myb5 and myb14 mutants have darker seed coats than the wild type, with myb5 having the stronger phenotypes.
Abstract
In Arabidopsis (Arabidopsis thaliana), the major MYB protein regulating proanthocyanidin (PA) biosynthesis is TT2, named for the transparent testa phenotype of tt2 mutant seeds that lack PAs in their coats. In contrast, the MYB5 transcription factor mainly regulates seed mucilage biosynthesis and trichome branching, with only a minor role in PA biosynthesis. We here characterize MYB5 and MYB14 (a TT2 homolog) in the model legume Medicago truncatula. Overexpression of MtMYB5 or MtMYB14 strongly induces PA accumulation in M. truncatula hairy roots, and both myb5 and myb14 mutants of M. truncatula exhibit darker seed coat color than wild-type plants, with myb5 also showing deficiency in mucilage biosynthesis. myb5 mutant seeds have a much stronger seed color phenotype than myb14. The myb5 and myb14 mutants accumulate, respectively, about 30% and 50% of the PA content of wild-type plants, and PA levels are reduced further in myb5 myb14 double mutants. Transcriptome analyses of overexpressing hairy roots and knockout mutants of MtMYB5 and MtMYB14 indicate that MtMYB5 regulates a broader set of genes than MtMYB14. Moreover, we demonstrate that MtMYB5 and MtMYB14 physically interact and synergistically activate the promoters of anthocyanidin reductase and leucoanthocyanidin reductase, the key structural genes leading to PA biosynthesis, in the presence of MtTT8 and MtWD40-1. Our results provide new insights into the complex regulation of PA and mucilage biosynthesis in M. truncatula.
Flavonoids are ubiquitous secondary metabolites synthesized by the general phenylpropanoid pathway. Flavonols, anthocyanins, and proanthocyanidins (PAs) are the three major classes of flavonoids widely distributed in the plant kingdom. Flavonols function as UV light protectants (Li et al., 1993), can be required for pollen fertility (Mo et al., 1992), and act as copigments with anthocyanins to modify flower and leaf color (Bloor, 1997). Anthocyanins are responsible for the red, purple, and blue color in various plant organs, especially in flowers, leaves, fruits, and seeds. PAs (also called condensed tannins) are present in the seed coats of many plants; they confer the astringency of fruits, leaves, stems, and seeds and hence function as herbivore feeding deterrents. PAs also contribute to seed coat color, becoming brown when oxidized (Albert et al., 1997). Besides their important roles in plant physiological processes, PAs also have pharmaceutical and agricultural value. PA monomers and their derivatives (epicatechin, catechin, epigallocatechin, gallocatechin, epicatechin-gallate, and epigallocatechin-gallate) in green tea have been shown to have numerous biological activities, including antibacterial, antioxidant, antitumor, and cancer preventive activities (Molan et al., 2009). Moderate PA content in the diet of ruminant animals can prevent potentially lethal pasture bloat (Li et al., 1996) and improve the availability of bypass protein out of the rumen (Naumann et al., 2013). Unfortunately, alfalfa (Medicago sativa), the most widely grown forage legume, contains almost no PAs in leaves and stems. Therefore, there is great interest to develop PA-containing alfalfa beneficial for grazing animals.
Most of the steps in the biosynthetic pathways to anthocyanin and PAs are shared (Supplemental Fig. S1). Leucoanthocyanidins are the first branch point between the anthocyanin and PA biosynthesis pathways. They can be oxidized to anthocyanidins by leucocyanidin dioxygenase (LDOX) or reduced to flavan-3-ols (catechins) by leucoanthocyanidin reductase (LAR; Tanner et al., 2003). The second branch point between the anthocyanin and PA biosynthesis pathways is anthocyanidin, which can be glycosylated by UDP glycosyltransferases (UGTs) to produce anthocyanins or reduced to (epi)flavan-3-ols (e.g. epicatechin) by anthocyanidin reductase (ANR; Xie et al., 2003, 2004). Epicatechin can also be glycosylated and then transported to the vacuole by a multidrug and toxic compound extrusion transporter (Pang et al., 2008; Zhao and Dixon, 2009), where it is believed to be polymerized by a laccase to form PA oligomers (Pourcel et al., 2005).
The regulation of anthocyanin and PA biosynthesis is controlled by a ternary complex of transcription factors, called the MBW complex, consisting of MYB, basic helix-loop-helix (bHLH), and WD40 transcription factors (Baudry et al., 2004). Among these, WD40 repeat transcription factors are required for both anthocyanin and PA biosynthesis. It is the MYBs, and to a lesser extent the bHLHs, that determine the specificity of gene activation for anthocyanin and PA biosynthesis. In Arabidopsis (Arabidopsis thaliana), a trichome-specific bHLH (ENHANCER OF GLABRA3 [EGL3]) and a PA-specific bHLH (TRANSPARENT TESTA8 [TT8]) have redundant functions in PA biosynthesis (Zhang et al., 2003). The MYBs specific to the anthocyanin pathway are PRODUCTION OF ANTHOCYANIN PIGMENTATION1 (PAP1; MYB75) in Arabidopsis and LEGUME ANTHOCYANIN PRODUCTION1 (LAP1) in Medicago truncatula (Borevitz et al., 2000; Peel et al., 2009). Homologs of PAP1 and LAP1 have been discovered in various plant species (Espley et al., 2007; Azuma et al., 2008; Lin-Wang et al., 2010). In Arabidopsis, TT2 (AtMYB123) is the major MYB transcription factor regulating the PA biosynthesis pathway. tt2 mutants almost completely lose PA accumulation in the seed coat (Nesi et al., 2001). TT2-related PA-regulating MYB transcription factors have also been discovered in Lotus japonicus (LjTT2a, LjTT2b, and LjTT2c; Yoshida et al., 2008), poplar (Populus spp.; MYB134; Mellway et al., 2009), and clover (Trifolium arvense; TaMYB14; Hancock et al., 2012). AtMYB5, a MYB transcription factor regulating trichome branching in leaves and mucilage accumulation in seed coats, plays only a minor role in PA biosynthesis in Arabidopsis (Gonzalez et al., 2009; Li et al., 2009). However, overexpression of VvMYB5a and VvMYB5b (the homologs of AtMYB5 in grape [Vitis vinifera]) in tobacco (Nicotiana tabacum) induces PA accumulation in flowers (Deluc et al., 2006, 2008). DkMYB4 (the homolog of AtMYB5 in persimmon [Diospyros kaki]) has been demonstrated to regulate PA accumulation in persimmon fruits, and overexpression or knockdown of DkMYB4 induces or reduces, respectively, PA accumulation in persimmon callus. Moreover, the PA accumulation deficiency in a nonastringent persimmon variety was shown to be associated with the low expression level of DkMYB4 (Akagi et al., 2009). These results indicate that MYB5-like transcription factors may play more important roles in PA biosynthesis in some plants than in Arabidopsis. However, there are no direct genetic data to support the relative importance of MYB5-related transcription factors, compared with TT2 homologs, in these plants.
Here, we provide genetic evidence to demonstrate that MtMYB5, the homolog of AtMYB5 in M. truncatula, plays a pivotal role in PA biosynthesis. We isolated mutants of MtMYB5 and MtMYB14 (a close ortholog of the TT2-related TaMYB14) and characterized their phenotypes. Both myb5 and myb14 mutants exhibit darker seed coats than the wild type. Strikingly, myb5 mutants have a stronger phenotype than myb14 mutants. M. truncatula MYB5 and MYB14 physically interact and synergistically activate transcription from the ANR and LAR promoters in the presence of MtTT8 and MtWD40-1, providing a revised model for the regulation of the PA biosynthesis pathway.
RESULTS
Identification of MtMYB5 and MtMYB14
A WD-40 protein is a key component of the transcriptional complex that activates PA biosynthetic genes (Lepiniec et al., 2006; Pang et al., 2009), and we recently reported that the MYB transcription factor MtPAR (for proanthocyanidin regulator) can activate the promoter of M. truncatula WD40-1 in yeast (Saccharomyces cerevisiae; Verdier et al., 2012). To search for new positive regulators of WD40-1, which may also play a role in PA biosynthesis, we performed a yeast one-hybrid (Y1H) screen against a complementary DNA (cDNA) library of young seeds harvested at 10 d after pollination (DAP). We found five MYB transcription factors able to activate the WD40-1 promoter in this assay. The activation activities of these MYB proteins were confirmed by using the full-length cDNAs to perform the Y1H assay a second time. Among these MYB transcription factors, two show similarity to Arabidopsis mucilage regulators, two show similarity to Arabidopsis MYB4, and one (Medtr3g083540) shows high similarity to known transcription factors activating PA biosynthesis. The fact that most of the genes identified by Y1H screening are related to known functions of WD40-1 proteins in plants suggests that these transcription factors are probably true regulators of Medicago spp. WD40-1. Here, we report the detailed characterization of the PA biosynthesis regulator encoded by Medtr3g083540.
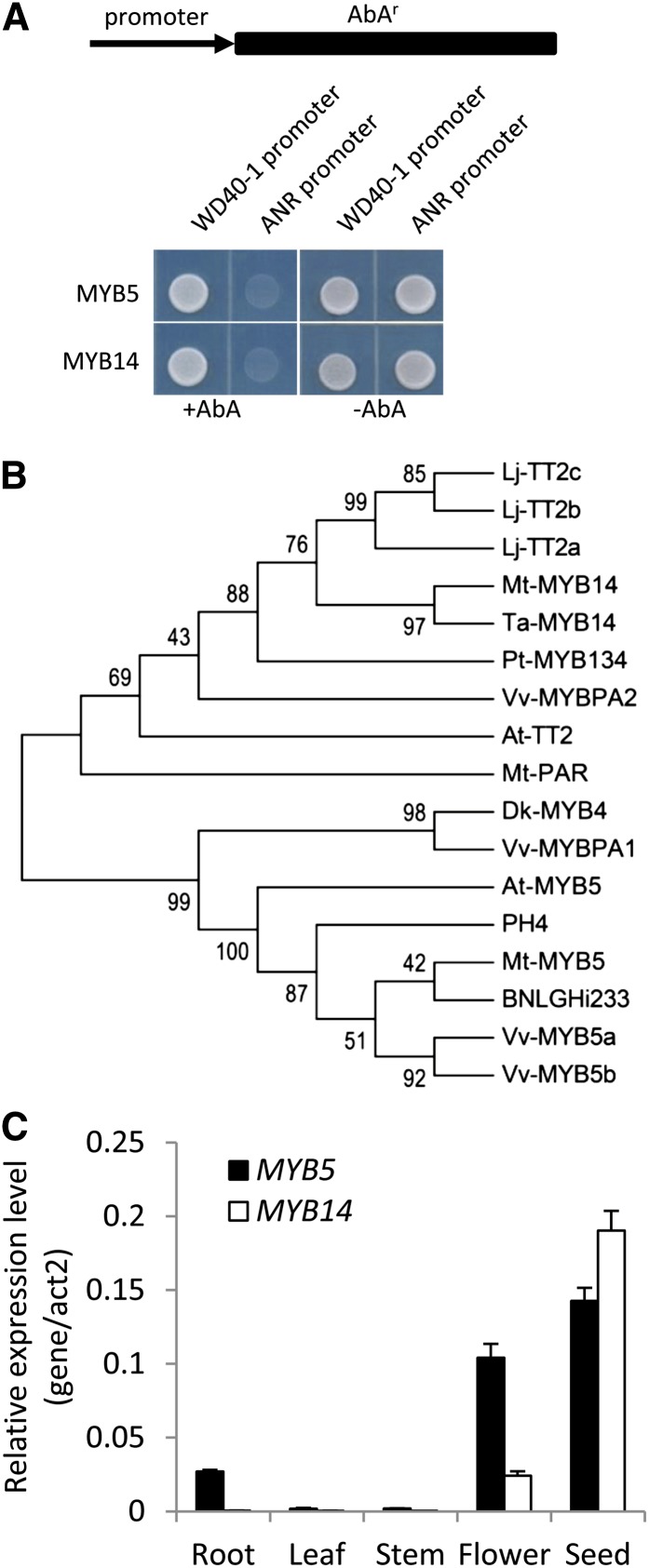
Medtr3g083540 encodes a 304-amino acid R2R3 MYB transcription factor that belongs to a MYB family that includes AtMYB5, VvMYB5a and VvMYB5b, and DkMYB4. Alignment of Medtr3g083540 with these MYBs indicated that the R2R3 domain is highly conserved among this family (Supplemental Fig. S2; Supplemental FASTA S1). More importantly, Medtr3g083540 contains the C1 and C3 C-terminal motifs shared by all MYB5-like proteins except DkMYB4, which lacks the C3 motif (Supplemental Fig. S2; Supplemental FASTA S1). Hence, we name Medtr3g083540 as MtMYB5. MYB5 activated the M. truncatula WD40-1 promoter in the Y1H assay but not the promoter of the PA biosynthetic pathway gene ANR, activation of which may require additional factors that are not present in the yeast assay (see below; Fig. 1A).
Figure 1.
Identification and expression of MtMYB5 and MtMYB14. A, MYB5 and MYB14 activate the MtWD40-1 promoter, but not the ANR promoter, in Y1H assays. The promoters of WD40-1 and ANR were cloned in front of the aureobasidin A resistance (AbAr) gene and integrated into Y1G Gold yeast (Clontech) to generate reporter strains. The reporter strains were transformed with the effector transcription factors (MYB5 and MYB14), and growth was recorded in the presence or absence of aureobasidin A. B, Phylogenetic tree of MYB transcription factors related to AtMYB5 and AtTT2. Protein sequences were obtained from GenBank with the following accession numbers: AtMYB5 (NP_187963.1), DkMYB4 (BAI49721.1), VvMYB5a (NP_001268108.1), VvMYB5b (NP_001267854.1), MtMYB5 (XP_003601609.1), PH4 (AAY51377.1), BNLGHi233 (AAK19611.1), AtTT2 (NP_198405.1), LjTT2a (BAG12893.1), LjTT2b (BAG12894.1), LjTT2c (BAG12895.1), TaMYB14-2 (AFJ53054.1), MtMYB14-2 (Mt3.5v4 contig_238935_1, Mt4.0v1 Medtr4g125520.1), MtPAR (XP_003627264.1), PtMYB134 (ACR83705.1), VvPA1 (NP_001268160.1), and VvPA2 (NP_001267953.1). The phylogenetic tree was constructed by the neighbor-joining method with 1,000 bootstrap replicates by the software MEGA6.0 (http://www.megasoftware.net/). Numbers indicate confidence percentages. C, Transcript levels of MYB5 and MYB14 in different organs of M. truncatula as determined by qRT-PCR. Reactions were performed as technical triplicates, and error bars indicate sd.
Arabidopsis TT2 is the best studied MYB transcription factor regulating PA biosynthesis (Nesi et al., 2001). No M. truncatula homolog of TT2 could be found in earlier versions of the M. truncatula genome database. However, a homology search using the AtTT2 protein sequence to blast the M. truncatula genome version Mt3.5v4 (Young et al., 2011) resulted in the positive identification of contig_238935_1 as the closest homolog of AtTT2 in M. truncatula. (Supplemental Fig. S3; Supplemental FASTA S2; in the most recent M. truncatula genome Mt4.0v1, contig_238935 is assembled as Medtr4g125520.1). Contig_238935_1 has only one amino acid difference from MtMYB14-2 (AFJ53058.1), which is the M. truncatula ortholog of T. arvense MYB14, a protein that induces PA accumulation in white clover (Trifolium repens) and alfalfa (Hancock et al., 2012). We conclude that contig_238935_1 is a new allele of MtMYB14-2, the single amino acid difference probably being due to the fact that the two genes were cloned from different ecotypes of M. truncatula. The Y1H assay indicated that, similar to MtPAR and MtMYB5, MtMYB14 is also able to activate the promoter of MtWD40-1, but not ANR, in yeast (Fig. 1A).
As a first step to analyze the functions of MtMYB5 and MtMYB14, we performed a phylogenetic analysis of MYBs known to regulate PA biosynthesis. In general, these MYBs form two clades in the phylogenetic tree. One includes MtMYB5-related proteins, and the other includes AtTT2, MtPAR, VvMYBPA2, LjTT2s, and MYB14-like proteins (Fig. 1B; Supplemental FASTA S3). Among these TT2-related MYBs, MYB14s and LjTT2s are closely clustered. Inspection of the multiple alignment of these proteins reveals that, besides the high homology at their N termini, these MYBs also share considerable amino acid identity at their C-terminal regions (Supplemental Fig. S3).
To experimentally demonstrate that MtMYB14 and MtPAR are functional orthologs of AtTT2, we complemented Arabidopsis tt2 mutant plants with MtPAR and MtMYB14 driven by the Arabidopsis ANR promoter (ProBan). For comparison, MtMYB5 was also included in the complementation experiments. As shown in Supplemental Figure S4, both MtPAR and MtMYB14 rescued the Arabidopsis tt2 phenotype (leading to dark seed coats), supporting the conclusion that MtPAR and MtMYB14 are functional orthologs of Arabidopsis TT2. Although Xu et al. (2014) observed that AtMYB5 could partially rescue the tt2 phenotype, we did not observe such rescue with MtMYB5.
We next performed quantitative reverse transcription (qRT)-PCR to check the transcript levels of MYB5 and MYB14 in different M. truncatula tissues. Both MYB5 and MYB14 are highly expressed in seeds (Fig. 1C). MYB5 is also highly expressed in flowers and moderately expressed in roots. There is a moderate MYB14 expression level in flowers. These different expression patterns suggest that MYB5 might have broader roles than MYB14 in Medicago spp.
MYB5 Promotes PA Accumulation in M. truncatula Hairy Roots
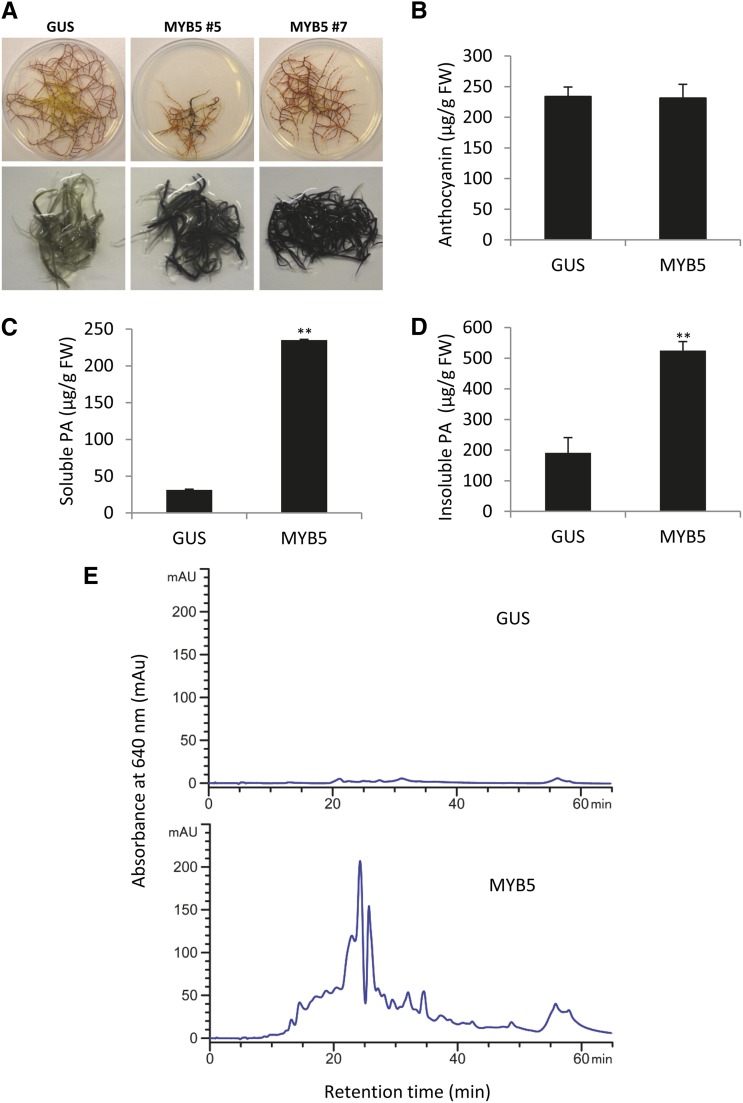
To demonstrate that MtMYB5 can promote PA accumulation in planta, we generated MYB5-overexpressing transgenic hairy roots in M. truncatula ecotype A17. Control roots were transformed with a GUS construct. Some lines of MYB5-transformed hairy roots showed retarded growth (Fig. 2A), whereas growth was unaffected in other lines. Staining with dimethylaminocinnamaldehyde (DMACA) suggested that both the retarded and normally growing hairy roots accumulated large amounts of PAs (Fig. 2A, bottom row). To avoid negative growth effects, we selected normally growing roots (derived from line 7) for further analysis. There was no significant difference in anthocyanin accumulation between GUS control and MYB5-overexpressing roots (Fig. 2B). In contrast, both soluble and insoluble PA levels were strongly induced in MYB5-overexpressing roots, reaching 250 and 500 μg g−1 fresh weight, respectively (Fig. 2, C and D).
Figure 2.
Overexpressing MtMYB5 in M. truncatula A17 hairy roots induces PA accumulation. A, Unstained (top row) and DMACA-stained (bottom row) GUS (transformed with vector harboring the GUS gene) and two independent transgenic MYB5-overexpressing hairy root lines. B, Anthocyanin levels quantified after extraction by measurement of A550 and expressed as cyanidin-3-O-glucoside equivalents. C, Soluble PA levels as quantified with DMACA reagent and expressed as epicatechin equivalents. D, Insoluble PA levels as quantified by the butanol-HCl method and expressed as procyanindin B1 equivalents. E, Soluble PA profiles as determined by normal-phase HPLC with postcolumn DMACA derivatization. GUS, Control lines overexpressing GUS; MYB5, MtMYB5-overexpressing line 7. Three independent biological samples of line 7 were analyzed in B to D. Error bars denote sd. **P < 0.01 by Student’s t test (n = 3). FW, Fresh weight.
PA profiling by normal-phase HPLC coupled with postcolumn DMACA derivatization indicated that most of the soluble PA-like DMACA-positive material accumulating in MYB5-overexpressing lines was of relatively low Mr (Fig. 2E); epicatechin monomer elutes at a retention time of approximately 15 min in this HPLC system (Pang et al., 2013). Since DMACA can react with other unknown metabolites, we then performed the more diagnostic phloroglucinolysis analysis to determine the nature and composition of putative PAs in MYB5-overexpressing lines after purification of the soluble PAs on Sephadex LH20 resin. Our results indicated that epicatechin is the main component of the PAs accumulating in MYB5-overexpressing hairy roots, both as starter and extension unit (Supplemental Fig. S5). The average degree of polymerization of the soluble PAs calculated by this method (ratio of extension unit to starter unit peak areas) was determined to be approximately 31. Taken together, our data indicate that the high-Mr DMACA-positive metabolites accumulating in MYB5-overexpressing lines are authentic PAs and similar to those occurring naturally in the M. truncatula seed coat (Pang et al., 2007).
We utilized ultra-performance liquid chromatography (UPLC)-mass spectrometry (MS) to determine whether the monomer flavan-3-ols also accumulate in MYB5-expressing hairy roots. This analysis revealed significant levels of epicatechin and epicatechin-3′-O-glucoside in MYB5-overexpressing lines; these compounds were barely detectable in GUS controls (Supplemental Fig. S6).
MYB14 Promotes PA Accumulation in M. truncatula Hairy Roots
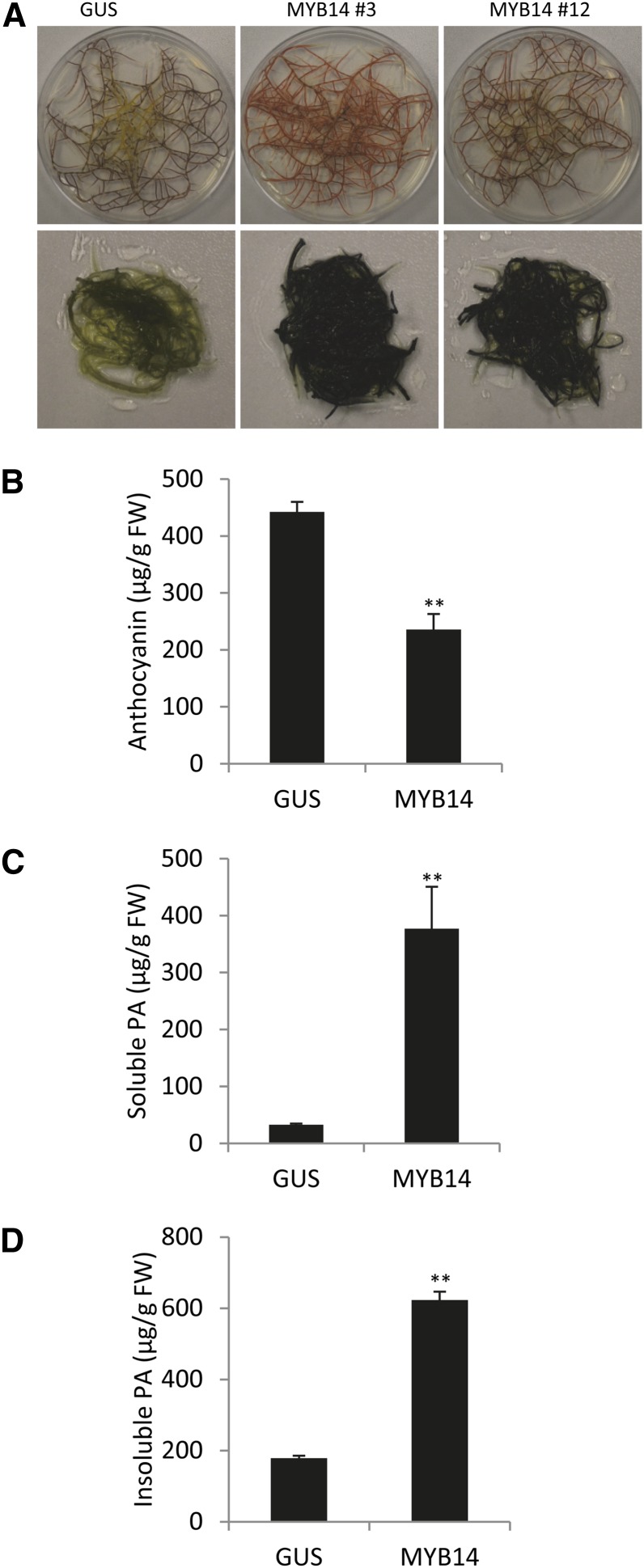
To confirm the function of MtMYB14 as a PA regulator, we generated MYB14-overexpressing hairy roots in M. truncatula A17. Unlike MYB5, overexpression of MYB14 did not result in growth retardation of hairy roots (Fig. 3A). Overexpression of MYB14 reduced the anthocyanin content to about half that of GUS control lines (Fig. 3B). DMACA staining and quantification of soluble and insoluble PAs demonstrated that MYB14 strongly induced PA accumulation in hairy roots (Fig. 3, A, C, and D). Phloroglucinolysis and UPLC-MS analyses confirmed the presence of similar types of epicatechin-rich PAs and similar accumulation of free epicatechin and epicatechin-3′-O-glucoside, as observed in MYB5-overexpressing hairy roots (Supplemental Figs. S5 and S7). The mean degree of polymerization of the PAs was somewhat higher in the MYB14 lines than in the MYB5 lines (Supplemental Fig. S5, A and B).
Figure 3.
Overexpressing MtMYB14 in M. truncatula A17 hairy roots induces PA accumulation. A, Unstained (top row) and DMACA-stained (bottom row) GUS (transformed with vector harboring the GUS gene) and two independent transgenic MYB14-overexpressing hairy roots. B, Anthocyanin levels. C, Soluble PA levels. D, Insoluble PA levels. Each was determined as described in the legend to Figure 2. GUS, Control lines overexpressing GUS; MYB14, MtMYB14-overexpressing line 3. Three independent biological samples of line 3 were analyzed in B to D. Error bars denote sd. **P < 0.01 by Student’s t test (n = 3). FW, Fresh weight.
Phenotypes of M. truncatula myb5 Mutants
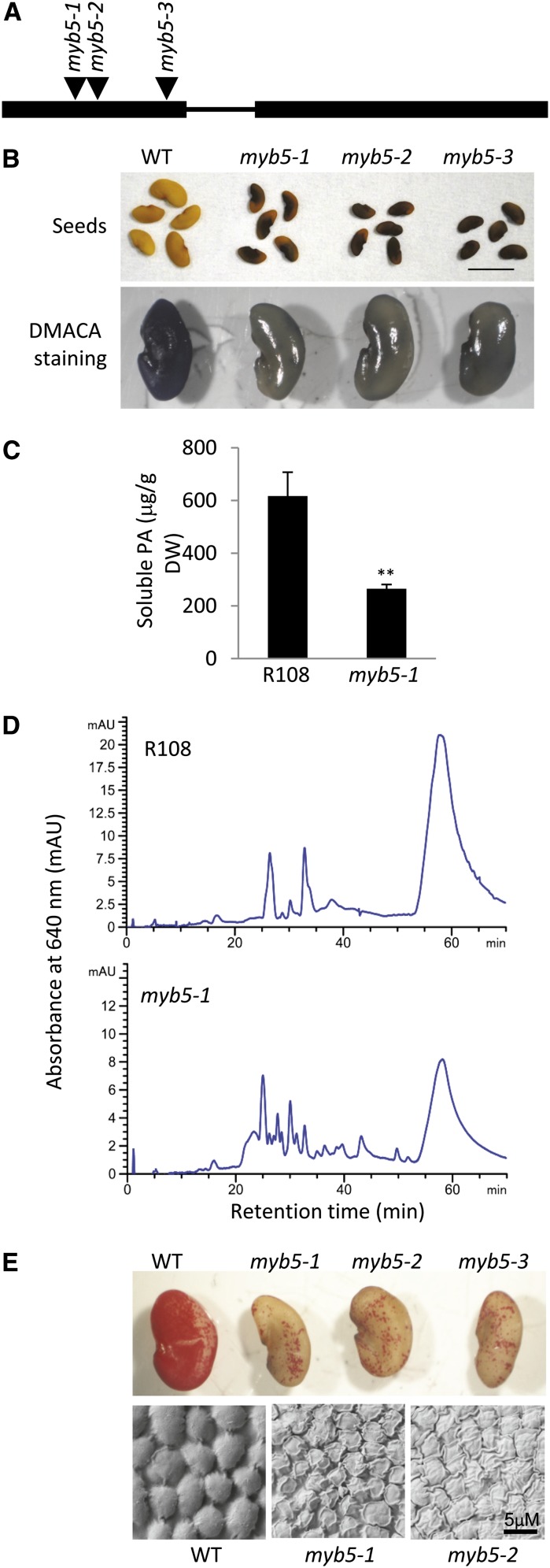
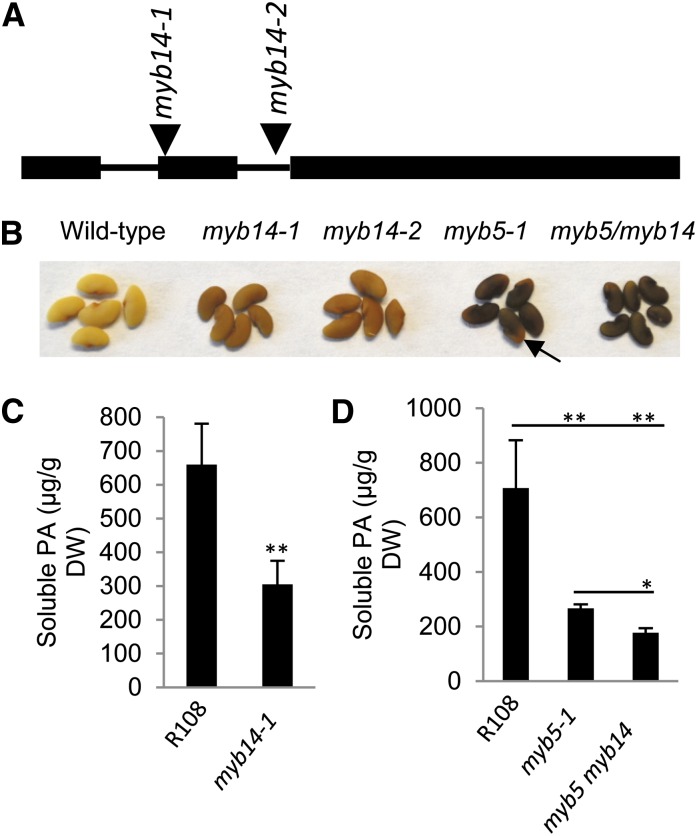
To better understand the functions of MtMYB5, we screened the Tnt1 (for the transposable element of tobacco [N. tabacum] cell type1) retrotransposon-mutagenized M. truncatula R108 population (Tadege et al., 2008) to isolate myb5 mutants. In total, we obtained three independent lines of MYB5 Tnt1 insertion mutants. All three Tnt1 insertions were located in the first exon of the MYB5 gene (Fig. 4A). Reverse transcription (RT)-PCR could not detect the full-length MYB5 transcript in any of the three lines, indicating that all three were null mutants (Supplemental Fig. S8A). There were no visible phenotypic differences in the vegetative organs between mutant and wild-type plants during the whole developmental period under our growth conditions. However, striking differences were observed in mature seeds, which exhibited a conspicuous dark-red color, whereas wild-type seeds are light yellow (Fig. 4B, top row). The dark-red pigments in mutant seeds are not extractable by acidic methanol or 70% acetone solutions, suggesting that these pigments are cross-linked in the seed coat (see “Discussion”). DMACA-stained mutant seeds displayed a much lighter blue color than wild-type seeds, suggesting that the PA content in mutant seeds was reduced (Fig. 4B, bottom row). Quantification of soluble PAs by the DMACA method indicated that mutant seeds accumulated approximately 30% of the PA content of wild-type seeds (Fig. 4C). However, quantification of insoluble PAs by the butanol-HCl method did not detect differences between the mutant and the wild type (Supplemental Fig. S9). The butanol-HCl method relies on the hydrolysis of PAs into anthocyanidin monomers; therefore, it cannot distinguish anthocyanidin released from true PAs and anthocyanidin released from non-PA complexes, the presence of which is suggested by the dark-red color of the myb5 mutant seeds. Profiling of the soluble PA fraction by normal-phase HPLC coupled with DMACA staining indicated that the myb5 mutant seeds produce, besides a lower amount of total soluble PAs, a more diverse range of small PA oligomers (Fig. 4D). Phloroglucinolysis analysis did not detect any difference in PA monomer composition or mean degree of polymerization between mutant and wild-type seeds (Supplemental Fig. S10).
Figure 4.
Phenotypes of myb5 null mutants. A, Positions of the Tnt1 insertions in the MYB5 gene in the mutant lines myb5-1 (NF11932), myb5-2 (NF12338), and myb5-3 (NF13006). Boxes indicate exons, and the line indicates the intron. B, Seed coat phenotypes of myb5 null mutants. myb5 mutant seed coats display a dark reddish color (top row). DMACA staining (bottom row) indicates that myb5 mutants accumulate less PA than the wild type (WT). C, Soluble PA levels in mature dry seeds of wild-type and myb5 mutant seeds. Three biological replicates were analyzed. Error bars indicate sd. **P < 0.01 by Student’s t test (n = 3). DW, Dry weight. D, Normal-phase HPLC analysis, with postcolumn derivatization with DMACA, of soluble PA fractions from the wild type (R108) and the myb5-1 mutant. E, Ruthenium Red staining (top row) and scanning electron microscopy images (bottom row; all three images are at the same magnification) of seeds of ecotype R108 (WT) and myb5 mutants.
AtMYB5 has been demonstrated to positively regulate mucilage biosynthesis in the Arabidopsis seed. To test whether MtMYB5 is also involved in mucilage biosynthesis in M. truncatula we stained the myb5 mutant seeds with Ruthenium Red, a dye used to detect mucilage (Western et al., 2001). Similar to the Arabidopsis myb5 mutant, M. truncatula myb5 mutant seeds exhibited much lighter Ruthenium Red staining than wild-type seeds, indicating that the mutant seeds accumulate much less mucilage in the seed coat (Fig. 4E, top row).
To determine if there are any differences in seed coat morphology between the myb5 mutant and the wild type, we examined the seed coat epidermal cells by scanning electron microscopy. In wild-type seeds, the epidermal cells were tightly arranged to form a smooth dome-like surface, with the cell walls between neighboring cells tightly sealed. In contrast, in mutant seeds, there were many cracks between the epidermal cells, and many epidermal cells were wrinkled with irregular shape (Fig. 4E, bottom row). The cells were also generally somewhat smaller than in the wild type, reflecting the overall somewhat smaller size of the seeds themselves (Fig. 4, B and E).
Phenotypes of M. truncatula myb14 Mutants
We isolated two independent myb14 mutants from the M. truncatula Tnt1 insertion mutant population (Fig. 5A). myb14-1 and myb14-2 harbor Tnt1 insertions in the second exon and the second intron, respectively (Fig. 5A). MYB14 transcripts could not be detected in line myb14-1, whereas very low levels of MYB14 transcripts were detected in line myb14-2, indicating that myb14-1 is a null mutant and myb14-2 is a hypomorphic mutant (Supplemental Fig. S8, B and C). Both myb14-1 and myb14-2 seeds display slightly but noticeably darker red-brown color than wild-type seeds, suggesting that the reduction of MYB14 transcripts in hypomorphic myb14-2 is sufficient to cause a similar phenotype to that in the null mutant (Fig. 5B). Compared with the myb5 mutant, the seed color of myb14 is closer to that of the wild type. To exclude the possibility that the different phenotype of M. truncatula myb14 is the result of gene redundancy, we used qRT-PCR with two pairs of primers, one primer pair located in the conserved R2R3 domain and the other located in the divergent C-terminal region, to measure the M. truncatula MYB14 gene copy number. Results from both primer pairs indicated that MYB14 is a single-copy gene in the M. truncatula genome (Supplemental Fig. S11).
Figure 5.
Seed phenotypes of myb14 and the myb5 myb14 double mutant. A, Tnt1 retrotransposon insertion positions in the MYB14 gene. Arrowheads indicate the insertion positions in myb14-1 (NF14565) and myb14-2 (NF84). Boxes indicate exons, and the lines indicate introns. B, Seed color phenotypes of myb14 and myb5 single mutants and the myb5 myb14 double mutant. Note the lighter color at the end of the seed (arrow). C and D, Soluble PA levels in myb14 -1, myb5-1, and myb5-1 myb14-1 (myb5 myb14) mutants as determined by the DMACA method. All quantifications used three independent biological replicates. Error bars indicate sd. **P < 0.01, *P < 0.05 by Student’s t test (n = 3). DW, Dry weight.
myb14 mutants accumulate approximately 50% of the PA level of wild-type seeds (Fig. 5C). Similar to myb5 mutants, we could not detect differences in the levels of insoluble PAs between myb14 mutant and wild-type seeds by the butanol-HCl method (Supplemental Fig. S7). Based on the same rationale, we conclude that insoluble PA levels in myb14 mutant seed cannot be determined by the butanol-HCl method due to the presence of insoluble anthocyanin moieties. No differences in mucilage staining were observed in myb14 mutant seeds compared with the wild type (Supplemental Fig. S12).
To dissect the genetic interaction between MYB5 and MYB14 in M. truncatula, we generated the myb5 myb14 double-knockout mutant by crossing myb5-1 and myb14-1. The seed coat color of the double mutant exhibited a more uniform dark red-brown color than the single mutant of myb5, which typically exhibits a lighter red-brown color at one end of the seed (Fig. 5B, arrow). These results suggest that MtMYB5 and MtMYB14 act redundantly to regulate PA biosynthesis. The soluble PA content in the double mutant was reduced to approximately 20% of that of the wild type (Fig. 5D).
Gene Expression Profiling of MtMYB5- and MtMYB14-Overexpressing Hairy Roots and Mutant Seeds
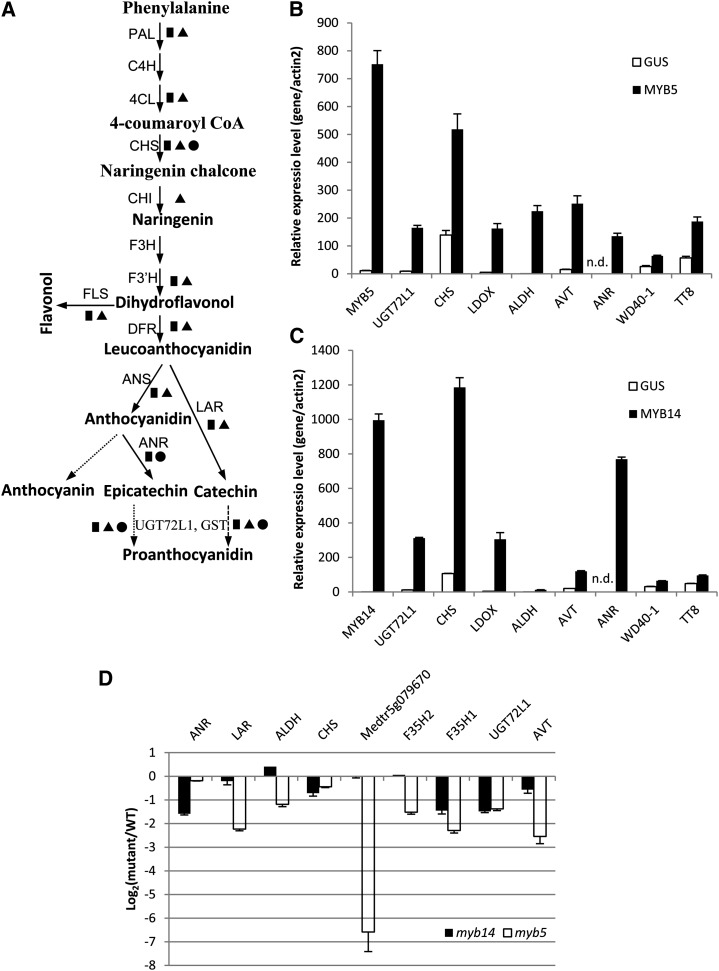
To gain insights into MYB5- and MYB14-regulated genes in M. truncatula, we performed transcriptome analysis of hairy roots by Affymetrix microarray analysis. We considered genes for which the transcript level changed by more than 2-fold, and with a statistical significance of P < 0.05, as differentially expressed genes. In total, 628 and 460 probe sets were induced in MYB5- and MYB14-overexpressing hairy roots, respectively (Supplemental Data Sets S1 and S2). qRT-PCR analysis was performed on a set of selected genes to confirm the microarray results (Fig. 6, B and C). Genes involved in the PA and anthocyanin biosynthesis pathways were highly induced (Fig. 6, A and B). Noticeably, ANR, the gene responsible for epicatechin synthesis, was induced by 255-fold (second strongest induced) and 985-fold (strongest induced) in MYB5- and MYB14-overexpressing hairy roots, respectively, consistent with the observation that these hairy roots accumulate large amounts of PAs. One hundred forty-nine probe sets were induced more than 2-fold by both MYB5 and MYB14 (Supplemental Data Set S3). Known PA and flavonoid biosynthesis genes, such as ANR, LAR, UGT72L1, LDOX, CHS, and F3′H, were among the common induced genes. Interestingly, an aldehyde dehydrogenase gene and a vacuolar acid invertase gene were also highly induced by both MYB5 and MYB14, possibly indicating that sugar metabolism is altered to accommodate the high level of PA biosynthesis in these roots.
Figure 6.
Genes regulated by MYB5 and MYB14. A, Scheme of the flavonoid pathway leading to PA production. Squares denote genes that are up-regulated in MYB5- and MYB14-overexpressing hairy roots based on Affymetrix microarray analysis. Triangles denote genes that are down-regulated in myb5-1 mutant seeds. Circles denote genes that are down-regulated in myb14-1 mutant seeds. B and C, qRT-PCR analysis to confirm the up-regulation of genes in MYB5-overexpressing (B) and MYB14-overexpressing (C) hairy roots. n.d., Not detected. Transcript levels were normalized to MtActin2 transcript levels. D, qRT-PCR to measure the transcript levels of down-regulated genes in seeds (12 DAP) of the myb5-1 and myb14-1 mutants. ALDH, Aldehyde dehydrogenase; ANS, anthocyanidin synthase (leucoanthocyanidin dioxygenase [LDOX]); AVT, acid vacuolar invertase; C4H, cinnamate 4-hydroxylase; CHI, chalcone isomerase, 4CL, 4-coumarate coenzyme A ligase; CHS, chalcone synthase; DFR, dihydroflavanol reductase; F3H, flavonoid 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; GST, glutathione S-transferase; F35H, flavonoid 3′,5′-hydroxylase; TT8, MtTT8; WD40-1, MtWD40-1; WT, wild type. All qRT-PCR analyses were performed on one independent biological replicate (separate from those used for the microarray analysis), giving technical triplicates; error bars indicate sd.
Transcripts encoding the transcription factors MtTT8 (Medtr1g072320.1, the putative homolog of Arabidopsis TT8) and MtWD40-1 (Pang et al., 2009), which are the integral components of the MBW complex and essential for the activation of PA pathway structural genes, were also up-regulated in transgenic hairy roots. MtTT8 (probe set Mtr.22479.1.S1_at) was induced 2.6- and 2.1-fold by MtMYB5 and MtMYB14, respectively. MtWD40-1 (Mtr.39774.1.S1_at) was induced 2.6-fold by MtMYB14 but less than 2-fold by MtMYB5. Because of the relatively low level of induction of these genes compared to that of the PA pathway structural genes, qRT-PCR was performed to quantify the expression levels of MtTT8 and MtWD40-1 in hairy roots. The results confirmed that both MtMYB5 and MtMYB14 can induce MtTT8 and MtWD40-1 (Fig. 6, B and C).
One MYB transcription factor gene, Medtr5g079670.1, represented by two probe sets (Mtr.16432.1.S1_at and Mtr.34401.1.S1_s_at), was induced 53- to 80-fold and 10-fold by MYB5 and MYB14, respectively (Supplemental Data Set S3). BLAST analysis of the GenBank database indicated that Medtr5g079670.1 belongs to the class of Arabidopsis MYB4-like proteins. Proteins in this family typically are transcriptional repressors, and this particular M. truncatula gene shows high sequence similarity to the known anthocyanin biosynthesis suppressor, AtMYBL2.
To identify transcript changes underlying the phenotypes of the myb5 and myb14 mutant seeds, we performed Affymetrix microarray analysis. Seeds of mutant and wild-type plants at 12 DAP were dissected from pods for the isolation of RNA. Two biological replicates were collected, and transcript levels were compared with those of wild-type plants. Nine hundred ninety-one genes (500 down-regulated and 491 up-regulated more than 2-fold) were differentially expressed in myb5 seeds (Supplemental Data Set S4), whereas only 151 genes were differentially expressed (100 down-regulated and 51 up-regulated more than 2-fold) in myb14 seeds (Supplemental Data Set S5).
We analyzed the 500 down-regulated genes in myb5 seeds by PathExpress (Goffard and Weiller, 2007) to identify the potential metabolic pathways associated with these genes. This indicated that flavonoid biosynthesis and sugar metabolism are the main pathways down-regulated in the myb5 mutant seeds (P < 0.01; Supplemental Table S1), consistent with the low PA and mucilage phenotypes of myb5 mutant seeds. The flavonoid pathway genes PAL, CHS, F3′H, DFR, LDOX, LAR, UGT72L1, and GST were all down-regulated in myb5 seeds. ANR, which is highly induced by MYB5 overexpression in hairy roots, was only slightly down-regulated in the myb5 seeds. However, LAR, another key structural gene for PA biosynthesis, was down-regulated more than 6-fold. The down-regulation of the above genes was subsequently confirmed by qRT-PCR from independent biological samples (Fig. 6D). It is noteworthy that most of the above genes belong to multigene families in the M. truncatula genome but only a subset of each gene family was down-regulated in myb5 seeds, suggesting that different gene family members might be involved in different flavonoid biosynthetic pathways. Medtr5g079670.1, the putative repressor highly induced in hairy roots, was the most strongly down-regulated gene in myb5 seeds (Fig. 6D; Supplemental Data Set S4). The strong down-regulation of an anthocyanin repressor was associated with the up-regulation of certain anthocyanin biosynthesis genes; two genes encoding anthocyanin acyltransferase and three genes encoding DFR were up-regulated more than 2-fold in myb5 seeds (Supplemental Data Set S4).
Genes encoding glycosyl hydrolase and α/β-fold hydrolase are down-regulated in Arabidopsis myb5 mutant seeds and were assumed to be associated with mucilage biosynthesis (Gonzalez et al., 2009; Li et al., 2009). Similarly, many genes encoding glycoside hydrolases and other cell wall carbohydrate-modifying enzymes are down-regulated in M. truncatula myb5 mutant seeds (Supplemental Table S2). The homolog of one gene in Arabidopsis that has been directly demonstrated to be involved in mucilage biosynthesis (MUCILAGE-MODIFIED2 [MUM2]; Supplemental Table S2) was down-regulated in myb5 seeds. Interestingly, genes encoding the aldehyde dehydrogenase and vacuolar acid invertase identified in MYB5-overexpressing hairy roots were also down-regulated in myb5 seeds, suggesting that they are probable targets of MYB5 (Fig. 6D). These enzymes are typically involved in carbohydrate and/or lipid metabolism, suggesting that they are probably associated with mucilage biosynthesis in the M. truncatula seed coat.
Analysis of 100 down-regulated genes in myb14 seeds by PathExpress indicated that flavonoid biosynthesis was likely the only pathway significantly affected (Supplemental Table S3), with ANR, UGT72L1, and CHS being down-regulated more than 2-fold in myb14 seed. Subsequent qRT-PCR analysis confirmed the microarray data (Fig. 6D). One gene encoding flavonoid 3′,5′-hydroxylase, which was down-regulated slightly less than 2-fold in myb5 and myb14 seeds (54% of the wild type) on the basis of microarray analysis, was found to be down-regulated by more than 2-fold by qRT-PCR analysis. The results show that loss of function of MYB14 only affects a small set of genes in the PA biosynthesis pathway, consistent with its less severe phenotypic effect than loss of function of MYB5. Unlike myb5 mutant seeds, no gene implicated in mucilage biosynthesis was down-regulated in myb14 seeds.
MYB5 and MYB14 Physically Interact and Synergistically Activate the ANR and LAR Promoters in the Presence of MtTT8 and MtWD40-1
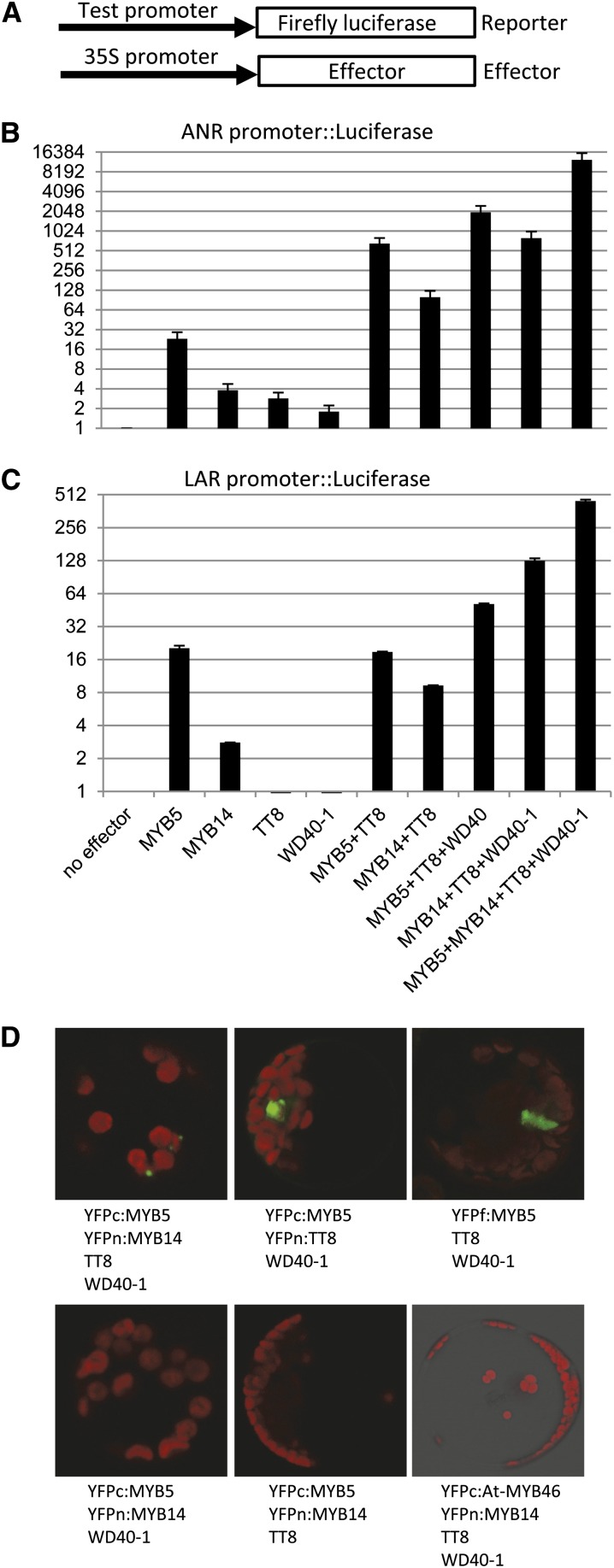
MYB transcription factors interact with bHLH and WD40 transcription factors to form the MBW complex (Lepiniec et al., 2006). To test the hypothesis that MYB5 and MYB14 form complexes with M. truncatula bHLH and WD40 transcription factors to regulate the PA biosynthesis pathway, we conducted transient promoter activation assays in Arabidopsis protoplasts. The promoters of M. truncatula ANR and LAR were cloned in front of the firefly luciferase reporter gene (Fig. 7A). The reporter constructs were cotransfected into the protoplasts with various effectors driven by the constitutive cauliflower mosaic virus 35S promoter (Fig. 7A). In addition to MYB5 and MYB14, the effectors included one bHLH transcription factor, MtTT8, and MtWD40-1 (Pang et al., 2009).
Figure 7.
MYB5 and MYB14 synergistically activate the ANR and LAR promoters in the presence of MtWD40-1 and MtTT8 in Arabidopsis protoplasts. A, Schematic diagram showing the structures of reporter and effector constructs. B and C, Activation of the ANR and LAR promoters in transient assays in Arabidopsis protoplasts. Various combinations of M. truncatula effectors (MYB5, MYB14, TT8, and WD40-1) were used to transfect Arabidopsis protoplasts along with ANR promoter (B) or LAR promoter (C) reporter constructs. Firefly luciferase activities were quantified and normalized to Renilla luciferase (transfection efficiency control) activities. The vertical axis is plotted as log2-fold to better show the smaller effects of single effectors. These effects may, in part, involve complexes with endogenous Arabidopsis effectors. D, BiFC assay showing the physical interaction between MYB5 and MYB14. The EYFP C-terminal half (YFPc) and EYFP N-terminal half (YFPn) were fused with the indicated genes and used to transfect Arabidopsis protoplasts. As positive controls, the interaction between MYB5 and TT8 was confirmed by BiFC (top row, middle), and the full-length EYFP (YFPf) fused with MYB5 was also included. As a negative control, Arabidopsis AtMYB46 fused with YFPc was used to cotransfect protoplasts with YFPn:MYB14 in the presence of TT8 and WD40-1 (bottom row, right). EYFP signals were rendered as green, and chloroplast autofluorescence was rendered as red.
Both MYB5 and MYB14 alone were able to weakly activate the ANR promoter (Fig. 7B); this contrasts with their inability to interact with the promoter in the Y1H assay (Fig. 1). MYB5 alone had higher activation activity than MYB14 alone (23-fold versus 3.8-fold). When MYB5 and MYB14 were cotransfected with TT8 and WD40-1, the ANR and LAR promoter activities were strongly activated. We observed a 1,965-fold induction when MYB5 was cotransfected with TT8 and WD40-1 and a 794-fold induction when MYB14 was cotransfected with TT8 and WD40-1. These results support the hypothesis that MYB5 and MYB14 are independently able to form a complex with TT8 and WD40-1 to activate the ANR promoter. We also observed low transactivation, 2.8- and 1.8-fold, for TT8 and WD40-1 alone, respectively, which, as with the low level of transactivation by MYB5 and MYB14 alone, likely reflects the nonspecific formation of complexes with endogenous Arabidopsis activators. All these data are consistent with the operation of a classical MBW ternary complex.
To test whether MYB5 and MYB14 act redundantly in separate ternary complexes, we cotransfected the protoplasts with all four transcription factors, resulting in a 12,400-fold activation of the ANR promoter (Fig. 7B). Because the induction activity of the combination of all four transcription factors is more than four times higher than the sum of the activation by MYB5 and MYB14 ternary complexes with TT8 and WD40-1, we conclude that MYB5 and MYB14 are able to activate the ANR promoter synergistically.
We next tested the activation activities of these transcription factors on the promoter of LAR (Fig. 7C). Similar results were obtained to those with the ANR promoter. A weak activation activity was observed for MYB5 alone (20-fold) and MYB14 alone (2.8-fold). When TT8 and WD40-1 were cotransfected with MYB5 or MYB14, we observed 51- or 127-fold induction activity, respectively. Similar to the results with the ANR promoter, the highest induction activity (448-fold) was observed when all four transcription factors were cotransfected. This activation is 2.5 times higher than the sum of the transactivation activities of the MYB5 and MYB14 complexes with TT8 and WD40-1. Therefore, we conclude that MYB5 and MYB14 also synergistically activate the LAR promoter.
To determine whether the apparent synergistic action between MYB5 and MYB14 reflects the existence of a MYB5/MYB14 complex on the same promoter, we performed bimolecular fluorescence complementation (BiFC) assays between MYB5 and MYB14 in the presence of WD40-1, TT8, and the ANR promoter. We transfected the Arabidopsis protoplasts with MYB5 fused with the C-terminal half of enhanced yellow fluorescent protein (EYFP; YFPc:MYB5) and MYB14 fused with the N-terminal half (YFPn:MYB14) along with TT8, WD40-1, and the ANR promoter. The complementary YFP signals between MYB5 and MYB14 were readily observed when TT8 and WD40-1 were cotransfected with the BiFC plasmids (Fig. 7D, top left). In contrast, when TT8 or WD40-1 was omitted, no complementary YFP signal could be observed (Fig. 7D, bottom left and middle, respectively), indicating that MYB5 and MYB14 form the complex in a TT8- and WD40-1-dependent manner. It is noteworthy that the complementary YFP signals between MYB5 and MYB14 are multiple spotted signals, suggesting that MYB5 and MYB14 are parts of multiple transcription foci. The complementary YFP signals between MYB5 and TT8 were more diffuse (Fig. 7D, top middle), similar to those of full-length EYFP fused with MYB5 (YFPf:MYB5; Fig. 7D, top right), suggesting that the interaction between MYB5 and TT8 is not dependent on transcription foci. Taken together, our results strongly suggest that MYB5 indeed forms a complex with MYB14 and that this is part of a quaternary complex with TT8 and WD40-1 for the high-level transactivation of PA biosynthesis.
DISCUSSION
Both MYB5 and MYB14 Are PA Regulators in M. truncatula
M. truncatula MYB5 belongs to a small family of MYB transcription factors characterized by their highly conserved R2R3 domains at the N terminus and two other highly conserved motifs, C1 and C3, at the C terminus. Although members of this family have been shown to be involved in PA biosynthesis, loss-of-function mutants of these transcription factors result in quite different phenotypes in different species, suggesting that the mechanisms underlying the transcriptional control of PA biosynthesis may be different in different species. In Arabidopsis, loss of function of myb5 does not affect the expression level of genes related to PA biosynthesis. The involvement of AtMYB5 in PA biosynthesis is only apparent when Arabidopsis myb5 and tt2 are both knocked out, indicating that AtMYB5 plays only a minor role in PA biosynthesis (Gonzalez et al., 2009). In persimmon, complete loss of PA in nonastringent-type fruit is associated with mutation of myb4, and most of the structural genes in PA biosynthesis are turned off (Akagi et al., 2009), suggesting that DkMYB4 is the major regulator, if not the sole regulator, of PA biosynthesis in persimmon. Interestingly, in petunia (Petunia hybrida), the MYB transcription factor PH4 regulates vacuolar acidification but has no effect on anthocyanin content (Quattrocchio et al., 2006). No PA-related phenotype was reported in ph4 mutants, but mutation of the gene encoding PH5, a proton pump that is the direct target of PH4, drastically reduces the PA content in seeds and produces the typical lighter seed color of PA-deficient mutants (Verweij et al., 2008). In contrast to these observations, M. truncatula myb5 mutants accumulate significantly reduced levels of soluble PAs in the seed coats but exhibit a dark-red seed coat color (presumably arising from an anthocyanin-related compound; see below), suggesting that the regulatory architecture at the anthocyanin/PA branch in M. truncatula is distinctly different from that in the other systems described above.
TT2 is the major MYB regulator of PA biosynthesis in Arabidopsis. MYB14 is the most closely related MYB to TT2 in M. truncatula, but it was only identified after a reannotation of the genome sequence MT3.5v4 (Young et al., 2011). Hancock et al. (2012) reported that overexpression of TaMYB14 (the homolog of MtMYB14 in T. arvense) could induce PA accumulation in leaf tissues of Trifolium spp. and alfalfa, consistent with our result that MtMYB14 is able to promote PA accumulation in M. truncatula hairy roots. MtPAR, another TT2-related MYB transcription factor in M. truncatula, also promotes PA accumulation in M. truncatula hairy roots and alfalfa plants (Verdier et al., 2012). PA accumulation is also induced by strong expression of Arabidopsis TT2 in M. truncatula hairy roots (Pang et al., 2008). These results indicate that there is some degree of promiscuity with regard to the MYB components of the MBW complex, at least in heterologous systems.
In many plants, the color of the seed coat is mainly determined by its anthocyanin and PA contents. Seed coats with high PA and low anthocyanin contents exhibit bright brown colors due to PA oxidation. In contrast, seed coats with high anthocyanin content exhibit a dark-red color. In soybean (Glycine max), the recessive i allele, which releases CHS from silencing, results in high anthocyanin accumulation in the seed coat, which can be dark brown or even black depending on other loci such as R and T (Tuteja et al., 2004; Yang et al., 2010). In contrast, the dominant I allele, which silences CHS expression and hence blocks both anthocyanin and PA biosynthesis, results in a completely colorless seed coat. In Arabidopsis, most of the mutants isolated from screening for seed coat color display various degrees of reduced color to colorless phenotypes (tt mutants), indicating that these mutations abolish or reduce both anthocyanin and PA biosynthesis. So far in Arabidopsis, only the banyuls (ban) mutant, which harbors a mutation in the ANR gene, contains high anthocyanin content in the seed coat and hence exhibits a dark-red seed coat phenotype (Devic et al., 1999; Xie et al., 2003). The ban phenotype supports the exclusive role of ANR in PA biosynthesis. In soybean, three varieties with red-brown seed coats are associated with reduced ANR mRNA levels (Kovinich et al., 2012).
The tt2 mutant of Arabidopsis exhibits a strong transparent testa phenotype, suggesting that TT2 regulates both the anthocyanin and PA pathways (Nesi et al., 2001). Mutation of AtMYB5 does not result in a transparent testa phenotype, although the tt2 myb5 double mutant exhibits a slightly stronger transparent testa phenotype than tt2 alone (Gonzalez et al., 2009), suggesting that AtMYB5 can regulate both anthocyanin and PA pathways in Arabidopsis. Unlike mutants of their homologs in Arabidopsis, both M. truncatula myb5 and myb14 mutant seeds exhibit darker color than wild-type M. truncatula seeds, with M. truncatula myb5 exhibiting the stronger phenotype in terms of seed color. Due to the fact that these pigments are resistant to acid methanol and aqueous acetone extraction, their identities cannot be determined at present. However, given the fact that anthocyanin and PA biosynthesis share most of the biosynthetic steps, it is reasonable to assume that these pigments are some kind of cross-linked anthocyanin-related compound. One plausible hypothesis to interpret the seed color phenotypes is that both MYB5 and MYB14 mainly regulate PA biosynthesis with little or no effect on anthocyanin biosynthesis, allowing for the accumulation of anthocyanins (which are then further processed, for example by oxidation) in the seed coat and hence the darker seed color.
Downstream Targets of MYB5 and MYB14 in M. truncatula
Gene expression profiling of MYB5- or MYB14-overexpressing hairy roots revealed that both genes strongly induce the expression of structural genes specific to the PA biosynthesis pathway. ANR was induced more than 250-fold in MYB5-overexpressing hairy roots and 985-fold in MYB14-overexpressing hairy roots. These extremely high values mirror the effects of high overexpression of AtTT2 in M. truncatula hairy roots (Pang et al., 2008) and are, in part, explicable in terms of extremely low ANR expression in control tissues. As in AtTT2-expressing M. truncatula hairy roots, UGT72L1, a glycosyltransferase specific for epicatechin (Pang et al., 2008), was also highly induced in MYB5- and MYB14-expressing hairy roots. Besides PA pathway-specific genes, genes encoding enzymes functioning upstream of ANR or LAR, such as CHS, F3′H, and DFR, which could potentially be involved in both anthocyanin and PA biosynthesis, were also induced by MYB5 and MYB14. However, compared with ANR, these genes are induced with much lower fold increase, suggesting that they might be induced through a secondary, feedback/feed-forward mechanism. MtTT8 and MtWD40-1, the two essential components of the MBW complex, are up-regulated around 2- to 3-fold, suggesting that there is a self-activating feedback regulation for the complex. In petunia, two MYB anthocyanin activators (Deep Purple and Purple Haze) also up-regulate Anthocyanin1 (AN1; TT8 homolog) and AN11 (WD40 homolog; Albert et al., 2014), consistent with our results here.
In addition to structural and regulatory genes, both MYB5 (strongly) and MYB14 (weakly) induced the expression of the MYB transcription factor Medtr5g079670 in hairy roots. Moreover, Medtr5g079670 is down-regulated in myb5 mutant seed, suggesting that MYB5 is a major regulator of this gene. Medtr5g079670 belongs to the MYB4-like transcription factor family, members of which have been characterized as transcriptional repressors (Jin et al., 2000). Interestingly, overexpression of AtMYB5 in Arabidopsis also induces an anthocyanin biosynthesis-suppressing MYB transcription factor, AtMYBL2 (Dubos et al., 2008; Matsui et al., 2008). Similar to Medtr5g079670, AtMYBL2 is also significantly down-regulated in the Arabidopsis myb5 mutant (Li et al., 2009). Medtr5g079670 is the most similar gene to AtMYBL2 in the M. truncatula genome and therefore might also function as an anthocyanin suppressor in M. truncatula. Unlike AtMYBL2, which has a truncated R2 domain, Medtr5g079670 has a more typical complete R2R3 domain, suggesting that structurally different MYB transcription factors might fulfill similar functions in different species. One MYB suppressor from petunia, PhMYB27, which shows high homology to Medtr5g079670, was recently reported to suppress anthocyanin biosynthesis (Albert et al., 2014). Therefore, one possible role for Medtr5g079670 in PA biosynthesis could be to suppress the anthocyanin-specific pathway so that metabolic flux can be more efficiently directed to PA biosynthesis.
Similar to the loss of function of AtMYB5, the M. truncatula myb5 mutant also produces less seed mucilage. Microarray gene expression profiling of the mutant seeds revealed that many genes encoding glycoside hydrolase and cell wall carbohydrate metabolism are down-regulated in the mutant seeds. One of the glycoside hydrolases, Medtr3g088520 (Mtr.17284.1.S1_at), shows high similarity with Arabidopsis MUM2. The mum2 mutant has defects in mucilage exclusion upon seed hydration (Huang et al., 2011). A gene annotated as encoding a putative aldehyde dehydrogenase is the most strongly induced gene in MYB5-overexpressing hairy roots, although the function of this gene (flavonoid or mucilage biosynthesis) and its potential aldehyde substrate in Medicago spp. are currently unclear.
Striking observations from this study are that myb5 mutant seeds exhibit a more conspicuous dark-red seed color than myb14 seeds and that less soluble PA can be extracted from myb5 seeds. In contrast to the minor role of MYB5 in PA biosynthesis in Arabidopsis, these results suggest that MtMYB5 plays at least as important a role as MtMYB14 (orthologous to AtTT2) in PA biosynthesis in Medicago spp. However, examination of the expression levels of major PA pathway structural genes did not reveal a significant difference between myb5 and myb14 seeds. Instead, a putative anthocyanin repressor, Medtr5g079670, appeared to be exclusively down-regulated in myb5 seeds. One hypothesis to explain the more conspicuous seed coat color of the myb5 mutant is that the anthocyanin repressor is essential to direct the metabolic flux to PA biosynthesis. In this way, knockout of myb5 will cause metabolic flux to be redirected to anthocyanin biosynthesis as well as via the down-regulation of the PA pathway structural genes. The combination of these two effects results in the stronger phenotype seen in the myb5 seeds. Another possible hypothesis is that myb5 predominantly regulates some unknown steps, such as epicatechin transport and condensation. The petunia homolog of MtMYB5 directly regulates a vacuolar proton pump (PH5) that is essential for PA transport (Verweij et al., 2008). It will be interesting to investigate whether a similar proton pump exists in Medicago spp. An acid vacuolar invertase is strongly down-regulated in myb5 seeds; this might possibly affect the vacuolar osmotic pressure and consequently affect PA accumulation in the vacuoles.
A Model for the Regulation of PA Biosynthesis in M. truncatula
The current model for the regulation of PA biosynthesis is mainly based on results from Arabidopsis, where TT2, TT8, and TTG1 form a ternary complex to activate PA biosynthesis genes (Baudry et al., 2004). In grape, VvMYB5a and VvMYB5b can activate the VvANR promoter in the presence of AtEGL3, suggesting that MYB5 proteins are also part of a similar ternary complex (Deluc et al., 2008). In our transactivation assays using Arabidopsis protoplasts, we observed that MtMYB5 and MtMYB14 alone can weakly activate both the ANR and LAR promoters, with MYB5 showing somewhat higher activity than MYB14. When MtTT8 and MtWD40-1 were included in the assay, the activation by MtMYB5 and MtMYB14 increased dramatically, supporting the ternary complex model. However, the highest transactivation activity was observed when all four transcription factors were cotransfected, and the effects of MYB5 and MYB14 were synergistic rather than additive. That this involves a previously unsuspected quaternary complex is suggested by the observation that BiFC analysis revealed a physical interaction between MYB5 and MYB14, but only in the presence of TT8 and WD40-1.
It was recently reported that petunia MYB27 functions as a cosuppressor by forming a quaternary complex with a second activator MYB and bHLH and WD40 transcription factors (Albert et al., 2014). This parallels our model in which MYB5 and MYB14 can form a quaternary complex with MtTT8 and MtWD40-1. Previous work has suggested the presence of two MYB molecules in transcriptional complexes for the activation of anthocyanin biosynthesis. For example, Kong et al. (2012) proposed a model to explain how different configurations of bHLH protein (maize [Zea mays] R) dimerization can function as a switch that directs the MBW complex to be tethered to different promoters. In either of the two configurations, there are two MYB molecules (maize C1) in the complex. It is tempting to speculate that the two MYB molecules can be present as a homodimer or, as suggested by this work and that of Albert et al. (2014), as a heterodimer. Such heterodimers may consist of two transcriptional activators with synergistic activities or an activator and a repressor. The promoters of ANR and LAR have both MYB and bHLH binding sites; it remains to be determined exactly how the transcriptional complex is assembled on these promoters.
Recently, we discovered that another M. truncatula MYB transcription factor, PAR, also regulates PA biosynthesis in the M. truncatula seed coat (Verdier et al., 2012). Unlike loss of function of MYB5 and MYB14, the par mutant exhibits a lighter seed coat color than the wild type, indicating that PAR either regulates both anthocyanin and PA biosynthesis or does not negatively regulate the anthocyanin pathway. The mechanism by which PAR, MYB5, and MYB14 together orchestrate PA biosynthesis is currently under investigation.
MATERIALS AND METHODS
Plant Materials
Medicago truncatula ecotype R108 was used as the wild type for comparison with Tnt1 insertion mutants. The tobacco (Nicotiana tabacum) Tnt1 insertion-mutagenized M. truncatula population was screened to identify MYB5 and MYB14 mutants as described (Tadege et al., 2008). Seeds were scarified with concentrated sulfuric acid for 10 min and then washed with a large amount of water five times to remove sulfuric acid. Scarified seeds were sterilized with 10% (v/v) bleach for 10 min and then rinsed five times with sterile water. myb5 mutant seeds were sterilized with 10% (v/v) bleach without sulfuric acid scarification. Sterilized seeds were vernalized at 4°C for 4 d on moist, sterile filter paper. Vernalized seeds were germinated on filter paper for 5 d before transfer to soil in pots. The plants were grown in a greenhouse set at a 12-h/12-h day/night cycle at 24°C.
Y1H Screening and Assays
The M. truncatula WD40-1 promoter reporter yeast (Saccharomyces cerevisiae) strain was generated as described by Verdier et al. (2012). A cDNA library was constructed from RNAs of 10-DAP seeds of M. truncatula A17 using the CloneMiner II cDNA Library Construction Kit (Invitrogen). The cDNA library was then cloned into pAG425GPD-ccdB (Alberti et al., 2007) to generate the library for yeast screening. The library plasmid DNA was used to transform the WD40-1 promoter reporter strain. Positive colonies were selected on yeast Leu dropout medium supplemented with 200 ng mL−1 aureobasidin A. To reconfirm the Y1H results from screening, full-length cDNAs of putative effectors were cloned into pAG425GPD-ccdB and Y1H assays were performed as described (Verdier et al., 2012).
Complementation of Arabidopsis tt2 with MtMYB5, MtPAR, and MtMYB14
The Arabidopsis (Arabidopsis thaliana) ANR (BAN) promoter was amplified by the Ban-pro-F/Ban-pro-R primer pair from genomic DNA and cloned into pMDC32 (Curtis and Grossniklaus, 2003) at the HindIII and KpnI sites to replace the 35S promoter. The resulting plasmid, pMDC32-ProBan, was used to generate pMDC32-ProBan:MYB5, pMDC32-ProBan:PAR, and pMDC32-ProBan:MYB14 by using Gateway LR Clonase (Invitrogen). The resulting binary vectors were used to transform the Arabidopsis tt2 mutant (Salk_005260) by the floral dip method (Clough and Bent, 1998).
Plasmid Construction for Generating MYB5- and MYB14-Overexpressing Plants
To generate binary vectors for MYB5 and MYB14 overexpression, the cDNA sequences were amplified by RT-PCR from seed RNA using the primer pairs MtMYB5-F/MtMYB5-R and MtMYB14-F/MtMYB14-R, respectively (for all oligonucleotide primers used in this work, see Supplemental Table S4). Amplified cDNAs were first cloned into pENTR/D TOPO vectors and then cloned into the pB7GW2D binary vector by LR recombination reaction to generate MtMYB5-pB7GW2D and MtMYB14-pB7GW2D binary vectors. These binary vectors were transformed into Agrobacterium rhizogenes strain ARqua1, and hairy roots were generated by transforming M. truncatula ecotype A17 with A. rhizogenes as described in the Medicago Handbook (http://www.noble.org/Global/Medicagohandbook/pdf/AgrobacteriumRhizogenes.pdf).
RNA Isolation, qRT-PCR, and DNA Microarray Analysis
RNAs from plant tissues except seeds were isolated using the RNeasy Plant Mini Kit (Qiagen). Except for root samples, which were collected from 1-week-old seedlings grown on one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc, all other tissues were collected from soil-grown plants. RNAs from seeds were isolated from 12-DAP seeds using Plant RNA Reagent (Invitrogen). Isolated RNAs were treated with DNase I and then purified by the RNeasy MinElute Cleanup Kit (Qiagen). Cleaned RNAs were used to perform RT with SuperScript III Reverse Transcriptase (Invitrogen). qRT-PCR analysis was performed on an ABI 7900HT quantitative PCR machine according to the manufacturer’s instructions. Three technical replicates were used for each gene, and statistics were performed with Excel. Three biological replicates for hairy root samples and two biological replicates for 12-DAP seeds were used to perform microarray analyses using the Medicago GeneChip (Affymetrix) according to the manufacturer’s instructions. Raw data were normalized by the robust multichip averaging method. Presence and absence calls for probe sets were obtained by using the dCHIP algorithm (Li and Wong, 2001). The type I family-wise error rate was calculated by using a Bonferroni-corrected P value (threshold of 0.05). To identify differentially regulated probe sets, we used a P value threshold of 5% and at least a 2-fold difference between transformant/mutant lines and their respective controls.
DMACA and Ruthenium Red Staining
To visualize PAs, seeds were soaked in water for 2 h and then stained with 1% DMACA solution (1% [w/v] DMACA in 1:1 methanol:concentrated HCl) for 1 h. Stained seeds were then washed in 70% (v/v) ethanol for 1 h. To visualize mucilage, seeds were stained with 0.01% (w/v) aqueous Ruthenium Red solution for 10 min and then washed in water.
Quantification of PAs
For hairy roots, approximately 0.2 g of fresh tissue was used for extraction of the soluble PA fraction. For seeds, plant materials were first ground to a powder in liquid nitrogen and then freeze dried for 16 h. Approximately 50 mg of freeze-dried materials was then extracted. Extraction was three times with 1 mL of 70% (v/v) acetone and 0.5% (v/v) acetic acid, as described by Pang et al. (2008). Assay of soluble PAs (with DMACA reagent) and insoluble PAs (with butanol-HCl) was performed as described previously (Pang et al., 2008).
Determination of PA Composition by Normal and Reverse-Phase HPLC
All analyses, including size estimation by HPLC followed by postcolumn derivatization with DMACA and phloroglucinolysis, were performed as described by Pang et al. (2008).
UPLC Analysis of PA Monomers
Ten milligrams of lyophilized tissue powder was extracted with 80% (v/v) methanol containing 18 μg mL−1 umbelliferone (internal standard) for 4 h at 4°C with constant agitation. Samples were then centrifuged at 3,000g for 15 min. The supernatants were transferred to liquid chromatography vials and analyzed by UPLC-electrospray ionization-quadropole time of flight-MS (Waters ACQUITY UPLC device) according to the procedure described (Pang et al., 2009).
Promoter Transactivation Assays
To construct luciferase reporter constructs, approximately 2-kb genomic sequences upstream of the translation start codons of the ANR and LAR genes were PCR amplified from M. truncatula A17 genomic DNA using the primer pairs ANRP2K-F/ANRP2K-R and LARP2k-F/LARP2K-R, respectively (Supplemental Table S4) and cloned into p2GW7 to replace the 35S promoter in the vector. A firefly luciferase gene was then cloned into this vector to generate the promoter::luciferase reporter construct. The cDNAs of the effector transcription factors were first cloned into pENTRD vector and then cloned into p2GW7 by LR reaction to form the 35S::effector constructs. A Renilla luciferase gene was also cloned into p2GW7 to form the reference gene construct. Arabidopsis protoplasts were isolated and transformed as described by Sheen et al. (http://molbio.mgh.harvard.edu/sheenweb/protocols_reg.html). Two micrograms of each plasmid for reporters and effectors and 100 ng of Renilla luciferase-expressing vector (internal control for transfection efficiency) were used to transform 100-μL batches of protoplasts (approximately 1 × 106 cells mL−1). The Dual-Luciferase Reporter Assay Kit (Promega) was used to quantify the luciferase activities according to the manufacturer’s instructions. The GloMax 96 Microplate Luminometer (Promega) was used to read the luciferase activities.
BiFC
The constructs for BiFC assays were constructed by cloning the indicated genes into Gateway destination vectors pSAT4-DEST-nEYFP-C1 and pSAT5-DEST-cEYFP-C1 (https://www.bio.purdue.edu/people/faculty/gelvin/nsf/protocols_vectors.htm). Arabidopsis protoplasts were isolated and transfected as described above. Images were acquired with the Zeiss LCM 710 microscope.
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MYB5 (XM_003601561, Medtr3g083540.1), MYB14 (Mt3.5v4 contig_238935_1, Mt4.0v1 Medtr4g125520.1), and TT8 (XM_003590608, Medtr1g072320.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Biosynthesis pathway of PA.
Supplemental Figure S2. MtMYB5 is the homolog of AtMYB5.
Supplemental Figure S3. Multiple alignment of the homologs of MtMYB14.
Supplemental Figure S4. Genetic complementation of Arabidopsis tt2 with MtMyb14, MtPAR, and MtMyb5.
Supplemental Figure S5. Phloroglucinolysis analysis of PAs in MYB5- and MYB14-overexpressing hairy roots.
Supplemental Figure S6. UPLC-MS detection of epicatechin and epicatechin-3′-O-glucoside ions in hairy roots overexpressing MYB5.
Supplemental Figure S7. UPLC-MS detection of epicatechin and epicatechin-3′-O-glucoside ions in hairy roots overexpressing MYB14.
Supplemental Figure S8. RT-PCR to detect MYB5 and MYB14 transcripts in myb5 and myb14 mutant seeds.
Supplemental Figure S9. Butanol-HCl analysis of insoluble PA-like material in R108, myb5, and myb14 mutant seeds.
Supplemental Figure S10. Phloroglucinolysis analysis of soluble PAs in seeds of the wild type (R108), myb5-1, and myb14-1.
Supplemental Figure S11. Measurement of MYB14 copy number in the M. truncatula genome by quantitative PCR.
Supplemental Figure S12. Ruthenium Red staining showing that mucilage levels in myb14 are similar to those in wild-type R108.
Supplemental Table S1. PathExpress analysis of genes down-regulated in myb5 mutant seeds (pathways with P < 0.01 are highlighted).
Supplemental Table S2. Genes putatively involved in mucilage biosynthesis that are down-regulated in seeds of the M. truncatula myb5 mutant.
Supplemental Table S3. PathExpress analysis of genes down-regulated in myb14 mutant seeds (the pathway with P < 0.01 is highlighted).
Supplemental Table S4. List of oligonucleotide sequences used in this study.
Supplemental Data Set S1. Differentially expressed genes in MYB5-overexpressing hairy roots.
Supplemental Data Set S2. Differentially expressed genes in MYB14-overexpressing hairy roots.
Supplemental Data Set S3. Genes up-regulated more than 2-fold by both MYB5 and MYB14.
Supplemental Data Set S4. Differentially expressed genes in myb5 mutant seeds.
Supplemental Data Set S5. Differentially expressed genes in myb14 mutant seeds.
Supplemental FASTA S1. Protein sequences of MYB5-like proteins.
Supplemental FASTA S2. Protein sequences of TT2-like proteins.
Supplemental FASTA S3. Protein sequences for phylogenetic tree construction.
Supplementary Material
Acknowledgments
We thank Dr. Xianzhi He for generating transgenic hairy roots, Drs. Jiangqi Wen and Xiaofei Chen for screening for M. truncatula Tnt1 insertion mutants, Dr. Yuhong Tang for microarray analysis, and Dr. Stephen Temple for critical reading of the article.
Glossary
- PA
proanthocyanidin
- bHLH
basic helix-loop-helix
- Y1H
yeast one-hybrid
- DAP
days after pollination
- cDNA
complementary DNA
- DMACA
dimethylaminocinnamaldehyde
- BiFC
bimolecular fluorescence complementation
- qRT
quantitative reverse transcription
- UPLC
ultra-performance liquid chromatography
- MS
mass spectrometry
- RT
reverse transcription
Footnotes
This work was supported by the U.S. Department of Agriculture/National Institute of Food and Agriculture (grant no. 2010–65115–20373), Forage Genetics International, and the Samuel Roberts Noble Foundation.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K. (2009) DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol 151: 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, Delseny M, Devic M. (1997) BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J 11: 289–299 [DOI] [PubMed] [Google Scholar]
- Alberti S, Gitler AD, Lindquist S. (2007) A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma A, Kobayashi S, Mitani N, Shiraishi M, Yamada M, Ueno T, Kono A, Yakushiji H, Koshita Y. (2008) Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor Appl Genet 117: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Bloor SJ. (1997) Blue flower colour derived from flavonol: anthocyanin co-pigmentation in Ceanothus papillosus. Phytochemistry 45: 1399–1405 [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S. (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140: 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F. (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147: 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M. (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19: 387–398 [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffard N, Weiller G. (2007) PathExpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res 35: W176–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev Biol 325: 412–421 [DOI] [PubMed] [Google Scholar]
- Hancock KR, Collette V, Fraser K, Greig M, Xue H, Richardson K, Jones C, Rasmussen S. (2012) Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol 159: 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, DeBowles D, Esfandiari E, Dean G, Carpita NC, Haughn GW. (2011) The Arabidopsis transcription factor LUH/MUM1 is required for extrusion of seed coat mucilage. Plant Physiol 156: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Pattanaik S, Feller A, Werkman JR, Chai C, Wang Y, Grotewold E, Yuan L. (2012) Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc Natl Acad Sci USA 109: E2091–E2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovinich N, Saleem A, Arnason JT, Miki B. (2012) Identification of two anthocyanidin reductase genes and three red-brown soybean accessions with reduced anthocyanidin reductase 1 mRNA, activity, and seed coat proanthocyanidin amounts. J Agric Food Chem 60: 574–584 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW. (2009) The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YG, Tanner G, Larkin P. (1996) The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric 70: 89–101 [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP. (2009) The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci USA 89: 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molan AL, De S, Meagher L. (2009) Antioxidant activity and polyphenol content of green tea flavan-3-ols and oligomeric proanthocyanidins. Int J Food Sci Nutr 60: 497–506 [DOI] [PubMed] [Google Scholar]
- Naumann HD, Muir JP, Lambert BD, Tedeschi LO, Kothmann MM. (2013) Condensed tannins in the ruminant environment: a perspective on biological activity. J Agric Sci 1: 8–20 [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Cheng X, Huhman DV, Ma J, Peel GJ, Yonekura-Sakakibara K, Saito K, Shen G, Sumner LW, Tang Y, et al. (2013) Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta 238: 139–154 [DOI] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA. (2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105: 14210–14215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Wright E, Wang ZY, Dixon RA. (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Wenger JP, Saathoff K, Peel GJ, Wen J, Huhman D, Allen SN, Tang Y, Cheng X, Tadege M, et al. (2009) A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol 151: 1114–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel GJ, Pang Y, Modolo LV, Dixon RA. (2009) The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J 59: 136–149 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I. (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. (2006) PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large scale insertional mutagenesis using Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR. (2003) Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278: 31647–31656 [DOI] [PubMed] [Google Scholar]
- Tuteja JH, Clough SJ, Chan WC, Vodkin LO. (2004) Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J, Zhao J, Torres-Jerez I, Ge S, Liu C, He X, Mysore KS, Dixon RA, Udvardi MK. (2012) MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc Natl Acad Sci USA 109: 1766–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F. (2008) An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat Cell Biol 10: 1456–1462 [DOI] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW. (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Dixon RA. (2004) Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch Biochem Biophys 422: 91–102 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C. (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol 202: 132–144 [DOI] [PubMed] [Google Scholar]
- Yang K, Jeong N, Moon JK, Lee YH, Lee SH, Kim HM, Hwang CH, Back K, Palmer RG, Jeong SC. (2010) Genetic analysis of genes controlling natural variation of seed coat and flower colors in soybean. J Hered 101: 757–768 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Iwasaka R, Kaneko T, Sato S, Tabata S, Sakuta M. (2008) Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol 49: 157–169 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21: 2323–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.