments, this number of WGCW sites must be located within an R-loop forming region.
EXPERIMENTAL PROCEDURES
Cell Culture and CSR Assay
CH12F3.2a and its derivative cells were cultured in 10%FCS RPMI medium supplemented with 50 μM β-mercaptoethanol. As for CSR assay, healthy cells in log phase were seeded at 5×104 cells/ml in medium with 1μg/ml anti-CD40 (eBioscience #16-0404-86), 5ng/ml IL-4 (R&D #404-ML-010) and 0.5ng/ml TGF- β1 (R&D #240-B-002), and grown for 72h. Cells were stained with FITC-conjugated anti-mouse IgA antibody (BD #559354) and analyzed by flow cytometry. CSR efficiency was determined by the percentage of IgA+ cells.
Plasmid Construction
The 80 nt oligonucleotides were 5′-phosphorylated and purified on a denaturing polyacrylamide gel. A pair of complementary oligonucleotides was annealed and self-ligated at a final concentration of 4μM. Ligation products were resolved on agarose gel, and DNA with expected length was recovered, blunted with Klenow and cloned into the exchange vector backbone. The sequence was confirmed by DNA sequencing.
Cellular Targeting and Screening
Five micrograms exchange vector and one microgram Cre-expression vector were cotransfected into 1F7 cells by electroporation (Lonza). Transfected cells were serially diluted and seeded in 96-well plates. After 72h, ganciclovir (Sigma-Aldrich #G2536-100MG) was added at a final concentration of 2μg/ml. At 7 days after transfection, single clones were picked for puromycin sensitity test at a final concentration of 1μg/ml. Puromycin-sensitive clones were screened by PCR around upstream and downstream boundaries of LoxP sites and across the entire switch region. A subset of clones were also examined by Southern blot. At least five clones were used for CSR assay.
Germ-Line Transcript Quantification
Two million healthy cells at a density around 1×106 cells/ml were supplemented with anti-CD40, IL-4 and TGF-β1 for 6h, and total RNA was extracted with GenElute™ Mammalian Total RNA Kit (Sigma-Aldrich #RTN350). 10% of RNA was reverse transcribed into cDNA with M-MuLV Reverse transcriptase (NEB M0253S), and 10% of the RT products were analyzed with real-time PCR. β-Actin was used as an internal control. Each sample was done in duplicates, and at least three independent cellular clones were analyzed for each construct.
S9.6 Purification
TCC HB-8730 hybridoma line (generously provided by Bradley Cairns) was cultured in a CELLine 1000 bioreactor (Satorius Biotech, NY) according to manufacturer’s instructions. Harvested antibody (culture supernatant) was purified on a column packed with Protein G Sepharose 4 Fast Flow (GE Healthcare) equilibrated with 1x phosphate buffered saline (PBS).
S9.6 Immunoprecipitation
Healthy cells in log phase were seeded at 3×105 cells/ml in medium with anti-CD40, IL-4 and TGF-β1 and grown for 24h. Genomic DNA was prepared by overnight proteinase K digestion, phenol-chloroform extraction and ethanol precipitation. Genomic DNA was fragmented with EcoRI; importantly, RNase A was added at this step to prevent S9.6 antibody binding to RNA species in subsequent steps (Phillips et al., 2013). Five microgram fragmented genomic DNA was incubated with 5μg S9.6 antibody in 400μl IP buffer (10mM sodium phosphate (pH 7.0)[0.195ml 1M NaH2PO4, 0.61ml 0.5M Na2HPO4/50ml], 140mM NaCl [1.4ml NaCl/50ml], 0.1% Tween 20[250μl 20%/50ml]) for 2h at 4 °C. Ten microliter pre-blocked Dynabeads (Invitrogen 10004D) were added into the mixture and gently rotated at 4°C. After 2h, beads were washed with IP buffer three times, and treated with proteinase K overnight. DNA bound to the beads was recovered by phenol-chloroform extraction, and quantified by real-time PCR.
Bisulfite Sequencing
One million healthy cells at a density around 1×105 cells/ml were supplemented with anti-CD40, IL-4 and TGF-β1 for 48h, and genomic DNA was extracted. Five micrograms genomic DNA was incubated with the bisulfite solution (Lightning Conversion Reagent by Zymo Research D5030) for 16h at 37°C, and recovered as described by the manufacturer. The switch regions were amplified with one converted primer and one native primer by PCR, and subcloned into the pGEM-T Easy vector (Promega A1360). At least twelve clones from each switch region were randomly chosen and analyzed.
Supplementary Material
Highlights.
Ig switch region strength is dependent on R-loop formation.
Ig switch region strength is dependent on the total number of WGCW sites.
R-loop formation requires a nontemplate DNA strand that is rich in G-clusters.
G-clusters can be at the upstream edge of or interspersed throughout the switch region.
Acknowledgments
We thank former lab member, Dr. Deepankar Roy (Genentech), for initial advice and guidance, and Dr. Ray Mosteller for comments on the manuscript. This work was supported by NIH grants to MRL and KY. ZZZ was supported initially by a USC Provost’s Fellowship. NRP is supported in part by The John H. Richardson Fellowship through the ARCS Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baaklini I, Usongo V, Nolent F, Sanscartier P, Hraiky C, Drlica K, Drolet M. Hypernegative supercoiling inhibits growth by causing RNA degradation. J Bacteriol. 2008;190:7346–7356. doi: 10.1128/JB.00680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli S, Rallu F, Sanscartier P, Cerritelli SM, Crouch RJ, Drolet M. Effects of RNA polymerase modifications on transcription-induced negative supercoiling and associated R-loop formation. Mol Microbiol. 2004;52:1769–1779. doi: 10.1111/j.1365-2958.2004.04092.x. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucl Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNoia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Ann Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci. 2003;8:d210–221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Hertz GZ, Scappino L, Gritzmacher C. DNA sequence at immunoglobulin switch region recombination sites. Nucl Acid Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick WA, Shi J, Graves KA, Collins JT. Germline transcription and switch recombination of a transgene containing the entire H chain constant region locus: effect of a mutation in a STAT6 binding site in the gamma 1 promoter. J Immunol. 2004;173:5531–5539. doi: 10.4049/jimmunol.173.9.5531. [DOI] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney JA, Misaghi S, Senger K, Garris C, Sun Y, Lorenzo MN, Zarrin AA. DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination. Adv Immunol. 2009;101:163–189. doi: 10.1016/S0065-2776(08)01005-5. [DOI] [PubMed] [Google Scholar]
- Han L, Masani S, Yu K. Overlapping activation-induced cytidine deaminase hotspot motifs in Ig class-switch recombination. Proc Natl Acad Sci U S A. 2011;108:11584–11589. doi: 10.1073/pnas.1018726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, Hsieh CL, Lieber MR. Sequence-dependence of chromosomal R-loops at the immunoglobulin heavy chain Smu class switch region. Mol Cell Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FT, Yu K, Hsieh CL, Lieber MR. The downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci. 2006;103:5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masani S, Han L, Yu K. Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol Cell Biol. 2013;33:1468–1473. doi: 10.1128/MCB.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata H, Tomizawa J. A mechanism of formation of a persistent hybrid betwen elongating RNA and template DNA. Cell. 1990;62:331–338. doi: 10.1016/0092-8674(90)90370-t. [DOI] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand V, Anant S, Sugai M, Kinoshita K, Davidson N, Honjo T. Specific Expression of Activation-Induced Cytidine Deaminase (AID), a Novel Member of the RNA-Editing Deaminase Family in Germinal Center B Cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Murphy KM. Janeway’s Immunobiology. 8. Garland Science; 2012. [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes to cells: devoted to molecular & cellular mechanisms. 2012;17:218–233. doi: 10.1111/j.1365-2443.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, Lingner J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013;32:2861–2871. doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Calabrese P, Park SJ, Goodman MF. Analysis of a single-stranded DNA-scanning process in which activation-induced deoxycytidine deaminase (AID) deaminates C to U haphazardly and inefficiently to ensure mutational diversity. J Biol Chem. 2011;286:24931–24942. doi: 10.1074/jbc.M111.241208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, Leysath CE. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit. 2013;26:376–381. doi: 10.1002/jmr.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR. Competition Between the RNA Transcript and the Nontemplate DNA Strand During R-Loop Formation In Vitro: A Nick Can Serve as a Strong R-loop Initiation Site. Mol Cell Biol. 2010;30:146–159. doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR. DNA Structure and Function. San Diego: Academic Press; 1994. [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Unequal crossover and the evolution of multigene families. Cold Spring Harb Symp Quant Biol. 1974;38:507–513. doi: 10.1101/sqb.1974.038.01.055. [DOI] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Snapper CM, Finkelman FD. Immunoglobulin Class Switching. In: Paul WE, editor. Fundamental Immunology. Philadelphia: Lippincott-Raven; 1999. pp. 831–861. [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachen T, Read AP. Human Molecular Genetics. 2. New York: Wiley-Liss; 1999. [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabl M, Steinberg C. Affinity maturation and class switching. Curr Opin Immun. 1996;8:89–92. doi: 10.1016/s0952-7915(96)80110-5. [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsurawat T, Jenjaroenpun P, Kwoh CK, Kuznetsov V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012;40:e16. doi: 10.1093/nar/gkr1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol Cell. 2014;53:484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation induced deaminase activity at cytidine. J Biol Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- Yu K, Lieber MR. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair. 2003;2:1163–1174. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Yu K, Roy D, Bayramyan M, Haworth IS, Lieber MR. Fine-structure analysis of activation-induced deaminase accessbility to class switch region R-loops. Mol Cell Biol. 2005;25:1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin AA, Tian M, Wang J, Borjeson T, Alt FW. Influence of switch region length on immunoglobulin class switch recombination. Proc Natl Acad Sci U S A. 2005;102:2466–2470. doi: 10.1073/pnas.0409847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, Hsu E. Origin of immunoglobulin isotype switching. Curr Biol. 2012;22:872–880. doi: 10.1016/j.cub.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.