A predicted xyloglucan endotransglucosylase (XET) protein interacts with a predicted xyloglucan endohydrolase protein to confer in vivo XET action and change the amount of aluminum retained in hemicellulose and aluminum sensitivity in Arabidopsis.
Abstract
Previously, we reported that although the Arabidopsis (Arabidopsis thaliana) Xyloglucan Endotransglucosylase-Hydrolase31 (XTH31) has predominately xyloglucan endohydrolase activity in vitro, loss of XTH31 results in remarkably reduced in vivo xyloglucan endotransglucosylase (XET) action and enhanced Al resistance. Here, we report that XTH17, predicted to have XET activity, binds XTH31 in yeast (Saccharomyces cerevisiae) two-hybrid and coimmunoprecipitations assays and that this interaction may be required for XTH17 XET activity in planta. XTH17 and XTH31 may be colocalized in plant cells because tagged XTH17 fusion proteins, like XTH31 fusion proteins, appear to target to the plasma membrane. XTH17 expression, like that of XTH31, was substantially reduced in the presence of aluminum (Al), even at concentrations as low as 10 µm for 24 h or 25 µm for just 30 min. Agrobacterium tumefaciens-mediated transfer DNA insertion mutant of XTH17, xth17, showed low XET action and had moderately shorter roots than the wild type but was more Al resistant than the wild type. Similar to xth31, xth17 had low hemicellulose content and retained less Al in the cell wall. These data suggest a model whereby XTH17 and XTH31 may exist as a dimer at the plasma membrane to confer in vivo XET action, which modulates cell wall Al-binding capacity and thereby affects Al sensitivity in Arabidopsis.
Soil acidity (pH < 5.5) affects about 40% of the world’s arable land (von Uexküll and Mutert, 1995) and more than 50% of land that is potentially arable, particularly in the tropics and subtropics (Kochian et al., 2004; Eticha et al., 2010). Al is the most growth-limiting factor for crop production on acid soils worldwide (Foy, 1988; Kochian, 1995), especially when the pH drops below 5 (Eswaran et al., 1997).
To survive in an Al-toxic environment, Al-resistant plants have evolved two mechanisms to cope with Al toxicity. One is to restrict Al uptake from the root, while the other is to cope with internalized Al (Taylor, 1991; Kochian et al., 2004). The latter is usually employed by Al-accumulating species such as Hydrangea macrophylla (Ma et al., 1997a) and buckwheat (Fagopyrum esculentum; Ma et al., 1997b). In most cases, secretion of the organic acid anions is the most important Al exclusion mechanism (Kochian, 1995; Ryan et al., 2001; Ma and Furukawa, 2003), although it does not explain all the Al resistance in some plants such as signalgrass (Brachiaria decumbens Stapf cv Basilisk; Wenzl et al., 2001), maize (Zea mays; Piñeros et al., 2005), buckwheat (Zheng et al., 2005), rice (Oryza sativa; Ma et al., 2005; Yang et al., 2008), or Fagopyrum tataricum (Yang et al., 2011a). Therefore, it is possible that for some plant species (such as buckwheat), Al tolerance is a combination of mechanisms including organic anion efflux.
Recently, evidence has accumulated that the cell wall, especially the hemicellulose component, may impact Al resistance. For example, Al induces significant changes in the hemicellulose fraction of wheat (Triticum aestivum; Tabuchi and Matsumoto, 2001), triticale (× Triticosecale Wittmack; Liu et al., 2008), rice (Yang et al., 2008), and Arabidopsis (Arabidopsis thaliana; Zhu et al., 2012), especially the Al-sensitive cultivars. Moreover, we found that Arabidopsis hemicellulose is not only very sensitive to Al stress (the content of hemicellulose increased quickly under Al stress), but is also the principal binding site for Al (Yang et al., 2011b). Furthermore, loss of Xyloglucan Endotransglucosylase-Hydrolase31 (XTH31) function resulted in lower xyloglucan content and cell wall Al-binding capacity in Arabidopsis (Zhu et al., 2012). Thus, xyloglucan may be a major Al-binding site in Arabidopsis, and any effects leading to xyloglucan modifications may regulate Al sensitivity.
XTHs are a family of enzymes that play principal roles in the construction and restructuring of the load-bearing cross links among cellulose microfibrils (Osato et al., 2006) through catalyzing the molecular grafting or hydrolyzing of the xyloglucans to form the framework (Fry et al., 1992; Nishitani and Tominaga, 1992; Okazawa et al., 1993; Rose et al., 2002). There are 33 identified XTH genes in the Arabidopsis genome, and one-third occur as clusters resulting from genome duplication (Blanc et al., 2000; Yokoyama and Nishitani, 2001); XTH1-11 are classified in subgroup 1, XTH12-26 are in subgroup 2, and XTH27-33 are in subgroup 3 (Rose et al., 2002). Each member of the XTH gene family is likely regulated by specific cues and committed to cell wall dynamics specific to certain tissues or cell types (Nishitani, 2002; Becnel et al., 2006; Osato et al., 2006). For example, XTH27 is involved in the cell wall modification of tracheary elements at a specific stage of rosette leaf development and is essential for tertiary vein development (Matsui et al., 2005), whereas XTH31 is involved in cell wall modification and cell elongation through modulating xyloglucan endotransglucosylase (XET) action under Al stress (Zhu et al., 2012). However, XTH31 is an XTH for which xyloglucan endohydrolase (XEH) activity has been predicted (Baumann et al., 2007), and in our previous report, we demonstrated that XTH31 produced heterologously in Pichia pastoris has high XEH activity but low XET activity in vitro (Zhu et al., 2012), which is in accordance with Kaewthai et al. (2013), who reported that XTH31 is a predominant hydrolase using the in vitro activity assays and enzyme product analysis, as well as the use of a fluorogenic substrate in vivo. Unexpectedly, however, the xth31 mutant has very low XET action and activity (Zhu et al., 2012). One possible explanation for this result is that XTH31 may interact with and be required for activity of XET-active XTHs.
In this study, we demonstrate that XTH17 can bind to XTH31 in vitro and in vivo and that a transfer DNA (T-DNA) insertional mutant of XTH17 has elevated Al resistance and exhibits a phenotype very similar to xth31. Together, these data are consistent with the interpretation that XTH17 and XTH31 may interact with each other and thereby contribute to Al-inhibited XET action in Arabidopsis.
RESULTS
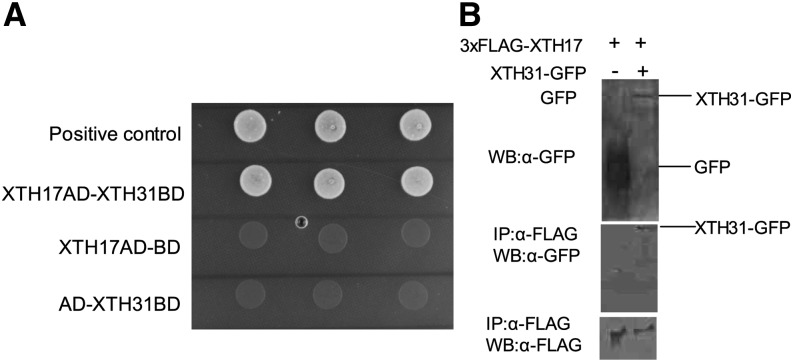
XTH17 Can Bind XTH31 in Vitro and in Vivo
In our previous study, we found that xth31 mutant is compromised in in situ XET action and in extractable XET activity. Because XTH31 produced in Pichia spp. presents predominantly XEH activity, we proposed that XTH31 in the Arabidopsis root may undergo synergistic or direct protein-protein interactions with other XTHs and that loss of XTH31 function diminishes the action and activity of other XTH proteins with XET activity (Zhu et al., 2012). As 11 XTH genes are predominately expressed in the roots (Yang et al., 2011b), we used the yeast (Saccharomyces cerevisiae) two-hybrid system to screen for XTHs that can bind XTH31 and found that only XTH17 showed strong interaction with XTH31 (Fig. 1A). XTH17 (At1g65310) is classified, together with XTH12 to XTH16 and XTH18 to XTH26 in subgroup 2, while XTH31 belongs to the XTH subgroup 3 (Rose et al., 2002). To confirm the interaction between XTH17 and XTH31 in plant cells, we performed coimmunoprecipitation assays. Transient expression of XTH31-GFP proteins in tobacco (Nicotiana benthamiana) was detected after coimmunoprecipitation experiments with 3XFlag-XTH17 using anti-Flag M2 affinity gel (Fig. 1B). Lack of 3XFlag-tagged XTH17 detected when assayed in the presence of control protein (35S-driven GFP) indicates that interaction requires XTH31 (Fig. 1B). These results are strong evidence that 3XFlag-XTH17 can bind with XTH31-GFP in plant cells.
Figure 1.
Physical interaction between XTH17 and XTH31. A, Yeast two-hybrid analysis of XTH17 and XTH31. XTH31 (with DNA-binding domain [BD]) and XTH17 (with activation domain [AD]) were introduced into yeast cells as indicated. Colony growth indicates binding domain and activation domain proximity enabled through interaction of proteins. The empty vector of binding domain with XTH17-AD and the empty vector of activation domain with XTH31-BD are represented as negative control. Three colonies for each line means replicates. B, Coimmunoprecipitation of 3XFlag-XTH17 and XTH31-GFP using tobacco. Total protein extracts (top) or proteins immunoprecipitated (IP) using anti-GFP (α-GFP) antibody from the solubilized microsomal and cytosolic fractions (middle and bottom) were analyzed by western blotting (WB) using α-GFP or α-FLAG antibodies.
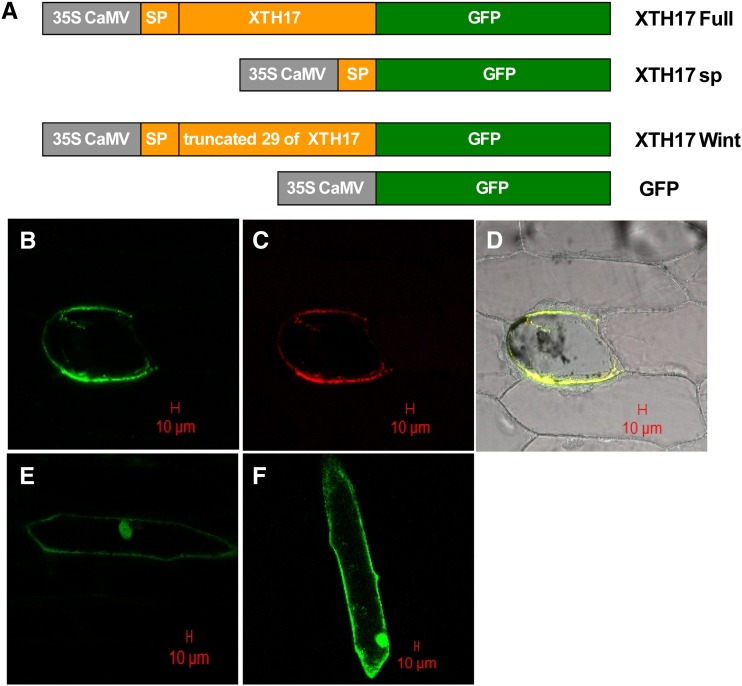
The Cellular Localization of the XTH17 Protein
Similar to XTH31, XTH17 harbors a potential signal peptide at the N terminus. To elucidate the subcellular localization of XTH17, we produced three different constructs fused to GFP (XTH17 Full-GFP: the full XTH17 coding sequence fused in the 3' region with the GFP, XTH17 sp-GFP: the XTH17 signal peptide fused in the 3' region with the GFP, and XTH17 Wint-GFP: only the XTH17 signal peptide fused in the 3' region with the GFP; Fig. 2A) and used these to transform onion (Allium cepa) cells. The full-length XTH17 fusion protein (XTH17 Full-GFP) was found around the cell periphery but proximal to and distinct from the cell wall and colocalized with a plasma membrane marker, suggesting plasma membrane localization (Fig. 2, B, C, and D). A XTH17 Wint-GFP fusion protein that lacks the N-terminal 26 amino acids predicted to compose the signal peptide showed similar localization to that of GFP alone (Fig. 2, E and F). We also attempted to assay the putative signal peptide function by fusing only the first 26 amino acids of XTH17 to GFP (XTH17 sp-GFP; Fig. 2A), but we were unable to determine the subcellular localization, as we could not detect the XTH17 sp-GFP protein, which may be due to the reason that these amino-terminal amino acids are not sufficient to confer localization of GFP.
Figure 2.
The subcellular localization of XTH17 by transient expression in onion epidermal cells. A, Schematic representation of XTH17 constructs fused to GFP. sp indicates the putative signal peptide (26 amino acids at the N terminus of the predicted XTH17 protein; see “Materials and Methods” for details). XTH17 Wint indicates the predicted XTH17 lacking the 26 putative signal peptide. B, XTH17 Full-GFP localization in a plasmolyzed cell. C, The localization of plasma membrane marker (pm-rk CD-1007) in the same plasmolyzed cell as in B. D, XTH17 Full-GFP colocalized with a plasma membrane marker (pm-rk CD-1007). E, XTH17 Wint-GFP localization in a nonplasmolyzed cell. F, 35S:GFP localization in a nonplasmolyzed cell. The red scale bars indicate 20 µm.
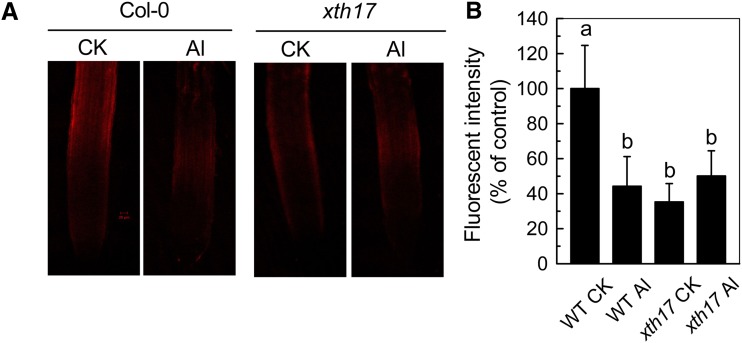
XTH17 Contributes to XET Action
In our previous study, we demonstrated that in situ XET action was reduced remarkably in xth31 (Zhu et al., 2012). However, as Pichia spp.-derived XTH31 has strong XEH activity and poor XET activity, the reduction of XET action in the xth31 mutant remained unexplained. Using endogenous xyloglucan as the donor substrate, we found that XET action was also sharply decreased in the xth17 mutant under control growth conditions (Fig. 3, A and C). Furthermore, in contrast to ecotype Columbia (Col-0; wild type; Fig. 3, A and B; Yang et al., 2011b), xth17 failed to show a dramatic reduction in XET action upon Al stress (Fig. 3, C and D), indicating that XTH17 is required for a major proportion of the assayable XET action in the root tip.
Figure 3.
Effect of Al on XET action in the wild type (WT) and xth17 mutant. Incubation without (control check [CK]) or with Al (Al) for 24 h corresponds to CK and Al, respectively. Seedlings with approximately 1-cm roots were grown on plates without or with 50 µm Al for 24 h. Roots were subjected to cytochemical assays of XET action (see “Materials and Methods” for details). A, Pictures of XET action shown as orange fluorescence in representative roots after 0 or 24 h treatment with 50 µm Al. The red bar in A represents 20 µm. B, XET action expressed as fluorescence relative to the untreated wild type. Values are mean ± sd (n = 6). Columns with different letters are significantly different at P < 0.05.
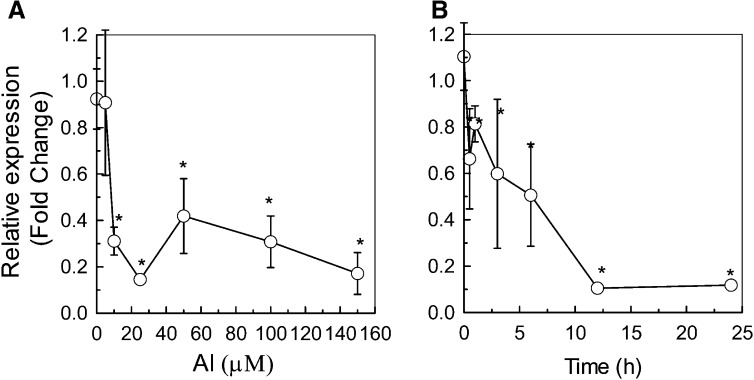
The Dose and Time Response of XTH17 Expression to Al
In our previous report, time course and dose response experiments indicated that XTH31 transcript accumulation is suppressed by 50 µm Al within 30 min of Al treatment, and a significant change in XTH31 RNA abundance can be detected even at an Al concentration as low as 5 µm (Yang et al., 2011b; Zhu et al., 2012). To examine whether XTH17 transcript accumulation is also sensitive to Al stress, we conducted both the time course and dose response experiments and found that XTH17 transcript levels was significantly reduced at Al concentrations higher than 10 µm for 24 h and the largest inhibition of RNA accumulation occurred at 25 µm (Fig. 4A). We then used this Al concentration to conduct a time course analysis and found that a significant down-regulation of XTH17 expression could be detected even within 30 min (Fig. 4B). These data indicate that XTH17 expression, like that of XTH31, is sensitive to Al stress.
Figure 4.
Dose and time response of XTH17 RNA levels to Al in roots measured by quantitative RT-PCR analysis. A, XTH17 transcript levels in 6-week-old seedlings after exposure to 0 to 150 µm Al3+ for 24 h. B, XTH17 transcript levels after exposure to 25 µm Al for 0, 0.5, 1, 6, 12, and 24 h. The y axis shows XTH17 RNA levels normalized to that of the control (0 µm Al). Values are mean ± sd (n = 3). The asterisks show significant differences between control and Al treatments at P < 0.05 by Student’s t test.
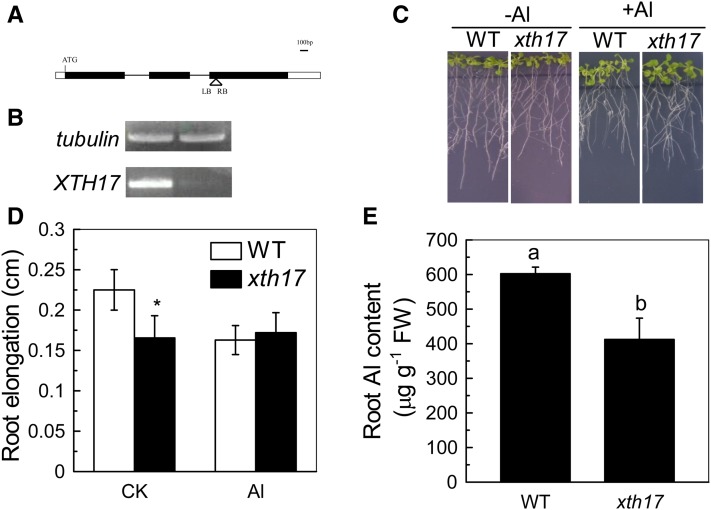
An XTH17 T-DNA Insertional Mutant Has Increased Al Resistance
In our previous study, we found that XTH31 is required to confer Al sensitivity in Arabidopsis by controlling the xyloglucan incorporated into the cell wall and Al-binding capacity of the cell wall (Zhu et al., 2012). To determine whether XTH17 function is also related to Al stress responses, we characterized a T-DNA insertional line of XTH17 (CS16535; Fig. 5A). The XTH17 transcripts were down-regulated in the homozygous line (Fig. 5B), indicating that the T-DNA insertion leads to the knockdown of the XTH17 function. The roots of xth17 were shorter than the wild type under normal growth conditions (Fig. 5C), suggesting that XTH17 function is required for normal elongation; a similar phenotype was found for xth31 (Zhu et al., 2012). Moreover, although the root elongation of the wild type was decreased from an average of 0.22 cm when grown on agar medium without Al3+ to an average of 0.16 cm when grown with 50 µm Al3+ for 24 h, the same Al treatment had no significant effect on xth17 seedling root growth (Fig. 5, C and D). Strikingly, Al-exposed xth17 roots accumulated significantly less Al than that in wild-type roots grown under similar conditions (Fig. 5E). These data strongly suggest that XTH17 impacts Al sensitivity by affecting Al accumulation in the roots.
Figure 5.
The xth17 mutant has altered root lengths, reduced Al sensitivity, and reduced Al accumulation. A, Schematic structure of the xth17 mutant locus carrying a T-DNA insertion in the third exon. The black and white boxes represent the coding and untranslated regions, respectively. B Semiquantitative RT-PCR study of XTH17 transcripts. C, The wild type (WT) and xth17 grown on MS plates without (–Al) or with 50 µm Al (+Al). D, Root elongation of the wild type and xth17 without (control check [CK]) or with (Al) 50 µm Al. Plants with approximately 1-cm roots were grown in agar medium containing 0 or 50 µm Al for 24 h. Root elongation was measured before and after treatment. Data are means ± sd (n = 10). The asterisk shows a significant difference at P < 0.05 by Student’s t test. E, Root Al content of the wild type and xth17. Data are means ± sd (n = 4). Columns with different letters are significantly different at P < 0.05. FW, Fresh weight.
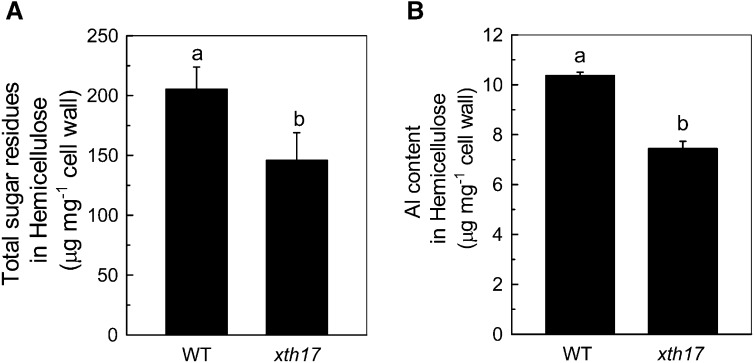
Lower Hemicellulose Content in xth17 Roots
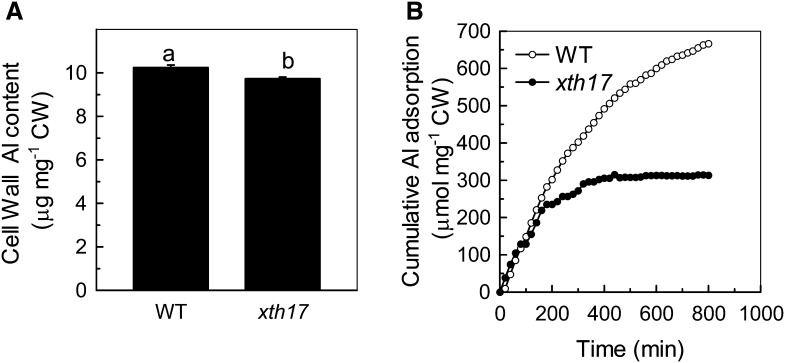
As hemicellulose is a major Al-binding site in Arabidopsis cell wall and XTH genes are involved in the modulation of hemicellulose (Fry et al., 1992; Zhu et al., 2012), we measured hemicellulose content in xth17 and the wild type and found that the total sugar and Al content in hemicellulose were both significantly lower in xth17 than in the wild type (Fig. 6, A and B). Furthermore, less Al was found in the cell wall of xth17 than in the wild type (Fig. 7A). Finally, the in vitro Al adsorption kinetics also showed that the extracted root cell walls of xth17 adsorbed significantly less Al than that of the wild type (Fig. 7B). These results indicate that the loss of XTH17 affects Al response association with cell wall components.
Figure 6.
Total sugar and Al content in hemicellulose. A, Total sugar in extracted hemicellulose of the wild type (WT) and xth17 mutant. Cell wall materials from Al-untreated roots were fractionated into different polysaccharide classes (see “Materials and Methods” for details). B, Al content in hemicellulose of the wild type and xth17 mutant. Cell wall hemicellulose from Al-treated roots was assayed for Al (see “Materials and Methods” for details). Data are means ± sd (n = 4). Columns with different letters show a significant difference between the wild type and xth17 mutant at P < 0.05 by Student’s t test.
Figure 7.
Al content in cell wall (CW) and Al adsorption kinetics in the extracted cell wall. A, Al content in cell wall of the Col-0 (wild type [WT]) and xth17 mutant. Al content in the cell wall from Al-treated roots was detected (see “Materials and Methods” for details). Data are means ± sd (n = 4). Columns with different letters show a significant difference between the wild type and xth17 mutant at P < 0.05 by Student’s t test. B, Al adsorption kinetics of cell walls in the wild type and xth17. Cell wall materials from Al-untreated roots were placed into a 2-mL column, and Al adsorption kinetics were monitored as previously described (Zheng et al., 2004).
DISCUSSION
In our previous study, we found that the XTH31 heterologously produced in Pichia spp. showed high XEH but low XET activity in vitro, which agrees with sequence-based predictions that XTH31 and XTH32 are the only two XTH members with XEH activity (Baumann et al., 2007). However, the xth31 mutant root has much reduced in situ XET action and extractable XET activity compared with the wild-type Col-0. Thus, we proposed that XTH31 in the root may undergo synergistic or direct protein-protein interactions with other XTHs to confer XET activity (Zhu et al., 2012). Here, by yeast two-hybrid and transient expression assays with Agrobacterium tumefaciens infiltration of tobacco leaves, we found that among the XTH genes expressed in roots, XTH17 could directly interact with XTH31 in vitro and in vivo (Fig. 1B). Moreover, using a XTH17-GFP fusion protein, we confirmed that XTH17 is localized at the plasma membrane (Fig. 2), which is in accordance with XTH31 (Zhu et al., 2012). Because xyloglucan is synthesized in the Golgi and undergoes transglycosylation immediately after release into the wall (Thompson and Fry, 2001), this membrane-localized XTH17 is well positioned for catalyzing this process. As XTH31 is also localized at the plasma membrane (Zhu et al., 2012), it is reasonable to hypothesize that XTH17 and XTH31 may form a dimer at the plasma membrane surface, where they could potentially function together to modify the newly secreted xyloglucans.
Xyloglucan is an important hemicellulose component of dicotyledonous plant cell walls, accounting for up to 20% of cell wall content (Fry, 1989; Hayashi, 1989). Thus, it is not surprising that plant genomes contain a large number of XET/XEHs to modify xyloglucans, with up to 33 genes in Arabidopsis (Campbell and Braam, 1999; Yokoyama and Nishitani, 2001), 29 in rice (Yokoyama et al., 2004), 25 in tomato (Solanum lycopersicum; Saladié et al., 2006), 41 in Populus spp. (Geisler-Lee et al., 2006), 22 in barley (Hordeum vulgare; Strohmeier et al., 2004), and 57 in wheat (Liu et al., 2007). All XTH enzymes studied to date display XET, XEH (Tabuchi et al., 2001; Rose et al., 2002), or both activities (de Silva et al., 1993; Fanutti et al., 1993), and their activity can loosen plant cell walls (Van Sandt et al., 2007). Recently, expression profiles of several XTH genes have provided evidence that distinct XTHs may have particular physiological roles in cell wall dynamics (Vissenberg et al., 2005; Becnel et al., 2006). Some XTH genes are prominently expressed in rapidly expanding tissues such as the elongation zone of roots, sites of future root hair initiation, which can be induced by Al in maize (Doncheva et al., 2005), growing root hairs (Vissenberg et al., 2000, 2001, 2003), growing internodes of adult stems (Potter and Fry, 1993; Uozu et al., 2000; Nakamura et al., 2003; Romo et al., 2005), and hypocotyls (Catalá et al., 1997; Yun et al., 2005). For instance, XTH19 is expressed in the apical dividing, elongating regions and the differentiation zone, while XTH20 is expressed specifically in vascular tissues in the basal mature region of the root (Vissenberg et al., 2005). XTH27 is expressed extensively during the development of tracheary elements, when the secondary wall undergoes thickening (Matsui et al., 2005). Furthermore, it was previously reported that XTH17 is expressed in all tissue types in the elongation and differentiation zone (Vissenberg et al., 2005), while XTH31 is predominantly expressed in the root tips, including the elongation zone (Zhu et al., 2012). The protein encoded by XTH17 belongs to the class II subfamily in the XTH protein family; members of this subfamily have been shown to exhibit exclusively XET activity (Maris et al., 2011). Thus, it is not unexpected that the xth17 mutant exhibited lower XET action (Fig. 3). However, as the xth31 mutant also harbors very low XET action/activity (Zhu et al., 2012), based on the data presented here, we now propose that XTH31 can confer its XET function only when both XTH17 and XTH31 coexist.
XET action/activity has been proposed to play important roles in the process of cell expansion (Smith and Fry, 1991; Fry et al., 1992; Van Sandt et al., 2007). For instance, extractable XET activity correlates well with growth rate (Fry et al., 1992; Hetherington and Fry, 1993; Potter and Fry, 1993, 1994; Pritchard et al., 1993; Zhu et al., 2012). Here, we found that the roots of xth17 grow shorter than the wild type (Fig. 4C), which is in accordance with the remarkable reduction of its XET action (Fig. 3), while the root growth of xth17 was less inhibited under Al stress (Fig. 4, C and D), being consistent with its almost unchanged XET action under Al (Fig. 3). As the in vivo XET action of XTH17 at or near the plasma membrane may modify the newly secreted xyloglucan to enhance further xyloglucan incorporation, the lower XET action may lead to the reduced cell wall polysaccharides (Fig. 6), which also result in the lower Al-binding capacity (Figs. 5 and 6B).
XTH genes have been shown to be responsive to diverse stimuli. As reported by Xu et al. (1995, 1996), expression of XTH genes is regulated by abiotic factors, such as touch, darkness, cold shock, heat shock, and others (Wu et al., 1994). Zurek and Clouse (1994) suggested that several hormones, e.g. abscisic acid, brassinosteroids, and GAs, can also regulate the XTH expression. Recently, Divol et al. (2007) and Maldonado-Mendoza et al. (2005) demonstrated that expression of XTH genes can be regulated by parasites or induced during arbuscular mycorrhizal symbiosis, respectively. In this study, we found that similar to XTH31, the expression of XTH17 was also very responsive to Al (Fig. 7).
Modification of the root cell wall’s binding properties has been taken as an important Al-resistant/-sensitive mechanism. Pectin content and degree of methylation have been implicated in cell wall Al-binding capacity due to pectin carboxylate groups (Horst et al., 2010). For example, Yang et al. (2008) found that in sensitive rice cultivar root tips, both the higher pectin content and the higher degree of demethylesterification in pectin result in the greater Al-binding capacity (Yang et al., 2008). However, we have recently found that hemicellulose binds much more Al than pectin, and XET action, along with the expression of XTH31, is significantly inhibited by Al stress; thus, we proposed that XTH function is a factor that predisposes plants to Al toxicity (Yang et al., 2011b; Zhu et al., 2012). In this study, we further investigated the function of XTH17 in its relationship with Al resistance and propose that XTH17 works with XTH31 through the formation of an XTH17-XTH31 dimer and that dimer formation is required for XTH17 XET action (Fig. 3) and, therefore, loss of XTH17 function results in lower Al accumulation in the cell wall (Fig. 5). As a consequence, xth17 is more Al resistant (Fig. 4). Altogether, these data indicate that XTH17 plays an important role in conferring Al sensitivity in Arabidopsis.
In conclusion, our results indicate that XTH17, together with XTH31, can form a complex required for XET action, thus modulating the cell wall Al-binding capacity in Arabidopsis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Both wild-type and xth17 (Arabidopsis [Arabidopsis thaliana]) plants used were in the Col-0 wild-type background. Seeds were surface sterilized and germinated on an agar-solidified nutrient medium in petri dishes. The nutrient medium was based on Murashige and Skoog (MS) salts (Murashige and Skoog, 1962) containing the macronutrients KNO3, 6.0 mm; Ca(NO3)2, 4.0 mm; MgSO4, 1 mm; and NH4H2PO4, 0.1 mm; and the micronutrients Fe(III)-EDTA, 50 μm; H3BO3, 12.5 μm; MnSO4, 1 μm; CuSO4, 0.5 μm; ZnSO4, 1 μm; H2MoO4, 0.1 μm; and NiSO4, 0.1 μm. The final pH was adjusted to 4.5. The seeds were vernalized at 4°C for 1 d. Petri dishes were placed into a growth chamber, positioned vertically, and kept under controlled environmental conditions at 24°C, 140 µmol m–2 s–1, and a 16-h/8-h day/night rhythm.
For hydroponic culture, seedlings were first aseptically germinated on the above solid nutrient medium. After 2 weeks, the young plantlets were placed on vermiculite for additional 3 weeks in an environmentally controlled growth chamber. Seedlings of similar rosette diameters were then transferred to the nutrient solution containing the above-mentioned nutrients for another 1 week. Then, the plants were subjected to the following treatments: control check (0.5 mm CaCl2, pH 4.5) and Al (50 μm Al in the 0.5 mm CaCl2, pH 4.5). After 24 h, the roots were excised for RNA extraction or for Al content analysis. For Al content analysis, the seedlings were washed three times with deionized water, and the fresh weight was recorded. For the dose response experiment, 6-week-old seedlings were exposed to 0.5 mm CaCl2 medium (pH 4.5) containing 0, 5, 10, 25, 50, 100, or 150 μm AlCl3 for 24 h. For the time response experiment, 6-week-old seedlings were exposed to 0.5 mm CaCl2 medium (pH 4.5) containing 50 μm AlCl3 for 0, 0.5, 1, 6, 12, and 24 h.
Effect of Al on Root Growth
Seedlings with a root length about 1 cm were selected and transferred to petri dishes containing agar-solidified CaCl2 (0.5 mm) medium with or without 50 µm Al in the form of AlCl3 for 24 h. Root morphology was recorded using a digital camera connected to a computer. Data were quantified and analyzed by Photoshop 7.0 (Adobe Systems). For the long-term treatment, seedlings about 1 cm long were selected and transferred to petri dishes containing agar-solidified MS medium with or without 50 µm Al in the form of AlCl3. Plants were grown vertically for an additional 7 d, at which point photographs were taken.
Cytochemical Assay
The XET action was determined according to Vissenberg et al. (2000). In brief, roots were incubated in a 6.5 µm sulphorhodamine-labelled oligosaccharides of xyloglucan (XGO-SRs) mixture according to Zhu et al. (2012). The assay was followed by a 10-min wash in ethanol/formic acid/water (15:1:4, v/v/v) to remove any remaining unreacted XGO-SRs; a further incubation overnight in 5% (v/v) formic acid removed apoplastic, non-wall-bound xyloglucan-SR. Samples were mounted on glass slides and inspected under a laser-scanning confocal microscope (LSM 510; Zeiss) using excitation light of 540 nm.
Gene Expression Analysis
Total RNA was isolated from root using TRIzol (Invitrogen). Complementary DNA was prepared from 1 µg of total RNA using the PrimeScript RT reagent kit (Takara). For real-time reverse transcription (RT)-PCR analysis, 1 µL of 10-fold-diluted complementary DNA was used for the quantitative analysis of gene expression performed with SYBR Premix ExTaq (Takara) with the following pairs of gene-specific primers (for tubulin: forward, 5′-AAGTTCTGGGAAGTGGTT-3′; reverse, 5′-CTCCCAATGAGTGACAAA-3′ and for XTH17: forward, 5′-AGTTCAAGAACCCCGAGGCGAT-3′; reverse, 5′-TTGCCCAATGCTCGGCTTCC-3′). Each complementary DNA sample was run in triplicate. Expression data were normalized with the expression level of the tubulin gene.
Cell Wall Extraction and Fractionation
Extraction of crude cell wall materials and subsequent fractionation of cell wall components were carried out according to Zhong and Lauchli (1993) with minor modifications. Roots were ground with a mortar and pestle in liquid nitrogen and then homogenized with 75% (v/v) ethanol for 20 min in an ice-cold water bath. The sample was then centrifuged at 8,000 rpm for 10 min, and the supernatant was removed. The pellets were homogenized and washed with acetone, methanol:chloroform at a ratio of 1:1, and methanol for 20 min each, with each supernatant removed after centrifugation between the washes. The remaining pellet, i.e. the cell wall material, was dried for further use.
Pectin was triple extracted by hot water for 1 h each and pooling the supernatants (pectin). The pellet was subjected to twice extraction with 24% (w/v) KOH for a total time of 24 h and pooling the supernatants (hemicelluloses).
Determination of Total Polysaccharide
The total polysaccharide contents in the hemicellulosic fractions were determined by the phenol-sulfuric acid method (Dubois et al., 1956) and expressed as Glc equivalents according to Zhu et al. (2012). Briefly, 200 μL of hemicellulose extracts was incubated with 1 mL of 98% (v/v) H2SO4 and 10 μL of 80% (v/v) phenol at room temperature for 15 min and then incubated at 100°C for 15 min. After cooling, the A490 was measured spectrophotometrically.
Al Content Measurement
Al content in each cell wall pellet was extracted by 2 n HCl for 24 h with occasional shaking. Al concentrations in the hemicellulose fraction were determined by inductively coupled plasma-atomic emission spectrometry (IRIS/AP optical emission spectrometer).
Adsorption Kinetics
To determine the ability of xth17 and Col-0 to adsorb Al, a total of 5 mg of cell wall materials was placed in a 2-mL column equipped with a filter at the bottom. The adsorption solution consisted of 20 µm AlCl3 in 0.5 mm CaCl2 at pH 4.5. The solution was passed through the bed of cell walls by a peristaltic pump at 12 mL h–1. The eluate was collected in 5-mL aliquots, which were assayed for Al spectrophotometrically with pyrocatechol violet according to Kerven et al. (1989) with some modification (Zhu et al., 2012). The kinetics study was carried out twice independently, and one set of adsorption curves is presented in “Results.”
Yeast Two-Hybrid Analysis
Yeast (Saccharomyces cerevisiae) two-hybrid analysis was performed using MatchMaker GAL4 Two-Hybrid System 3 (Clontech, http://www.clontech.com/) according to the manufacturer’s instructions. A yeast strain (AH109) was transformed with pairs of pGBKT7 vectors (Clontech) harboring XTH31 and pGADT7 vectors (Clontech) harboring XTH17. The transformants were tested on the screening medium.
Agrobacterium tumefaciens-Mediated Infiltration of Tobacco Leaves
The A. tumefaciens-mediated transient expression in tobacco (Nicotiana benthamiana) leaves was conducted as described by Liu et al. (2012). A preculture of the A. tumefaciens EHA105 strain harboring the constructs of 3XFlag-XTH17 and XTH31-GFP was prepared in Luria-Bertani medium with the proper antibiotics and incubated overnight with shaking at 28°C. A 1-mL aliquot of preculture was used to inoculate 50 mL of Luria-Bertani medium with the appropriate antibiotics, 10 mm MES, and 20 μm acetosyringone, and the bacteria were allowed to grow overnight. After centrifugation at 5,000 rpm for 10 min at 4°C, the cell pellet was resuspended in the infiltration medium (10 mm MgCl2, 10 mm MES, and 100 μm acetosyringone) to an optical density at 600 nm OD600 of 1.0. The cell suspension was then left standing at room temperature for 2 to 3 h before infiltration of tobacco leaves. A mix of cells containing the 3XFlag-XTH17 and XTH31-GFP was then prepared to infiltrate the second or third true leaves of 5-week-old tobacco plants. Infiltrated tobacco was grown for another 3 d before sample collection.
Protein Extraction and Protein Gel-Blot Analysis
To prepare total protein extracts, leaves of 5-week-old tobacco were ground into fine powder in liquid nitrogen and thawed in cold lysis buffer (50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 0.05%–0.1% [v/v] Tween 20, and a protease inhibitor mixture [Roche Applied Science, http://www.roche.com]) using a mortar and pestle according to Tanaka et al. (2012). The extracts were then centrifuged at 10,000g for 5 min at 4°C, and the cell debris was removed by centrifugation of the supernatant at 100,000g for 10 min at 4°C. The total protein concentration was determined by Bradford Protein Assay. Twenty microliters of Anti-Flag M2 Affinity Gel (Sigma, catalog no. A2220) was added to 500 to 1,000 µg of total soluble protein solution and incubated at 4°C for 2 to 3 h with a rotator, and then the antigen-Anti-Flag M2 Affinity Gel conjugates were precipitated by centrifugation and prior to washing. After being centrifuged at 3,000 rpm, pellets were washed twice with 1 mL of lysis buffer containing 250 mm NaCl. Finally, the pellets were resuspended in SDS protein loading buffer. The XTH31-GFP protein was detected using polyclonal anti-GFP antibody.
35S:XTH17–GFP Expression Constructs, Transient Onion Transformation, and Plasmolysis
The full XTH17 coding sequence (XTH17 Full), the same without the signal peptide (XTH17 Int), and only the XTH17 signal peptide (XTH17 sp) were cloned in pBI 221 vector under the control of a CaMV 35S promoter and fused in the 3' region with the GFP according to Zhu et al. (2012). Onion cells were bombarded at 900 psi with 5μg of DNA plasmids for expression of the fusion with or without plasma membrane marker pm-rk CD-1007, or GFP alone as a control using a biolistic PSD-1000/He particle delivery system (Bio-Rad). After particle bombardment, the samples were incubated for 24–60 h at 25°C in the dark. Samples were mounted on glass slides and inspected under a laser-scanning confocal microscope (LSM 510; Zeiss). When indicated, cells were plasmolysed in saturated Suc for 15 min.
Statistical Analysis
Each experiment was repeated independently at least two times, and one set of representative data are shown in the results. Data were analyzed by one-way ANOVA, and the means were compared by Student’s t test. Different letters and asterisks on the histograms indicate statistical differences at the P < 0.05 level.
Acknowledgments
We thank Stephen C. Fry (Institute of Cell and Molecular Biology, University of Edinburgh) for kindly donating the XGO-SRs.
Glossary
- XEH
xyloglucan endohydrolase
- XET
xyloglucan endotransglucosylase
- T-DNA
transfer DNA
- Col-0
ecotype Columbia
- MS
Murashige and Skoog
- cDNA
complementary DNA
- RT
reverse transcription
Footnotes
This work was supported by the Natural Science Foundation of China (grant no. 31370294), the 973 program (grant no. 2014CB441002), the National High-Tech Research and Development Program of China (grant no. 2012AA101101), the Program for Innovative Research Team in Universities (grant no. IRT1185), and the Fundamental Research Funds for Central Universities.
Articles can be viewed online without a subscription.
References
- Baumann MJ, Eklöf JM, Michel G, Kallas ÅM, Teeri TT, Czjzek M, Brumer H., III (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J. (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Braam J. (1999) Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 4: 361–366 [DOI] [PubMed] [Google Scholar]
- Catalá C, Rose JKC, Bennett AB. (1997) Auxin regulation and spatial localization of an endo-1,4-β-d-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. Plant J 12: 417–426 [DOI] [PubMed] [Google Scholar]
- de Silva J, Jarman CD, Arrowsmith DA, Stronach MS, Chengappa S, Sidebottom C, Reid JSG. (1993) Molecular characterization of a xyloglucan-specific endo-(1→4)-β- d-glucanase (xyloglucan endo-transglycosylase) from nasturtium seeds. Plant J 3: 701–711 [PubMed] [Google Scholar]
- Divol F, Vilaine F, Thibivilliers S, Kusiak C, Sauge MH, Dinant S. (2007) Involvement of the xyloglucan endotransglycosylase/hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant Cell Environ 30: 187–201 [DOI] [PubMed] [Google Scholar]
- Doncheva S, Amenós M, Poschenrieder C, Barceló J. (2005) Root cell patterning: a primary target for aluminium toxicity in maize. J Exp Bot 56: 1213–1220 [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Eswaran H, Reich P, Beinroth F (1997) Global distribution of soils with acidity. In AC Moniz, AMC Furlani, RE Schaffert, NK Fageria, CA Rosolem, H Cantarella, eds, Plant–Soil Interactions at Low pH. Brazil Society of Soil Science, Campinas, Brazil, pp 159–164 [Google Scholar]
- Eticha D, Zahn M, Bremer M, Yang Z, Rangel AF, Rao IM, Horst WJ. (2010) Transcriptomic analysis reveals differential gene expression in response to aluminium in common bean (Phaseolus vulgaris) genotypes. Ann Bot (Lond) 105: 1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid JSG. (1993) Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1→4)-β-d-glucanase) from the cotyledons of germinated nasturtium seeds. Plant J 3: 691–700 [DOI] [PubMed] [Google Scholar]
- Foy CD. (1988) Plant adaptation to acid, aluminum-toxic soils. Commun Soil Sci Plant 19: 959–987 [Google Scholar]
- Fry SC. (1989) The structure and functions of xyloglucan. J Exp Bot 40: 1–11 [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunnerås S, et al. (2006) Poplar carbohydrate-active enzymes: gene identification and expression analyses. Plant Physiol 140: 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol 40: 139–168 [Google Scholar]
- Hetherington PR, Fry SC. (1993) Xyloglucan endotransglycosylase activity in carrot cell suspensions during cell elongation and somatic embryogenesis. Plant Physiol 103: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst WJ, Wang Y, Eticha D. (2010) The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: a review. Ann Bot (Lond) 106: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewthai N, Gendre D, Eklöf JM, Ibatullin FM, Ezcurra I, Bhalerao RP, Brumer H. (2013) Group III-A XTH genes of Arabidopsis thaliana encode predominant xyloglucan endo-hydrolases that are dispensable for normal growth. Plant Physiol 161: 440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerven GL, Edwards DG, Asher CJ, Halman PS, Kokot S. (1989) Aluminium determination in soil solution: II. Short-term colorimetric procedures for the measurement of inorganic monomeric aluminium in the presence of organic ligands. Aust J Soil Res 27: 91–102 [Google Scholar]
- Kochian LV. (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46: 237–260 [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA. (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Liu Q, Yang JL, He LS, Li YY, Zheng SJ. (2008) Effect of aluminum on cell wall, plasma membrane, antioxidants and root elongation in triticale. Biol Plant 52: 87–92 [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ. (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu D, Zhang H, Gao H, Guo X, Wang D, Zhang X, Zhang A. (2007) The α- and β-expansin and xyloglucan endotransglucosylase/hydrolase gene families of wheat: molecular cloning, gene expression, and EST data mining. Genomics 90: 516–529 [DOI] [PubMed] [Google Scholar]
- Ma JF, Furukawa J. (2003) Recent progress in the research of external Al detoxification in higher plants: a minireview. J Inorg Biochem 97: 46–51 [DOI] [PubMed] [Google Scholar]
- Ma JF, Hiradate S, Nomoto K, Iwashita T, Matsumoto H. (1997a) Internal detoxification mechanism of Al in hydrangea: Identification of Al form in the leaves. Plant Physiol 113: 1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Nagao S, Huang CF, Nishimura M. (2005) Isolation and characterization of a rice mutant hypersensitive to Al. Plant Cell Physiol 46: 1054–1061 [DOI] [PubMed] [Google Scholar]
- Ma JF, Zheng SJ, Hiradate S, Matsumoto H. (1997b) Detoxifying aluminium with buckwheat. Nature 390: 569–5709403684 [Google Scholar]
- Maldonado-Mendoza IE, Dewbre GR, Blaylock L, Harrison MJ. (2005) Expression of a xyloglucan endotransglucosylase/hydrolase gene, Mt-XTH1, from Medicago truncatula is induced systemically in mycorrhizal roots. Gene 345: 191–197 [DOI] [PubMed] [Google Scholar]
- Maris A, Kaewthai N, Eklöf JM, Miller JG, Brumer H, Fry SC, Verbelen JP, Vissenberg K. (2011) Differences in enzymic properties of five recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis thaliana. J Exp Bot 62: 261–271 [DOI] [PubMed] [Google Scholar]
- Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K. (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42: 525–534 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–496 [Google Scholar]
- Nakamura T, Yokoyama R, Tomita E, Nishitani K. (2003) Two azuki bean XTH genes, VaXTH1 and VaXTH2, with similar tissue-specific expression profiles, are differently regulated by auxin. Plant Cell Physiol 44: 16–24 [DOI] [PubMed] [Google Scholar]
- Nishitani K. (2002) A genome-based approach to study the mechanisms by which cell-wall type is defined and constructed by the collaborative actions of cell-wall-related enzymes. J Plant Res 115: 303–307 [DOI] [PubMed] [Google Scholar]
- Nishitani K, Tominaga R. (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058–21064 [PubMed] [Google Scholar]
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, Nishitani K. (1993) Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. J Biol Chem 268: 25364–25368 [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119: 153–162 [DOI] [PubMed] [Google Scholar]
- Piñeros MA, Shaff JE, Manslank HS, Alves VMC, Kochian LV. (2005) Aluminum resistance in maize cannot be solely explained by root organic acid exudation: a comparative physiological study. Plant Physiol 137: 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter I, Fry SC. (1993) Xyloglucan endotransglycosylase activity in pea internodes: effects of applied gibberellic acid. Plant Physiol 103: 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter I, Fry SC. (1994) Changes in xyloglucan endotransglycosylase (XET) activity during hormone-induced growth in lettuce and cucumber hypocotyls and spinach cell suspension cultures. J Exp Bot 45: 1703–1710 [Google Scholar]
- Pritchard J, Hetherington PR, Fry SC, Tomos AD. (1993) Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. J Exp Bot 44: 1281–1289 [Google Scholar]
- Romo S, Jiménez T, Labrador E, Dopico B. (2005) The gene for a xyloglucan endotransglucosylase/hydrolase from Cicer arietinum is strongly expressed in elongating tissues. Plant Physiol Biochem 43: 169–176 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Ryan P, Delhaize E, Jones D. (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52: 527–560 [DOI] [PubMed] [Google Scholar]
- Saladié M, Rose JKC, Cosgrove DJ, Catalá C. (2006) Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J 47: 282–295 [DOI] [PubMed] [Google Scholar]
- Smith RC, Fry SC. (1991) Endotransglycosylation of xyloglucans in plant cell suspension cultures. Biochem J 279: 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier M, Hrmova M, Fischer M, Harvey AJ, Fincher GB, Pleiss J. (2004) Molecular modeling of family GH16 glycoside hydrolases: potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Sci 13: 3200–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi A, Matsumoto H. (2001) Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiol Plant 112: 353–358 [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Mori H, Kamisaka S, Hoson T. (2001) A new type of endo-xyloglucan transferase devoted to xyloglucan hydrolysis in the cell wall of azuki bean epicotyls. Plant Cell Physiol 42: 154–161 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2012) Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J 70: 599–613 [DOI] [PubMed] [Google Scholar]
- Taylor GJ. (1991) Current views of the aluminum stress response: the physiological basis of tolerance. Curr Top Plant Biochem Physiol 10: 57–93 [Google Scholar]
- Thompson JE, Fry SC. (2001) Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J 26: 23–34 [DOI] [PubMed] [Google Scholar]
- Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M. (2000) Characterization of XET-related genes of rice. Plant Physiol 122: 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt VS, Suslov D, Verbelen JP, Vissenberg K. (2007) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot (Lond) 100: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Fry SC, Verbelen JP. (2001) Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiol 127: 1125–1135 [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Martinez-Vilchez IM, Verbelen JP, Miller JG, Fry SC. (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell 12: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato Y, Yokoyama R, Verbelen JP, Nishitani K. (2005) Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots. Physiological roles in specification in cell wall construction. Plant Cell Physiol 46: 192–200 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Van Sandt V, Fry SC, Verbelen JP. (2003) Xyloglucan endotransglucosylase action is high in the root elongation zone and in the trichoblasts of all vascular plants from Selaginella to Zea mays. J Exp Bot 54: 335–344 [DOI] [PubMed] [Google Scholar]
- von Uexküll HR, Mutert E (1995) Global extent, development and economic impact of acid soils. In RA Date, NJ Grundon, GE Raymet, ME Probert, eds, Plant-Soil Interactions at Low pH: Principles and Management. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 5–19 [Google Scholar]
- Wenzl P, Patiño GM, Chaves AL, Mayer JE, Rao IM. (2001) The high level of aluminum resistance in signalgrass is not associated with known mechanisms of external aluminum detoxification in root apices. Plant Physiol 125: 1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC. (1994) Root growth maintenance at low water potentials: increased activity of xyloglucan endotransglucosylase and its possible regulation by abscisic acid. Plant Physiol 106: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J. (1996) The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J 9: 879–889 [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Li YY, Zhang YJ, Zhang SS, Wu YR, Wu P, Zheng SJ. (2008) Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol 146: 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Zhu XF, Peng YX, Zheng C, Li GX, Liu Y, Shi YZ, Zheng SJ. (2011b) Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol 155: 1885–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Zhu XF, Zheng C, Zhang YJ, Zheng SJ. (2011a) Genotypic differences in Al resistance and the role of cell-wall pectin in Al exclusion from the root apex in Fagopyrum tataricum. Ann Bot (Lond) 107: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JKC, Nishitani K. (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice: classification and expression analysis. Plant Physiol 134: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun HS, Kwon C, Kang BG, Lee JS, Han TJ, Chang SC. (2005) A xyloglucan endotransglycosylase/hydrolase1, VrXTH1, is associated with cell elongation in mungbean hypocotyls. Physiol Plant 125: 106–113 [Google Scholar]
- Zheng SJ, Lin XY, Yang JL, Liu Q, Tang C. (2004) The kinetics of aluminum adsorption and desorption by root cell walls of an aluminum resistance wheat (Triticum aestivum L.) cultivar.. Plant Soil 261: 85–90 [Google Scholar]
- Zheng SJ, Yang JL, He YF, Yu XH, Zhang L, You JF, Shen RF, Matsumoto H. (2005) Immobilization of aluminum with phosphorus in roots is associated with high aluminum resistance in buckwheat. Plant Physiol 138: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Lauchli A. (1993) Changes of cell wall component and polymer size in primary roots of cotton seedlings under high salinity. J Exp Bot 44: 773–778 [Google Scholar]
- Zhu XF, Shi YZ, Lei GJ, Fry SC, Zhang BC, Zhou YH, Braam J, Jiang T, Xu XY, Mao CZ, et al. (2012) XTH31, encoding an in vitro XEH/XET-active enzyme, controls Al sensitivity by modulating in vivo XET action, cell wall xyloglucan content and Al binding capacity in Arabidopsis. Plant Cell 24: 4731–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD. (1994) Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol 104: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]