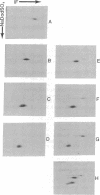
Abstract
A mitochondrial mutation that genetically maps in the middle of the gene coding cytochrome c oxidase subunit II has been found to be a single-base-pair deletion. Three independently isolated spontaneous revertants of this mutant have different single-base-pair insertions within 15 nucleotides of the mutation. These findings clearly identify the location of the gene and suggest that the mutation causes a frame-shift. The sequence of about 900 base pairs surrounding the mutation has been determined and found to have several chain termination codons in every possible reading frame. The sequence can, however, be translated in one frame by assuming that the codon TGA does not cause chain termination in yeast mitochondira, as was recently suggested for the human organelle [Barrell, B. G., Bankier, A. T. & Drouin, J. (1979) Nature (London), in press]. If TGA codes for tryptophan residues, as is apparently the case in human mitochondria, a polypeptide can be read from the yeast mtDNA that is identical to bovine cytochrome oxidase subunit II at 37.8% of its residues. Furthermore, the DNA sequences of the frame-shift revertants discussed above predict relative isolectric point differences between the wild-type and various revertant forms of the polypeptide. The detection of these isolectric point differences by two-dimensional electrophoresis of subunit II from the various strains independently confirms the presumed reading frame of the gene. It is concluded that TGA is translated in yeast mitochondria, most probably as tryptophan.
Keywords: frame-shift revertants, DNA sequence, genetic code, protein sequence homology
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borst P., Grivell L. A. The mitochondrial genome of yeast. Cell. 1978 Nov;15(3):705–723. doi: 10.1016/0092-8674(78)90257-x. [DOI] [PubMed] [Google Scholar]
- Brenner S., Barnett L., Katz E. R., Crick F. H. UGA: a third nonsense triplet in the genetic code. Nature. 1967 Feb 4;213(5075):449–450. doi: 10.1038/213449a0. [DOI] [PubMed] [Google Scholar]
- Cabral F., Schatz G. Identification of cytochrome c oxidase subunits in nuclear yeast mutants lacking the functional enzyme. J Biol Chem. 1978 Jun 25;253(12):4396–4401. [PubMed] [Google Scholar]
- Cabral F., Solioz M., Rudin Y., Schatz G., Clavilier L., Slonimski P. P. Identification of the structural gene for yeast cytochrome c oxidase subunit II on mitochondrial DNA. J Biol Chem. 1978 Jan 10;253(1):297–304. [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Bedbrook J. R., Bogorad L., Rich A. Maize chloroplast DNA fragment encoding the large subunit of ribulosebisphosphate carboxylase. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5487–5491. doi: 10.1073/pnas.74.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G., Tzagoloff A. Assembly of the mitochondrial membrane system. DNA sequence of subunit 2 of yeast cytochrome oxidase. J Biol Chem. 1979 Sep 25;254(18):9324–9330. [PubMed] [Google Scholar]
- Douglas M. G., Butow R. A. Variant forms of mitochondrial translation products in yeast: evidence for location of determinants on mitochondrial DNA. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1083–1086. doi: 10.1073/pnas.73.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Kafatos F. C., Maniatis T. The primary structure of rabbit beta-globin mRNA as determined from cloned DNA. Cell. 1977 Apr;10(4):571–585. doi: 10.1016/0092-8674(77)90090-3. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Sequence of the lacI gene. Nature. 1978 Aug 24;274(5673):765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Genetic and physical analysis of the mitochondrial gene for subunit II of yeast cytochrome c oxidase. J Mol Biol. 1979 May 5;130(1):63–82. doi: 10.1016/0022-2836(79)90552-7. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Identification of phage SP01 proteins coded by regulatory genes 33 and 34. Nature. 1976 Aug 26;262(5571):748–753. doi: 10.1038/262748a0. [DOI] [PubMed] [Google Scholar]
- Hartley M. R., Wheeler A., Ellis R. J. Protein synthesis in chloroplasts. V. Translation of messenger RNA for the large subunit of fraction I protein in a heterologous cell-free system. J Mol Biol. 1975 Jan 5;91(1):67–77. doi: 10.1016/0022-2836(75)90372-1. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C. UGA mutations and UGA suppressors in yeast. Biochimie. 1976;58(1-2):179–182. doi: 10.1016/s0300-9084(76)80368-9. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Grivell L. A., Borst P., Bos J. L. Nucleotide sequence of the mitochondrial structural gene for subunit 9 of yeast ATPase complex. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1663–1667. doi: 10.1073/pnas.76.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol. 1971 Jun 14;58(2):439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Macino G., Coruzzi G., Nobrega F. G., Li M., Tzagoloff A. Use of the UGA terminator as a tryptophan codon in yeast mitochondria. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3784–3785. doi: 10.1073/pnas.76.8.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system: partial sequence of a mitochondrial ATPase gene in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979 Jan;76(1):131–135. doi: 10.1073/pnas.76.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman R. B., Tzagoloff A. Breakage of yeast: a method for processing multiple samples. Anal Biochem. 1975 Apr;64(2):545–549. doi: 10.1016/0003-2697(75)90466-2. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Poyton R. O., Schatz G. Cytochrome c oxidase from bakers' yeast. III. Physical characterization of isolated subunits and chemical evidence for two different classes of polypeptides. J Biol Chem. 1975 Jan 25;250(2):752–761. [PubMed] [Google Scholar]
- Prunell A., Bernardi G. The mitochondrial genome of wild-type yeast cells. VI. Genome organization. J Mol Biol. 1977 Feb 15;110(1):53–74. doi: 10.1016/s0022-2836(77)80098-3. [DOI] [PubMed] [Google Scholar]
- Roth J. R. UGA nonsense mutations in Salmonella typhimurium. J Bacteriol. 1970 May;102(2):467–475. doi: 10.1128/jb.102.2.467-475.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonimski P. P., Tzagoloff A. Localization in yeast mitochondrial DNA of mutations expressed in a deficiency of cytochrome oxidase and/or coenzyme QH2-cytochrome c reductase. Eur J Biochem. 1976 Jan 2;61(1):27–41. doi: 10.1111/j.1432-1033.1976.tb09994.x. [DOI] [PubMed] [Google Scholar]
- Steffens G. J., Buse G. Studies on cytochrome c oxidase, IV[1--3]. Primary structure and function of subunit II. Hoppe Seylers Z Physiol Chem. 1979 Apr;360(4):613–619. [PubMed] [Google Scholar]
- Stewart J. W., Sherman F., Jackson M., Thomas F. L., Shipman N. Demonstration of the UAA ochre codon in bakers yeast by amino-acid replacements in iso-1-cytochrome c. J Mol Biol. 1972 Jul 14;68(1):83–96. doi: 10.1016/0022-2836(72)90264-1. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Weber K. A single UGA codon functions as a natural termination signal in the coliphage q beta coat protein cistron. J Mol Biol. 1973 Nov 15;80(4):837–855. doi: 10.1016/0022-2836(73)90213-1. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Gette W. R., Furth M. E., Nomura M. Effects of ribosomal mutations on the read-through of a chain termination signal: studies on the synthesis of bacteriophage lambda O gene protein in vitro. Proc Natl Acad Sci U S A. 1977 Feb;74(2):689–693. doi: 10.1073/pnas.74.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]