Elimination of four MADS-box genes leads to premature expression of floral genes in leaves.
Abstract
Multiple factors, including the MADS-domain proteins AGAMOUS-LIKE15 (AGL15) and AGL18, contribute to the regulation of the transition from vegetative to reproductive growth. AGL15 and AGL18 were previously shown to act redundantly as floral repressors and upstream of FLOWERING LOCUS T (FT) in Arabidopsis (Arabidopsis thaliana). A series of genetic and molecular experiments, primarily focused on AGL15, was performed to more clearly define their role. agl15 agl18 mutations fail to suppress ft mutations but show additive interactions with short vegetative phase (svp) mutations in ft and suppressor of constans1 (soc1) backgrounds. Chromatin immunoprecipitation analyses with AGL15-specific antibodies indicate that AGL15 binds directly to the FT locus at sites that partially overlap those bound by SVP and FLOWERING LOCUS C. In addition, expression of AGL15 in the phloem effectively restores wild-type flowering times in agl15 agl18 mutants. When agl15 agl18 mutations are combined with agl24 svp mutations, the plants show upward curling of rosette and cauline leaves, in addition to early flowering. The change in leaf morphology is associated with elevated levels of FT and ectopic expression of SEPALLATA3 (SEP3), leading to ectopic expression of floral genes. Leaf curling is suppressed by sep3 and ft mutations and enhanced by soc1 mutations. Thus, AGL15 and AGL18, along with SVP and AGL24, are necessary to block initiation of floral programs in vegetative organs.
Appropriate timing of the shift from vegetative to reproductive growth is an important determinant of plant fitness. The time at which a plant flowers is determined through integration of signals reflecting extrinsic and intrinsic conditions, such as photoperiod, the duration of cold, plant health, and age (for review, see Amasino, 2010). One of the most important pathways regulating the timing of the floral transition is the photoperiod pathway (for review, see Imaizumi and Kay, 2006). Under long-day (LD) inductive conditions in Arabidopsis (Arabidopsis thaliana), photoperiod pathway components act to promote flowering by inducing CONSTANS (CO) and downstream genes. The floral integrator FLOWERING LOCUS T (FT) is a major target of multiple flowering pathways and the photoperiod pathway in particular. It is directly activated by CO (Samach et al., 2000). Under LD conditions, the peak of CO expression is coincident with the presence of light, and CO activates FT expression in the leaf vascular system (Yanovsky and Kay, 2003). FT travels through the phloem to the shoot apex (Corbesier et al., 2007), where, together with FLOWERING LOCUS D (Abe et al., 2005; Wigge et al., 2005), it activates APETALA1 (AP1) and other floral meristem identity genes, starting the flowering process. Other flowering time pathways converge on FT and/or directly impact gene expression in the meristem. The changes in gene expression that accompany the floral transition must be rapid, robust, largely irreversible, and strictly controlled spatially. This is achieved through positive feed-forward and negative feedback loops involving multiple regulatory factors (for recent review, see Kaufmann et al., 2010).
Members of the MADS-box family of regulatory factors are central players in the regulatory loops controlling the floral transition (for a recent review, see Smaczniak et al., 2012a). MADS-domain factors typically act in large multimeric complexes and are well suited for regulation that involves combinatorial action. During the floral transition, MADS-domain proteins can act either as repressors or activators. In Arabidopsis, important floral repressors include SHORT VEGETATIVE PHASE (SVP) and members of the FLOWERING LOCUS C (FLC)-like group, including FLC, FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING1 (MAF1), and MAF2 to MAF5. Promoters of flowering include such MADS-domain factors as SUPPRESSOR OF CONSTANS1 (SOC1) and AGAMOUS-LIKE24 (AGL24). Together with non-MADS-box proteins FT and TWIN SISTER OF FT, SOC1 and AGL24 function as floral integrators. These operate downstream of the flowering time pathways but upstream of the meristem identity regulators such as LEAFY (LFY) and the MADS-domain factor AP1.
The MADS-domain factors AGL15 and AGL18 also contribute to regulation of the floral transition in Arabidopsis. While single mutants have no phenotype, agl15 agl18 double mutants flower earlier than the wild type (Adamczyk et al., 2007). Therefore, AGL15 and AGL18 appear to act in a redundant fashion in seedlings, and like SVP, FLC, and MAF1 to MAF5, they act as floral repressors. The contributions of AGL15 and AGL18 are most apparent in the absence of strong photoperiodic induction: the agl15 agl18 double mutant combination partially suppresses the delay in flowering observed in co mutants, as well as the flowering delay associated with growth under short-day (SD) noninductive conditions. The earlier flowering in agl15 agl18 mutants under these conditions is associated with up-regulation of FT, and both AGL15 and AGL18 are expressed in the vascular system and shoot apex of young seedlings (Adamczyk et al., 2007), raising the possibility that AGL15 and AGL18 act directly on FT in leaves, as well as other targets in the meristem.
AGL15, and to a lesser extent AGL18, have been further implicated in the networks that control flowering through molecular studies. Zheng et al. (2009) performed a chromatin immunoprecipitation (ChIP) analysis using AGL15-specific antibodies, tissue derived from embryo cultures, and a tiling array. Floral repressors (SVP and FLC), floral integrators (FT and SOC1), and a microRNA targeting AP2-like factors (miR172a) were identified as possible AGL15 targets (Zheng et al., 2009), suggesting that AGL15 may contribute to regulation through multiple avenues during the floral transition. AGL15 itself is directly bound and activated by AP2, which is both an A-class floral identity gene and a floral repressor (Yant et al., 2010). AGL15 is down-regulated in ap2 mutants, which are early flowering, while AGL18 is the nearest locus to multiple AP2-bound sites (Yant et al., 2010). Both AGL15 and AGL18 were identified as SOC1 targets through ChIP analyses (Immink et al., 2009; Tao et al., 2012). In yeast (Saccharomyces cerevisiae) two-hybrid assays, AGL15 interacts with a number of other MADS-domain proteins (de Folter et al., 2005), and in a one-hybrid study based on the SOC1 promoter, AGL15-SVP, AGL15-AGL24, and AGL15-SOC1 heterodimers were shown to bind to regions containing CArG boxes (Immink et al., 2012). AGL18 may act redundantly to AGL15 in these contexts. However, AGL18 either does not interact or only interacts weakly with other proteins in yeast two-hybrid assays (de Folter et al., 2005; Hill et al., 2008; Causier et al., 2012). It remains to be determined whether this truly reflects weaker or nonredundant in planta interactions or a technical problem in the artificial yeast system.
Guided by the knowledge gained about AGL15 targets and interactions from molecular studies, we asked the following question: what is the functional significance of these molecular relationships in the context of the floral transition? We performed a series of genetic experiments combining agl15 agl18 mutations and mutations in interacting factors such as SVP, AGL24, and SOC1, as well as targets such as FT and SOC1. We also performed further molecular experiments focused on AGL15, for which a variety of tools are available. Among other things, we show that AGL15 and AGL18, along with AGL24 and SVP, play a role in blocking expression of the floral MADS-domain factor SEPALLATA3 (SEP3) during the vegetative phase. In the absence of these four factors, reproductive programs are initiated early, and floral genes are expressed in the youngest rosette leaf and cauline leaves.
RESULTS
Genetic Interactions with SOC1 and FT
AGL15 and AGL18 contribute to flowering time regulation, but their effects can only be easily measured under noninductive conditions. We reasoned that their contributions might be partially obscured by the contributions of SVP and members of the FLC clade, which have large quantitative effects on flowering time in Arabidopsis (for review, see Amasino, 2010). svp mutations result in early flowering under both inductive and noninductive conditions and are largely epistatic to flc and flm mutations but show additive interactions with agl15 agl18 mutations (Adamczyk et al., 2007). SVP has been previously shown to repress FT and SOC1 through direct binding (Lee et al., 2007; Li et al., 2008). Therefore, genetic interactions between AGL15, AGL18, and their putative targets FT and SOC1 were examined in the presence and absence of SVP.
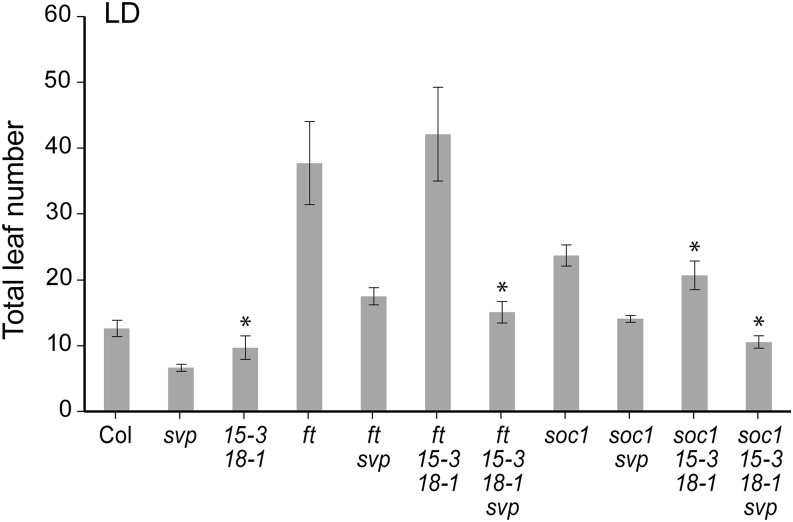
The agl15 agl18 double mutant combination and svp mutations were introduced into a background carrying the ft-1 mutation, and the impact on flowering time was assessed (Fig. 1). Because the FT mobile signal is decreased or eliminated in the leaves of ft-1 plants, they are late flowering under LD conditions. The agl15 agl18 ft-1 plants flower at the same time as ft-1 plants. By contrast, svp ft-1 plants flower significantly earlier than ft-1 plants. When agl15, agl18, and svp mutations are combined in the ft-1 background, flowering time is further accelerated relative to the svp ft-1 plants. In fact, the plants flower only a few leaves later than the wild type (Fig. 1). Therefore, agl15 agl18 and svp mutations show an additive relationship even in the absence of FT, suggesting that AGL15 and AGL18 act on additional targets that impact flowering.
Figure 1.
Genetic interactions between agl15, agl18, and svp mutations in backgrounds containing ft and soc1 mutations. Flowering time was measured under LD conditions. Asterisk indicates mutant combinations where the addition of agl15 agl18 mutations results in statistically significant differences (P < 0.01) in the means. The means ± 1 sd are shown (n ≥ 17 plants). Col, Columbia.
When FT arrives at the meristem, multiple changes in gene expression occur, including up-regulation of SOC1 and AP1. Because AGL15 has been shown to bind to the SOC1 promoter in yeast one-hybrid assays (Immink et al., 2012) and is physically associated with the SOC1 locus in seedlings according to ChIP-chip assays (Zheng et al., 2009), we tested whether agl15 agl18 effects are SOC1 dependent. Mutations in SOC1 produce plants that flower later than the wild type but not as late as ft mutants. The agl15 agl18 double mutant combination weakly suppresses soc1 mutations. svp mutations almost completely suppress soc1 mutations, and svp soc1 plants flower only slightly later than wild-type plants. When svp and agl15 agl18 mutations are combined in a soc1 background, the plants flower significantly earlier than svp soc1 or wild-type plants and at approximately the same time as agl15 agl18 mutants. Therefore, AGL15 and AGL18 have effects on flowering time that are independent of SOC1.
AGL15 Associates with Putative Regulatory Regions of FT in Vivo
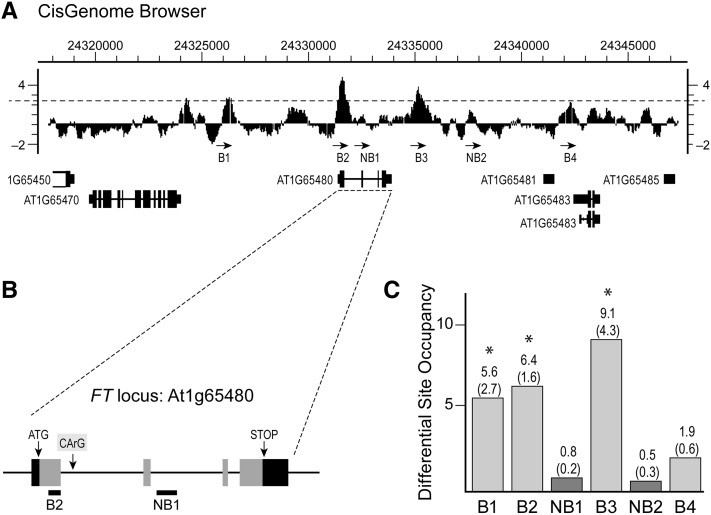
Previous work suggested that AGL15 regulates SOC1 via sequences in its promoter and the 5′ untranslated region (Immink et al., 2012); however, the location of AGL15-binding sites at the FT locus and their relationship to other known regulatory sites were unknown. To define these sites, we used ChIP followed by quantitative PCR (qPCR) and primer pairs spanning the FT locus in enrichment tests. First, we determined regions of interest based on data from a previously published ChIP-chip analysis using AGL15-specific antibodies (Zheng et al., 2009). Two peaks were identified in the vicinity of the FT locus using CisGenome (Ji et al., 2008). Default parameters were used except that the peak moving average cutoff was decreased from at least 3-fold to at least 2.5-fold change (immune versus preimmune) and the required number of continuous probes passing the cutoff for the peak was increased from 5 to 7. The former was done to allow a false discovery rate that was not zero for at least some peaks and the latter to decrease the number of considered peaks. As shown in Figure 2A, one peak corresponds to the 5′ region of the gene (position 24331418–24331713, The Arabidopsis Information Resource 10 annotation) and includes part of the first intron that contains a binding site for MADS-domain proteins (CArG box) that has been previously identified as important for SVP- (Lee et al., 2007) and FLC-mediated (Helliwell et al., 2006; Searle et al., 2006) regulation of FT (peak B2 in Fig. 2A; CArG shown in Fig. 2B). A second CArG 5′ to the ATG (CArG V in Lee et al. (2007) and present in peak B2 has a form of C(A/T)8G that is preferentially bound in vitro by AGL15 (Tang and Perry, 2003). This region also showed significant binding by SVP (Lee et al., 2007). A second peak was identified by CisGenome in the 3′ intergenic region (B3 in Fig. 2A, position 24335034–24335291). Two additional regions that did not meet the cutoffs used for CisGenome and two contiguous nonbound regions were also selected for further analysis (B1, B4, NB1, and NB2; Fig. 2A).
Figure 2.
AGL15 associates with regions near/in FT in vivo. A, The moving average CisGenome track of the region encompassing FT (At1g65480) is shown. The dashed line indicates the cutoff of 2.5 used to detect peaks (B2, B3) in Cisgenome. Other peaks of potential interest (B1, B4) are marked. Two contiguous nonbound regions (NB1, NB2) were also analyzed. B, Locus map showing the introns, exons, location of a previously identified CArG motif important for regulation of FT by MADS-domain proteins in the first intron, and the location of sites amplified in the ChIP analysis. C, The DSO calculations from qPCR on three independent ChIP experiments. Recovery of target by coimmunoprecipitation with anti-AGL15 antiserum was compared with recovery of a nonbound control (TUA3) in the same immune precipitation. The averages (sds) are shown. Asterisk indicates values that indicate significant enrichment relative to TUA3.
After regions of interest were identified, three additional independent ChIP assays were performed to isolate AGL15-DNA complexes, and qPCR with specific primers was used to test for association of AGL15 with these regions. The amount of amplicon corresponding to each B or NB fragment was expressed relative to the amount of amplicon corresponding to a nonbound control fragment (tubulin alpha-3 [TUA3], At5g19770) in the same immune precipitation. This yielded a differential site occupancy (DSO) value for each fragment. As shown in Figure 2C, the B1, B2, and B3 regions were significantly enriched and confirmed as being associated with AGL15-containing complexes. B4, NB1, and NB2 did not show significant enrichment.
To further confirm specific association of AGL15 with select DNA fragments, the immunoprecipitation was performed independent of the AGL15 antisera. Instead, tissue accumulating AGL15 with a C-terminal tandem affinity purification (TAP) tag was used. The TAP tag consists of a calmodulin-binding peptide, protease cleavage site, and two protein A domains (Puig et al., 2001). This later part allows precipitation of TAP-tagged proteins using IgG Sepharose. The control tissue accumulated AGL15 lacking the TAP tag. As shown in Supplemental Figure S1, the regions B1 to B4 are coprecipitated with AGL15-TAP but not in the untagged control tissue.
AGL15 Contributes to Flowering Time Regulation in Both the Leaves and Meristem
If FT and SOC1 are direct targets of AGL15, then AGL15 activity may be required both in leaf vascular tissue and the meristem. To test this, we used a similar strategy to that used for tissue-specific FLC expression (Searle et al., 2006). AGL15 complementary DNA (cDNA) was expressed under the control of a phloem-specific promoter (SUCROSE-PROTON SYMPORTER2 [SUC2]) or a meristem-specific promoter (KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 [KNAT1]) in agl15 agl18 plants.
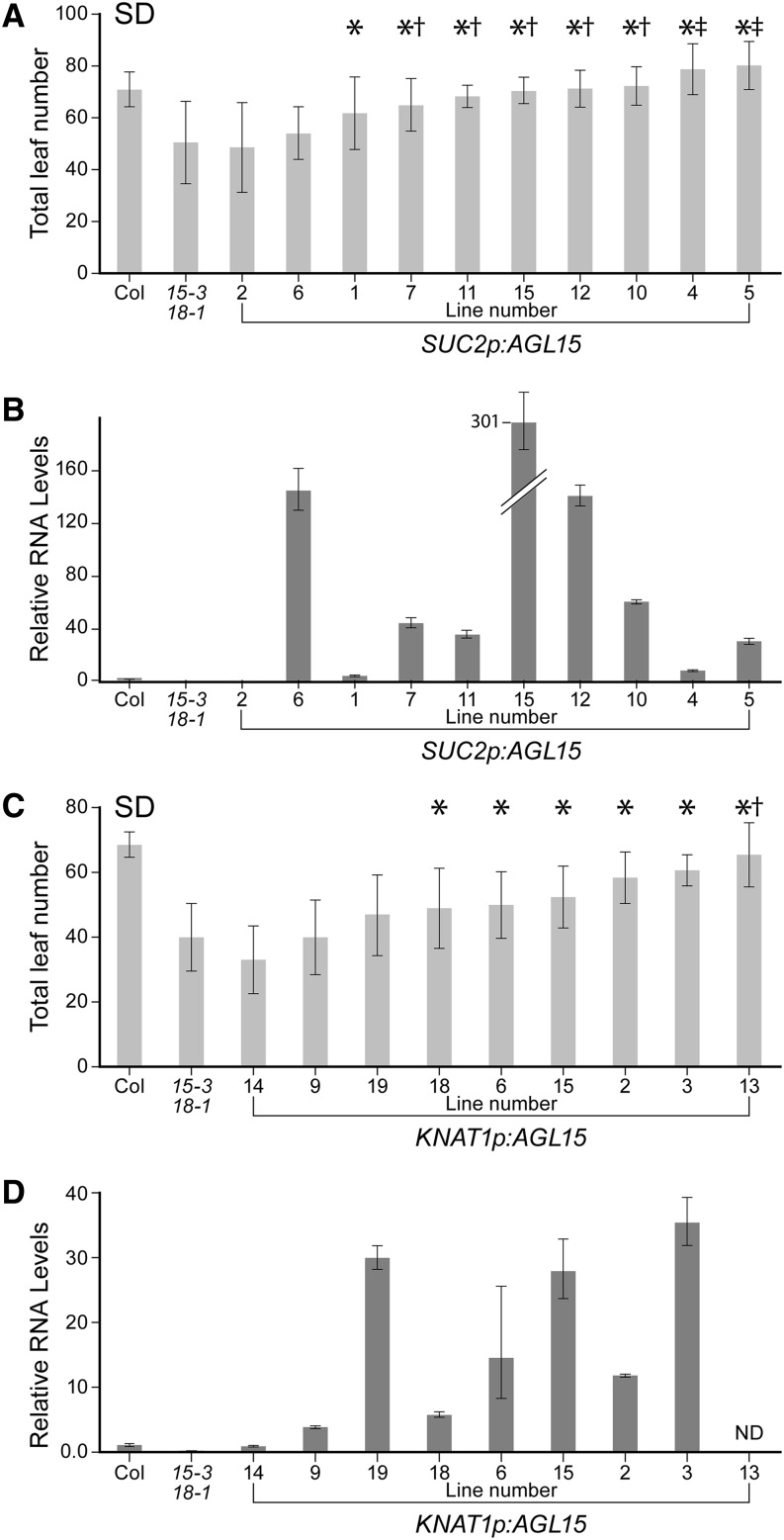
When AGL15 was expressed at high levels in the phloem, eight out of 10 independent lines had average flowering times that were significantly later than the agl15 agl18 mutant when grown under SD conditions (Fig. 3A). In five of those lines, flowering times were statistically indistinguishable from the wild type; and in two lines, flowering was delayed relative to the wild type. AGL15 RNA levels were also measured in 8-d-old seedlings grown under SD conditions for each line. Recovery of wild-type (or later) flowering times was consistently associated with elevated AGL15 transcript levels, although a strong correlation between levels of transcript accumulation and the magnitude of the flowering time change was not observed. In the seven lines with wild-type (or later) flowering times, AGL15 RNA levels varied from 8.7-fold to 300-fold higher than wild-type levels (Fig. 3B). In the two lines that did not show changes in flowering time, one had almost undetectable levels of AGL15 transcripts and the other had almost 145-fold higher levels than the wild type. We speculate that, in the latter, AGL15 protein is either not accumulating, not localized in the nucleus, or not incorporated into active regulatory complexes. Alternatively, the transcripts may not be expressed in the correct cell types in this line.
Figure 3.
Analysis of transgenic plants with AGL15 expressed in specific tissues. Flowering time (A and C) and AGL15 transcript levels (B and D) under SD conditions in independent lines of transformed agl15 agl18 plants expressing AGL15 in the phloem under the control of the SUC2 promoter (A and B) or in the meristem under the control of the KNAT1 promoter (C and D) are shown. For flowering times, means ± 1 sd are shown (n ≥ 9 plants). Asterisk indicates values that are significantly different from the agl15 agl18 control. Single dagger indicates values that are not significantly different from the Columbia (Col) wild type. Double dagger indicates values that are significantly different and greater than the Columbia wild type. For RNA, error bars indicate 1 sd.
When AGL15 was expressed in the meristem, six out of nine independent lines flowered significantly later than the agl15 agl18 mutant, but only one of these lines had a flowering time that was statistically indistinguishable from the wild type (Fig. 3C). In lines showing delayed flowering, expression levels ranged from 5.7-fold to 35-fold higher than the wild type (Fig. 3D). The measurements were based on whole seedling samples; therefore, we suspect that levels were considerably higher in individual cells in the meristem. As with the SUC2p:AGL15 constructs, one line with a relatively high level of expression (30-fold, line 19) flowered earlier than the wild type.
The results of these experiments indicate that AGL15 activity in either the leaves or the meristem can impact flowering time. When a phloem-specific promoter was used, 70% (7/10) of the lines flowered at the same time or later than the wild type. When a meristem-specific promoter was used, only 11% (1/9) of the lines had flowering times similar to the wild type. This is consistent with the molecular and genetic results that link AGL15 and AGL18 to the regulation of FT in the phloem and SOC1 in the meristem. Expression of AGL15 in the phloem is most effective in restoring wild-type flowering times in agl15 agl18 mutants, as we might expect given that FT functions upstream of SOC1.
Phenotypic and Molecular Changes in agl15 agl18 agl24 svp Mutants
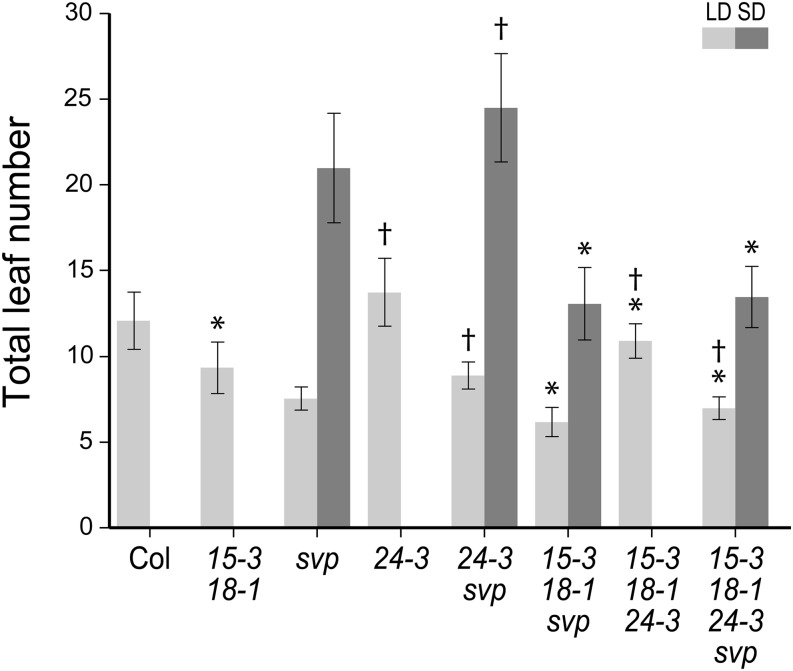
Next, we considered the relationship between AGL15, AGL18, SVP, and SVP’s closest relative AGL24. Together, these form two distinct two-member clades within the MADS-domain family in Arabidopsis (Parenicová et al., 2003). While AGL15, AGL18, and SVP act as floral repressors in seedlings, AGL24 is up-regulated at the floral transition and acts to promote inflorescence fate (Yu et al., 2002, 2004). Because most of the AGL24 alleles in current use are transposon alleles that can be unstable, we isolated and characterized a transfer DNA (T-DNA) allele of AGL24 for our experiments. In the allele we are designating as agl24-3, the T-DNA has inserted in the 4th exon (Supplemental Fig. S2A), and no full-length transcript can be detected in homozygous mutant plants (Supplemental Fig. S2B). agl24-3 plants flower later than wild-type Columbia (Supplemental Fig. S2C), which is consistent with the behavior of previously studied loss-of-function alleles and AGL24’s proposed function as a floral activator and promoter of inflorescence fate (Michaels et al., 2003; Yu et al., 2004).
When agl24-3 mutations are combined with agl15, agl18, and/or svp mutations, flowering time changes in a manner that is consistent with AGL24’s role as a floral activator and AGL15 and AGL18’s roles as floral repressors. Under SD conditions, addition of the agl24 mutation has no effect on the flowering time of agl15 agl18 svp plants; however, in all other cases, addition of either agl15 agl18 or agl24 mutations causes statistically significant changes in flowering time (Fig. 4A). agl24 mutations delay flowering, while the agl15 agl18 mutant combination consistently accelerates flowering. svp agl24 plants and agl15 agl18 agl24 plants flower later than svp and agl15 agl18 plants, respectively. svp agl15 agl18 and agl24 agl15 agl18 plants flower earlier than svp and agl24 plants, respectively. Finally, agl15 agl18 agl24 svp plants flower earlier than 24 svp plants but slightly later than agl15 agl18 svp plants. As expected, agl24 and agl15 agl18 mutations act antagonistically on the timing of the floral transition.
Figure 4.
Genetic interactions between agl15, agl18, agl24, and svp mutations. Flowering time under LD (light gray) and SD (dark gray) conditions. Asterisk indicates mutant combinations where the addition of agl15 agl18 mutations results in statistically significant differences (P < 0.01) in the means. Single dagger indicates mutant combinations where the addition of agl24 mutations results in statistically significant differences (P < 0.01) in the means. The means ± 1 sd are shown (n ≥ 20 plants). Col, Columbia.
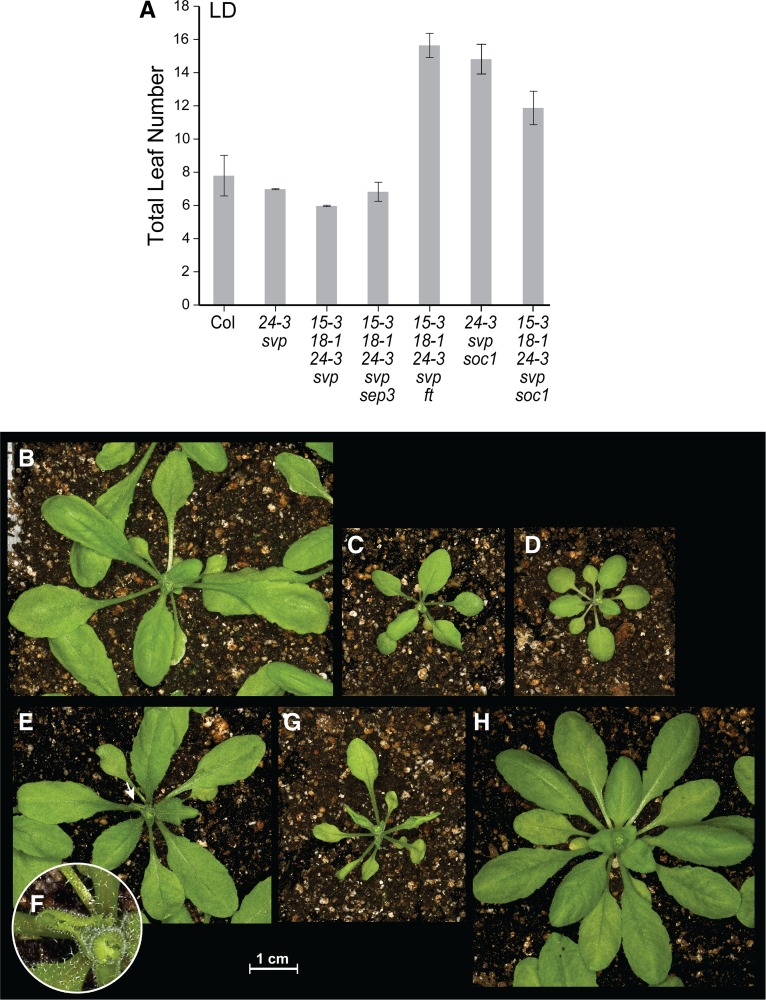
In addition to altered flowering times, the agl15 agl18 agl24 svp mutants show position-dependent changes in leaf morphology, which was an unexpected phenotype. The plants are small relative to the wild type (Fig. 5A), and the blades of the youngest rosette leaf as well as any cauline leaves (typically 1–2) curl tightly in an upward direction (Fig. 5, E and F). agl15 agl18 agl24 plants (Fig. 5B) do not show leaf curling; in agl15 agl18 svp (Fig. 5C) and agl24-3 svp (Fig. 5D) plants, leaf curling is sometimes observed but occurs inconsistently and is less pronounced. The robust leaf curling in the quadruple mutants suggest that AGL24, SVP, AGL15, and AGL18 act in a partially redundant or additive fashion on a program that impacts leaf morphogenesis.
Figure 5.
Phenotype of wild-type and mutant plants, grown under LD conditions. The appearance of the rosette just before bolting is shown. A, The Columbia wild type. B, agl15 agl18 agl24. C, agl15 agl18 svp. D, agl24 svp. E, agl15 agl18 agl24 svp. The small arrow indicates the youngest rosette leaf, which is tightly curled. F, Higher magnification view of the curled leaf shown in E.
Next, the molecular basis of the change in leaf morphology was investigated. Similar leaf curling has been reported in association with ectopic expression of FT (Teper-Bamnolker and Samach, 2005) or ectopic expression of MADS-domain proteins involved in floral development, either through viral (35S) promoters (Mizukami and Ma, 1992; Krizek and Meyerowitz, 1996; Honma and Goto, 2001; Pelaz et al., 2001) or derepression, as in curly leaf mutants (Goodrich et al., 1997). To identify candidate genes that show consistent changes in the agl15 agl18 agl24 mutants, we performed a microarray analysis. FT RNA levels are low in both mutant and wild-type plants at 6 d (Fig. 6A); therefore, we used RNAs isolated from 6-d-old seedlings to investigate gene expression patterns before the floral transition. We used RNAs isolated from cauline leaves to compare expression patterns in flat (wild-type) versus curled (mutant) leaves. cDNA was prepared and hybridized to NimbleGen whole-genome microarray chips, and a statistical analysis was performed on the data as described previously (Adamczyk and Fernandez, 2009). In mutant seedlings, SHATTERPROOF2 and SEP3 were the only MADS-domain genes to be significantly up-regulated (greater than 2-fold) relative to the wild type. In the curled cauline leaves of the mutants, multiple MADS-domain genes that are typically expressed during the reproductive phase were up-regulated, including AP3, PISTILLATA (PI), AP1, and SEP3. Based on this analysis, we focused on SEP3, which showed the earliest and most consistent effect, as a candidate regulatory target. SEP3 functions as a coregulator with LFY and activates B and C class organ identity genes in flowers. It also acts in ternary complexes with meristem and organ identity factors (Honma and Goto, 2001; Immink et al., 2009; Liu et al., 2009). In plants carrying 35S:FT constructs and grown in blue light-enriched environments, the curled leaf phenotype was shown to be associated with ectopic expression of the MADS-domain factor SEP3 (Teper-Bamnolker and Samach, 2005). Ectopic expression of SEP3 leads, in turn, to expression of other floral genes in the curled leaves (Castillejo et al., 2005; Teper-Bamnolker and Samach, 2005).
Figure 6.
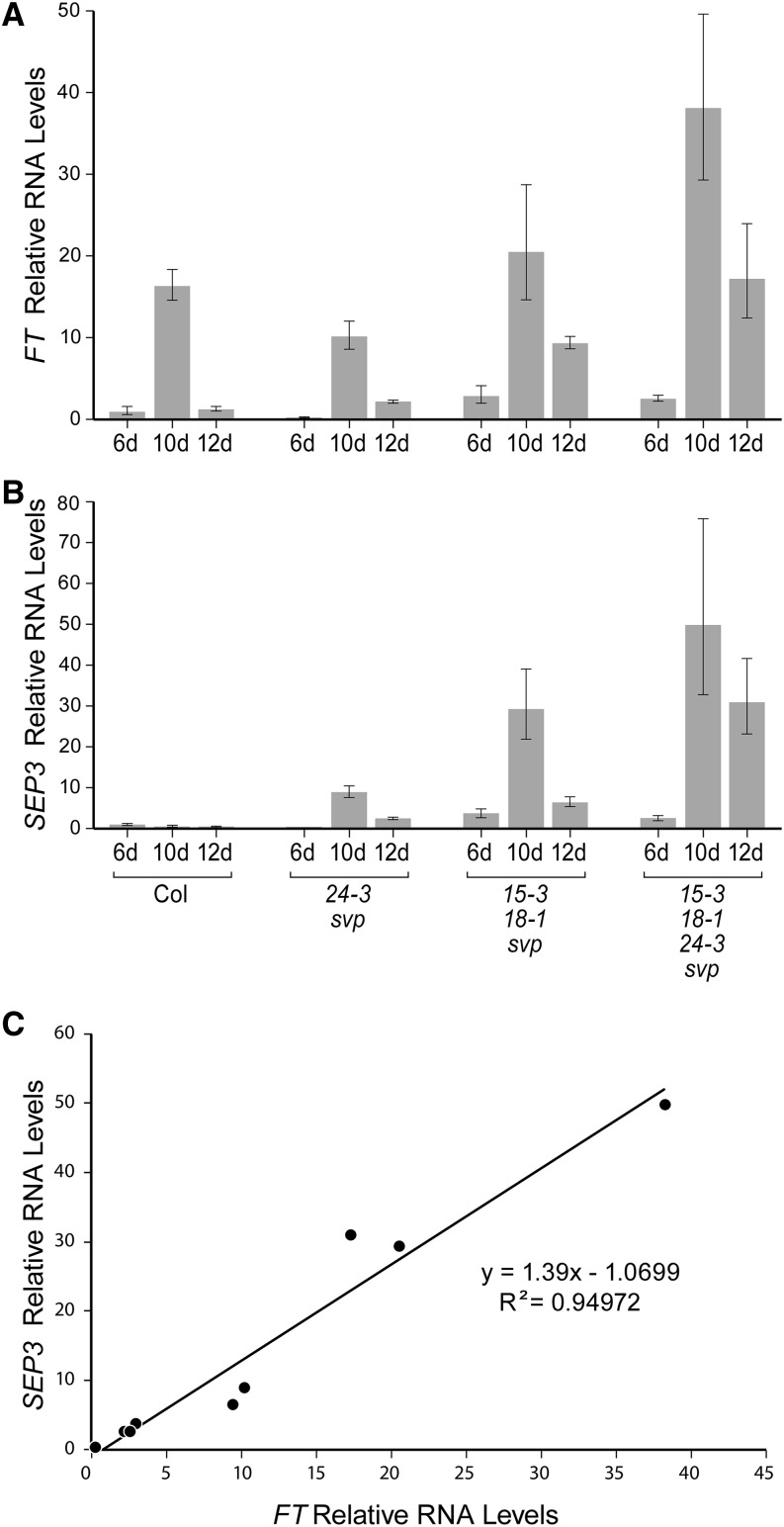
qPCR analysis of transcript accumulation during the floral transition period. Relative levels of FT RNA (A) and SEP3 RNA (B) in wild-type and mutant seedlings, grown for different lengths of time under LD conditions, are shown. The floral transition occurs between 6 and 10 d in wild-type plants under these conditions. RNA amounts were normalized to β-tubulin and expressed relative to the 6-d Columbia (Col) value. Error bars indicate 1 sd. C, Graph showing the relationship between FT and SEP3 relative RNA levels in the mutants.
To investigate the expression changes further, qPCR was performed to compare changes in relative levels of FT and SEP3 transcripts over time in wild-type and mutant seedlings grown under LD conditions (Fig. 6). Because FT RNA levels vary throughout the day, care was taken to collect tissue samples at the same time each day. In 6-d-old seedlings, FT RNA levels were approximately 2.5- to 3-fold higher in the early flowering agl15 agl18 svp and agl15 agl18 agl24 svp plants than in wild-type Columbia or agl24 svp plants (Fig. 6A). In all genotypes, FT transcript levels increased between 6 and 10 d, which corresponds to the period of floral induction under our growth conditions. At 10 d, FT RNA levels were highest in agl15 agl18 agl24 svp plants; but in all genotypes, the 10-d levels were more than 10-fold higher than wild-type levels at 6 d. FT RNA levels were lower overall in the 12-d-old samples, but agl15 agl18 agl24 svp and agl15 agl18 svp plants had higher levels than wild-type or agl24 svp plants. The overall pattern of expression changes was similar in all the seedlings. We concluded that FT is not constitutively expressed at high levels in the agl15 agl18 agl24 svp seedlings as it is in 35S:FT plants nor is induction of FT severely perturbed in any of the mutants.
The accumulation of SEP3 transcripts, on the other hand, differed significantly in wild-type and mutant seedlings (Fig. 6B). In wild-type seedlings, SEP3 RNA levels were low throughout the period from 6 to 12 d. In mutant seedlings, SEP3 RNA levels increased during the period of floral induction. At 10 d, SEP3 RNA levels were elevated by approximately 9-fold (agl24 svp), 29-fold (agl15 agl18 svp), or 50-fold (agl15 agl18 agl24 svp), relative to the wild type at 6 d. SEP3 RNA levels were lower in 12-d-old samples but were still relatively high. When we compared SEP3 and FT RNAs levels in the mutants, a strong linear relationship was seen (Fig. 6C). At the time of the floral transition then, there are relatively high FT and SEP3 RNA levels in agl15 agl18 agl24 svp plants, intermediate levels in agl15 agl18 svp plants, and lower levels in agl24 svp plants. In wild-type plants, on the other hand, FT RNAs are at an intermediate level at 10 d, but there is little or no accumulation of SEP3 RNA.
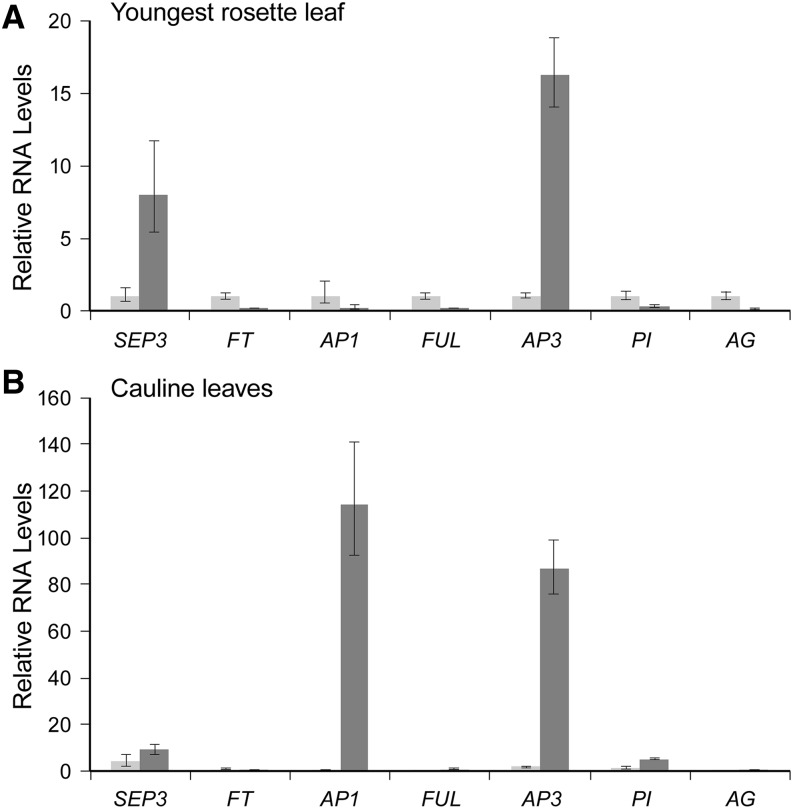
The association of elevated SEP3 RNA levels with the change in leaf morphology was confirmed by sampling the youngest rosette leaves of wild-type plants (typically leaf 9 or 10) and agl15 agl18 agl24 svp plants (typically leaf 4 or 5) at 31 d. This is approximately 1 week after bolting for the wild type and approximately 2 weeks after bolting for agl15 agl18 agl24 svp plants. SEP3 RNA levels were approximately 8-fold higher in the curled leaves of agl15 agl18 agl24 svp plants than the flat leaves of the wild type (Fig. 7A). FT RNA levels were lower in the curled leaves, possibly because these plants are further past the floral transition. Elevated SEP3 can lead to activation of floral programs. To confirm that this is happening in the curled leaves, we compared the RNA levels of various MADS-domain factors expressed during the reproductive phase in the youngest rosette leaves of mutant and wild-type plants (Fig. 7A) and, in a separate experiment, in the cauline leaves of mutant and wild-type plants (Fig. 7B). In the curled rosette leaves of agl15 agl18 agl24 svp plants, AP3 RNA levels were elevated 16-fold, while AP1, FRUITFUL (FUL), PI, and AGAMOUS (AG) RNA levels were as low or lower than they are in the wild type. In the curled cauline leaves of agl15 agl18 agl24 svp plants, AP1 and AP3 RNAs accumulated to very high relative levels, and SEP3 and PI RNA levels were also elevated relative to the levels in wild-type rosette and cauline leaves. On the other hand, FUL and AG RNA levels were no higher than they are in the youngest wild-type rosette leaf.
Figure 7.
qPCR analysis of transcript accumulation in rosette and cauline leaves of wild-type and agl15 agl18 agl24 svp mutant plants. Relative transcript levels in the youngest rosette leaf (A) and cauline leaves (B) of flowering plants are shown for different genes associated with flowering. The leaves from wild-type plants (light gray) were flat, while the leaves from mutant plants (dark gray) were curled. For each transcript, RNA amounts were normalized to β-tubulin and expressed relative to the level in the youngest rosette leaf of the Columbia wild type. Error bars indicate 1 sd.
Genetic and Environmental Modifiers of the Curled Leaf Phenotype
If leaf curling in agl15 agl18 agl24 svp plants is mediated by FT and/or SEP3 expression in young seedlings, we might expect ft and sep3 mutations to suppress this phenotype. The introduction of the ft-1 mutation into the agl15 agl18 agl24 svp background resulted in an increase in flowering time from six to almost 16 leaves (Fig. 8A) and a complete suppression of leaf curling (Fig. 8B). The introduction of the sep3-2 mutation resulted in relatively little change in flowering time (Fig. 8A) but also completely suppressed the leaf curling phenotype (Fig. 8C). By analyzing populations fixed for agl15 agl18 agl24 svp and segregating for sep3, we found that leaf curling is sensitive to the dosage of SEP3. As in 35S:FT plants (Teper-Bamnolker and Samach, 2005), the curled leaf phenotype was suppressed in plants that were heterozygous for sep3-2 (data not shown). Leaf curling was also affected by environmental conditions. When agl15 agl18 agl24 svp plants were grown in SD, the plants flowered with 12 to 14 leaves and leaf curling was completely suppressed (Fig. 8D). In 35S:FT plants, on the other hand, growth in SD conditions leads to intermediate curling, i.e. the phenotype is attenuated but not eliminated (Teper-Bamnolker and Samach, 2005).
Figure 8.
Phenotype of plants carrying agl15 agl18 agl24 svp mutations, grown under different conditions or in the presence of genetic modifiers. A, Flowering time under LD conditions. The means ± 1 sd are shown (n ≥ 9 plants). B to G, The appearance of rosettes just before bolting, for agl15 agl18 agl24 svp ft mutant grown under LD conditions (B), agl15 agl18 agl24 svp sep3 mutant grown under LD conditions (C), agl15 agl18 agl24 svp mutant grown under SD conditions (D), and agl24 svp soc1 mutant grown under LD conditions (E). The small arrow indicates a curled leaf. F, Higher magnification view of the curled leaf shown in E. G, agl15 agl18 agl24 svp soc1 mutant grown under LD conditions. Several curled leaves are visible. H, agl15 agl18 agl24 svp soc1 mutant grown under SD conditions. Col, Columbia.
Because SOC1 is proposed to act along with AGL24 and SVP to regulate SEP3 during early stages of floral development (Liu et al., 2009), we also tested the effect of soc1-2 mutations. agl24 svp soc1 plants are larger than agl24 svp plants but resemble them in that leaf curling is less pronounced and appears less consistently than in agl15 agl18 agl24 svp plants (Fig. 8, E and F). When agl15 agl18 mutations are introduced, the agl15 agl18 agl24 svp soc1 plants flower earlier than agl24 svp soc1 plants but later than wild-type plants (Fig. 8A). Both the plants and the leaves are larger at the floral transition than with agl15 agl18 agl24 svp plants, and an increased number of rosette and cauline leaves show curling (Fig. 8G). Following the floral transition, the phenotype of agl15 agl18 agl24 svp soc1 plants resembles that of agl24 svp soc1 plants, with all of the phenotypic changes, such as bract development and floral abnormalities (not shown), that were described in previous studies (Liu et al., 2009). Leaf curling, but not bract formation or floral abnormalities, is suppressed by growth of agl15 agl18 agl24 svp soc1 plants under SD conditions (Fig. 8H).
DISCUSSION
Role of AGL15 in Flowering Time Regulation
We sought to more clearly define the role of AGL15 in regulation of the floral transition. By expressing AGL15 separately and specifically in the leaf vasculature and the meristem, we showed that AGL15 is likely to contribute to regulation in both tissues and at different stages in the floral induction process. Expression of AGL15 in the phloem of agl15 agl18 mutants was sufficient to restore wild-type flowering times, which is consistent with a role for AGL15 in regulating FT or another gene of major effect in the leaf vasculature. Expression of AGL15 in the meristem via the KNAT1 promoter resulted in delayed flowering relative to the agl15 agl18 mutant. This is consistent with repression of genes contributing to flowering in the meristem, such as the known target SOC1. However, because wild-type flowering time was only restored in one line, it appears that AGL15 activity, when restricted to the meristem, is typically not sufficient to overcome the effects of a strong inductive signal coming from the leaves. A model summarizing the effect of agl15 agl18 and svp mutations on flowering time via regulation of FT and SOC1 is shown in Supplemental Figure S3.
By mapping the sites where complexes containing AGL15 bind to the FT locus, we sought to determine the relationship between these complexes and those containing the repressors SVP or FLC. The largest peak (B2) of AGL15 binding is centered over the first exon and intron, where the major binding sites for FLC and SVP are also located (Helliwell et al., 2006; Searle et al., 2006; Lee et al., 2007). FLC and SVP are known to be present in the developmental context (embryonic tissue) where the ChIP analysis was performed (Lehti-Shiu et al., 2005), and FLC-containing repressor complexes have been shown to be quite large (800 kD; Helliwell et al., 2006). AGL15 may be a component of these complexes and add to their effectiveness. However, AGL15 and AGL18 also contribute to repression of flowering under conditions where SVP and FLC are absent (Adamczyk et al., 2007). Additional ChIP experiments in svp flc backgrounds would be needed to test whether AGL15’s association with this region is reduced or otherwise altered under these conditions. AGL15 shows associations with more distal regions both upstream (B1 peak) and downstream (B3 peak) of the FT coding region. The importance of distal upstream regions to the regulation of FT was recently demonstrated (Adrian et al., 2010). The downstream region is also likely to contain important regulatory elements. ChIP-chip analysis indicated that the AP2-like factor SCHLAFMüTZE (SMZ) binds approximately 1.5 kb downstream of the coding sequence (Mathieu et al., 2009). SMZ acts as repressor of FT, and this activity is dependent on the presence of the MADS-domain factor FLM (Mathieu et al., 2009). It is not known whether AGL15 is important for SMZ-mediated FT repression; however, it is intriguing that AGL15 binds to sites near this important control region (approximately 1.2 kb downstream).
From molecular analyses, it is clear that SVP and AGL15 each affect a large number of genes other than FT and SOC1 within the flowering networks. Through ChIP analyses, 333 enriched regions, corresponding to 328 genes, have been identified as possible direct targets of SVP (Tao et al., 2012). Two hundred thirty-one or 69% of these regions also appear on the list of possible AGL15 targets, which includes approximately 2,000 genes (Zheng et al., 2009). The AP2-like transcription factors and microRNAs targeting SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes (miR156e) and AP2-like genes (miR172a) have been highlighted as particularly relevant SVP targets with regard to flowering time regulation (Tao et al., 2012). AP2 and the AP2-like factors TEMPRANILLO1, TARGET OF EAT1 (TOE1), and TOE3, as well as miR172a, SOC1, and the miRNA156 target SPL11 appear both on the SVP and AGL15 lists. A comparison of the binding-site distribution profiles generated by CisGenome suggests that the SVP- and AGL15-binding sites may overlap or be in close proximity at these loci as well. Other possible direct targets of AGL15 include genes encoding flowering time regulators (FLC, SVP, AGL18, MAF3, MAF5, and AGL19) and meristem and organ identity genes (LFY, FUL, AG, AGL24, AP3, and SEP2).
The large number of shared targets and the close proximity of SVP- and AGL15-binding sites is intriguing and raises the possibility that these factors may act in a partially redundant way in the context of large repressor complexes. AGL15 could contribute to repression by increasing the efficiency of complex formation or recruitment or by enhancing corepressor recruitment. AGL15, AGL18, AGL24, and SVP all contain ethylene response factor-associated amphiphilic repression (EAR) motifs (LxLxL) in their C-terminal domains (Hill et al., 2008; Kagale et al., 2010). EAR motifs are involved in recruiting corepressors, and, through them, histone deacetylase complexes, to genetic loci (Ohta et al., 2001; for review, see Kagale and Rozwadowski, 2011). Alternate means of recruiting corepressors might become especially important in situations where SVP protein levels are low, as at high ambient temperatures (Lee et al., 2013). Future experiments may show that AGL15 and AGL18 play critical roles under particular, as yet undetermined, environmental conditions or in other species, particularly those that lack the FLC clade (Ruelens et al., 2013; supplemental material).
Leaf Curling Is Associated with SEP3 Activation and Floral Gene Expression
The agl15 agl18 agl24 svp mutant combination leads to elevated expression of SEP3 in young seedlings. SEP3 is normally repressed during vegetative growth and the early stages of reproductive growth. In young floral meristems, repression is mediated by a combination of SVP, AGL24, and SOC1 (Liu et al., 2009). In agl24 svp soc1 plants, SEP3 is also up-regulated in 6-d-old seedlings; however, leaf curling was not reported (Liu et al., 2009). We found only mild leaf curling in the agl24 svp and agl24 svp soc1 mutants we created using a strong agl24 T-DNA allele. However, if agl15 agl18 mutations are added, to create agl15 agl18 agl24 svp or agl15 agl18 agl24 svp soc plants, the youngest rosette leaf and the cauline leaves are tightly curled. Therefore, the agl15 agl18 mutant combination is acting as an enhancer of the change in leaf morphology. As in other studies, we find that leaf curling is correlated with expression of floral programs in leaf tissues. We found elevated levels of AP3 transcripts in curled rosette leaves and elevated levels of AP1, AP3, and PI in curled cauline leaves. The cauline leaves maintain leaf identity and produce branched trichomes. No obvious signs of cell type changes could be seen in scanning electron micrographs of the exposed abaxial surfaces (data not shown). The leaf curling effect appears to be dependent on high SEP3 levels because sep3 mutations suppress this phenotype in a dose-dependent manner.
Only the youngest rosette leaf and cauline leaves curl in agl15 agl18 agl24 svp and agl15 agl18 agl24 svp soc plants. Therefore, there is a temporal and spatial pattern to changes in leaf morphology in the mutants that is not seen in 35S:FT and 35S:SEP3 plants, where all leaves curl (Teper-Bamnolker and Samach, 2005). The onset of leaf curling coincides with the dramatic increase in SEP3 RNA levels that occurs during the floral induction period in the mutants. During the 6- to 12-d window, SEP3 and FT RNA levels are directly proportional. Therefore, the early and strong induction of FT in the mutant plants would appear to be an important factor leading to SEP3 accumulation above the threshold levels needed to induce leaf curling. This is consistent with previous work showing that SEP3 activation is FT dependent in transgenic plants (Teper-Bamnolker and Samach, 2005). Conditions that eliminate strong induction through FT, such as ft mutations or SD conditions, suppress leaf curl. In their study of 35S:FT plants, Teper-Bamnolker and Samach (2005) suggest that organ fate depends on the developmental stage of the organ at the time when FT (and subsequently SEP3) is activated. According to this view, the youngest rosette leaf and cauline leaves are specifically affected because they are the only organs in a relatively immature state when SEP3 levels reach the critical threshold in the mutants.
If AGL15 is involved in direct regulation of SEP3, as well as indirect regulation through FT, we might expect to see a physical association between AGL15 and the SEP3 locus, and ChIP-chip analyses support this (Zheng et al., 2009). CisGenome shows a peak of binding in the distal SEP3 promoter (data not shown), which contains CArG boxes that are required for SEP3 autoregulation and enhancement of expression in floral tissues. Complexes containing AP1, SEP3, FUL, and AG have been shown to bind to CArG box pairs in this region (Smaczniak et al., 2012b) and presumably contribute to activation. In young floral meristems, repression is mediated by a combination of SVP, AGL24, and SOC1, which also directly bind to regions upstream of the translational start site upstream region (Liu et al., 2009). AGL24 and SOC1, along with AGL15, have been shown to interact with the corepressor SIN3-ASSOCIATED POLYPEPTIDE18 (Hill et al., 2008; Liu et al., 2009) and with TOPLESS (TPL) and TOPLESS-RELATED (TPR) corepressors in yeast two-hybrid assays (Causier et al., 2012). AGL18 has also been shown to interact with TPL/TPR corepressors (Causier et al., 2012). Based on these interactions, AGL15, AGL18, AGL24, and SOC1 potentially act in a partially redundant fashion. The corepressors contribute to repression by recruiting histone deacetylase complexes (Liu et al., 2009; Krogan et al., 2012). We propose that AGL15 and AGL18 play a role in recruiting histone deacetylases to the SEP3 locus in embryos or young seedlings, where levels of AGL24 and SOC1 are relatively low and AGL15 and AGL18 levels are relatively high. At the floral transition, AGL24 and SOC1 are up-regulated and take over this important role as the levels of AGL15 and AGL18 decline.
agl15 agl18 agl24 svp Mutations Affect Coordination of the Floral Transition
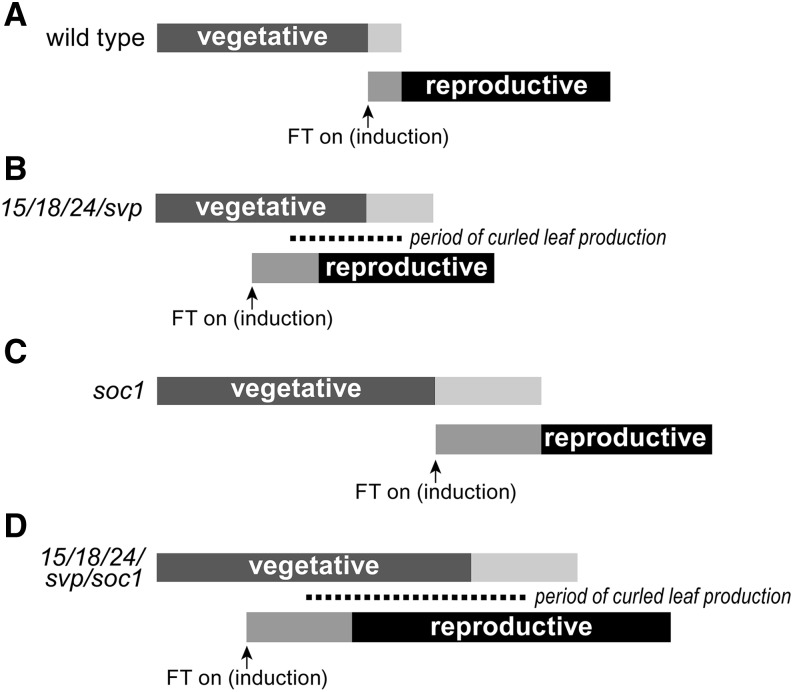
The following simple conceptual model was developed to explain the effects observed in agl15 agl18 agl24 svp and agl15 agl18 agl24 svp soc1 mutants. In the wild type, silencing of the vegetative-phase programs is coupled to activation of reproductive-phase programs, as marked by inflorescence and floral bud development, such that there is minimal overlap between them (Fig. 9A). A smooth transition over a relatively short period of time (from floral induction to floral determination) depends on the operation of positive feed-forward and negative feedback loops that include a number of MADS-domain proteins. AGL24, SVP, and SOC1 have been shown previously to be integral players in such loops (Immink et al., 2012; Tao et al., 2012). AGL15 potentially forms heterodimers in vivo with all three of these factors (Immink et al., 2012). AGL18 may as well, because it is closely related to AGL15 and shows complete redundancy with regard to flowering time (Adamczyk et al., 2007), but it has been more difficult to work with experimentally. In agl15 agl18 agl24 svp plants, both positive and negative feedback loops may be disrupted, resulting in a longer transition period. With the removal of repressors from the system, activation of FT and SEP3 is both early and strong. This would result in a period of overlap between the vegetative and reproductive phases (Fig. 9B). As a consequence, organs developing during this period retain vegetative identity and develop as leaves, but floral programs are also initiated, which results in leaf curling.
Figure 9.
Models of the floral transition in wild-type and mutant plants. During the floral transition in the wild type (A), developmental programs that mark the vegetative phase are down-regulated and developmental programs that mark the reproductive phase are up-regulated. Lighter colors indicate the transition period when gene expression patterns are changing in the shoot apex. Leaf primordia that form during the vegetative phase but mature after floral induction (marked by up-regulation of FT) develop into cauline leaves. B, In agl15 agl18 agl24 svp mutants, expression of floral programs starts prematurely in developing leaves, resulting in leaf curl. C, In soc1 mutants, flowering is delayed and the transition period is longer. D, In agl15 agl18 agl24 svp soc1 mutants, expression of floral programs starts prematurely and the period of leaf production is extended, so many curled leaves are produced.
As a floral integrator, SOC1 plays a pivotal role in the transition from vegetative to reproductive growth. Its regulatory targets include both upstream (flowering time) and downstream (meristem and organ identity) genes. As a consequence, in soc1 plants, the vegetative phase is prolonged (more rosette leaves are produced) and the transition to reproductive growth occurs more slowly (Fig. 9C). In agl15 agl18 agl24 svp soc1 plants, the two effects are combined. Floral induction and expression of floral programs are early, but the transition is slowed and more organs develop during the period of overlap (Fig. 9D). The result is strong leaf curling that affects multiple rosette and cauline leaves.
In conclusion, with these experiments, we provide additional evidence that AGL15 and AGL18 make important contributions to control of the transition from the vegetative to reproductive phase. This is mediated, at least partially, through regulation of FT in the phloem. AGL15 and AGL18, along with SVP and AGL24, are necessary to block premature activation of SEP3 and expression of reproductive programs during the vegetative phase.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) plants were grown on plates from sterile seed and transplanted to soil after 7 d, as described previously (Lehti-Shiu et al., 2005). Plants were grown in growth chambers at 22°C under either inductive LD conditions (16-h days, 8-h nights; Econair Ecological Chambers) or noninductive SD conditions (8-h days, 16-h nights; Conviron). Lighting was provided by a mix of incandescent and cool fluorescent bulbs, with levels set at approximately 125 μE m–2 s–1. For flowering time experiments, the total number of leaves produced on the main axis was counted. Differences between the means were tested using a two-tailed Student’s t test. Mean values were considered statistically different when P < 0.01. For seedling RNA isolations, seedlings were grown on plates with germination media containing 0.5× Murashige and Skoog salts and vitamins (Murashige and Skoog, 1962) supplemented with 10 g L–1 Suc, 0.5 g L–1 MES, and 7 g L–1 agar, pH 5.7. For rosette leaf RNA isolations, the youngest rosette leaf was harvested after bolting from mutant and wild-type plants grown for 31 d under LD conditions. Cauline leaves were harvested in a different experiment, and because of the difference in flowering time, mutant cauline leaves were collected 8 d earlier than wild-type cauline leaves. In each experiment, wild-type and mutant plants were grown in parallel, and tissues were harvested at the same time of day, frozen in liquid nitrogen, and stored at –80°C.
All experiments were conducted using mutant alleles in the Columbia-0 background. With the exception of agl24-3, the mutant alleles were described previously: agl15-3 (SALK_093946), agl18-1 (SALK_083061), svp-32 (SALK_072930), sep3-2, ft-1, and soc1-2 (Kardailsky et al., 1999; Pelaz et al., 2000; Moon et al., 2003; Lehti-Shiu et al., 2005; Lee et al., 2007). A seed pool (SALK_095007) containing the agl24-3 allele was obtained from the Arabidopsis Biological Resource Center at The Ohio State University (Alonso et al., 2003). Plants carrying the T-DNA allele were backcrossed five times to Columbia-0 to establish the agl24-3 line. For genotyping, DNA was isolated from leaf samples as described previously (Adamczyk et al., 2007). Mutant alleles were identified by PCR genotyping with gene-specific primers (Supplemental Table S1) and ExTaq polymerase (TaKaRa Bio), with the exception of ft-1. For ft-1 alleles, homozygous plants were identified based on the late-flowering phenotype and progeny testing.
Generation and Analysis of Transgenic Plants
For expression of AGL15 in the phloem, the SUC2p:AGL15 construct (DF422), with the SUC2 promoter fused to full-length AGL15 cDNA, was generated. The SUC2 promoter consisted of approximately 2 kb of sequence upstream of the ATG and was amplified from Columbia genomic DNA using oligo 944 (5′-TTCTGCAGAAAATCTGGTTTCATATTAATTTCA-3′), which introduced a PstI site, and oligo 945 (5′-TTGGATCCATTTGACAAACCAAGAAAGTAAGA-3′), which introduced a BamHI site. AGL15 cDNA sequence was amplified using oligo 953 (TTGGATCCATGGGTCGTGGAAAAATCGAG), which introduced a BamHI site, and oligo 954 (TTGGTACCCTAAACAGAGAACCTTTGTCTT), which introduced a KpnI site. The SUC2 and AGL15 sequences were introduced into a modified PZP221 vector (DF264) that contained the NOPALINE SYNTHASE terminator.
For expression of AGL15 in the meristem, the KNAT1p:AGL15 construct (DF421), with the KNAT1 promoter fused to full-length AGL15 cDNA, was generated. The KNAT1 promoter consisted of approximately 1.5 kb of sequence upstream of the ATG and was amplified from Columbia genomic DNA using oligo 946 (TTGTCGACAGAGCCCTAGGATTTGACGAT-3′), which introduced a SalI site, and oligo 947 (TTTCTAGAACCCAGATGAGTAAAGATTTGAG-3′), which introduced a XbaI site. AGL15 cDNA sequence was amplified as described for the SUC2p:AGL15 construct. The KNAT1 and AGL15 sequences were introduced into a modified PZP221 vector (DF375) that contained the NOPALINE SYNTHASE terminator.
Constructs were introduced into Agrobacterium tumefaciens strain GV3101 and then agl15-3 agl18-1 plants using the floral dip method (Clough and Bent, 1998). Transformants were selected on germination media supplemented with 100 μg mL–1 gentamycin. At least nine independent lines were analyzed in detail for each construct. For each line, the average flowering time of at least nine T2 progeny carrying at least one copy of the transgene was determined under SD conditions. Each transgenic line was compared with the agl15 agl18 control using Dunnett’s method for comparison of means (Dunnett, 1955). Differences were considered statistically different when P < 0.05. RNA was isolated from T2 seedlings grown on plates for 8 d under SD conditions, and qPCR analysis was conducted to determine the average relative level of AGL15 transcripts accumulating in each line.
Enrichment Test and qPCR
ChIP was performed as described previously (Zheng et al., 2009). In brief, AGL15-specific antiserum and preimmune serum (as a control) were used to immunoprecipitate AGL15-DNA complexes from an embryonic culture tissue expressing 35S:AGL15. Floral repressors are highly expressed in this tissue (Lehti-Shiu et al., 2005) and are likely to contribute to repression of floral programs before and immediately after germination. For enrichment tests, oligonucleotides for regions identified as bound (B2, B3) and possibly bound (B1, B4) and contiguous regions that are not bound (NB1, NB2) as well as oligonucleotides that amplify a nonbound control remote from FT (TUA3, At5g19770) were used in qPCR reactions with independently generated ChIP populations. Recovered DNA from ChIP (0.5 µL) or controls or 1 µL of input DNA diluted 125-fold was added to a reaction consisting of 40,000× diluted SYBR Green I (Invitrogen), 50 mm KCl, 0.2 mm deoxynucleotide triphosphate, 0.5 µm of each oligonucleotide, and 2 to 2.4 units Klentaq in 1× PC2 buffer (Ab Peptides). PCR was performed in an iCycler (Bio-Rad) with an initial 2-min denaturation at 95°C followed by 40 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30s, and, finally, 72°C for 5 min, followed by a melt curve determination. Oligonucleotides used for these experiments are listed in Supplemental Table S2. Quantitation was as described previously (Zheng et al., 2009). DSO indicates binding relative to control fragments (Haring et al., 2007; Mukhopadhyay et al., 2008) and was calculated using the delta delta cycle threshold (δδCT) method (Livak and Schmittgen, 2001), where δδCT = experimental (B or NB) δCT – control (TUA3) δCT, in samples derived from a single immune precipitation.
For gene expression analyses, RNA was isolated and cDNA was synthesized from 1 μg of total RNA as described previously (Lehti-Shiu et al., 2005). qPCR was performed in a 16-μL final volume containing 8 μL of SYBR-Green PCR master mix (Stratagene), 0.5 μL each of forward and reverse primers (12 μm), and 5 μL of a 1:10 dilution of the cDNA reaction mixture as template. Reactions were performed on an MX3000P QPCR System (Agilent). Calculations were based on two to three technical replicates. The amplification efficiency was tested for each primer pair by preparing a standard dose response curve. In addition, in the transgenic plant experiments, LinReg software, which takes the amplification efficiency into consideration while calculating the CT values (Ruijter et al., 2009), was used. CT values were normalized to levels of either β-tubulin (mutant studies) or CN1 (At2g28390, transgenic plants), and relative quantities were determined using the δδCT method (Livak and Schmittgen, 2001). RNA quantities in transgenic plants or mutants are expressed relative to wild-type values. Primer sequences are listed in Supplemental Table S2.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Coimmunoprecipitation of selected DNA fragments at the FT locus in plants carrying 35S:AGL15-TAP constructs.
Supplemental Figure S2. Analysis of the agl24-3 mutant allele.
Supplemental Figure S3. Model summarizing the effects of agl15, agl18, and svp mutations on FT and SOC1 expression and flowering time under inductive conditions.
Supplemental Table S1. Oligonucleotides used for PCR genotyping.
Supplemental Table S2. Oligonucleotides used for qPCR analyses.
Supplementary Material
Acknowledgments
We thank Drs. Richard Amasino, Scott Michaels, and Hao Yu for seed stocks; members of the Amasino lab for helpful discussions of flowering time regulation; Kaija Goodman, Patrick McMinn, David Merline, Vanessa Nurmi, and Cullen Vens for help with clone construction and expression analyses; and Sarah Friedrich and Kandis Elliot for plant photos and figure preparation.
Glossary
- ChIP
chromatin immunoprecipitation
- LD
long-day
- SD
short-day
- qPCR
quantitative PCR
- DSO
differential site occupancy
- cDNA
complementary DNA
- T-DNA
transfer DNA
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS–0718598 to D.E.F. and IOS–0922845 to S.E.P.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Adamczyk BJ, Fernandez DE. (2009) MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol 149: 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Castillejo C, Romera-Branchat M, Pelaz S. (2005) A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J 43: 586–596 [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RGH, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnett CW. (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50: 1096–1121 [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE. (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Immink RGH, Posé D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk AD, Schmid M, et al. (2012) Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RGH, Tonaco IAN, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC. (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC. (2010) Regulation of transcription in plants: mechanisms controlling developmental switches. Nat Rev Genet 11: 830–842 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM. (1996) The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22 [DOI] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. (2012) APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ryu HS, Chung KS, Posé D, Kim S, Schmid M, Ahn JH. (2013) Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Adamczyk BJ, Fernandez DE. (2005) Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol Biol 58: 89–107 [DOI] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H. (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33: 867–874 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJM, Tissenbaum HA. (2008) Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc 3: 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF. (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K. (2013) FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4: 2280. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Angenent GC, Kaufmann K. (2012a) Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139: 3081–3098 [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. (2012b) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Perry SE. (2003) Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J Biol Chem 278: 28154–28159 [DOI] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. (2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. (2003) Living by the calendar: how plants know when to flower. Nat Rev Mol Cell Biol 4: 265–275 [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM. (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36: 157–161 [DOI] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP. (2002) AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci USA 99: 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.