Abstract
The binding of the TRIM5α restriction factor to the HIV capsid is mediated by the C-terminal SPRY domain of TRIM5α. Atomic-level details of this host-pathogen interaction, which involves mobile variable loops of the SPRY domain, remain unclear. Some of the key determinants of restriction are encompassed by the long and disordered v1 loop of the SPRY domain. We applied molecular modeling to elucidate the conformational repertoire of the v1 loop and its role in the interaction with the capsid. All-atom replica exchange molecular dynamics revealed multiple transient, interconverting states of the v1 loop consistent with the intrinsic disorder observed experimentally. The docking of the SPRY conformations representing 10 most populated states onto the high-resolution model of the assembled HIV-1 capsid revealed that a subset of v1 conformations produced plausible binding poses, in which the SPRY domain binds close to the pseudo-2-fold symmetry axis and the v1 loop spans the interhexamer gap. Such binding mode is well supported by the NMR binding data and known escape mutants. We speculate that the binding mode that involves interaction of the capsid with a subset of preexisting SPRY conformations arising from the intrinsic disorder of the v1 loop may explain the remarkable ability of TRIM5α to resist viral evasion by mutagenesis and to restrict divergent retroviruses.
TRIM5α is a restriction factor that blocks retroviral replication following entry of the virus into the cytoplasm of the target cell.1 TRIM5α binds to the assembled capsid protein of the mature viral core, and this molecular recognition event determines the specificity of TRIM5α-mediated restriction and contributes to the host tropism of primate immunodeficiency viruses.2–6 The human variant of TRIM5α has low affinity for the HIV capsid and is a poor restrictor of the HIV retrovirus, whereas TRIM5α of the rhesus monkey binds HIV capsid with higher affinity and displays very potent HIV restriction.3,5 The host–pathogen interface formed by TRIM5α and the retroviral capsid is subject to strong positive selection resulting from antagonistic coevolution of retroviruses and their primate hosts.7 The protein segments under positive selection map onto the C-terminal SPRY domain, which is the capsid-binding module of TRIM5α.2–4,7
The individual SPRY domains bind the capsid with low affinity, and capsid recognition depends on the avidity effect arising from TRIM5α oligomerization.8–13 High-avidity protein-protein interactions are refractory to structural studies and pose significant experimental challenges because contributions of oligomerization and individual epitope binding to the overall binding affinity cannot be easily separated. Here, we focus on the interaction of the isolated SPRY domains with the capsid, which, albeit weak, are key determinants of the specificity and affinity of the TRIM5α-capsid binding. The structure of the rhesus TRIM5α SPRY domain has recently been determined by X-ray crystallography and NMR,14,15 and two atomic-resolution reconstructions of the fully assembled HIV capsid are also available.16,17 However, it remains poorly understood how the two proteins come together to form the restriction-competent complex because the interfaces of the two proteins contain flexible segments, making it difficult to predict the relative orientation of the binding partners and to interpret mutagenesis data. Particularly, the long v1 loop within the SPRY domain, which displays very high variability in primates and is critical for SPRY–capsid interactions, is highly mobile.14
The long and mobile v1 loop of SPRY displays significant NMR chemical shift perturbations upon binding to the capsid,8,14 but detailed understanding of the v1 conformational change upon binding is lacking. The question of the v1 involvement in the SPRY-capsid interactions is probably best approached in the framework developed for studies of intrinsically disordered proteins (IDPs), proteins which do not possess a well-defined fold but instead can exist in multiple structural states with certain probabilities and, thus, are better described as ensembles of structures.18-21 Disordered protein regions commonly undergo a disorder-to-order transition and adopt defined conformations upon binding to their interaction partners, but this is not a strict requirement, and some proteins can remain largely unstructured in the bound state.22,23 For weak protein-protein interactions steric and electrostatic complementarity becomes less stringent and the complex may no longer be represented by a single well-defined structure but should instead be described as an ensemble of bound states.24 Such binding can be determined by a property not attributable to specific amino acid residues, as observed, for example, in the interaction of the intrinsically disordered transactivation domain of the Ewing's sarcoma oncoprotein (EAD) with its target. 25 Interestingly, no single-residue mutation within the v1 loop of the rhesus monkey SPRY domain has been shown to eliminate binding to the HIV-1 capsid, which may be a direct consequence of intrinsic disorder at the SPRY–capsid interface. Gaining mechanistic insight into the mode of SPRY-capsid interaction and understanding where it fits on the spectrum of binding modes observed for IDPs may facilitate interpretation of the mutagenesis data and inform future functional studies.
In this study, we investigate conformational properties of the mobile capsid-binding surface of the rhesus TRIM5α SPRY using replica exchange molecular dynamics (REMD) simulations, an extension of the conventional molecular dynamics technique that allows for accelerated and more complete coverage of the conformational space of a protein.26,27 For example, conformational sampling of a 21 residue-long peptide Fs-21 was 35.1 times faster with REMD over conventional MD.28 REMD has become the method of choice for problems involving major conformational rearrangements and disorder-to-order transitions, such as chaperone-assisted folding,29 coupled folding and binding of intrinsically disordered proteins,30 and binding of proteins to short disordered peptides.31 REMD may prove particularly powerful for the SPRY domain modeling because, on the one hand, the resulting trajectories can be validated using available NMR-derived relaxation and nuclear Overhauser effect (NOE) data, whereas on the other, REMD trajectories can provide a much more exhaustive picture of the conformational repertoire of the SPRY domain than the conventional simulated-annealing approaches. Simulated-annealing calculations that use NMR-derived distance and angle restraints are not optimal in the regions with significant disorder because the NOE restrains are usually sparse in the mobile protein segments and may arise from distinct protein conformations. In this work, the conformational space sampled by the SPRY domain was explored by calculating 100 replica trajectories of 100 ns in a 200 K temperature window in explicit solvent. The derived conformational repertoire of the intrinsically disordered v1 loop was then evaluated by docking of the 10 most-populated states onto the surface of the assembled HIV capsid. We find that a subset of transient conformations adopted by the free SPRY domain in solution can produce plausible complexes that are in good agreement with experimental data. We suggest that the remarkable resilience of the SPRY domain to capsid mutations may arise from its ability to bind to the capsid using several distinct conformations of the v1 loop.
Materials and Methods
MD and REMD
Molecular dynamics calculations were performed with GROMACS 4.5.3 software using resources of the University of Texas Health Science Center at San Antonio (UTHSCSA) and the Texas Advanced Computing Center (TACC)(Alamo and Lone Star clusters, respectively). We selected the ff99sb-ildn-NMR32 variant of the original Amber ff99 force filed33 with improved the φ/ψ dihedral 32,34,35 and side chain torsion potentials for Ile, Leu, Asp, and Asn residues.35 This force field scored high when tested against a suite of 524 chemical shift and J coupling ((3)JH(N)H(α), (3)JH(N)C(/β), (3)JH(α)C′, (3)JH(N)C′, and (3)JH(α)N) measurements on dipeptides, tripeptides, tetra-alanine, and ubiquitin. The lowest-energy conformation from the family of NMR structures (PDB ID: 2LM3)14 was used as the starting structure for the simulation. The 205-residue protein contains 3206 atoms. The protein was immersed into a triclinic box with a minimal distance of 1 nm to box wall. Initially, 500 steps of steepest descent energy minimization were conducted to remove steric clashes. The box was filled with 8291 SPC water molecules and 26 pairs of Na+ and Cl− ions to mimic physiological conditions. The resulting system of 28 131 atoms was energetically minimized for additional 500 steps. The system's temperature was then gradually raised to 300 K using 1 K per 1 ps step. Solvent was equilibrated in a 5 ns molecular dynamics calculation with the protein-heavy atoms harmonically restrained to their positions in NPT ensemble. Pressure of 1 atm was controlled with Parinello-Rahman algorithm and temperature kept fixed with v-rescale. The PME method was used to treat electrostatics beyond the cutoff of 0.9 nm, and grid spacing of 0.16 nm was applied. For van der Waals interactions the same 0.9 nm cutoff was applied and the neighbor list was updated every 5 steps. The stochastic integrator with a 2 fs time step was used in all MD calculation. The resulting SPRY conformation was used as input for both, the conventional and replica exchange MD. Conventional molecular dynamics simulations were performed as 10 independent trajectories of 100 ns each and snapshots were saved every 2 ps. For the replica exchange calculation we selected a temperature window of 200 K. T-REMD server37 was used to generate the temperature distribution of the 100 replica within the 300-501.18 K temperature window. Exchange probability was set to 0.25 and exchange attempts were made every 1000 steps (2 ps). Trajectories of 100 ns were calculated for each of the 100 replicas, totaling in 10 μs of the cumulative REMD trajectory.
Docking
The HIV capsid lattice of the entire conical mature viral core is heterogeneous, consisting of hexamers and pentamers that form surfaces of distinct curvature. 38,39,16,17 In vitro, the capsid protein can be assembled into cylindrical structures devoid of pentamers with the surface curvature similar to the one observed on the lateral surfaces of the conical viral cores. In the cosedimentation studies, the TRIM5α binding to the cylindrical in vitro assemblies correlates with the TRIM5α restriction activity.3,5 Therefore, we used the relatively low curvature of the lateral surface of the cone for our docking studies.
The SPRY domain was docked onto the segment of the assembled capsid surface formed by three neighboring capsid hexamers related by the pseudo-3-fold local symmetry axis in the assembled structure. The trimer of hexamers contains all possible relative arrangements of epitopes on the capsid surface that could potentially be recognized by the SPRY domain. We used the crystal structure40 of the capsid hexamer (PDB ID: 3H4E, chains A-F) for each of the three hexamers. The trimer of hexamers was constructed by fitting Cα atoms of the prepared hexamer described above to the coordinates of three hexamers on the lateral surface of the all atom model of the fully assembled HIV-1 cone.16
Protein–protein docking that accounts for backbone and side chain flexibility is beyond current computational capabilities for systems of this size, so the capsid surface had to be kept rigid in the docking calculations. Thus, the main challenge in constructing the assembled capsid surface for the docking studies was posed by the most flexible element of the HIV-1 capsid: the cyclophilin A (CypA) binding loop located between helices 4 and 5 of the N-terminal domain.41,42 TRIM5α SPRY domain could theoretically be in contact with as many as 9 cyclophilin-binding loops on the surface of the assembled capsid model, so accounting for multiple different loop conformations was not possible and we were limited to a single conformation. Inspection of the available X-ray structures of the HIV-1 CA NTD in which the loop electron density is resolved revealed that there is one conformation that is adopted most frequently (Supporting Information Figure S1).40,43-45 This observation suggested that this conformation, even if only marginally stable, is predominant on the surface of the assembled capsid, and it was selected for our docking studies. Therefore, the same conformation of the CypA binding loop (3H4E, chain A) was used in every CA monomer within the hexamer.
The SIVmac239 capsid surface was constructed using the crystal structure of the SIVmac239 N-terminal domain.46 Electron density for the P88 in the CypA binding loop is missing in the SIVmac239 CA-NTD crystal, and both possible P88 isomers were modeled. The Schrodinger Maestro program was used to build the missing proline residue into the loop in the cis and trans conformations and an energy minimization was performed. Superposition on the original conformation from the crystal structure revealed that only the trans conformation was consistent with the electron density observed in the crystal and it was used to construct the capsid surface. Outside of the cyclophilin binding loop, the SIVmac239 capsid is virtually identical to the HIV ortholog, so we used the assembled HIV capsid as the template and only replaced the N-terminal domains with the SIVmac239 CA-NTD structure.
Rigid protein–protein docking was performed with Piper47 through Bioluminate interface of the Schrodinger suite. A total of 70 000 orientations were sampled, and 30 poses per ligand were saved. The poses were scored using the following criteria: (1) SPRY domain has to be oriented relative to the capsid in such a way that the interaction is primarily mediated by the variable loops v1 through v3, (2) the SPRY domain has to interact with more than one hexamer, (3) the v1 loop has to span the interhexamer gap, (4) the pose is scored depending on how well it agrees with mutagenesis and NMR titration data, and (5) the pose is scored depending on how frequently it occurs.
Results and Discussion
Amino Acid Composition of the v1 Loop Is Not Typical for Flexible Protein Loops
NMR relaxation properties of the v1 loop (S324–C349) of SPRY domain from rhesus TRIM5α indicate a high degree of intrinsic disorder and mobility.14 However, the amino acid composition of the SPRY v1 is distinct from what is usually observed in disordered regions: the 5 aromatic residues (Y331, F338, F340, F346, and Y348) located in the loop constitute 19% of its total amino acid content (Figure 1, bottom panel). Such large aromatic content is characteristic for stable protein folds48 and, in contrast, not commonly observed in the intrinsically disordered proteins or regions.49 Histidine 312 is sterically adjacent to Y348 and is located between v1 stems and points its side chain into the loop interior. Comparison of the v1 loop composition to the amino acid content in disordered regions of proteins50 reveals that 35% of amino acids in the v1 loop are order promoters, whereas only 38% are disorder inducers (Figure 1). Distribution of the order-promoting residues is not uniform. With the exception of I329 and Y331, all order-promoting residues are located in the C-terminal segment of the loop (P334–C349), whereas in the N-terminal part disorder-promoting residues dominate. This observation agrees with the intrinsic disorder prediction algorithms, which return relatively low disorder probabilities for v1, with higher values observed within N-terminal portion of the loop (Supporting Information Figure S2).
Figure 1.
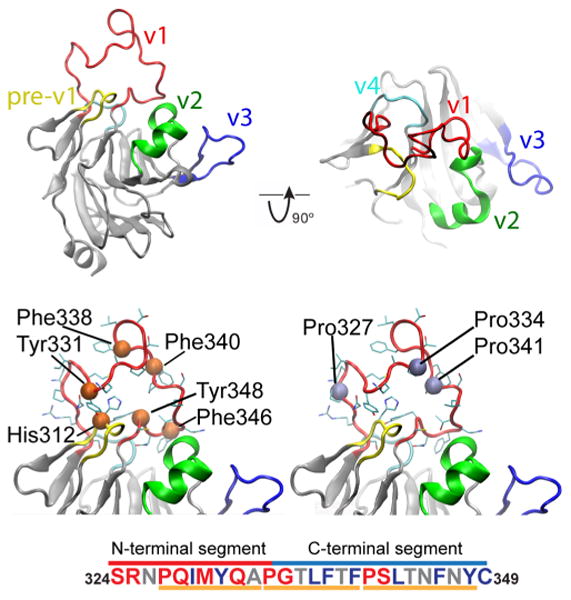
The structure of the rhTRIM5α SPRY domain. (a) Cartoon representation of the rhesus SPRY domain structure with the variable capsid-interacting loops highlighted. The close-up box shows one representative v1 conformation from the family of NMR structures. (b) Location of aromatic and prolines residues within v1 loop. With the exception of Y331, all aromatic residues are located in the C-terminal segment of the loop. Residue H312 from the pre-v1 segment is located in-between the v1 loop stems, adjacent to Y348, and acts as a key interfacial residue between the v1 loop and the core of the SPRY domain. (c) Amino acid composition of the v1 loop. Residues are colored according to their contribution to order/disorder.50 Red, promoting disorder; blue, promoting order; gray, low promoting capability. N-terminal part is enriched with residues that confer disorder, whereas C-terminal is abundant with order promoting residues. The three segments that can undergo unfold-to-fold transitions are denoted below sequence.
The length of the v1 loop (26aa) is within limits of minimal folds (e.g., Trp-cage, BBA5, WW domain, etc.), so theoretically, a metastable v1 conformation is feasible through ordered π–π interactions or hydrophobic collapse. The stems of the loop are located adjacent to each other (less than 10 Å apart), which provides high conformational freedom with many possibilities for backbone folding and side chain packing. Three v1 proline residues (P327, P334, and P341) divide the loop into three almost equal segments (P327– A333, P334–F340, and P341 – Y349) and render formation of extended secondary structure elements within v1 unfavorable. The length of these segments is similar to that of short linear motifs (SLiMs)—peptide stretches implicated in binding of IDPs to their targets.51,52 Notably, the v1 loop of a related TRIM21 protein adopts a well-defined and stable conformation determined by a network of hydrogen bonds and hydrophobic interactions with the adjacent v2 loop.53 In summary, the analysis of the v1 amino acid content suggests that, despite the observed disorder, the v1 loop may be capable of forming defined transient conformations, which can be further stabilized by the interaction with the binding partner.
Replica Exchange Molecular Dynamics Reveals Multiple Shallow Energetic Minima on the Free Energy Surface of V1 Conformations
In order to gain insight into the nature of v1 motions and to characterize the ensemble of v1 conformations, we performed replica exchange molecular dynamics (REMD) simulations in explicit solvent. To accomplish this, we calculated 100 ns REMD trajectories for 100 replica within the 200 K temperature window (see Materials and Methods). The observed random walk of the replica within the temperature window indicates sufficiently high exchange rate needed for efficient energy barrier crossing on the free energy landscape (Supporting Information Figure S3). For further analysis we used the lowest temperature (300 K) REMD trajectory.
Dihedral principal component analysis (dPCA)54,55 is a convenient approach to analyze conformational distribution in internal coordinates as it naturally separates internal from overall motion. This is of particular importance to study free energy landscapes of biomolecules undergoing large structural rearrangements. We applied dPCA to derive free energy profile of the v1 loop by projecting of the trajectory onto its first two principal components (Figure 2). The map reveals multiple energetic minima separated by continuous shallow areas. The basins are only up to ∼2.5 kcal/mol deep, which is not sufficient to confer stability to those states. From the derived free energy map, the conformational dynamics of the v1 loop can be interpreted as fluctuations within these basins with frequent transitions between them. This pattern seems to be indicative of structural disorder as has been observed with other intrinsically disordered proteins studied with REMD. 56-58
Figure 2.
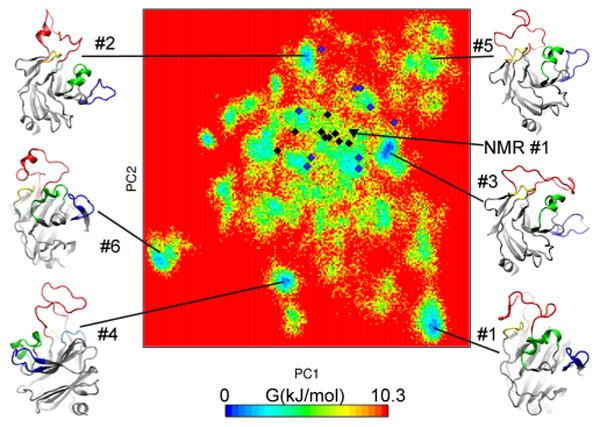
Free energy landscape of the v1 loop projected into first two main principal components. Conformations of the first six centroids are are linked to corresponding energetic basins. Location of NMR conformations (2LM3) shown in black diamonds and the starting NMR conformation is black triangle. Final conformations from 10 conventional MD simulations are in purple diamonds. Conventional MD SPRY has explored only the shallow part of the free energy surface.
In order to analyze population distribution, we performed structural clusterization of the v1 loop conformations with a cutoff 0.25 nm. Distribution of cluster sizes did not reveal a prevalent conformation: the top five clusters accommodated 7.37%, 4.94%, 4.90%, 4.69%, and 4.64% of total conformations (Supporting Information Figure S4a). A total of 38 clusters had sizes of at least 10% of the top one, accumulating together 80.7% of all conformations. Similarly, the distribution of eigenvalues from the dPCA analysis reveals that 22 eigenvectors have eigenvalues at least 10% of that of the first vector (Supporting Information Figure S4b). Collectively these data further support the intrinsically disordered nature of the v1 loop.
In order to evaluate the convergence of the trajectory, we analyzed the distribution of a set of interatomic distances within the v1 loop throughout the simulation. (Supporting Information Figure S4c). Similar distance distribution profiles in the initial and the final halves of the full trajectory are indicative of acceptable convergence of the REMD simulation. Furthemore, analysis of conformations in the most populated clusters reveals that these energetic basins were revisited multiple times along the trajectory (Supporting Information Figure S4d), another indicator of good convergence.
v1 Loop in Populated Clusters Is Stabilized by Hydrogen Bonding and Hydrophobic Interactions
Figure 2 shows the representative conformations (centroids) of the top six structural clusters. The backbone conformations and the side chain packing are markedly different in the six centroid structures consistent with the significant separation of the clusters on the free energy map. Inspection of the structures reveals that the conformations are stabilized either through secondary structural elements or the hydrophobic collapse of the side chains.
Among the top 10 clusters, four (#2, #6, #9, and #10) contain elements with helix-like hydrogen bonding of consecutive residues (Supporting Information Figure S5a). Residual secondary structure was observed in three distinct v1 segments: P327–A333, P334–F340, and P341–Y348, with the highest probability detected in the central segment P334–F340 (Figure 3). Disorder-to-order transition is commonly observed in intrinsically disordered proteins upon association with their binding partners. For example, intrinsically disordered nuclear coactivator binding domain (NCBD) of the transcriptional activator CBP has three regions with residual α-helical signatures, albeit largely disordered in the unbound state.57 Upon interaction with either of the two structurally distinct NCBD binding partners, CABD and IRF-3, all three helices are fully stabilized.59,60 Thus, we speculate that residual secondary structure observed in the REMD simulation of the free SPRY domain may be important for recognition of the HIV capsid.
Figure 3.
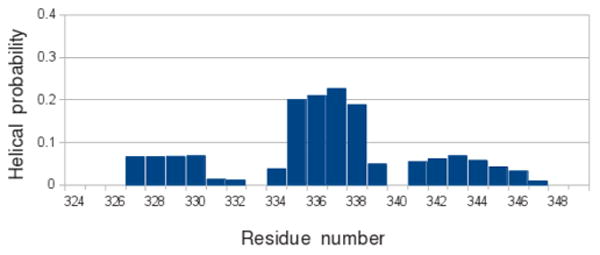
Helix propensity of the v1 loop residues. The prolines of the v1 loop divide it into three segments of equal size: P327–A333, P334–F340 and P341–Y348. The central segment P334–F340 displays the highest helix propensity.
Another observed stabilization mechanism in the v1 loop is the formation of a local hydrophobic core in which aromatic residues act as key contributors. 61,62 Side chains of H312 and Y348 are located at the interface between the loop and the protein and nucleate the hydrophobic collapse. Depending on particular cluster representatives, contributing amino acids varied to include other aromatic (Y331, F340), apolar (I329, M330), and positively charged residues (R325) to introduce π–π, liphophilic, or cation–π interactions into the core, respectively. The participating amino acids are distributed throughout the v1 loop; therefore, their involvement in the core prevent it from adopting extended conformations. For example, in cluster #1, the aromatic core includes residues H312, R325, Y331, and Y348, which constrains most of the v1 loop (Supporting Information Figure S5b). Additionally, the tip of the loop (residues adjacent to P334) is locked through hydrogen bonding with N309 in the pre-v1 segment. H312, located between v1 loop stems, is particularly important as it is observed to nucleate interactions in most clusters with hydrophobic collapse. The majority of its interactions is with aromatic residues, but hydrogen bonding is also observed in one of the structures. H312 was identified as one of the residues undergoing positive selection in TRIM5α evolution,7 consistent with its contribution to the v1 conformational repertoire that we observe in our study. Other residues commonly observed in this position in primate TRIM5α variants are cysteine and tyrosine, both of which are similar to histidine in their ability to mediate hydrophobic interactions as well as participate in hydrogen bonding.
v1 Loop Is Not Uniformly Flexible
The stems of the v1 loop are in close proximity to each other; thus, the loop flexibility is unrestrained by the attachment points and is solely determined by its amino acid composition. In order to characterize the v1 disorder and its propensity for hydrophobic collapse, we looked at the radial distribution function (RDF) commonly used in the statistical analysis of chaotic systems. We used the three proline residues (P327, P334, and P341) as reference points in the v1 loop and calculated the RDFs between Cγ atoms each of the three prolines and the loop base (geomteric center of Cγ atoms of H312 and Y348) (Figure 4). The RDFs display two major peaks at r1 = 0.44 nm and r2 = 0.9 nm, which can be interpreted as direct van der Waals contact or through another atoms layer, respectively. RDF of the N-terminal P327 reveals nearly equal probability of the r1 and r2 states, whereas the C-terminal P341 displays a marked preference for the fully collapsed state. These observations are consistent with the distribution of aromatic residues within the v1 and with the disorder probabilities predicted by the disorder-prediction algorithms (Supporting Information Figure S2). The strong tendency toward hydrophobic collapse observed in the C-terminal segment of v1 may be functionally important because mutation of five consecutive residues (NFNYC) in the C-terminus of v1 to alanine displays a very strong defect in HIV restriction and capsid binding.8,14
Figure 4.
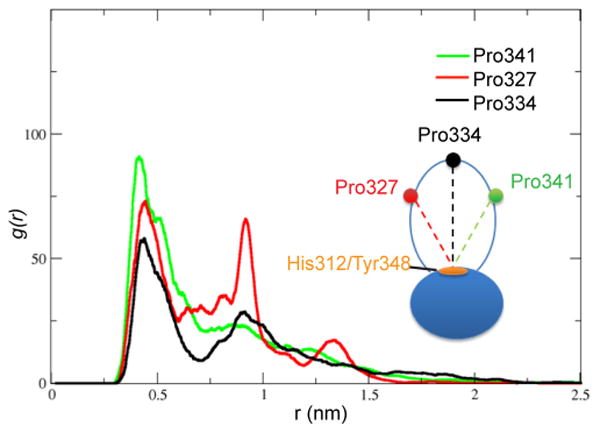
Radial distribution functions of P327 (red), P334 (black), and P341 (green) with respect to H312/Y348. P327, P341, and P334 represent the N terminus, the C terminus, and the tip of the v1 loop respectively. Fluctuations are higher in the N-terminal segment than in the C-terminal one.
Evaluation of the Energy Function Using X-ray and NMR Data
We performed 10 100 ns-long MD simulations of the same system that was used for the replica exchange calculations to assess flexibility of the v1 loop under conventional regime. We observed dissimilar dynamics in all 10 simulations, which is reflected in root-mean-square fluctuations profiles of Cα atoms (Figure 5a). In some systems (#1 or #6), v1 has reached metastable states, whereas most others showed unconstrained fluctuations. Such v1 behavior agrees with nearly radial distribution of the final conformations with respect to the starting NMR state on the FES map (Figure 2).
Figure 5.
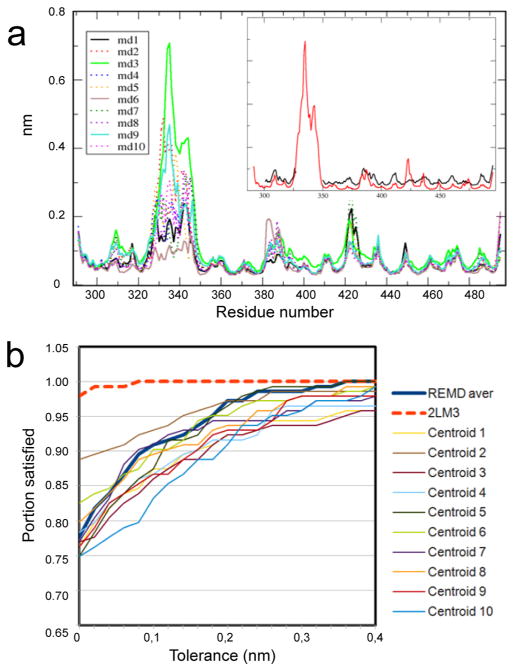
MD and REMD trajectories display good agreement with experimental B factors and NOE measurements. (a) Root mean square fluctuations (nanometers) of Cα atoms of rhSPRY observed in the 10 conventional MD simulations. The inset panel shows B factor calculated by averaging over the MD trajectories (red) compared to the values of obtained from the X-ray data (black). (b) NOE violations within v1 loop observed in the 10 centroid structures of the REMD simulation.
Despite high flexibility in conventional MD simulations, the trajectories provide only sparse coverage of the energy surface (see legend of the Figure 2). The v1 loop of SPRY is a particularly challenging system because the abundance of aromatic side chains and hydrogen bond donors/acceptors results in multiple possibilities for intraloop contacts and slows down the sampling of the accessible free energy surface. The data illustrate advantages of the REMD approach for simulation of long and mobile protein segments.
We used trajectories from 10 conventional MD simulations to analyze per residue fluctuations and calculate average B factors in order to compare with B factors determined from the 1.55 Å resolution X-ray diffraction data set14 (Figure 5a). All residues except variable loops have exhibited similar flexibility. We observe a very good correspondence between experimental and theoretical B factors, which indicates appropriate selection of the force field parameters.32 The discrepancy in the 419–425 region is caused by the changes in the hydrogen bonding involving the backbone amide of G420 that we observe in the simulation, which is probably impeded in the crystal.
The conformational ensemble derived from the REMD simulation was evaluated using experimental pairwise distance restrains derived from nuclear Overhauser effect (NOE) NMR spectroscopy. We used distance restraints obtained from NOE measurements for V1 protons located at least one residue apart (NOE (i, i + n) with n > 1) and compared them to the distances between the same protons derived from the REMD simulations (Figure 5b). The pairwise distances averaged over 10 centroids showed good correspondence to experimental data: 78% of the NOE contacts are satisfied, and 91% fall within 1 Å tolerance.
Then, we analyzed the NOE violations (Supporting Information Supplementary Table 1). Of the 143 long-range NOE contacts, 13 are violated by more than 1 Å on average over the 10 centroid structures. However, when these NOEs are analyzed for each centroid, only one NOE is violated by more than 1 Å in all 10 centroids, whereas the other NOEs are within 1 Å in at least one centroid. The most likely explanation of this observation is that these NOEs arise from just one particular transient conformation of v1. This is a well-known phenomenon frequently observed in the NMR studies of short peptides in solution, which complicates interpretation of NOE data because NOE contacts that arise from distinct conformations should not be used simultaneously in a structural calculation. Notably, one of the three most violated averaged NOEs (L337HD2–T339HG2) is only satisfied in the first, most populated centroid. Another indication that NOE signals may arise from distinct v1 conformations comes from the distribution of the calculated NMR structures on the free energy surface map (Figure 2). Taking into account that principal component metrics on the dPCA map are conformationally linked,54,55 a structure that is calculated to satisfy NOE contacts arising from distinct conformations are expected to be located close to the geometric center of the projections of these conformations onto the dPCA map. This is roughly what we observe for the v1 loop of the SPRY domain. The possibility of distinct conformations observed in our computational analysis should be taken into account when analyzing the NOE data for the SPRY domain variants. Analysis of REMD trajectories may provide a valuable tool for sorting NOE signals into sets that correspond to distinct conformations.
Docking of the SPRY Domain onto the HIV-1 CA Surface
Our next goal was to evaluate conformations of the SPRY domain obtained from the REMD studies for their complementarity with the surface of the assembled capsid using molecular docking. Computational protein–protein docking provides an unbiased approach for scoring, sorting, and identifying most compatible structures from an ensemble of conformations. However, the limitations of the force fields, pose scoring, and computational capabilities make accurate docking of weak protein–protein interactions challenging, and the predicted binding poses can by no means be considered high-resolution binding models. The main limitation of this study is the inability to fully account for protein flexibility at the binding interface. The conformational repertoire of the SPRY domain was represented by the 10 centroid structures representing the most populated energetic basins derived from REMD. However, the capsid surface was represented by a single model. The most commonly occurring conformation of the flexible cyclophilin-binding loop was used for all capsid monomers in the assembled capsid structure (see Materials and Methods). The SPRY conformers were docked onto the segment of CA surface formed by three neighboring CA hexamers that was constructed to approximate the curvature observed on the lateral surface of the conical HIV capsid. The docked poses were evaluated using five criteria based on existing experimental data for SPRY–capsid interactions (see Materials and Methods). A total of 33 out of the 300 poses satisfied the first four criteria. Among these, we found 17 complexes in which the orientation of the SPRY domain was similar, with multiple poses observed for three SPRY centroids (#1, #6, and #10). This binding mode is analyzed in more detail below.
The gaps between neighboring CA-NTD hexamers in the assembled capsid form deep crevices in the outer surface of the mature viral core. There are two major openings in the interhexamer space, one centered at the 2-fold symmetry axis of the hexagonal lattice (2-fold gap) and the other at the 3-fold symmetry axis (3-fold gap). The 2-fold gap is a deep, tunnellike opening, whereas the 3-fold gap is more shallow and rounded with its three sides lined by residues of the C-terminal end of helix 4 and the N-terminal end of the CypA-binding loop (Figure 6a). In the selected docked poses the SPRY domain binds such that the v2 and v3 loops are positioned in the 2-fold gap making contacts with two neighboring CA-NTD units within one hexamer and the v1 loop spans the 3-fold gap making contacts with the CypA-binding loop of the neighboring hexamers (Figure 6b). Superposition of the poses from the three centroids shows that the difference between the docked poses is mainly in the conformation of the v1 loop, as v2 and v3 loops occupy similar positions in the 2-fold gap.
Figure 6.
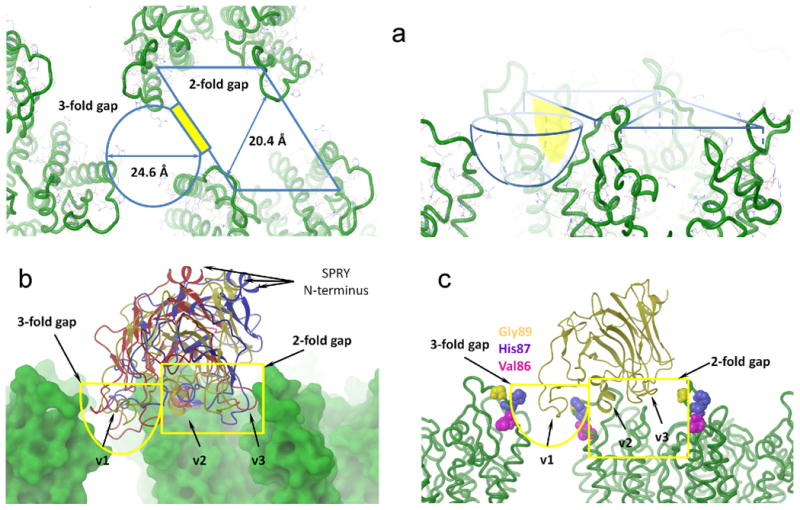
Docking of the SPRY domain onto the capsid surface. (a) Crevices on the surface of the HIV-1 CA lattice. The 2-fold gap has rhombic shape while the 3-fold gap is conical. There is an opening between the two gaps highlighted in yellow. (b) The selected binding mode of SPRY. Surfaces of two HIV-1 hexamers are shown in green. Centroids #1, #6, and #10 are shown as cartoons. The v1 loop bound to the 3-fold gap, whereas the v2 and v3 loops fit to 2-the fold gap. (c) Solvent exposed residues, mutations of which in the HIV-1 CA compromise restriction of the rhesus TRIM5α.
The observed poses agree well with the models of SPRY– capsid binding based on the NMR titration data,8 which suggests that the v1 loop is most likely involved in forming interhexamer contacts. They are also consistent with mutagenesis data available for the capsid–TRIM5α interaction from a number of studies.46,63–72 The CypA-binding loop lines the surfaces of the 2-fold and 3-fold gaps occupied by the SPRY domain in the docked poses described above, which explains the deleterious effect of mutations in the loop on TRIM5α restriction. For example, residues V8663,69 and H8766,69 from the CypA-biniding loop were found to be important for resistance emerging (Figure 6c). Another class of resistant mutations identified includes residues in helix 6 and its immediate vicinity.69,72 For example, a recently published selected evolution study describes HIV-1 CA variants that escape TRIM5α restriction without an associated fitness defect.72 G116E and M96I mutations identified in this study are of particular interest because these residues also display enhanced NMR broadening in the in vitro titration studies.8 These mutations are expected to affect relative orientation of the cyclophilin-binding loop and helix 6 of the capsid. In our docked poses, helix 6 makes contacts with the v2 loop of the SPRY domain, so any relative movement of these two distinct CA epitopes involved in the SPRY-CA interaction would compromise binding affinity.
Docking of the SPRY Domain onto the SIVmac CA Surface
To further evaluate the utility of protein–protein docking for studies of TRIM5α–capsid binding we performed docking of the rhesus SPRY domain onto the SIVmac239 capsid. SIVmac239 CA was selected because it is not restricted by the rhesus TRIM5α, and the high resolution structure of SIVmac239 CA-NTD has recently been determined.46 The model of the SIVmac239 lattice was constructed by replacing the CA-NTD domain in the HIV-1 model with the SIVmac structure (see Materials and Methods). The backbone conformations of the two CA-NTD domains are virtually identical with the exception of the cyclophilin-binding loop, the amino acid composition of which is significantly different in the two proteins (SIVmac239, 85QPAPQQGQLREPS97 and HIV-1, 86VHAGPIAPGQMREPR100). Docking of the same 10 most-populated conformers of the rhesus SPRY that were used for HIV-1 CA docking returned only four poses that satisfied the first four selection criteria (see Materials and Methods). Three out of the four poses were formed by the centroid #4 and represent the same binding mode, which is somewhat similar to the one observed with the HIV-1 capsid. The significant difference in the occurrence of the plausible binding poses between the SIVmac239 capsid (1%) and the HIV-1 capsid (5.6%) suggests that protein–protein docking can distinguish between restricted and nonrestricted capsids despite the limitations of the rigid-body approximation. There are two factors that contribute to the different outputs of the docking calculation for the two capsids. First is the different conformation of the cyclophilin-binding loop in the two capsids, which brings about differences in the overall steric complementarity between the CypA binding loop and the SPRY. The correlation between restriction and the number of plausible binding poses may suggest that, despite the mobility of the cyclophilin-binding loop its conformations found in the crystals are the more stable ones and are most abundant on the surface of the assembled capsid. The second factor is the electrostatic complementarity between the SPRY domain and the capsid surface, which may contribute to the selectivity of TRIM5α restriction. The outer surface of the HIV-1 capsid in the vicinity of the suggested binding site is populated with hydrophobic and charged amino acids (Supporting Information Figure S6). The corresponding patch on the SIVmac239 variant is enriched with glutamines, which do not strongly contribute to neither hydrophobic nor to charge interactions. V86Q and H87Q mutations in the HIV-1 capsid contribute to the escape from the restriction by the rhesus TRIM5α,69 whereas the 89QQ90 to LPA substitution in the SIVmac239 capsid sensitizes it to restriction by the rhesus TRIM5αTFP variant.46,71 We speculate that the glutamines can attenuate contacts with the largely hydrophobic v1 loop of the rhesus SPRY domain, thus weakening the interaction.
Interactions of IDPs with their binding partners can be divided into two broad classes based on their mechanism: the conformational selection, where the bound-like conformation preexists in the ensemble of conformations of the free protein,73–75 and the coupled folding and binding, where the bound conformation is largely determined by the interaction with the binding partner.76 Our data suggest that the interaction with the capsid mediated by the intrinsically disordered surface of the SPRY domain is a variant of conformational selection, because several preexisting SPRY conformations produce docked poses that are in good agreement with experimental data. Such a binding mechanism may explain some of the functional features of TRIM5α. First, the existence of several distinct conformations that are complementary to the capsid surface, but bind somewhat differently, may explain the ability of TRIM5α to resist viral evasion: a mutation on the capsid surface may disrupt binding of one preexisting SPRY conformation from the binding subset but is not likely to disrupt binding of all of them (Figure 7). Second, the mobility of the v1 loop that results in multiple divergent conformations of the interaction interface may allow recognition of distinct retroviral capsids because one subset of SPRY conformations can bind to one retroviral capsid, whereas a distinct subset of conformations can recognize a different capsid. The hypothesis that the SPRY–capsid complex can be formed with distinct conformations of the SPRY domain is attractive because it may explain some of the tantalizing properties of TRIM5α, but given the limitations of our computational study, it is nevertheless possible that the distinct poses we observe are just intermediates in the SPRY–capsid binding, which eventually coalesces into a single well-defined bound conformation.
Figure 7.
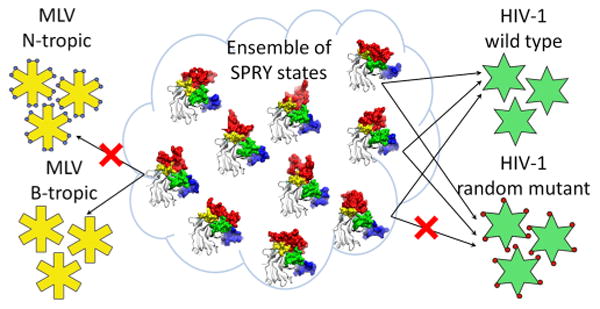
The model of capsid recognition. Ensemble of unbound states of rhesus SPRY is comprised of multiple interconverting conformations, a subset of which have good complementarity with the surface of the assembled HIV-1 capsid. SPRY conformers from this subset can all bind to the capsid, but the bound forms are distinct in their v1 conformations and the contacts they make with the CA. Random mutations on the CA surface may disrupt interaction with some of the SPRY conformers, but are not likely to disrupt interaction with all of them. A different subset of SPRY conformers from the ensemble may recognize retroviral capsids that share little amino acid similarity with the HIV capsid.
Conclusions
Rapid progress in computational power, algorithms and force fields is extending the applicability of computational tools to biological systems of ever increasing complexity. In this study, we used REMD simulations to study the conformational repertoire of the capsid-binding SPRY domain of the rhesus monkey TRIM5α restriction factor that potently blocks HIV-1 replication. It is a challenging system for exhaustive molecular dynamics calculations because the capsid-binding surface of the SPRY domain is formed by variable backbone segments of irregular structure, which include a 26 residue-long, highly mobile v1 loop. The results strongly support intrinsically disordered nature of the v1 loop. The calculated REMD trajectories and the observed conformational repertoire are in very good agreement with experimental data obtained from X-ray crystallography and NMR spectroscopy.
The docking of the most populated conformations onto the surface of the assembled HIV-1 capsid allowed identification of a plausible binding mode that is in a good agreement with mutagenesis and NMR titration data. Remarkably, the outcome of the docking calculation correlated with the known capsid selectivity of the rhesus TRIM5α SPRY domain, as the docking of the SPRY onto the nonrestricted SIVmac239 capsid returned very few plausible binding poses. Our findings are consistent with the conformational selection mechanism of the capsid recognition by the TRIM5α SPRY domains. Although molecular docking remains tentative for weakly interacting proteins, we show that it provides a useful tool for evaluating the orientation of the SPRY domain relative to distinct capsid epitopes and is useful for interpretation of the mutagenesis data. Our findings suggest that REMD may become a powerful method in the efforts to elucidate this host-pathogen interface and the molecular mechanism of TRIM5α-mediated contribution to the host-tropism of primate immunodeficiency viruses.
Supplementary Material
Acknowledgments
Authors acknowledge Dr. Nikolaos Biris for helpful discussions and Texas Advanced Computing Center (TACC) for the computational resources. This work was supported in part by the Scholar Award from the Cancer Prevention and Research Institute of Texas (CPRIT) to D.I.
Abbreviations
- TRIM
tripartite motif
- NMR
nuclear magnetic resonance
- MD
molecular dynamics
- REMD
replica exchange molecular dynamics
- HIV
human immunodeficiency virus
- CA
capsid
- IDP
intrinsically disordered proteins
- NOE
nuclear Overhauser effect
Footnotes
Notes The authors declare no competing financial interest.
Supporting Information: Figure S1 shows similarity between CypA binding loops conformations of HIV-1 capsid found in X-ray structures. Figures S2 and S3 provide additional details on the IDP-like properties of the variable loops and v1 in particular. Figure S4 provides analysis of REMD calculations. Figure S5 describes interactions that stabilize conformations of the v1 loop. Figure S6 compares exposed segments of the HIV-1 and SIMmac239 capsid monomers. The table contains NOE violation predicted from REMD simulations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5 alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song BW, Gold B, O'hUigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5 alpha exhibits line age-specific length and sequence variation in primates. J Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biris N, Tomashevski A, Bhattacharya A, Diaz-Griffero F, Ivanov DN. Rhesus Monkey TRIM5alpha SPRY Domain Recognizes Multiple Epitopes That Span Several Capsid Monomers on the Surface of the HIV-1 Mature Viral Core. J Mol Biol. 2013;425(24):5032–5044. doi: 10.1016/j.jmb.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz-Griffero F, Kar A, Perron M, Xiang SH, Javanbakht H, Li X, Sodroski J. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J Virol. 2007;81:10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J Virol. 2008;82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, Sundquist WI. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biris N, Yang Y, Taylor AB, Tomashevski A, Guo M, Hart PJ, Diaz-Griffero F, Ivanov DN. Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module. Proc Natl Acad Sci USA. 2012;109:13278–13283. doi: 10.1073/pnas.1203536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Ji X, Zhao G, Ning J, Zhao Q, Aiken C, Gronenborn AM, Zhang P, Xiong Y. Structural insight into HIV-1 capsid recognition by rhesus TRIM5alpha. Proc Natl Acad Sci USA. 2012;109:18372–18377. doi: 10.1073/pnas.1210903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graphics Modell. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 19.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 20.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature reviews Molecular cell biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 21.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourhis JM, Receveur-Brechot V, Oglesbee M, Zhang X, Buccellato M, Darbon H, Canard B, Finet S, Longhi S. The intrinsically disordered C-terminal domain of the measles virus nucleoprotein interacts with the C-terminal domain of the phosphoprotein via two distinct sites and remains predominantly unfolded. Protein science: a publication of the Protein Society. 2005;14:1975–1992. doi: 10.1110/ps.051411805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sigalov AB, Kim WM, Saline M, Stern LJ. The intrinsically disordered cytoplasmic domain of the T cell receptor zeta chain binds to the nef protein of simian immunodeficiency virus without a disorder-to-order transition. Biochemistry. 2008;47:12942–12944. doi: 10.1021/bi801602p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuxreiter M, Tompa P. Fuzzy complexes: a more stochastic view of protein function. Adv Exp Med Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 25.Song J, Ng SC, Tompa P, Lee KA, Chan HS. Polycation–π interactions are a driving force for molecular recognition by an intrinsically disordered oncoprotein family. PLoS Comput Biol. 2013;9:e1003239. doi: 10.1371/journal.pcbi.1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsutake A, Mori Y, Okamoto Y. Enhanced sampling algorithms. Methods Mol Biol. 2013;924:153–195. doi: 10.1007/978-1-62703-017-5_7. [DOI] [PubMed] [Google Scholar]
- 27.Swendsen RH, Wang JS. Replica Monte Carlo simulation of spin glasses. Phys Rev Lett. 1986;57:2607–2609. doi: 10.1103/PhysRevLett.57.2607. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Wu C, Duan Y. Convergence of replica exchange molecular dynamics. J Chem Phys. 2005;123:154105. doi: 10.1063/1.2056540. [DOI] [PubMed] [Google Scholar]
- 29.Fan H, Periole X, Mark AE. Mimicking the action of folding chaperones by Hamiltonian replica-exchange molecular dynamics simulations: application in the refinement of de novo models. Proteins. 2012;80:1744–1754. doi: 10.1002/prot.24068. [DOI] [PubMed] [Google Scholar]
- 30.Higo J, Nishimura Y, Nakamura H. A free-energy landscape for coupled folding and binding of an intrinsically disordered protein in explicit solvent from detailed all-atom computations. J Am Chem Soc. 2011;133:10448–10458. doi: 10.1021/ja110338e. [DOI] [PubMed] [Google Scholar]
- 31.Lockhart C, Klimov DK. Molecular interactions of Alzheimer's biomarker FDDNP with Abeta peptide. Biophysical J. 2012;103:2341–2351. doi: 10.1016/j.bpj.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li DW, Bruschweiler R. NMR-based protein potentials. Angew Chem. 2010;49:6778–6780. doi: 10.1002/anie.201001898. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 34.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beauchamp KA, Lin YS, Das R, Pande VS. Are Protein Force Fields Getting Better? A Systematic Benchmark on 524 Diverse NMR Measurements. J Chem Theory Comput. 2012;8:1409–1414. doi: 10.1021/ct2007814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patriksson A, van der Spoel D. A temperature predictor for parallel tempering simulations. Phys Chem Chem Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- 38.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 39.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 40.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin R, Tzou YM, Krishna NR. Structure of a monomeric mutant of the HIV-1 capsid protein. Biochemistry. 2011;50:9457–9467. doi: 10.1021/bi2011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 43.Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 44.Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Structural insights into the catalytic mechanism of cyclophilin A. Nat Struct Biol. 2003;10:475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 45.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373:355–366. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy KR, Schmidt AG, Kirmaier A, Wyand AL, Newman RM, Johnson WE. Gain-of-sensitivity mutations in a Trim5-resistant primary isolate of pathogenic SIV identify two independent conserved determinants of Trim5alpha specificity. PLoS Pathog. 2013;9:e1003352. doi: 10.1371/journal.ppat.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 48.Carugo O. Amino acid composition and protein dimension. Protein science: a publication of the Protein Society. 2008;17:2187–2191. doi: 10.1110/ps.037762.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 50.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 51.Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Mol BioSyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen BA AN, Yeh BJ, van Dyk D, Davidson AR, Andrews BJ, Weiss EL, Moses AM. Proteome-wide discovery of evolutionary conserved sequences in disordered regions. Sci Signaling. 2012;5:rs1. doi: 10.1126/scisignal.2002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci USA. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altis A, Nguyen PH, Hegger R, Stock G. Dihedral angle principal component analysis of molecular dynamics simulations. J Chem Phys. 2007;126:244111. doi: 10.1063/1.2746330. [DOI] [PubMed] [Google Scholar]
- 55.Mu Y, Nguyen PH, Stock G. Energy landscape of a small peptide revealed by dihedral angle principal component analysis. Proteins. 2005;58:45–52. doi: 10.1002/prot.20310. [DOI] [PubMed] [Google Scholar]
- 56.Espinoza-Fonseca LM, Ilizaliturri-Flores I, Correa-Basurto J. Backbone conformational preferences of an intrinsically disordered protein in solution. Mol BioSyst. 2012;8:1798–1805. doi: 10.1039/c2mb00004k. [DOI] [PubMed] [Google Scholar]
- 57.Knott M, Best RB. A preformed binding interface in the unbound ensemble of an intrinsically disordered protein: evidence from molecular simulations. PLoS Comput Biol. 2012;8:e1002605. doi: 10.1371/journal.pcbi.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potoyan DA, Papoian GA. Energy landscape analyses of disordered histone tails reveal special organization of their conformational dynamics. J Am Chem Soc. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 59.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 60.Qin BY, Liu C, Srinath H, Lam SS, Correia JJ, Derynck R, Lin K. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13:1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 61.Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 62.Salonen LM, Ellermann M, Diederich F. Aromatic rings in chemical and biological recognition: energetics and structures. Angew Chem. 2011;50:4808–4842. doi: 10.1002/anie.201007560. [DOI] [PubMed] [Google Scholar]
- 63.Chatterji U, Bobardt MD, Stanfield R, Ptak RG, Pallansch LA, Ward PA, Jones MJ, Stoddart CA, Scalfaro P, Dumont JM, Besseghir K, Rosenwirth B, Gallay PA. Naturally occurring capsid substitutions render HIV-1 cyclophilin A independent in human cells and TRIM-cyclophilin-resistant in Owl monkey cells. J Biol Chem. 2005;280:40293–40300. doi: 10.1074/jbc.M506314200. [DOI] [PubMed] [Google Scholar]
- 64.Ikeda Y, Ylinen LM, Kahar-Bador M, Towers GJ. Influence of gag on human immunodeficiency virus type 1 species-specific tropism. J Virol. 2004;78:11816–11822. doi: 10.1128/JVI.78.21.11816-11822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kootstra NA, Munk C, Tonnu N, Landau NR, Verma IM. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc Natl Acad Sci USA. 2003;100:1298–1303. doi: 10.1073/pnas.0337541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kootstra NA, Navis M, Beugeling C, van Dort KA, Schuitemaker H. The presence of the Trim5alpha escape mutation H87Q in the capsid of late stage HIV-1 variants is preceded by a prolonged asymptomatic infection phase. AIDS. 2007;21:2015–2023. doi: 10.1097/QAD.0b013e3282effa87. [DOI] [PubMed] [Google Scholar]
- 67.Maillard PV, Zoete V, Michielin O, Trono D. Homology-based identification of capsid determinants that protect HIV1 from human TRIM5alpha restriction. J Biol Chem. 2011;286:8128–8140. doi: 10.1074/jbc.M110.187609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagao T, Hatcho K, Doi N, Fujiwara S, Adachi A, Nomaguchi M. Amino acid alterations in Gag that confer the ability to grow in simian cells on HIV-1 are located at a narrow CA region. J Med Invest: JMI. 2009;56:21–25. doi: 10.2152/jmi.56.21. [DOI] [PubMed] [Google Scholar]
- 69.Owens CM, Song B, Perron MJ, Yang PC, Stremlau M, Sodroski J. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J Virol. 2004;78:5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacheco B, Finzi A, Stremlau M, Sodroski J. Adaptation of HIV-1 to cells expressing rhesus monkey TRIM5alpha. Virology. 2010;408:204–212. doi: 10.1016/j.virol.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O'Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, Kaur A, Hirsch VM, Johnson WE. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soll SJ, Wilson SJ, Kutluay SB, Hatziioannou T, Bieniasz PD. Assisted Evolution Enables HIV-1 to Overcome a High TRIM5alpha-Imposed Genetic Barrier to Rhesus Macaque Tropism. PLoS Pathog. 2013;9:e1003667. doi: 10.1371/journal.ppat.1003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nature Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein science: a publication of the Protein Society. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsai CJ, Ma B, Sham YY, Kumar S, Nussinov R. Structured disorder and conformational selection. Proteins. 2001;44:418–427. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- 76.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


