Chromatin from the rice egg nucleus initially moves into the sperm nucleus during karyogamy, which enlarges the latter before the male chromatin decondenses in nuclear fusion.
Abstract
In angiosperms, the conversion of an egg cell into a zygote involves two sequential gametic processes: plasmogamy, the fusion of the plasma membranes of male and female gametes, and karyogamy, the fusion of the gametic nuclei. In this study, the nuclei and nuclear membranes of rice (Oryza sativa) gametes were fluorescently labeled using histones 2B-green fluorescent protein/red fluorescent protein and Sad1/UNC-84-domain protein2-green fluorescent protein, respectively, which were heterologously expressed. These gametes were fused in vitro to produce zygotes, and the nuclei and nuclear membranes in the zygotes were observed during karyogamy. The results indicated that the sperm nucleus migrates adjacent to the egg nucleus 5 to 10 min after plasmogamy via an actin cytoskelton, and the egg chromatin then appears to move unidirectionally into the sperm nucleus through a possible nuclear connection. The enlargement of the sperm nucleus accompanies this possible chromatin remodeling. Then, 30 to 70 min after fusion, the sperm chromatin begins to decondense with the completion of karyogamy. Based on these observations, the development of early rice zygotes from plasmogamy to karyogamy was divided into eight stages, and using reverse transcription PCR analyses, paternal and de novo synthesized transcripts were separately detected in zygotes at early and late karyogamy stages, respectively.
In angiosperms, the sporophytic generation is initiated by double fertilization to form seeds (for review, see Raghavan, 2003). In double fertilization, one sperm cell from the pollen grain fuses with the egg cell, and the resultant zygote develops into an embryo that can transmit genetic material from the parents to the next generation. The central cell fuses with the second sperm cell to form a triploid primary endosperm cell, which develops into the endosperm that nourishes the developing embryo/seedling (Nawaschin, 1898; Guignard, 1899; Russell, 1992). The conversion of the egg cell into the zygote is completed by two sequential gametic processes: plasmogamy, the fusion of the male and female gametes’ plasma membranes, and karyogamy, the fusion of the male and female gametes’ nuclei.
GENERATIVE CELL SPECIFIC1/HAPLESS2 (GCS1/HAP2) and EGG CELL1 (EC1) have been identified as male and female, respectively, gamete factors for plasmogamy. GCS1/HAP2 was identified as a key male membrane protein with a single transmembrane domain and an His-rich domain in the extracellular region (Mori et al., 2006; von Besser et al., 2006). Recently, Sprunck et al. (2012) indicated that small Cys-rich EC1 proteins, which accumulated in the storage vesicles of Arabidopsis (Arabidopsis thaliana) egg cells, were secreted through exocytosis on sperm cell attachment to the egg cell, and the secreted EC1 proteins functioned in redistributing GCS1/HAP2 proteins to the sperm cell surface, resulting in successful gamete fusion.
Karyogamy involves the approach and fusion of male and female nuclei in a zygote. An intensively investigated karyogamic event is the fusion of two haploid nuclei during yeast mating, in which cytoskeleton-dependent nuclear migration, termed nuclear congression, and chaperon/endoplasmic reticulum (ER) protein-dependent nuclear fusion have been reported (Kurihara et al., 1994; Melloy et al., 2009; Tartakoff and Jaiswal, 2009; Gibeaux et al., 2013). Additionally, the cell cycle arrest and resumption induced by fertilization have been well studied using animal eggs (for review, see Kishimoto, 2003). Interestingly, in starfish eggs, Tachibana et al. (2008) indicated that cyclin B-cyclin-dependent kinase1 was required to form the sperm astral microtubules on which female and male pronuclei migrate during karyogamy. This suggested alternative cyclin B-cyclin-dependent kinase1 function in addition to its well-known role in mitotic spindle assembly during M phase (Karsenti and Vernos, 2001). In animals, the pronuclear migration in the fertilized gamete during karyogamy is dependent on the microtubule system. However, changes in the actin distribution by cytochalasin D, an inhibitor of actin polymerization, in mouse eggs is also reported to prevent pronuclear migration (Maro et al., 1984).
In angiosperms, the mechanisms responsible for the migration of the sperm nucleus toward the egg nucleus in the fertilized egg cell still remain undetermined, although the actin corona/patch is organized around or in female gametes and supposed to function in gamete fusion (Huang and Russell, 1994; Fu et al., 2000). Using transmission electron microscopy, nuclear fusion in angiosperm zygotes was first observed in the pollinated ovaries of cotton (Gossypium hirsutum; Jensen, 1964), and later, karyogamy in several kinds of plants, including wheat (Triticum aestivum) and barley (Hordeum vulgare), was reported (Van Went, 1970; Wilms, 1981; Mogensen, 1982; You and Jensen, 1985; Hause and Schröder, 1987; Tian and Shen, 1992). From these observations, three main steps for nuclear fusion have been described: (1) the external nuclear membranes become closely apposed or in contact through the ER and then fuse; (2) the inner nuclear membranes fuse, and bridges are formed between nuclei; and (3) the bridges enlarge and can entrap some cytoplasm. The male chromatin then begins to decondense.
To address the dynamics of karyogamy, Faure et al. (1993) obtained a precise timetable of the karyogamic progression by observing the nuclear approach and fusion in zygotes produced by the electrofusion of isolated maize (Zea mays) gametes. Faure et al. (1993) indicated that karyogamy in the zygotes occurred within 2 to 3 h after gamete fusion. 4′,6-diamino-phenylindole staining of barley zygotes isolated from pollinated ovaries also indicated the integration of male chromatin into the egg nucleus, and the decondensation of the male chromatin in the fused nucleus occurs a few hours after pollination (Mogensen and Holm, 1995). As for the membrane fusion of gamete nuclei, Maruyama et al. (2010) revealed that Bip, an ER-localized heat shock protein70 chaperon, is essential for the nuclear fusion of polar nuclei in the central cell. In addition, NUCLEAR FUSION DEFECTIVE1, a component of mitochondrial ribosomes, is also reported to be involved in the nuclear fusion of Arabidopsis karyogamy (Portereiko et al., 2006). After nuclear fusion, chromatin that was tightly packed in the sperm nucleus decondenses in the fused nucleus. Interestingly, nascent synthesis of the paternal mRNA and proteins in fused gametes are reported to coincide with male chromatin decondensation (Scholten et al., 2002), and the zygotic genome appears to switch on only hours after fertilization (Ingouff et al., 2007; Meyer and Scholten, 2007; Zhao et al., 2011; Nodine and Bartel, 2012). Moreover, it has been indicated that Histone3 (H3) variants contributed by paternal or maternal chromatin are diminished from the Arabidopsis zygotic nucleus within a few hours after fertilization, and these H3 variants are then de novo synthesized in the zygote, suggesting that the zygotic resetting of the H3 variants is part of the epigenetic reprogramming for zygotic development and embryogenesis (Ingouff et al., 2010). Although the mechanisms behind karyogamy and zygotic development have gradually become evident, how the male nucleus approaches and fuses with the female nucleus still remains unclear. This may be because of their location in zygote within the embryo sac, which is deeply embedded in the ovaries, making them difficult to trace.
To investigate the mechanisms of gametic and/or early zygotic development in angiosperms, we previously established a procedure to isolate rice (Oryza sativa) gametes and an in vitro fertilization (IVF) system to produce zygotes that can develop into fertile plants (Uchiumi et al., 2006, 2007). The rice IVF system has been used for analyzing gene/protein expression profiles in gametes/zygotes and polarity changes during zygotic development (Nakajima et al., 2010; Sato et al., 2010; Ohnishi et al., 2011; Abiko et al., 2013a, 2013b). In this study, zygotes were prepared by IVF of rice gametes heterologously expressing histone 2B (H2B)-GFP/red fluorescent protein (RFP) and Sad1/UNC-84-domain protein2 (SUN2)-GFP, which labeled the nuclei and nuclear membranes, respectively, and the dynamics of karyogamy in the rice zygotes were monitored. The results indicated that the sperm nucleus migrated adjacent to the egg nucleus in an actin filament-dependent manner within 5 to 10 min after gamete fusion, and then the egg chromatin became detectable in the sperm nucleus. On female chromatin transfer to inside the sperm nucleus through a possible connection between nuclei, the size of the nucleus was enlarged. Thereafter, according to the progression of nuclear fusion, sperm chromatin began to decondense 30 to 70 min after fusion. De novo gene expression was detectable when male chromatin had almost completely decondensed, and then, karyogamy was completed. In addition, based on the karyogamic progression, the precise staging of the early zygote was also presented for subsequent analyses on the early development of the fertilized egg/zygote.
RESULTS
Fluorescent Labeling of Chromatin and Nuclear Membrane in Rice Gametes
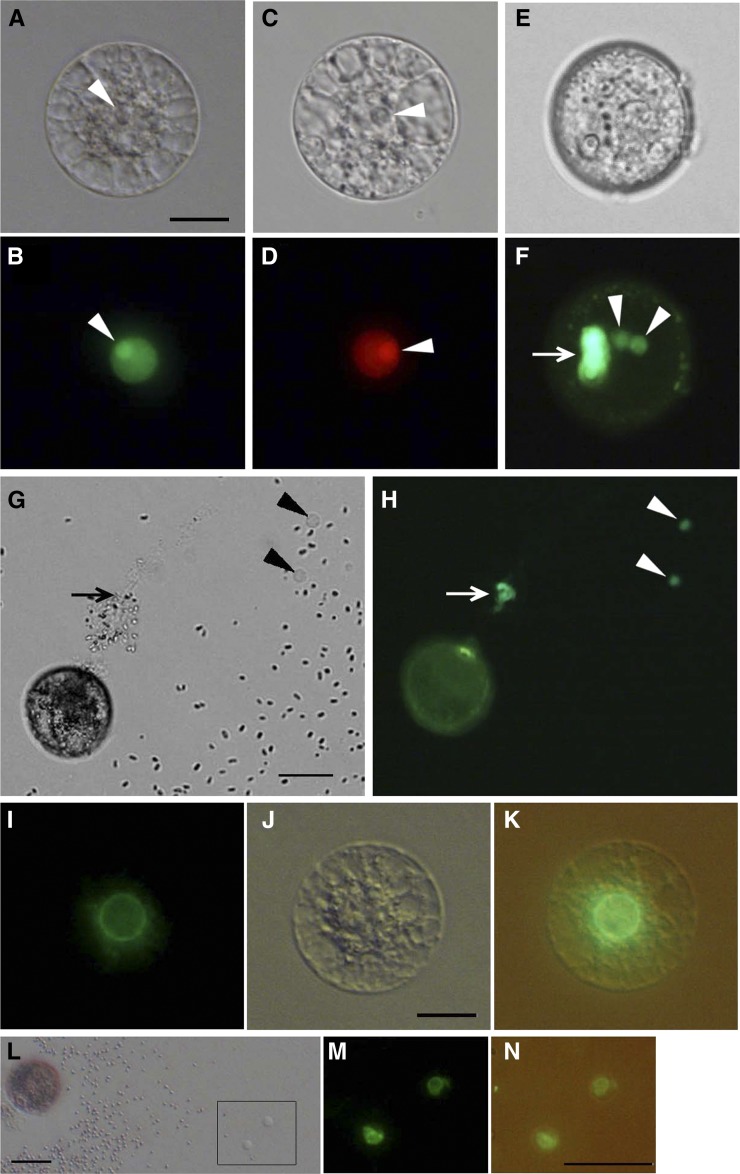
In egg cells prepared from transformed rice expressing H2B-GFP under the ubiquitin promoter, the nucleus was clearly visible (Fig. 1, A and B; Abiko et al., 2013b). In addition, observations for divisional profile of the zygotes, which were isolated from pollinated ovaries of the transformant, resulted in the possible identification of fluorescently labeled chromosomes during mitosis (Supplemental Fig. S1). These data are consistent with the report that H2B-GFP is incorporated into the chromatin and chromosome structures (Howe et al., 2012). In addition, nucleoli were strongly fluorescently labeled, although why such a strong signal was detected cannot be explained. A H2B-RFP fusion protein was heterologously expressed in rice plants under the DD45 promoter (Steffen et al., 2007), which is known to be egg cell specific in Arabidopsis. In egg cells isolated from the transformants, the nuclei were labeled by the fusion protein (Fig. 1, C and D), suggesting that the DD45 promoter could function as an active promoter in rice egg cells as well. In pollen grains from transformants expressing H2B-GFP, two putative sperm nuclei and a vegetative nucleus were visible (Fig. 1, E and F). When pollen grains were immersed in mannitol adjusted to 370 mosmol kg−1 of water, the pollen grains burst, releasing their contents, and the nuclei of the two released sperm cells and the vegetative nucleus were visible (Fig. 1, G and H).
Figure 1.
Rice gametes expressing the H2B-GFP, H2B-RFP, or SUN2-GFP fusion protein. A and B, An egg cell isolated from transgenic rice expressing H2B-GFP under control of the ubiquitin promoter visualized with bright-field (A) and fluorescence (B) microscopy. The arrowheads indicate nucleoli. C and D, An egg cell isolated from transgenic rice expressing H2B-RFP under control of the DD45 promoter visualized with bright-field (C) and fluorescence (D) microscopy. The arrowheads indicate nucleoli. E and F, A pollen grain expressing H2B-GFP under control of the ubiquitin promoter visualized with bright-field (E) and fluorescence (F) microscopy. Arrowheads and the arrow indicate sperm and vegetative nuclei, respectively. G and H, A pollen grain expressing H2B-GFP releasing its content in mannitol solution visualized with bright-field (G) and fluorescence (H) microscopy. Arrowheads indicate sperm cells, and the arrow indicates a possible vegetative nucleus. I to K, An egg cell expressing SUN2-GFP under control of the ubiquitin promoter visualized with fluorescence (I) and bright-field (J) microscopy. K contains the merged images of I and J. L to N, A pollen grain expressing SUN2-GFP releasing its content in mannitol solution (L). Two released sperm cells enclosed within the square in L were visualized with fluorescence microscopy (M). N contains the merged bright-field and fluorescent images. Bars = 20 μm. [See online article for color version of this figure.]
Arabidopsis SUN2 protein, a nuclear membrane protein (Graumann et al., 2010), was tagged with GFP, and the fusion protein was heterologously expressed in transformed rice under the ubiquitin promoter. The putative nuclear membrane was clearly visible in isolated egg cells (Fig. 1, I–K) and sperm cells (Fig. 1, L–N).
Male Chromatin Decondensation
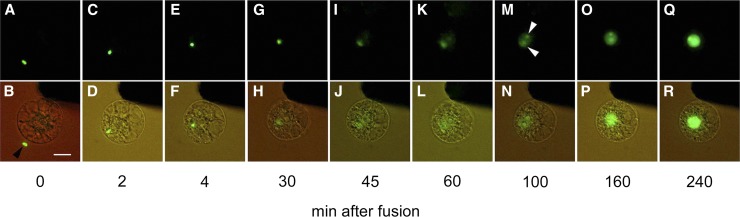
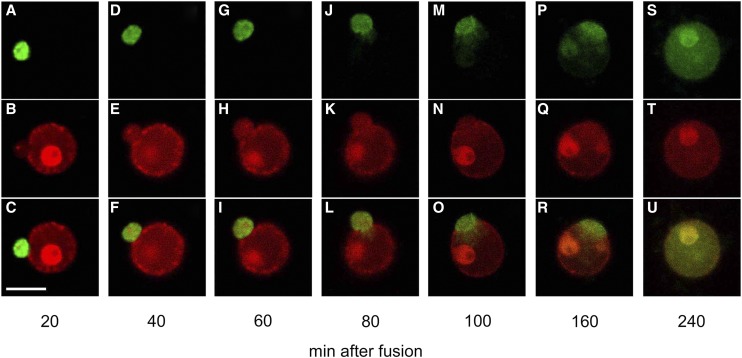
A sperm cell expressing H2B-GFP was electrofused with a wild-type egg cell, and fluorescence in the resulting zygote was observed (Fig. 2, A and B). The sperm nucleus migrated adjacent to the egg nucleus approximately 5 to 10 min after fusion (Fig. 2, C–F; Supplemental Fig. S2, A and B). Then, the sperm nucleus appeared to enlarge (Fig. 2, E–H; Supplemental Fig. S2, A–D), and the signal from the male chromatin began to distribute within the fused nucleus (Fig. 2, I–L; Supplemental Fig. S2, E–J). These observations suggested that nuclear fusion and the subsequent decondensation of sperm chromatin had occurred. After or during decondensation of the male chromatin, two nucleoli were detectable (Fig. 2, M and N; Supplemental Fig. S2, K and L), and then the nuclear signal intensity became progressively stronger (Fig. 2, M–R; Supplemental Fig. S3). This gradual increase in the H2B-GFP protein signal suggested that the H2B-GFP transcript and protein were de novo synthesized in the zygotes during or immediately after nuclear fusion, consistent with the report that nascent mRNA and protein synthesis coincided with the decondensation of male chromatin in fused nuclei of maize zygotes (Scholten et al., 2002).
Figure 2.
Karyogamy in rice zygotes. A and B, Alignment of a sperm cell expressing H2B-GFP with a wild-type egg cell on one of the electrodes under an alternating current field in a fusion droplet. C to R, Gametes in B were electrically fused, and the resulting fused cell (zygote) was observed at the time points indicated. Top shows fluorescent images, and bottom shows merged bright-field and fluorescent images. Black and white arrowheads indicate sperm cells expressing H2B-GFP and nucleoli, respectively. Bar = 20 μm. [See online article for color version of this figure.]
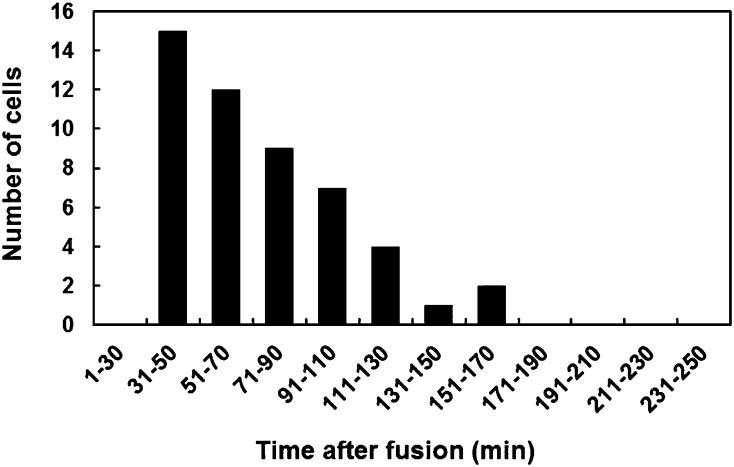
In this study, 50 independent zygotes produced by the fusion of a wild-type egg cell with a sperm cell expressing H2B-GFP were observed every 15 to 20 min after fusion to estimate the timetable from gamete fusion to the onset of sperm chromatin decondensation. Male chromatin decondensation was most prominent 31 to 70 min after fusion (Fig. 3), although the time ranged from 31 to 170 min after fusion. These results indicated that the time course from gamete fusion to decondensation in rice zygotes was almost equivalent to that in zygotes of maize (2 h after gamete fusion; Scholten et al., 2002), barley (80–100 min after pollination; Mogensen and Holm, 1995), and Arabidopsis (2 h after fertilization; Ingouff et al., 2010).
Figure 3.
Timetable of sperm chromatin decondensation in the fused nuclei of 50 independent rice zygotes. Zygotes were produced by in vitro fusion of a wild-type egg cell with a sperm cell expressing a H2B-GFP fusion protein, and the fluorescence signal was observed every 15 to 20 min after fusion. The y axis indicates the number of zygotes in which initial decondensation of sperm chromatin was observed.
Cytoskelton and Karyogamy
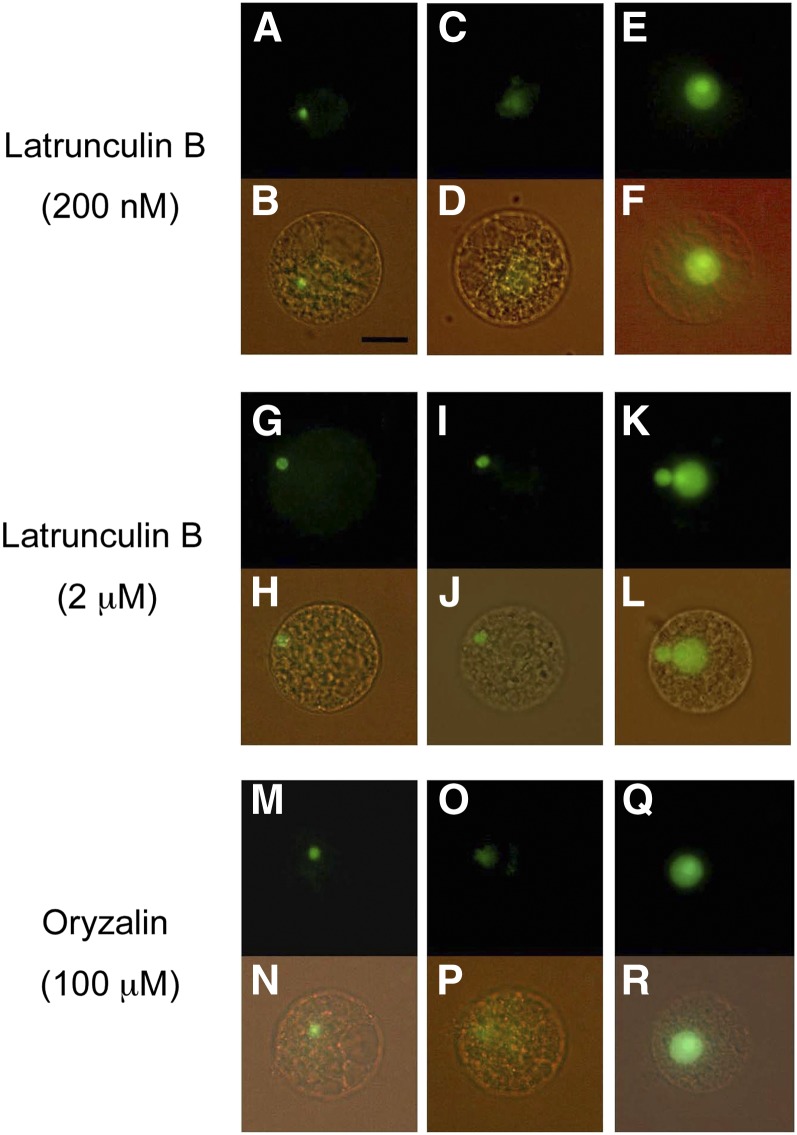
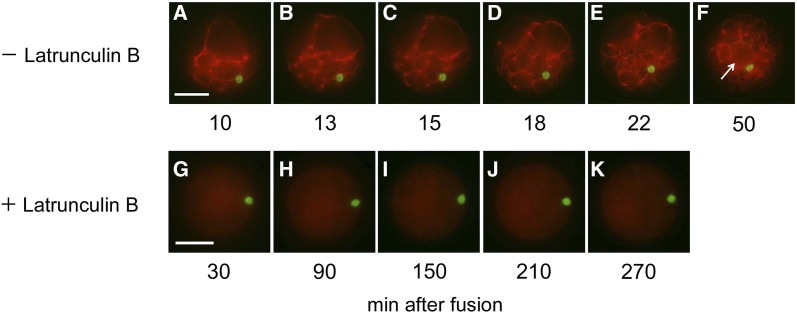
The effects of latrunculin B, an inhibitor of actin polymerization (Coué et al., 1987; Spector et al., 1989), and oryzalin, an inhibitor for tubulin assembly (Hugdahl and Morejohn, 1993), on karyogamy in rice zygotes were observed. In zygotes, which were prepared from the fusion of a sperm cell expressing H2B-GFP with an egg cell treated with 200 nm latrunculin B, karyogamy progressed normally (Fig. 4, A–D), and the zygotic nucleus was clearly visible after the completion of karyogamy (Fig. 4, E and F). When an egg cell treated with 2 μm latrunculin B was used for the production of a zygote, the sperm nucleus did not approach the egg nucleus (Fig. 4, G–J). In zygotes at 12 h after introduction, the sperm nucleus was still not fused with the egg nucleus, and interestingly, the fluorescent signal was detected in both the sperm and egg nuclei (Fig. 4, K and L). The effect of latrunculin B on actin organization in egg cells was also observed (Supplemental Fig. S4). In the egg cell, a mesh-like structure labeled by Lifeact-tagRFP was clearly visible, and the structure remained evident after treatment with 200 nm latrunculin B for 120 min. However, the structure was not visible in egg cells treated with 2 μm latrunculin B for 90 min. The difference in the effects of latrunculin B on karyogamy between the 200 nm and 2 µm concentrations (Fig. 4) may be caused by differences in the actin filament structure at the respective inhibitor concentrations. In contrast to latrunculin B, oryzalin showed no effect on karyogamy in rice zygotes at 10 (data not shown) and 100 µm (Fig. 4, M–R) concentrations.
Figure 4.
Effects of latrunculin B and oryzalin on karyogamy in fused rice gametes. Egg cells were treated with inhibitors and used for electrofusion with sperm cells expressing H2B-GFP. A to F, Egg cells and fused cells that were treated with 200 nm latrunculin B. G to L, Egg cells and fused cells that were treated with 2 μm latrunculin B. M to R, Egg cells and fused cells that were treated with 100 μm oryzalin. After in vitro fertilization, the fused cells were observed at 10 to 15 min (A, B, G, H, M, and N), 80 to 120 min (C, D, I, J, O, and P), and 10 to 12 h (E, F, K, L, Q, and R) after fusion. In zygotes treated with 2 μm latrunculin B, the decondensation of sperm chromatin was not observed for 2 h after fusion (I and J), and the signal from H2B-GFP was detected in both sperm and egg nuclei at 12 h after fusion. Bar = 20 μm. [See online article for color version of this figure.]
A sperm cell and an egg cell expressing H2B-GFP and Lifeact-tagRFP, respectively, were fused to trace the migration of the sperm nucleus and organization of actin filaments in a zygote. Actin filaments existed around or near sperm nucleus, and the sperm nucleus appeared to migrate along actin filaments (Fig. 5). In contrast, the sperm nucleus hardly migrated in the zygote when actin depolymerization was induced by latrunculin B treatment.
Figure 5.
Actin organization and sperm nucleus migration. Egg cells expressing Lifeact-tagRFP without (A–F) or treated with (G–K) 2 μm latrunculin B were fused with sperm cells expressing H2B-GFP, and resultant zygotes were observed at the time point indicated. An arrow indicates the nucleus surrounded by actin filaments. Bars = 20 μm.
Movement of Female Chromatin and Enlargement of the Sperm Nucleus
A wild-type sperm cell and an egg cell expressing H2B-GFP were fused. Although the sperm nucleus was not visible during intracellular migration or shortly after contacting the egg nucleus (Supplemental Fig. S2, M and N), interestingly, the fluorescent signal was detected in the sperm nucleus as well as in the egg nucleus 20 min after fusion (Supplemental Fig. S2, O and P). Thereafter, the nuclear fusion progressed, and karyogamy was completed (Supplemental Fig. S2, Q–X). In addition, the fusion process of the nuclear membranes was observed when male and female gametes, with nuclear membranes that were labeled with SUN2-GFP, were fused (Supplemental Fig. S5).
To visualize karyogamy in detail, zygotes produced by electrofusion of a sperm cell expressing H2B-GFP and an egg cell expressing H2B-RFP were observed using a confocal laser scanning (CLS) microscope. In zygotes, both the GFP-derived and RFP-derived signals from the sperm and egg nuclei, respectively, were observed, with the signal from the egg nucleus localizing inside the sperm nucleus at 20 min after fusion (Fig. 6, A–C). This suggests that female chromatins labeled by H2B-RFP or free H2B-RFP proteins in egg nucleus were distributed into the sperm nucleus through possible connection between male and female nuclear membranes. Alternatively, the possibility cannot be excluded that H2B-RFP proteins translated in the fused egg cell were transported into the sperm nucleus and that the H2B-RFP-derived signal became detectable. Therefore, an egg cell expressing H2B-RFP was treated with 2 μm latrunculin B and fused with a sperm cell expressing H2B-GFP to see whether H2B-RFP signal becomes detectable in a sperm nucleus that does not contact with egg nucleus. The results indicated that the H2B-RFP-derived signal was not detected in a sperm nucleus at least 150 min after fusion (Supplemental Fig. S6), suggesting that detection of the H2B-RFP in a sperm nucleus attaching with an egg nucleus within 20 min after fusion (Fig. 6, A–C) was not caused by transport of translational products in a fused egg but rather was the result of the migration of egg nucleus contents through possible connection between male and female nuclei. In addition, because most H2B-GFP proteins heterologously expressed in the rice egg cells and zygotes appear to be incorporated in chromatins and chromosomes (Supplemental Fig. S1), detected H2B-RFP signals would be from egg chromatin rather than free H2B-RFP proteins in the egg nucleus. However, a high-time resolution observation should be conducted for judging which possibility is plausible.
Figure 6.
Chromatin state during or after nuclear fusion in rice zygotes. Zygotes produced by electrofusion of a sperm cell expressing H2B-GFP and an egg cell expressing H2B-RFP were observed using a CLS microscope. The fluorescent signals from H2B-GFP and H2B-RFP are presented in top (A, D, G, J, M, P, and S) and middle (B, E, H, K, N, Q, and T), respectively, whereas bottom (C, F, I, L, O, R, and U) shows merged images. Bar = 10 μm.
The putative egg chromatin signal became more condensed in the sperm nucleus (Fig. 6, D–I), but sperm chromatin was not detected in the egg nucleus. This indicated that sperm chromatin did not migrate into the egg nucleus at this karyogamic stage. Thereafter, nuclear fusion and the decondensation of male chromatin progressed (Fig. 6, J–R), and finally, karyogamy was completed (Fig. 6, S–U).
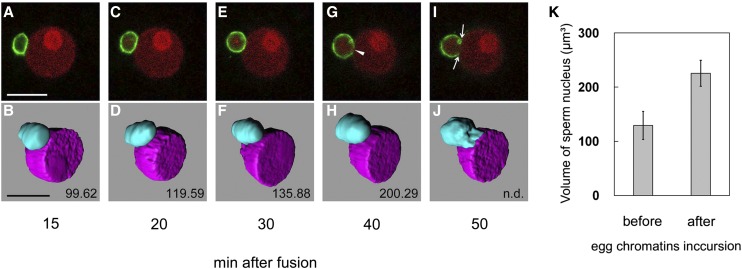
The possible one-way migration of egg chromatin into the sperm nucleus explains the enlargement of the sperm nucleus during or after contact with the egg nucleus (Fig. 2, E–H; Supplemental Fig. S2, A–D). Thus, a sperm cell expressing SUN2-GFP was fused with an egg cell expressing H2B-RFP, the serial sectional images of the nuclei in the fused cells were obtained with a CLS microscope, and the volume of the sperm nucleus before and after the incursion of female chromatin was measured. Female chromatin was not detectable in the sperm nucleus at 15 min after fusion, but the signal was observed at 20 min after fusion (Fig. 7, A and C). Although the detection of the egg nucleus contents in the sperm nucleus suggests that the connection between male and female nuclear membranes was formed 20 min after fusion, the SUN2-GFP signal was uniformly detected on the sperm nuclear membrane, and such possible connection could not be detected using fluorescent labeling of nuclear membrane by SUN2-GFP (Fig. 7C). To observe the initial connection between nuclear membranes, termed internuclear bridge, electron microscopic analysis is essential as reported (Jensen, 1964; Van Went, 1970; Wilms, 1981; Mogensen, 1982; You and Jensen, 1985; Hause and Schröder, 1987; Tian and Shen, 1992). When egg chromatin became detectable in the sperm nucleus, its volume was slightly larger than before the distribution of the egg chromatin (Fig. 7, B and D). The volume of the sperm nucleus continued to increase (Fig. 7, C–H), and a possible fusion point between the sperm and egg nuclei was observed (Fig. 7G). The fusing area enlarged as nuclear fusion progressed (Fig. 7, I and J). The volume of the sperm nucleus after the incursion of egg chromatin was approximately 1.75-fold larger compared with before egg chromatin incursion (Fig. 7K; before egg chromatin incursion, 129.34 ± 25.99 μm3; after egg chromatin incursion, 225.58 ± 24.08 μm3; n = 3). Statistical tests indicated that this difference was highly significant (P < 0.01).
Figure 7.
Changes over time in the volume of a rice sperm nucleus during contact with an egg nucleus. Zygotes produced by electrofusion of sperm cells expressing SUN2-GFP and an egg cell expressing H2B-RFP were observed using a CLS microscope. A, C, E, G, and I are CLS images, and B, D, F, H, and J are three-dimensional structure images. An arrowhead indicates the possible fusion point between the nuclear membranes. Arrows indicate the fusing area between sperm and egg nuclear membranes. Numbers in B, D, F, H, and J represent the volume of the sperm nucleus. Bars = 10 μm. K, Changes in the volume of the sperm nucleus before (A and B) or after (G and H) egg chromatin incursion. The data are means ± sds of three sperm nuclei.
DISCUSSION
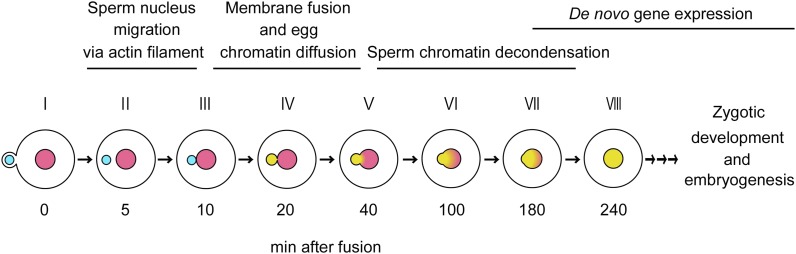
Based on these observations of karyogamy in rice zygotes, a schematic diagram of the progression of karyogamy is presented in Figure 8. After gamete fusion (stage I), the sperm nucleus migrates inside of the fused cell to appose the egg nucleus through an actin filament-dependent mechanism (stages II and III). In contrast to actin cytoskelton-dependent nuclear migration, in many animal species, it has been known that pronuclear congression in fused eggs depends on a microtubule aster, which is built from paternally inherited centrioles and maternally provided pericentriolar materials (Chambers, 1939; Schatten, 1994; Reinsch and Gönczy, 1998). The actin filament-dependent sperm nucleus migration in plant zygotes may be consistent with the characteristics of sperm cell, the male gamete in angiosperms, which does not possess the centrioles. After apposing of nuclei (stage III), female chromatin possibly moves into the sperm nucleus via putative internuclear connection, resulting in sperm nucleus enlargement, but the sperm chromatin remains tightly packed (stage IV). Male chromatin begins to decondense (stage V), and male chromatin further decondenses according to the progression of membrane fusion (stages VI and VII). The karyogamic event is finally completed at stage VIII.
Figure 8.
A schematic diagram of the progression of karyogamy in rice zygotes. After gamete fusion, the sperm nucleus migrates inside of the fused cell to appose the egg nucleus through possible actin filament-dependent machinery (stages I–III). Through a possible internuclear connection, female chromatin moves into the sperm nucleus, but sperm chromatin remains tightly packed (stage IV). Male chromatin begins decondensing (stage V), and male chromatin further uniformly distributes in accordance with the progression of nuclear fusion (stages VI and VII). The karyogamic event is finally completed at stage VIII, and thereafter, the zygote develops into an embryo. Light-blue and pink circles indicate sperm and egg nuclei/chromatin, respectively. Yellow indicates the merged sperm and egg chromatin.
The nascent synthesis of mRNAs and proteins from the parental genomes in zygotes was reported to be initiated either during karyogamy or within hours after fertilization in maize (Meyer and Scholten, 2007), Arabidopsis (Ingouff et al., 2010; Nodine and Bartel, 2012), and tobacco (Nicotiana tabacum; Zhao et al., 2011). However, a relationship between a precise karyogamic stage and the start of de novo gene expression in the zygote was not clearly presented. We recently conducted transcriptome analyses using isolated rice gametes and zygotes prepared from ovaries 2 to 4 h after pollination and globally identified genes that were up-regulated or down-regulated after fertilization (Abiko et al., 2013b). Therefore, in the study, the precise timing of the expression of several genes, which were known to be up-regulated after fertilization in rice zygotes, was monitored during karyogamy.
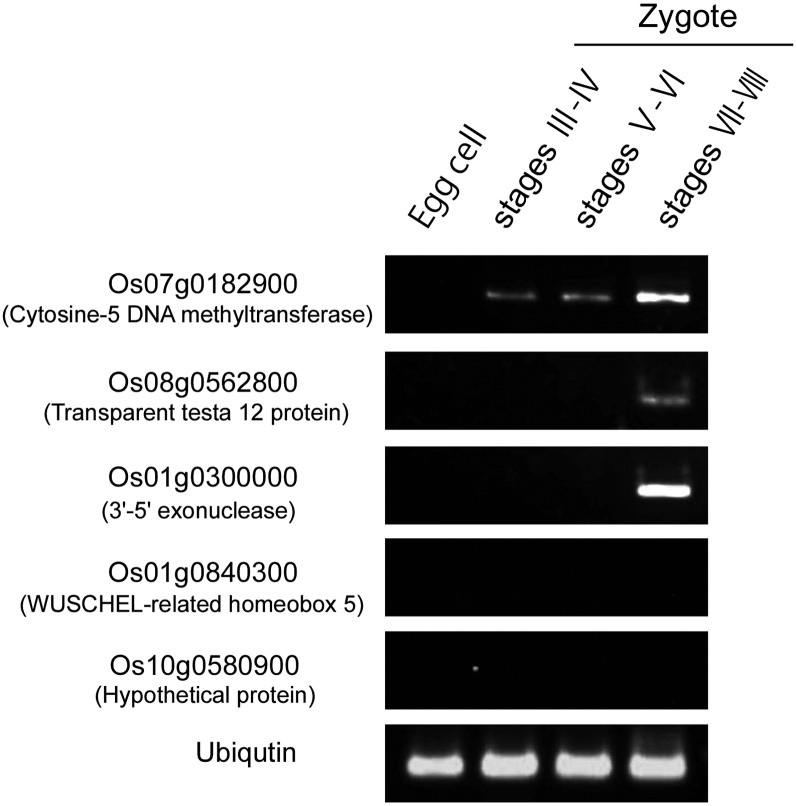
Zygotes produced by the fusion of sperm cells expressing H2B-GFP and wild-type egg cells were prepared and incubated. Then, zygotes that were in karyogamic stages III to IV, V to VI, and VII to VIII were selected for subsequent reverse transcription (RT)-PCR analyses. For the Os08g0562800 and Os01g0300000 genes, detectable expression levels occurred only at stages VII and VIII (Fig. 9). This indicates that the de novo expression of these genes in rice zygotes starts between stages VII and VIII, at which time the male chromatin in the fused nucleus has mostly decondensed, but there was no de novo gene expression during stages V and VI, at which time the initial decondensation of male chromatin occurs. For the Os07g0182900 gene, the transcript was detected from stages III to IV and V to VI, and the expression level increased during stages VII and VIII. The initial transcript detected at stages III to IV, approximately 10 to 20 min after fusion, will be derived from the paternal transcript, because the gene is not expressed in the egg cell before fusion (Fig. 9), and it was reported that the Os07g0182900 gene is highly expressed in sperm cells (Abiko et al., 2013b). Its increased expression level detected at stages VII and VIII will be a result of de novo zygotic gene expression. As for the Os01g0840300 and Os10g0580900 genes, no expression was detected, suggesting that the expression of these genes occurs in zygotes after the completion of karyogamy (stage VIII).
Figure 9.
Expression patterns of five genes putatively induced after fertilization in rice zygotes. Semiquantitative RT-PCR was performed on total RNAs isolated from 10 to 12 zygotes at the appropriate karyogamic stages. Ubiquitin mRNA was used as an internal control. Supplemental Table S1 shows primer sequences.
This study indicated that de novo zygotic gene expression starts in rice zygotes at stage VII to VIII of karyogamy, during which sperm chromatin has mostly decondensed in the fusing nucleus, and that paternally supplied transcripts were also detected in the zygote at stages III and IV of karyogamy before de novo expression started. In Arabidopsis, the contribution of such paternal mRNA for zygotic development is well known. After fertilization, vacuoles in Arabidopsis zygotes fragment, and the zygote elongates 2-fold to 3-fold before a large vacuole is reassembled (Mansfield and Briarty, 1991; Faure et al., 2002). The polarized zygote divides asymmetrically into a two-celled proembryo consisting of an apical cell and a basal cell, which develop into the embryo proper and the suspensor/hypophysis, respectively, suggesting that cellular polarity in the zygote is tightly linked to the establishment of the initial apical-basal axis in plants (Jeong et al., 2011; Zhang and Laux, 2011; Ueda and Laux, 2012). Bayer et al. (2009) indicated that transcripts of SHORT SUSPENSOR (SSP), encoding a member of the interleukin-1 receptor-associated kinase/Pelle-like kinase, are produced in sperm cells and delivered to the zygotes. Then, SSP proteins accumulate and activate the YODA-dependent mitogen-activated protein kinase kinase kinase signaling pathway that is thought to be crucial for the cell elongation of the zygote and the specified cell fate of the basal cell of Arabidopsis two-celled proembryo (Lukowitz et al., 2004). In addition, it has been reported that the WRKY2 transcription factor expressing in sperm cell as well as egg cell functions in establishment of early body plan in Arabidopsis zygote (Ueda et al., 2011). In the study, transcripts from the Os07g0182900 gene encoding cytosine-5 DNA methyltransferase1 detected at early karyogamy stage were considered as paternally supplied mRNA. Interestingly, it has been reported that the establishment of polarity or asymmetric cell division of rice zygotes is partly affected by treatment of zygotes with RG108, a specific inhibitor of methyltransferase1 (Abiko et al., 2013b). This could be another example of sperm mRNA involvement in zygotic development in angiosperms. Transcriptome analyses of rice zygotes at stages III to IV of karyogamy will result in the identification of genes with transcripts that are delivered from sperm cells and function in early zygotic development.
In rice zygote produced in vitro, de novo zygotic gene expression occurs during or after nuclear fusion in accordance with male chromatin decondensation in fused nucleus. However, in latrunculin B-treated zygotes, which are produced by fusion of a wild-type egg cell with a sperm cell expressing H2B-GFP, the H2B-GFP signal became detectable in an egg nucleus as well as a sperm nucleus, even if the sperm nucleus did not fuse with the egg nucleus (Fig. 4, K and L). This may indicate that egg activation and subsequent de novo gene expression progress in such a zygote without completion of karyogamy. Using a Ca2+-mediated IVF system with maize gametes, Antoine et al. (2000), (2001) indicated that gamete fusion induced egg cell activation, including rapid cell wall formation (Kranz et al., 1995), through a transitory increase of the Ca2+ levels in the fertilized egg cells. H2B-GFP expression in karyogamy-defected zygotes may suggest that the egg activation can be triggered by fusion stimuli and subsequent delivery of sperm cell cytoplasm into fertilized eggs and that initial zygotic events, including transcriptional/translational initiation, H3 replacement, and chromatin remodeling, toward de novo gene expression occur in each unfused male and female nucleus without karyogamy.
MATERIALS AND METHODS
Plant Materials, Isolation of Gametes and Zygotes, and the Electrofusion of Gametes
Rice (Oryza sativa ‘Nipponbare’) was grown in environmental chambers (K30-7248; Koito Industries) at 26°C with a 13-h/11-h light-dark cycle. The isolation of egg cells and sperm cells from rice flowers and the electrofusion of isolated gametes for zygote production were conducted as described by Uchiumi et al. (2006, 2007).
Vector Construction and Preparation of Transformants
Transformed rice expressing the H2B-GFP fusion protein was prepared as previously described (Abiko et al., 2013b). The plasmid vector, pENTRTM3C (Life Technologies), harboring Arabidopsis (Arabidopsis thaliana) DD45 promoter::H2B-TagRFP was a gift from Tomoko Igawa (Chiba University, Japan; Igawa et al., 2013). After the expression cassette was transferred to the destination vector pGWB1 (Nakagawa et al., 2007) using the Gateway LR Clonase Enzyme Mix (Invitrogen), the vector was used as the DD45 promoter::H2B-RFP vector to transform rice plants. For actin labeling using Lifeact (Era et al., 2009), pGWB1 binary vector, harboring Arabidopsis DD45 promoter::Lifeact-TagRFP, was a gift from Tomoko Igawa (Chiba University) and was used for rice transformation as below.
The promoter region of a ubiquitin gene (Os02g0161900) was subcloned into the HindIII and XbaI sites of pIG121-Hm, a binary plasmid vector harboring a GUS coding sequence, and the subsequent plasmid was used as Ubi promoter::GUS vector according to the work by Abiko et al. (2013b). The pGWB405 vector harboring a DNA fragment encoding the GFP-tagged Arabidopsis SUN2 (At5g22880) was a gift from Kentaro Tamura, (Kyoto University, Japan; Tamura et al., 2013). This vector was used as the template for PCR amplification of the DNA region encoding the H2B-GFP fusion protein using the specific primers 5′-GCTAGCATGTCGGCGTCAACGGTGT-3′ and 5′-GAGCTCTTACTTGTACAGCTCGTCCA-3′. The amplified PCR product was subcloned into the pGEM-T Easy vector (Promega), and the plasmid possessing the SUN2-GFP sequence was cut with NheI and SacI. Then, the excised DNA fragment was subcloned into the XbaI-SacI site of Ubi promoter::GUS vector, replacing the GUS sequence with SUN2-GFP. The vector was used as the Ubi promoter::SUN2-GFP vector to transform rice plants.
Agrobacterium tumefaciens LBA4404 was transformed with the above-mentioned vectors, and transgenic rice plants were prepared by cocultivation of scutellum tissue with A. tumefaciens according to Toki et al. (2006).
Microscopic Observation, Three-Dimensional Reconstruction, and Volume Measurements of Sperm Nuclei
Zygotes produced by IVF of isolated rice gametes were transferred into droplets of mannitol adjusted to 450 mosmol kg−1 of water. Gametes/zygotes expressing H2B-GFP or SUN2-GFP were observed with a BX-71 Inverted Fluorescence Microscope (Olympus) with 460- to 490-nm excitation and 510- to 550-nm emission wavelengths (U-MWIBA2 Mirror Unit; Olympus). For H2B-TagRFP proteins in gametes/zygotes, observations were conducted with 520- to 550-nm excitation and >580-nm emission wavelengths (U-MWIG Mirror Unit; Olympus). Digital images of gametes, zygotes, and their resulting embryos were obtained through a cooled CCD Camera (Penguin 600CL; Pixcera) and InStudio software (Pixcera). In addition to the BX-71 Inverted Fluorescence Microscope, zygotes were observed with a BZ9000 Microscope (Keyence) with 440- to 470-nm excitation and 535- to 550-nm emission wavelengths (OP-66836 GFP-BP filter set; Keyence) for H2B-GFP proteins and 525- to 540-nm excitation and 605- to 655-nm emission wavelengths (OP-66837 TRITC filter set; Keyence) for H2B-RFP and Lifeact-tagRFP proteins. IVF-produced zygotes were observed also with an LSM 710 CLS Microscope (Carl Zeiss) with 488-nm excitation and 505- to 530-nm emission wavelengths for H2B-GFP and SUN2-GFP and 543-nm excitation and >560-nm emission wavelengths for H2B-TagRFP.
The three-dimensional structure of the sperm nucleus in the zygote was constructed using IMARIS software (Bitplane) from a z series of CLS images in which sperm and egg nuclei were labeled with SUN2-GFP and H2B-TagRFP, respectively. The volume of the sperm nucleus was measured by surface rendering using the Surface function of IMARIS.
Inhibitor Treatment of Egg Cells and Zygotes
Latrunculin B (Wako) and oryzalin (Wako) were dissolved with methanol into stock solutions of 2 and 100 mm, respectively. Egg cells were incubated in droplets of mannitol solution adjusted to 370 mosmol kg−1 of water containing latrunculin B (200 nm and 2 μm) or oryzalin (10 and 100 μm) for 1.5 h. The egg cells were used for IVF with sperm cells, and the resultant zygotes were transferred into droplets of mannitol adjusted to 450 mosmol kg−1 of water containing the inhibitors for microscopic observation.
RT-PCR
Zygotes were produced by electrofusion of sperm cells expressing H2B-GFP with wild-type egg cells and incubated in droplets of mannitol solution adjusted to 450 mosmol kg−1 of water. The progression of karyogamy was observed by fluorescence microscopy as mentioned above, and the zygotes, at appropriate karyogamic stages, were selected and washed three times by transferring the cells into fresh droplets of mannitol solution on coverslips. The washed cells were submerged in 5 μL of extraction buffer supplied in a PicoPure RNA Isolation Kit (Life Technologies) and stored at −80°C until use. Complementary DNAs (cDNAs) were synthesized from total RNAs of 10 to 12 zygotes using the High-Capacity RNA-to-cDNA Kit (Life Technologies) according to the manufacturer’s instructions. For RT-PCR, 0.3 μL of first-strand cDNA was used as the template in a 20-μL PCR reaction with 0.3 μm primers using KOD-FX DNA Polymerase (Toyobo) as follows: 45 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 1 min. The expression of the ubiquitin gene (Os02g0161900) was monitored as an internal control. Primer sequences used for PCR analyses are listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mitotic division of an isolated rice zygote expressing H2B-GFP.
Supplemental Figure S2. Karyogamy in rice zygotes produced by in vitro fertilization.
Supplemental Figure S3. Karyogamy progression and de novo gene expression.
Supplemental Figure S4. Membrane fusion between rice sperm and egg nuclei.
Supplemental Figure S5. Effect of latrunculin B on the actin structure in rice egg cells.
Supplemental Figure S6. Male and female nuclei in zygote without karyogamy.
Supplemental Table S1. Nucleotide sequences of PCR primers.
Supplementary Material
Acknowledgments
We thank Hiroki Maeda (Tokyo Metropolitan University) for help in preparing transformed plants, Tomoko Mochizuki (Tokyo Metropolitan University) for isolating rice egg cells, Dr. Tomoko Igawa (Chiba University) for the DD45 promoter::H2B-TagRFP in pENTRTM3C and the DD45 promoter::Lifeact-TagRFP in pGWB1, Dr. Kentaro Tamura (Kyoto University) for the GFP–SUN2 in pGWB405, Dr. Tsuyoshi Nakagawa (Shimane University) for the pGWB1 and pGWB405 vectors, and the RIKEN Bio Resource Center (Tsukuba, Japan) for providing cultured rice cells (Oc line).
Glossary
- cDNA
complementary DNA
- CLS
confocal laser scanning
- ER
endoplasmic reticulum
- H3
Histone3
- IVF
in vitro fertilization
- RFP
red fluorescent protein
- RT
reverse transcription
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abiko M, Furuta K, Yamauchi Y, Fujita C, Taoka M, Isobe T, Okamoto T. (2013a) Identification of proteins enriched in rice egg or sperm cells by single-cell proteomics. PLoS ONE 8: e69578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abiko M, Maeda H, Tamura K, Hara-Nishimura I, Okamoto T. (2013b) Gene expression profiles in rice gametes and zygotes: identification of gamete-enriched genes and up- or down-regulated genes in zygotes after fertilization. J Exp Bot 64: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine AF, Faure JE, Cordeiro S, Dumas C, Rougier M, Feijó JA. (2000) A calcium influx is triggered and propagates in the zygote as a wavefront during in vitro fertilization of flowering plants. Proc Natl Acad Sci USA 97: 10643–10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine AF, Faure JE, Dumas C, Feijó JA. (2001) Differential contribution of cytoplasmic Ca2+ and Ca2+ influx to gamete fusion and egg activation in maize. Nat Cell Biol 3: 1120–1123 [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Chambers EL. (1939) The movement of the egg nucleus in relation to the sperm aster in the echinoderm egg. J Exp Biol 16: 409–424 [Google Scholar]
- Coué M, Brenner SL, Spector I, Korn ED. (1987) Inhibition of actin polymerization by latrunculin A. FEBS Lett 213: 316–318 [DOI] [PubMed] [Google Scholar]
- Era A, Tominaga M, Ebine K, Awai C, Saito C, Ishizaki K, Yamato KT, Kohchi T, Nakano A, Ueda T. (2009) Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure JE, Mogensen HL, Dumas C, Lörz H, Kranz E. (1993) Karyogamy after electrofusion of single egg and sperm cell protoplasts from maize: cytological evidence and time course. Plant Cell 5: 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure JE, Rotman N, Fortuné P, Dumas C. (2002) Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J 30: 481–488 [DOI] [PubMed] [Google Scholar]
- Fu Y, Yuan M, Huang BQ, Yang HY, Zee SY, O’Brien TP. (2000) Changes in actin organization in the living egg apparatus of Torenia fournieri during fertilization. Sex Plant Reprod 12: 315–322 [Google Scholar]
- Gibeaux R, Politi AZ, Nédélec F, Antony C, Knop M. (2013) Spindle pole body-anchored Kar3 drives the nucleus along microtubules from another nucleus in preparation for nuclear fusion during yeast karyogamy. Genes Dev 27: 335–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE. (2010) Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J 61: 134–144 [DOI] [PubMed] [Google Scholar]
- Guignard ML. (1899) Sur les anthérozoides et la double copulation sexuelle chez les végétaux angiosperms. Rev Gén de Bot 11: 129–135 [DOI] [PubMed] [Google Scholar]
- Hause G, Schröder MB. (1987) Reproduction in Triticale. 2. Karyogamy. Protoplasma 139: 100–104 [Google Scholar]
- Howe ES, Clemente TE, Bass HW. (2012) Maize histone H2B-mCherry: a new fluorescent chromatin marker for somatic and meiotic chromosome research. DNA Cell Biol 31: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang BQ, Russell SD. (1994) Fertilization in Nicotiana tabacum: cytoskeletal modifications in the embryo sac during synergid degeneration. Planta 194: 200–214 [Google Scholar]
- Hugdahl JD, Morejohn LC. (1993) Rapid and reversible high-affinity binding of the dinitroaniline herbicide oryzalin to tubulin from Zea mays L. Plant Physiol 102: 725–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Yanagawa Y, Miyagishima SY, Mori T. (2013) Analysis of gamete membrane dynamics during double fertilization of Arabidopsis. J Plant Res 126: 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. (2007) Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Soljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F. (2010) Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20: 2137–2143 [DOI] [PubMed] [Google Scholar]
- Jensen WA. (1964) Observations on the fusion of nuclei in plants. J Cell Biol 23: 669–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Bayer M, Lukowitz W. (2011) Taking the very first steps: from polarity to axial domains in the early Arabidopsis embryo. J Exp Bot 62: 1687–1697 [DOI] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. (2001) The mitotic spindle: a self-made machine. Science 294: 543–547 [DOI] [PubMed] [Google Scholar]
- Kishimoto T. (2003) Cell-cycle control during meiotic maturation. Curr Opin Cell Biol 15: 654–663 [DOI] [PubMed] [Google Scholar]
- Kranz E, von Wiegen P, Lörz H. (1995) Early cytological events after induction of cell division in egg cells and zygote development following in vitro fertilization with angiosperm gametes. Plant J 8: 9–23 [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. (1994) Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol 126: 911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C. (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–467 [Google Scholar]
- Maro B, Johnson MH, Pickering SJ, Flach G. (1984) Changes in actin distribution during fertilization of the mouse egg. J Embryol Exp Morphol 81: 211–237 [PubMed] [Google Scholar]
- Maruyama D, Endo T, Nishikawa S. (2010) BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 1684–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloy P, Shen S, White E, Rose MD. (2009) Distinct roles for key karyogamy proteins during yeast nuclear fusion. Mol Biol Cell 20: 3773–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Scholten S. (2007) Equivalent parental contribution to early plant zygotic development. Curr Biol 17: 1686–1691 [DOI] [PubMed] [Google Scholar]
- Mogensen HL. (1982) Double fertilization in barley and the cytological explanation for haploid embryo formation, embryo-less caryopses, and ovule abortion. Carlsberg Res Commun 47: 313–354 [Google Scholar]
- Mogensen HL, Holm PB. (1995) Dynamics of nuclear DNA quantities during zygote development in barley. Plant Cell 7: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. (2006) GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol 8: 64–71 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Uchiumi T, Okamoto T. (2010) Positional relationship between the gamete fusion site and the first division plane in the rice zygote. J Exp Bot 61: 3101–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaschin S. (1898) Resultate einer revision der befruchtungsvorgänge bei Lilium martagon und Fritillaria tenella. Bull Sci Acad Imp Sci Saint Pétersbourg 9: 377–382 [Google Scholar]
- Nodine MD, Bartel DP. (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Takanashi H, Mogi M, Takahashi H, Kikuchi S, Yano K, Okamoto T, Fujita M, Kurata N, Tsutsumi N. (2011) Distinct gene expression profiles in egg and synergid cells of rice as revealed by cell type-specific microarrays. Plant Physiol 155: 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko MF, Sandaklie-Nikolova L, Lloyd A, Dever CA, Otsuga D, Drews GN. (2006) NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol 141: 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan V. (2003) Some reflections on double fertilization, from its discovery to the present. New Phytol 159: 565–583 [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gönczy P. (1998) Mechanisms of nuclear positioning. J Cell Sci 111: 2283–2295 [DOI] [PubMed] [Google Scholar]
- Russell SD. (1992) Double fertilization. Int Rev Cytol 140: 357–390 [Google Scholar]
- Sato A, Toyooka K, Okamoto T. (2010) Asymmetric cell division of rice zygotes located in embryo sac and produced by in vitro fertilization. Sex Plant Reprod 23: 211–217 [DOI] [PubMed] [Google Scholar]
- Schatten G. (1994) The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol 165: 299–335 [DOI] [PubMed] [Google Scholar]
- Scholten S, Lörz H, Kranz E. (2002) Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J 32: 221–231 [DOI] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Blasberger D, Kashman Y. (1989) Latrunculins—novel marine macrolides that disrupt microfilament organization and affect cell growth. I. Comparison with cytochalasin D. Cell Motil Cytoskeleton 13: 127–144 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T. (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338: 1093–1097 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN. (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51: 281–292 [DOI] [PubMed] [Google Scholar]
- Tachibana K, Hara M, Hattori Y, Kishimoto T. (2008) Cyclin B-cdk1 controls pronuclear union in interphase. Curr Biol 18: 1308–1313 [DOI] [PubMed] [Google Scholar]
- Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. (2013) Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol 23: 1776–1781 [DOI] [PubMed] [Google Scholar]
- Tartakoff AM, Jaiswal P. (2009) Nuclear fusion and genome encounter during yeast zygote formation. Mol Biol Cell 20: 2932–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian GW, Shen JH. (1982) Ultrastructural observations of fertilization in wheat (Triticum aestivum). I. The fusion of the sperm and egg nuclei. Chin J Bot 4: 87–91 [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Komatsu S, Koshiba T, Okamoto T. (2006) Isolation of gametes and central cells from Oryza sativa L. Sex Plant Reprod 19: 37–45 [Google Scholar]
- Uchiumi T, Uemura I, Okamoto T. (2007) Establishment of an in vitro fertilization system in rice (Oryza sativa L.). Planta 226: 581–589 [DOI] [PubMed] [Google Scholar]
- Ueda M, Laux T. (2012) The origin of the plant body axis. Curr Opin Plant Biol 15: 578–584 [DOI] [PubMed] [Google Scholar]
- Ueda M, Zhang Z, Laux T. (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20: 264–270 [DOI] [PubMed] [Google Scholar]
- Van Went JL. (1970) The ultrastructure of the fertilized embryo sac of petunia. Acta Bot Neerl 19: 468–480 [Google Scholar]
- von Besser K, Frank AC, Johnson MA, Preuss D. (2006) Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Wilms HJ. (1981) Pollen tube penetration and fertilization in spinach. Acta Bot Neerl 30: 101–122 [Google Scholar]
- You R, Jensen WA. (1985) Ultrastructural observations of the mature megagametophyte and the fertilization in wheat (Triticum aestivum). Can J Bot 63: 163–170 [Google Scholar]
- Zhang Z, Laux T. (2011) The asymmetric division of the Arabidopsis zygote: from cell polarity to an embryo axis. Sex Plant Reprod 24: 161–169 [DOI] [PubMed] [Google Scholar]
- Zhao J, Xin H, Qu L, Ning J, Peng X, Yan T, Ma L, Li S, Sun MX. (2011) Dynamic changes of transcript profiles after fertilization are associated with de novo transcription and maternal elimination in tobacco zygote, and mark the onset of the maternal-to-zygotic transition. Plant J 65: 131–145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.