Rapid HR-like cell death confers plant resistance to aphids.
Abstract
BOTRYTIS-INDUCED KINASE1 (BIK1) plays important roles in induced defense against fungal and bacterial pathogens in Arabidopsis (Arabidopsis thaliana). Its tomato (Solanum lycopersicum) homolog is required for host plant resistance to a chewing insect herbivore. However, it remains unknown whether BIK1 functions in plant defense against aphids, a group of insects with a specialized phloem sap-feeding style. In this study, the potential role of BIK1 was investigated in Arabidopsis infested with the green peach aphid (Myzus persicae). In contrast to the previously reported positive role of intact BIK1 in defense response, loss of BIK1 function adversely impacted aphid settling, feeding, and reproduction. Relative to wild-type plants, bik1 displayed higher aphid-induced hydrogen peroxide accumulation and more severe lesions, resembling a hypersensitive response (HR) against pathogens. These symptoms were limited to the infested leaves. The bik1 mutant showed elevated basal as well as induced salicylic acid and ethylene accumulation. Intriguingly, elevated salicylic acid levels did not contribute to the HR-like symptoms or to the heightened aphid resistance associated with the bik1 mutant. Elevated ethylene levels in bik1 accounted for an initial, short-term repellence. Introducing a loss-of-function mutation in the aphid resistance and senescence-promoting gene PHYTOALEXIN DEFICIENT4 (PAD4) into the bik1 background blocked both aphid resistance and HR-like symptoms, indicating bik1-mediated resistance to aphids is PAD4 dependent. Taken together, Arabidopsis BIK1 confers susceptibility to aphid infestation through its suppression of PAD4 expression. Furthermore, the results underscore the role of reactive oxygen species and cell death in plant defense against phloem sap-feeding insects.
Aphids are specialized to feed and survive on phloem sap of their host plants. In contrast to chewing insects that cause extensive plant tissue damage, aphids have evolved to manipulate resource allocation within the host plant by converting the feeding site into a sink to deplete photoassimilates (Girousse et al., 2005). Their highly modified stylets navigate through plant tissues predominantly intercellularly before reaching phloem, causing very limited host cell damage. During probing and feeding, aphids secrete gelling and watery saliva (Tjallingii, 2006). Gelling saliva forms the sheath enveloping the stylet along the pathway leading to the vascular bundle. The sheath limits damage to plant cells and avoids triggering extracellular defenses. Watery saliva is thought not only to prevent clogging of phloem sieve elements and the food canal in aphid stylets due to protein coagulation, but also to modulate host cellular processes and mitigate host defense (Tjallingii, 2006; Will and van Bel, 2006; Will et al., 2007). Aphids make use of their stealthy feeding strategies and intimate associations with their hosts to disguise themselves and overcome plant defense, reminiscent of the deceptive strategies frequently employed by pathogens (Kaloshian, 2004; Walling, 2008).
During the long history of coevolution, plants have developed sophisticated means to protect themselves against assaults from various herbivorous insects. Most plants are equipped with constitutive and induced defense mechanisms, including physical barriers, such as trichomes and cell walls, and chemical defense, such as secondary metabolites. Despite the deceptive feeding style of aphids, the brief intracellular punctures along the stylet passage and secretions from salivation nevertheless can trigger responses in host plants (Tjallingii, 2006; Will and van Bel, 2006; De Vos and Jander, 2009; Bos et al., 2010). Plant defense responses can be classified as antibiosis, which curtails insect survival and reproduction, and/or antixenosis, which deters insect settling and herbivory. Transcriptomic studies suggest that phloem sap feeders modulate known defense signaling pathways, oxidative stress response, senescence, and plant metabolism and structure (Moran and Thompson, 2001; Zhu-Salzman et al., 2004; De Vos et al., 2005; Thompson and Goggin, 2006; Kuśnierczyk et al., 2008). Plant response to aphids involves genes regulated by the major plant hormones salicylic acid (SA), jasmonic acid (JA), ethylene (ET), and abscisic acid (ABA) and genes encoding transcriptional regulators. Exogenous JA application enhances plant resistance to aphids (Ellis et al., 2002; Zhu-Salzman et al., 2004; Cooper and Goggin, 2005). Furthermore, reduced population expansion was observed in green peach aphids (Myzus persicae) when raised on the Arabidopsis (Arabidopsis thaliana) constitutive expression of vegetative storage protein1 mutant constantly expressing JA responses, whereas the JA-insensitive mutant coronatine-insensitive1 supports more rapid growth of aphids than wild-type plants (Ellis et al., 2002; Mewis et al., 2005). Aphid infestation has been shown to trigger ET production (Mantelin et al., 2009). Elevated ET levels have been both positively and negatively correlated with plant resistance to aphids (Thompson and Goggin, 2006). In tomato (Solanum lycopersicum), ET biosynthesis renders plants more susceptible to potato aphids (Macrosiphum euphorbiae; Mantelin et al., 2009). However, the Arabidopsis ET-insensitive mutant ein2 promotes performance of green peach aphids (Kettles et al., 2013), indicating that ET plays a defensive role in Arabidopsis. Aphid feeding activates the SA signaling pathway in a number of plant species (Moran and Thompson, 2001; Moran et al., 2002; Zhu-Salzman et al., 2004). SA-mediated resistance to aphids has been observed on some occasions (Mohase and van der Westhuizen, 2002; Kaloshian, 2004), but SA does not seem to play a defensive role in Arabidopsis against aphids (Pegadaraju et al., 2005). ABA has also been implicated as a modulator of plant immunity via signaling cross talk (Fujita et al., 2006; Koornneef and Pieterse, 2008). Mutations in ABA biosynthesis and signaling have significant impacts on aphid population growth (Kerchev et al., 2013). Comparison of plant gene expression profiles reveals that aphid feeding and pathogen infection induce both similarly and differentially regulated gene sets (Barah et al., 2013).
The localized cell death elicited by microbial pathogens known as the hypersensitive response (HR) is considered a defense mechanism used by plants to prevent further spread of infection (Torres et al., 2006). A hallmark of hypersensitivity in many plants is local production of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2). HR-like symptoms, manifested as localized chlorotic and necrotic lesion spots, can also be detected in plants attacked by various insect herbivores. Strong HR-like symptoms, including rapid and prolonged accumulation of H2O2, were detected in lines of wheat (Triticum aestivum) resistant to Hessian fly (Mayetiola destructor) but not in the susceptible line (Liu et al., 2010). Enhanced resistance against phloem sap-sucking brown planthopper (Nilaparvata lugens) is accompanied by increased H2O2 levels as well as HR-like cell death in rice (Oryza sativa) expressing an antisense lipoxygenase (Zhou et al., 2009). Oxidative stress induced by insect herbivory is considered a component of soybean (Glycine max) resistance to invading corn earworm (Helicoverpa zea; Bi and Felton, 1995). Arabidopsis PHYTOALEXIN DEFICIENT4 (PAD4), a lipase-like protein essential for defense against microbial pathogens (Jirage et al., 1999), has been demonstrated to enhance plant resistance to green peach aphid by promoting premature leaf senescence and cell death (Pegadaraju et al., 2005, 2007). Functional dissection further revealed that the molecular mechanism of PAD4 resistance against aphids is distinct from that against pathogens (Louis et al., 2012).
Basal disease resistance, the first line of plant defense response, is elicited upon detection of pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs) by specific transmembrane pattern recognition receptors and is collectively termed PAMP-triggered immunity (Boller and Felix, 2009; Monaghan and Zipfel, 2012). Among the best characterized Arabidopsis PAMP/MAMP receptors are receptor-like kinases (RLKs) such as FLAGELLIN-SENSITIVE2 (FLS2) that recognizes bacterial flagellin and EF-TU RECEPTOR (EFR) that recognizes bacterial elongation factor EF-Tu (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). Upon binding to their cognate MAMPs, FLS2 or EFR associate with another RLK, BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE (BAK1; Chinchilla et al., 2007). BOTRYTIS-INDUCED KINASE1 (BIK1), a receptor-like cytoplasmic kinase (RLCK), is directly phosphorylated by BAK1 and associates with FLS2/BAK1 complex in modulating PAMP-mediated signaling (Lu et al., 2010; Zhang et al., 2010; Liu et al., 2013). Most recently, BAK1 is shown to be required for aphid elicitor-mediated ROS induction and plant innate immunity to aphids (Prince et al., 2014). Likewise, TOMATO PROTEIN KINASE1b (TPK1b), the tomato homolog of BIK1, plays an important role in plant resistance to a chewing insect herbivore (Abuqamar et al., 2008). The second layer of plant defense response is mediated by plant disease resistance (R) proteins, which recognize specific avirulence proteins from pathogens. R gene-mediated resistance to aphids has been reported, although the corresponding avirulence proteins from aphids remain unknown (Kaloshian, 2004). The tomato R gene Mi-1 confers resistance to some biotypes of potato aphids as well as to whiteflies (Bemisia tabaci) and root-knot nematodes (Meloidogyne incognita; Rossi et al., 1998; Vos et al., 1998; Nombela et al., 2003).
In this study, we examined the roles of several RL(C)Ks, including FLS2, EFR, BAK1, and BIK1, in Arabidopsis response to aphid infestation. We challenged these loss-of-function mutants with green peach aphids, a phloem sap-feeding generalist, to evaluate aphid performance and plant response. bik1 plants displayed heightened antibiosis and antixenosis toward aphids, which was correlated with pronounced aphid-induced HR-like cell death. Further exploration of potential interactions between BIK1 and known defense pathways revealed that BIK1 modulated plant response to aphid infestation through its control of PAD4 expression.
RESULTS
bik1 Exhibited Increased Resistance to Green Peach Aphids
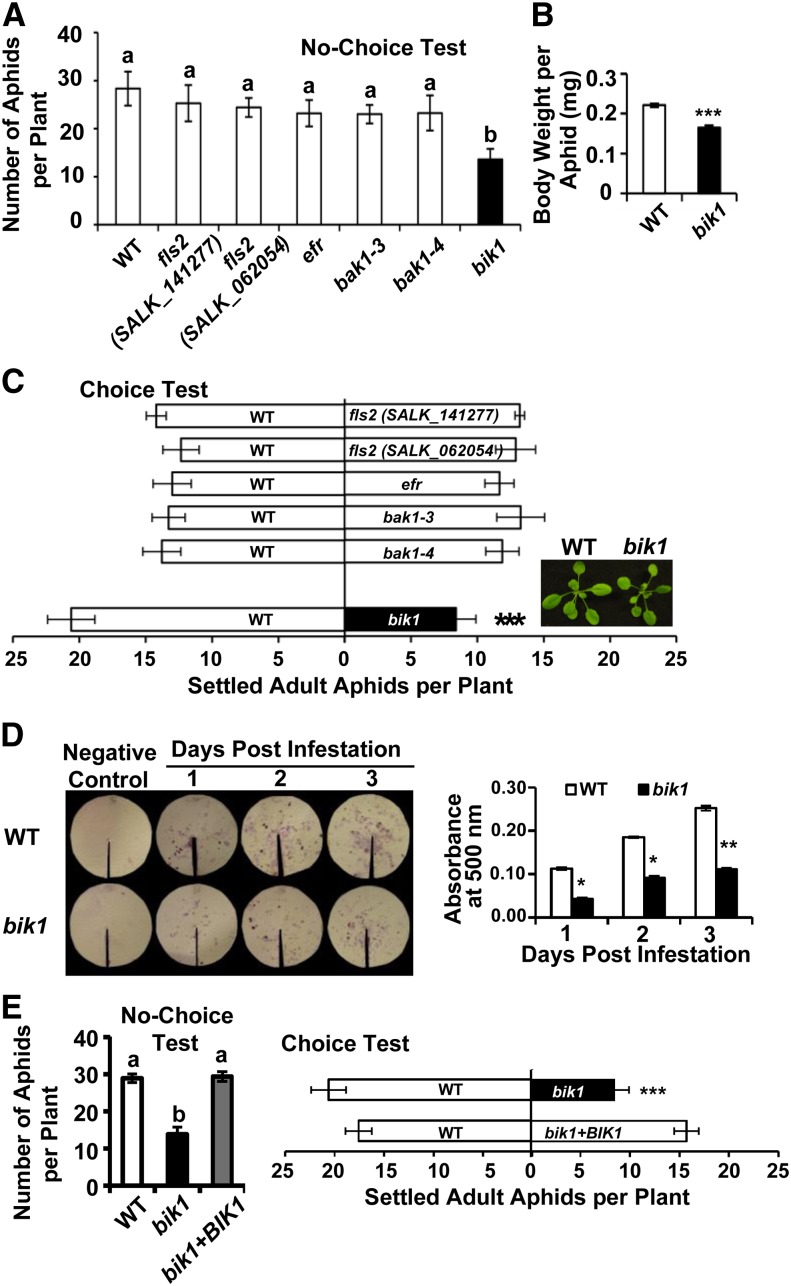
Plant defense response upon aphid infestation is often reflected by reduced offspring production (antibiosis) in a no-choice test with reduced feeding and body weight or by nonpreference (antixenosis) in a choice test. To determine whether the several known RL(C)Ks, which play important roles in PAMP-triggered immunity, extend their function to aphid-associated defense response, we evaluated aphid performance on the loss-of-function mutants (Fig. 1). Aphids infesting fls2, efr, and bak1 mutants had fecundities comparable to that on the wild-type plants (ecotype Columbia-0; Fig. 1A). Likewise, no particular preference was detected among them (Fig. 1C), suggesting that these RLKs may not play a major role in plant defense against aphids. Interestingly, on bik1, the amount of aphid progeny was, on average, about one-half that on wild-type plants (Fig. 1A). In agreement with this no-choice test result, aphids on bik1 excreted less honeydew (Fig. 1D), indicative of less food intake, and had less body weight (Fig. 1B) than those reared on the wild type. In the choice tests, approximately twice as many aphids preferred wild-type versus bik1 plants (Fig. 1C). Thus, BIK1 was a negative regulator of plant resistance to aphids. In addition, we confirmed that the heightened resistance in bik1 is due to loss of BIK1 function via complementation experiments. Transgenic plants expressing BIK1 complementary DNA (cDNA) in bik1 mutant recovered the susceptibility to aphids in both choice and no-choice tests (Fig. 1E), verifying that the observed aphid resistance in bik1 was due to loss of BIK1 function.
Figure 1.
Loss of BIK1 function confers resistance to green peach aphids. No-choice tests (A) and aphid body weight (B) of indicated genotypes. For no-choice tests, six second-instar nymphs were inoculated on each plant (4-5 weeks old). Total aphid numbers were recorded 7 d later. At least 10 replications were performed for each genotype. To obtain average body weight of adult aphids, neonates were reared on the wild type (WT) or bik1 for 10 d. Adults were then collected and were weighed as six groups of 10 aphids each. C, Choice tests. Three-week-old plants were used. At this developmental stage, no apparent size differences were observed between genotypes including the wild-type versus bik1 pair. Settled aphids were counted 6 h after releasing 35 adults in between two plants of the tested genotypes. Each test was comprised of 10 replicates. Inset image of the shoot phenotypes of the 3-week-old, uninfested wild type and bik1. D, Aphids on bik1 excreted less honeydew than those reared on the wild type. Quantity of honeydew secretion was correlated with the area and intensity of ninhydrin stains (left) and with optical density at 500 nm values (right). E, Expression of BIK1 cDNA confers wild-type levels of aphid susceptibility to bik1. One-way ANOVA was applied to no-choice tests, and the χ2 test was used to analyze data derived from choice tests. Body weight and honeydew secretion data were analyzed by independent samples’ Student’s t tests. Bars represent means ± se. Statistical significance for treatment effects is marked *P < 0.05, **P < 0.01, or ***P < 0.001. Means with different letters were significantly different (P < 0.05).
Notably, bik1 mutant showed comparable size and biomass during the first 3 weeks of growth (Fig. 1C; Supplemental Table S1), when choice tests were performed. Later, bik1 mutants exhibited growth defect and were smaller than the wild type (Supplemental Fig. S1; Supplemental Table S1). However, the antibiotic activity was unlikely due to their small stature, as inoculating six second-instar nymphs and rearing them for 7 d on 4- to 5-week-old plants would by no means result in a population limited by space or nutrients.
Aphids Induced HR-Like Lesions in bik1
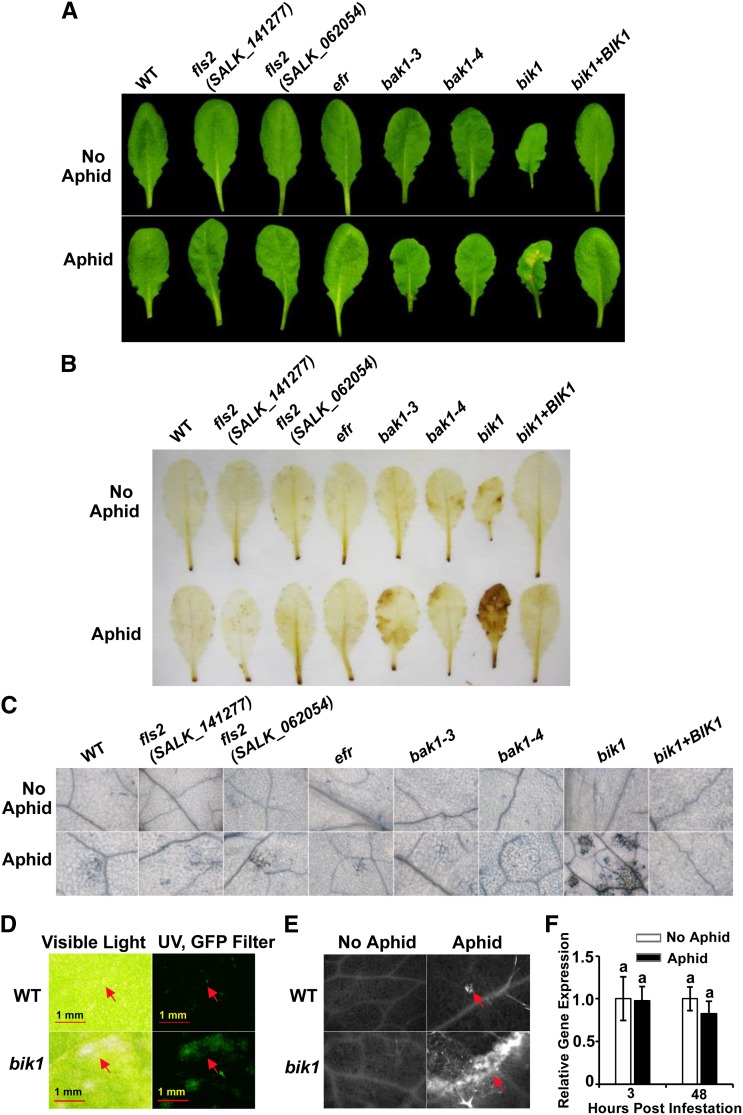
Despite an enhanced resistance to aphid infection, bik1 began to show apparent lesion spots approximately 5 d after aphid infestation, while no visible lesions were observed in fls2, efr, and bak1 mutants or in the wild type (Fig. 2A). With continued aphid infestation, all infested plants, regardless of the genotype, eventually displayed stunted growth, yellowing, and necrosis, with lesions spreading to the entire leaf and the whole plants. Notably, bik1 is not a lesion mimic mutant as no spontaneous lesions were observed without aphid infestation. Because bik1 plants are dwarfs, the number of aphids applied was adjusted by a ratio proportional to the rosette area. For plant symptom assessment, this ratio was applied for all genotypes exhibiting size differences relative to the wild type to exclude potential misjudgment due to size discrepancies.
Figure 2.
Aphid infestation induces a heightened HR in bik1. Representative leaf images of lesion formation (A), DAB staining (B; H2O2 indicator), and trypan blue staining (C; cell death indicator) prior to (top) or 6 d after (bottom) aphid infestation of genotypes indicated. D, Autofluorescence of aphid-induced lesion spots under UV excitation with GFP filter set (right). The same fields of view are shown under visible light (left). E, Callose deposition at lesion sites. Left, control leaves; right, callose deposition after aphid treatment. Arrows point to lesion sites. F, Relative expression of BIK1 in wild-type (WT) plants in the presence and absence of aphid infestation. Three-week-old plants were infested with aphids as described in “Materials and Methods.” Data were analyzed by independent samples’ Student’s t test. Means with different letters were significantly different (P < 0.05).
We further examined whether the aphid-induced lesion formation in the bik1 mutant resembles the features with an HR process that is often correlated with plant resistance against microbial pathogens (Lamb and Dixon, 1997; Heath, 2000). Using 3,3′-diaminobenzidine (DAB) staining, we observed that leaves of aphid-infested bik1 plants had much higher H2O2 accumulation than any other genotypes examined (Fig. 2B). Likewise, more severe cell death was shown in aphid-infested bik1 leaves compared with the wild type and the other mutants by the trypan blue staining assay (Fig. 2C). By contrast, fls2, efr, and bak1 mutants showed phenotypes similar to wild-type plants in either H2O2 or cell death assays. Furthermore, we detected accumulation of autofluorescent phenolic compounds and deposition of callose at necrotic spots in aphid-infested bik1 plants (Fig. 2, D and E), which are also HR lesion-associated histological markers (Hunt et al., 1997; Luna et al., 2011; Williams et al., 2011). Wild-type levels of H2O2 and lesions upon aphid infection were restored in the bik1 BIK1 complementation line (Fig. 2). Taken together, the data indicate that aphid-induced lesions in bik1 were an HR-like response.
Although BIK1 is highly induced by pathogens (Veronese et al., 2006), we did not detect a significant change in BIK1 expression upon aphid infestation (Fig. 2F). This is further supported by published microarray data (Couldridge et al., 2007; Kusnierczyk et al., 2007; Kuśnierczyk et al., 2008).
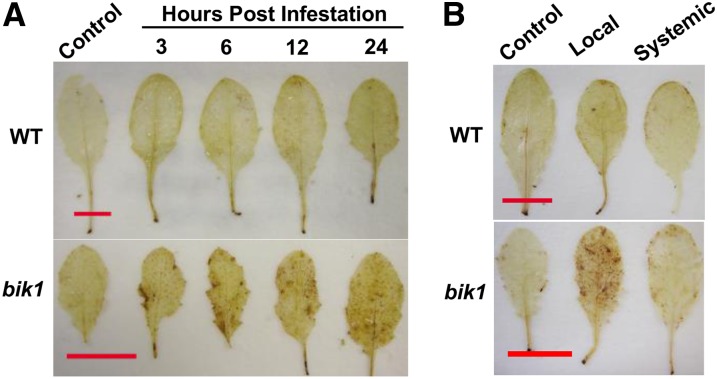
Because cellular H2O2 accumulation precedes cell death (Hoeberichts and Woltering, 2003), earlier time points were chosen for DAB staining. Staining became apparent within 3 h upon aphid infestation in bik1 leaves but was absent from the infested wild-type leaves over the 24-h course of the experiment (Fig. 3A). When aphids were caged on specific leaves, H2O2 could only be detected in infested local leaves, not in uninfested systemic leaves (Fig. 3B), supporting our conclusion that the lesion formation in bik1 is an HR rather than a constitutive plant damage phenotype. Correlation between plant symptoms and aphid performance suggests that elevated H2O2 accumulation and cell death in bik1 could be the defense mechanism compromising aphid fitness. BIK1 thus functions to counteract aphid-induced ROS production and cell death, distinct from its role in PAMP pathways.
Figure 3.
bik1 exhibits earlier and stronger ROS accumulation in locally infested leaves compared with the wild type (WT). A, DAB staining (H2O2 indicator) of aphid-infested leaves collected at 3, 6, 12, and 24 h post infestation. Four-week-old Arabidopsis plants were infested with aphids using the caged-leaf method as described in “Materials and Methods.” Caged (24 h) but uninfested leaves served as a control. B, DAB staining of local and infested, as well as systemic and uninfested, leaves of the same plant at 24 h post infestation. All leaves were caged. Controls were caged leaves from uninfested plants. Experiments were repeated three times. Bars = 1.0 cm.
Aphids Altered Phytohormone Contents and Gene Expression in bik1
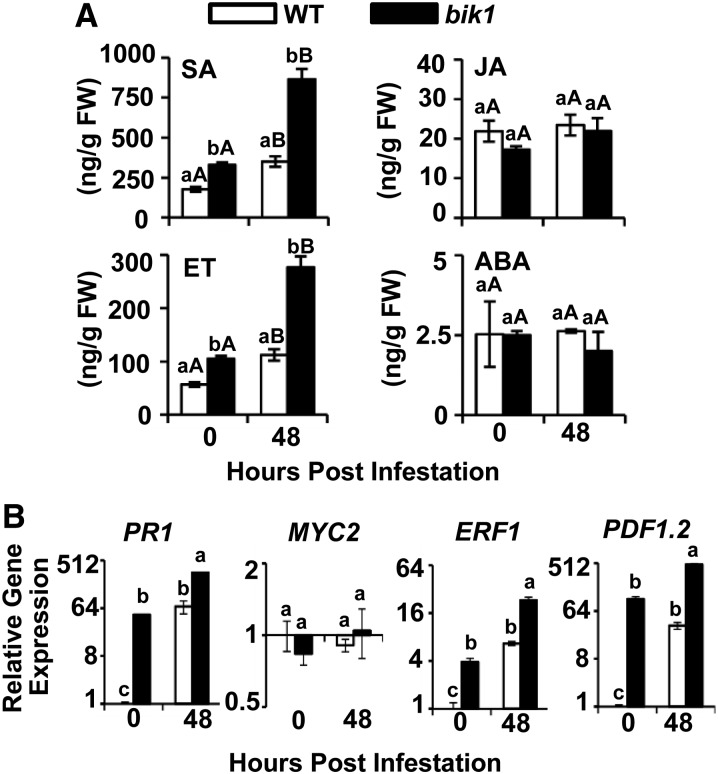
Aphid-induced plant defense and cell death pathways are often regulated by certain plant hormones (De Vos et al., 2005). To determine whether the resistance to aphids conferred by loss of BIK1 function involved defense-related plant hormones, we measured SA, JA, ET, and ABA levels in the presence and absence of aphid feeding in both wild-type and bik1 plants (Fig. 4A). Elevated basal SA, consistent with Veronese et al. (2006), and ET levels were detected in bik1, while JA and ABA contents were comparable in both genotypes. SA and ET levels increased in both the wild type and bik1 upon aphid infestation, and the levels of both hormones were higher in bik1 than in the wild type (Fig. 4A). No significant changes in JA and ABA were observed after aphid feeding. Basal expression levels of the SA-signaling marker gene PATHOGENESIS-RELATED PROTEIN1 (PR1) and the ET/JA marker genes ETHYLENE RESPONSE FACTOR1 (ERF1) and PLANT DEFENSIN1.2 (PDF1.2) were greater in bik1 compared with the wild type (Fig. 4B). Aphid infestation up-regulated expression of these genes in both wild-type and mutant plants. In comparison, basal expression of the JA-regulated transcription factor MYC2 was similar in both genotypes and was not altered by aphid infestation in either genotype (Fig. 4B). These data imply that BIK1 may function as a negative regulator of SA and ET accumulation both in the presence and absence of aphid infestation, thereby suppressing expression of their responsive genes.
Figure 4.
bik1 shows higher basal and induced levels of SA and ET and elevated expression of their marker genes during aphid infestation than the wild type (WT). A, SA, JA, ABA, and ET levels in the wild type and bik1 before and after aphid infestation. Three-week-old plants were infested with aphids for 48 h. Four replicates were used for each genotype. Hormone measurements were performed as described in “Materials and Methods.” Data were analyzed by the independent samples’ Student’s t test (P < 0.05). Different lowercase letters indicate significant differences between genotypes within the same treatment. Different uppercase letters indicate significant differences between treatments within the same genotype. B, Relative expression of SA, JA, and ET marker genes PR1, MYC2, ERF1, and PDF1.2 in response to aphid feeding at 0- and 48-h time points. Data were analyzed by one-way ANOVA. Tukey’s multiple range test analysis was used for pairwise comparisons of the difference between treatments for mean separation (P < 0.05). FW, Fresh weight.
Resistance to Aphids Conferred by Loss of BIK1 Function Was SA Independent
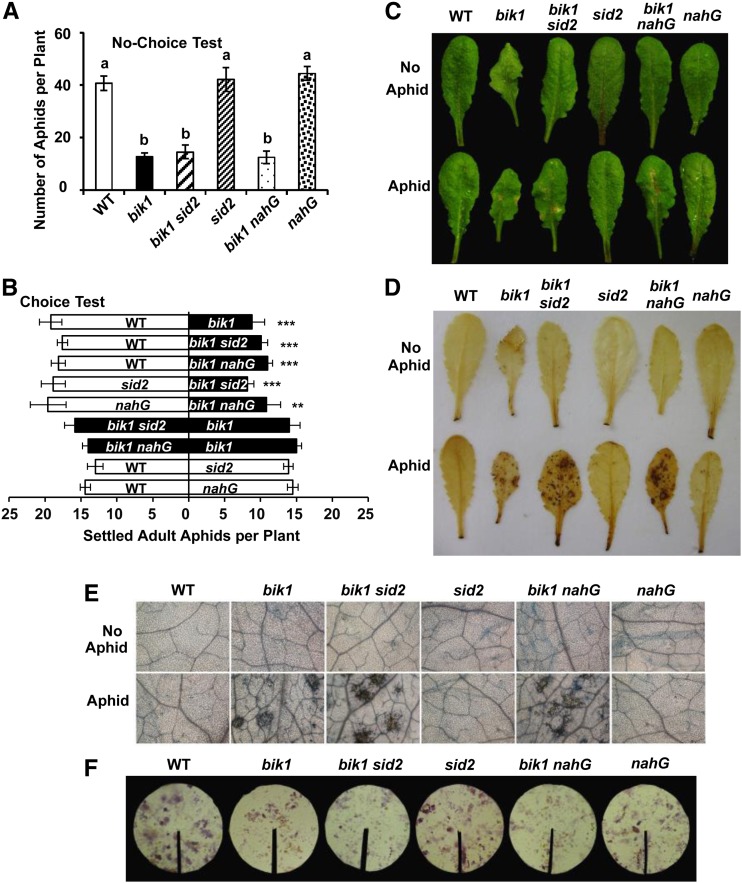
Loss of SALICYLIC ACID INDUCTION DEFICIENT2 (SID2) function blocks SA biosynthesis (Wildermuth et al., 2001), and nahG plants express the bacterial salicylate hydroxylase that degrades SA to catechol (Delaney et al., 1994). To assess the role that SA may play in bik1 resistance to aphids, bik1 sid2 and bik1 nahG plants were used for choice and no-choice tests (Fig. 5). In no-choice tests, the aphid numbers on bik1 sid2 or bik1 nahG plants paralleled those on bik1, and numbers on SA-deficient sid2 or nahG did not significantly differ from the wild type (Fig. 5A). Similar results were obtained in choice tests (Fig. 5B), as well as from honeydew excretion assays (Fig. 5F). Apparently, reducing the SA level did not weaken aphid resistance in bik1, nor did it influence aphid response in the wild type. Therefore, elevated SA accumulation was not required for bik1 resistance to the aphid, in contrast to its requirement for bik1’s resistance to a virulent strain of Pseudomonas syringae (Veronese et al., 2006).
Figure 5.
SA is not required for resistance to aphids and is not responsible for heightened HR in bik1. No-choice (A) and choice tests (B) on genotypes indicated. Representative leaf images of 4- to 5-week-old plants (C), DAB staining (D; H2O2 indicator), and trypan blue staining (E; cell death indicator) before (top) or after aphid infestation (bottom). F, Ninhydrin staining of honeydew after 48-h aphid feeding. All experiments were performed as described in in “Materials and Methods.” Bars represent means ± se. Statistical significance for treatment effects is marked *P < 0.05, **P < 0.01, or ***P < 0.001. Means with different letters were significantly different (P < 0.05).
To examine how SA impacted the aphid-triggered HR-like lesion formation, H2O2 production, and cell death in bik1, DAB and trypan blue staining were conducted on the SA-deficient plants. No correlations were observed between the SA status and lesion formation, H2O2 production, or cell death phenotypes (Fig. 5, C–E), a result supporting previous studies showing that SA is not essential for aphid defense in Arabidopsis (Pegadaraju et al., 2005). By contrast, a correlation was observed between resistance to aphids and H2O2 production as well as cell death occurrence. Notably, in terms of the plant size and morphology, bik1 sid2 and bik1 nahG were closer to the wild type than to bik1, yet they exhibited levels of H2O2 production, cell death, and aphid resistance comparable to bik1. Therefore, dwarfism was unlikely the cause of enhanced resistance to aphids in bik1. Heightened endogenous SA has been reported previously to confer bik1 with resistance to the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Veronese et al., 2006). Results from our study revealed differential function of SA in BIK1-mediated plant responses to bacterial pathogens versus phloem sap-feeding aphids.
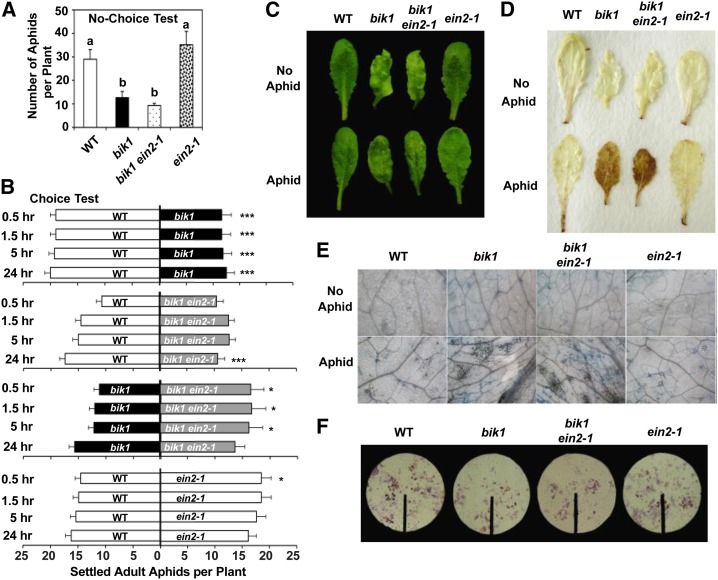
Elevated ET Signaling in bik1 Increased Aphid Repellence during Early Stages of Infestation
Like SA, ET is known to play a key role in cell death and plant response to pathogens and insects (Dong et al., 2004; Cohn and Martin, 2005; Bouchez et al., 2007). To examine whether elevated ET has a role in aphid resistance in bik1, we pretreated plants with 1-methylcyclopropene (1-MCP), an inhibitor of ET action that binds to the ET receptor. In choice tests, there was no significant difference in the number of aphids on 1-MCP-treated bik1 and wild-type plants 6 h after aphid inoculation (Fig. 6), suggesting that 1-MCP may have compromised resistance in bik1. As time went on, however, 1-MCP-treated bik1 gradually regained their aphid repellence, presumably due to loss of 1-MCP function.
Figure 6.
1-MCP temporarily attenuates bik1 deterrence of aphids. Choice tests between 3-week-old wild-type (WT) and bik1 plants in the presence and absence of 1-MCP. Settled aphids were recorded 6 and 12 h after aphid infestation. Application of 1-MCP began 5 d prior to choice tests and was reapplied every 12 h to prevent the loss of its effectiveness. Control plants were subjected to the same manipulation without 1-MCP. Statistical significance for treatment effects is marked *P < 0.05, **P < 0.01, or ***P < 0.001.
Because the 1-MCP effect was temporary, this pharmacological approach was limited to choice tests. To further investigate whether increased ET contributes to bik1 resistance to aphids, a genetic approach was used to impair ET signaling in bik1, and longer term no-choice tests were performed. The bik1 mutant was crossed with two ET-insensitive mutants, ein2-1 and ein3-1 (Guo and Ecker, 2004; Broekaert et al., 2006). EIN2 (a transducer of ET signaling) and EIN3 (a primary ET-responsive transcription factor) are essential components of the ET signaling pathway. In no-choice tests, the bik1 ein2-1 double mutant showed resistance comparable to bik1 (Fig. 7A), suggesting that ET was not important in suppressing aphid reproduction in bik1, in agreement with honeydew secretion data (Fig. 7F). However, in choice tests, blocking ET signaling in bik1 (i.e. bik1 ein2-1) increased plant attractiveness to aphids (Fig. 7B), implying that elevated ET in bik1 contributed to its aphid repellence. Interestingly, when compared with bik1, bik1 ein2-1 was preferred more by aphids early on. As experiments continued, the difference in the number of aphids on each genotype became nonsignificant. Thus, the overall effect of ET on bik1-mediated aphid resistance appeared to be only temporary and rather subtle.
Figure 7.
Elevated ET increases bik1 repellence against aphids but shows no effect on aphid reproduction or on aphid-induced plant HR. No-choice (A) and choice tests (B) on genotypes and at time points as indicated. Representative leaf images of 4- to 5-week-old plants (C), DAB staining (D; H2O2 indicator), and trypan blue staining (E; cell death indicator) before (top) or after aphid infestation (bottom). F, Ninhydrin staining of honeydew after 48-h aphid feeding. All experiments were performed as described in in “Materials and Methods.” Bars represent means ± se. Statistical significance for treatment effects is marked *P < 0.05, **P < 0.01, or ***P < 0.001. Means with different letters were significantly different (P < 0.05). WT, Wild type.
The bik1 ein2-1 double mutant maintained the small stature of the bik1 single mutant (Supplemental Fig. S1C). Feeding response in the bik1 ein2-1 double mutant, i.e. lesion formation, H2O2 production, and cell death upon aphid infestation, resembled that of bik1 (Fig. 7, C–E). Similar results were obtained with bik1 ein3-1 plants (Supplemental Fig. S2). Taken together, ET signaling in bik1 was mainly involved in aphid deterrence initially in choice tests but appeared to play little role in cell death-mediated defense in bik1.
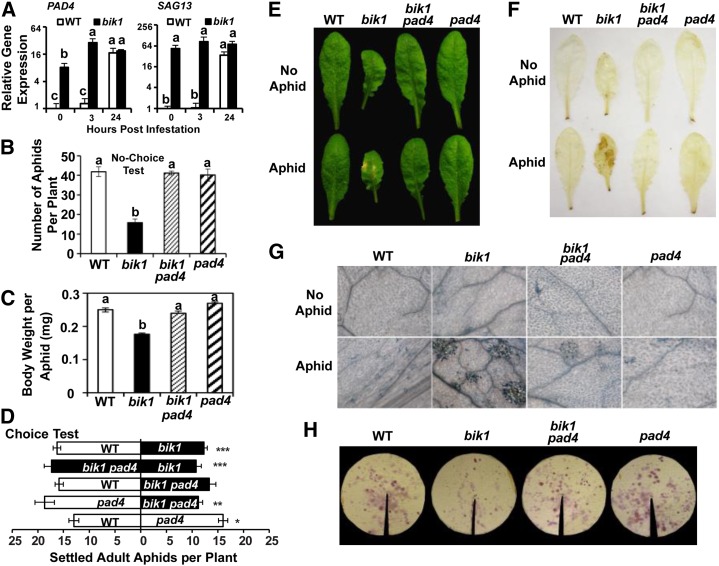
Aphid Resistance and HR-Like Cell Death in bik1 Is PAD4 Dependent
PAD4 is a lipase-like protein that, upon aphid feeding, promotes premature leaf senescence to suppress insect reproduction and colonization (Pegadaraju et al., 2005, 2007). Aphids induced PAD4 expression in both bik1 and the wild type (Fig. 8A). Compared with the wild-type plants, bik1 had much higher PAD4 basal expression. Consistently, a senescence marker gene, SENESCENCE ASSOCIATED GENE13 (SAG13), regulated by PAD4 during aphid infestation (Weaver et al., 1998; Pegadaraju et al., 2005) shared a similar expression pattern with PAD4 (Fig. 8A). These results indicated that BIK1 suppresses PAD4 and senescence gene expression.
Figure 8.
Resistance to aphids and aphid-induced HR in bik1 were PAD4 dependent. A, Relative expression of PAD4 and SAG13 in wild-type (WT) and bik1 plants in the presence and absence of aphid infestation. Three-week-old plants were infested with aphids as described in “Materials and Methods.” No-choice test (B), average aphid body weight (C), and choice tests (D) were performed on genotypes indicated. Representative leaf images of 4- to 5-week-old plants (E), DAB staining (F; H2O2 indicator), and trypan blue staining (G; cell death indicator) before (top) or after aphid infestation (bottom). H, Ninhydrin staining of honeydew after 48-h aphid feeding. Bars represent means ± se. Statistical significance for treatment effects is marked *P < 0.05, **P < 0.01, or ***P < 0.001. Means with different letters were significantly different (P < 0.05).
To learn whether potential interactions exist between BIK1 and PAD4 in cell death-mediated aphid resistance, we examined aphid performance on the bik1 pad4 double mutant. In no-choice tests, aphid numbers and body weight were both significantly higher on bik1 pad4 than on bik1 plants and were comparable to the wild type (Fig. 8, B and C). Honeydew excretion showed the same trend (Fig. 8H). Likewise, in choice tests, aphids showed a strong preference for bik1 pad4 when paired with bik1 (Fig. 8D). Apparently, the antibiosis and antixenosis observed in bik1 diminished when the pad4 mutation was introduced. The pad4 mutant did not support more aphid growth than the wild-type plant, although it attracted more aphids in the choice test. Therefore, the suppression of aphid performance in bik1 was dependent on elevated basal PAD4 expression.
Consistent with insect performance, bik1 pad4 plants displayed phenotypes similar to those of the wild type in terms of lesion formation, H2O2 accumulation, and cell death (Fig. 8, E–G). Inactivation of PAD4 in bik1 blocked the cell death, indicating that PAD4 was required for hypersensitivity and aphid resistance resulting from loss of BIK1 function.
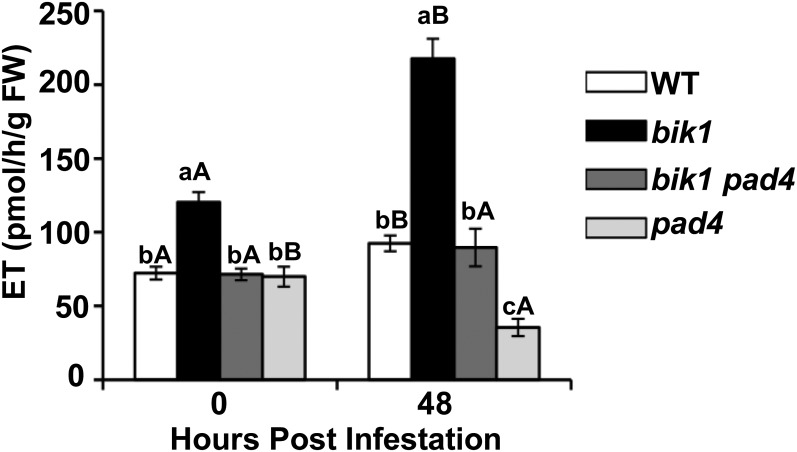
Interestingly, ET emission decreased in bik1 pad4 compared with bik1, both in the presence and absence of aphids (Fig. 9). This observation suggested that PAD4 may positively regulate ET accumulation.
Figure 9.
PAD4 potentially promotes ET production. ET production by wild-type (WT), bik1, bik1 pad4, and pad4 plants measured before or after 48-h aphid infestation as described in “Materials and Methods.” Bars represent means ± se from at least six individual plants. Different lowercase letters indicate significant differences between genotypes by one-way ANOVA and Tukey’s multiple range test (P < 0.05). Different uppercase letters indicate significant differences between treatments by an independent sample’s Student’s t test (P < 0.05). FW, Fresh weight.
Loss of BIK1 Function Did Not Confer Resistance to Chewing Insects
Unlike aphids, chewing insects massively damage the host cells during infestation. To assess the role of BIK1 in Arabidopsis defense against chewing insects, we performed bioassays using fall armyworm (Spodoptera frugiperda) neonate larvae placed on 4-week-old wild-type and bik1 plants (Supplemental Fig. S3). No significant weight and size differences were detected between larvae reared on the two genotypes (Supplemental Fig. S3, A and B). In addition, fall armyworm elicited comparable H2O2 production on wild-type and bik1 plants (Supplemental Fig. S3C). The data suggested that BIK1 has distinct roles in Arabidopsis response to two groups of insects that differ in their feeding behaviors. This observation is also different from a previous study showing that TPK1b, the tomato homolog of BIK1, enhances host plant resistance against tobacco hornworm (Manduca sexta; Abuqamar et al., 2008).
DISCUSSION
Plants in the natural environment are constantly challenged by insect herbivory and pathogen infection. As a result, they have developed a plethora of sophisticated means to cope with diverse biotic stresses. Given the common features between plant responses to phloem sap feeders and pathogens, we studied several PAMP/MAMP signal receptors for involvement in plant response to aphids using their loss-of-function lines. While FLS2, BAK1, and EFR did not seem to be associated with response to aphid infestation, BIK1 acted as a negative regulator of the defense response against aphids. This is in contrast to its positive role in resistance to fungal necrotrophs (Veronese et al., 2006) and flagellin-mediated immune responses (Lu et al., 2010). Thus, the PAMP recognition components did not seem to have a parallel role in perceiving or transmitting signals from invading aphids.
HR-Like Cell Death Could Be Pivotal for Aphid Resistance in bik1 Plants
The bik1 mutant exhibited heightened resistance to aphids as well as enhanced local H2O2 production and necrotic cell death upon aphid infestation (Figs. 1 and 2). As in plant-microbe interactions, cell death could be either considered a plant defense factor or viewed as an effect of aphid manipulation of host nutritional quality (Goggin, 2007). Although bik1 plants displayed severe lesion formation, this aphid-induced symptom correlated with impeded aphid colonization, growth, and reproduction. Thus, rather than a damage symptom, H2O2 accumulation and cell death represent a major defense mechanism in bik1 to enhance resistance to aphids. These features were limited to aphid-infested bik1 leaves (Fig. 3) and unrelated to dwarfism (Fig. 5; Supplemental Fig. S1B). Furthermore, SA, JA, ET, and ABA did not have major involvement.
Oxidative stress induced by insect feeding is believed to be an important component of plant resistance to invading insects. Detoxification of ROS may decrease antioxidant levels and increase toxic oxidation products in plants as shown in soybean following herbivory by corn earworm (Bi and Felton, 1995). In addition, increased H2O2 and other oxidative products in plants also directly damage the insect midgut and affect growth. Consumption of artificial diets containing even relatively low concentrations of H2O2 caused high mortality of insects (Liu et al., 2010). At high concentrations, ROS can react with almost all cellular macromolecules, including proteins, lipids, and DNAs (Van Breusegem and Dat, 2006). Accordingly, the elevated ROS generated in bik1 may result in decreased quantity and quality of nutrients and antioxidants, causing damage to aphid tissues and ultimately reducing their fitness. Furthermore, it is plausible that H2O2-potentiated HR in infected and adjacent cells could limit photoassimilate flow to the feeding sites, although it is questionable how effective such an approach can be, given that aphids can move away from their feeding sites before a sufficient defense response is mounted. Nevertheless, poor aphid performance on bik1 plants relative to the wild type supported the hypothesis that rapid and potent HR-like cell death placed limitations on aphid infestation.
ROS Production, Cell Death, and Defense against Aphids in bik1 Required Functional PAD4
While loss of BIK1 function promoted aphid-induced lesions, no lesions were formed without aphid infestation (Figs. 2 and 3). Furthermore, the spread of the aphid-induced lesions in bik1 required continued aphid feeding (data not shown). These data suggest that BIK1 does not directly repress but rather indirectly modulates a cell death pathway through an aphid-responsive component. We postulated that BIK1 may exert its negative regulation via PAD4, a lipase-like protein, for the following reasons. First, PAD4 regulates the activation of premature leaf senescence, i.e. a cell death-mediated resistance mechanism against aphids (Pegadaraju et al., 2005), consistent with the tight correlation between HR lesions and resistance we observed in bik1. Second, although PAD4 is involved in SA signaling, SA is not important for the defense against aphids conferred by PAD4, agreeing with our conclusion that bik1 resistance is SA independent. Third, expression of PAD4 is induced in response to aphid feeding (Pegadaraju et al., 2005), potentially furnishing an aphid-triggered control point downstream of BIK1. Experimental results demonstrated that PAD4 was required for bik1 resistance to aphids (Fig. 8). It should be noted that although more aphids preferred pad4 plants over the wild type in the choice tests (Fig. 8D), no obvious increase in insect reproduction was observed on pad4 in the no-choice tests (Fig. 8B). This is in contrast to the observations of Pegadaraju et al. (2005), who reported significantly higher population growth of green peach aphids on pad4 than on the wild type. Differences in plant growth conditions or in insect strain, age, and quantity used by the two laboratories could account for the different results. We witnessed relatively mild lesion formation in the wild type, which may explain the nonsignificant difference in aphid propagation on the wild type versus pad4. Furthermore, different conditions under which the ROS experiments were performed may explain the discrepancy in time needed for detection of ROS between different laboratories; in the current in vivo study, oral secretion was delivered via the aphid’s fine mouthpart and was only in contact with a very limited number of plant cells, probably making ROS hard to detect in the early stage. Prince et al. (2014), on the other hand, used leaf disks submerged in 5 mg mL–1 aphid-derived extract. It is possible that exposing the entire leaf tissue to a relatively high concentration of aphid elicitors permitted early ROS response. Alternatively, the early response could be triggered by factors in the aphid-derived extract that normally would not come into direct contact with the host cells.
We propose that BIK1 modulates cell death and resistance to aphids through its control of PAD4 (Fig. 10). Removal of PAD4 function was sufficient to eliminate the strong HR-like cell death of bik1 and restore its susceptibility to aphids. Ectopic expression of PAD4 triggered more rapid cell death in aphid-infested leaves and stronger resistance to aphids than in the wild type (Pegadaraju et al., 2007). Inactivation of BIK1 repression in a sense resembles overexpression of PAD4. On the other hand, although aphid feeding induced PAD4 expression and localized cell death in wild-type plants, DAB staining revealed only marginal differences in H2O2 production between the wild type and the pad4 mutant (Fig. 8). These data suggest that in wild-type plants, BIK1 suppression most likely is the dominant control factor for cell death, prevailing over the stimulus from aphid feeding. It should be pointed out that high basal PAD4 expression alone, i.e. in the bik1 mutant without aphid feeding, was insufficient to result in cell death. Contrasting results of DAB staining of the bik1 mutant with and without aphid treatment appeared to support this assumption. It is possible that PAD4-mediated cell death is initiated and propagated by aphid oral secretion-triggered signaling cascades, which are predominantly repressed by BIK1.
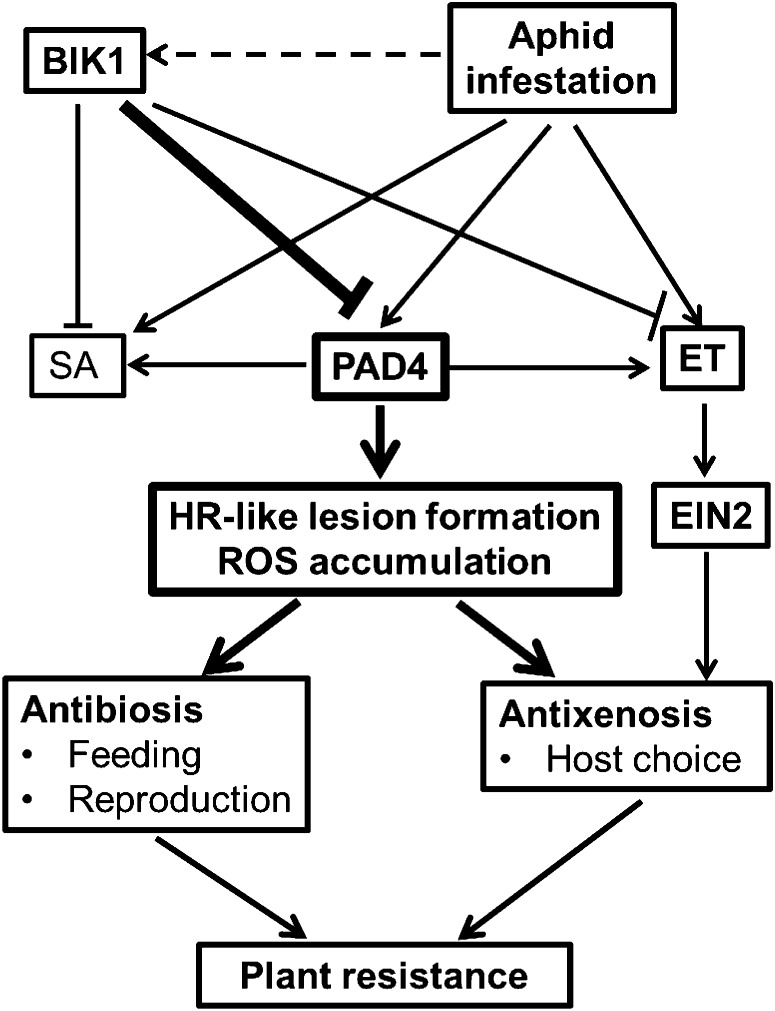
Figure 10.
Model depicting Arabidopsis resistance to aphids conferred by bik1 mutant. PAD4 is a positive regulator of aphid-induced plant antibiotic and antixenotic responses. PAD4-regulated defense, potentially resulting from ROS-mediated cell death, is suppressed by BIK1. Based on the intensity of DAB staining, the BIK1 suppression is presumably much stronger than the aphid induction, illustrated by thicker lines in the graph. BIK1 also suppresses SA and ET accumulation. SA has no direct influence on resistance to aphids. ET increased host repellence early on, possibly prior to significant ROS production.
It should be noted that bik1 is not the only mutant conferring PAD4-dependent aphid resistance. Loss of function of SUPPRESSOR OF SALICYLIC ACID INSENSITIVITY2 (SSI2), a desaturase, resulted in hyperresistance to aphids, and the resistance required PAD4 as well (Louis et al., 2012). As with bik1, ssi2 resistance diminished in the ssi2 pad4 double mutant. But unlike the bik1 mutant that expressed high basal PAD4 transcript, the ssi2 mutant did not show elevated PAD4 expression in the absence of aphid feeding. Thus, the role of PAD4 in aphid resistance could be regulated by distinct pathways; while bik1 may exert its resistance through releasing the suppression of PAD4 by BIK1, the interaction with SSI2 could be indirect.
Pleiotropic Effects of BIK1
It is rather counterintuitive, at first glance, that a gene like BIK1 that confers plant susceptibility to invaders exists. A logical explanation could be that it plays an indispensable role in other processes and/or is involved in multiple pathways in the plant where a balance has to be achieved through cross talk. Constitutive defense is often associated with fitness costs, e.g. altered leaf morphology, stunted growth, and decreased fertility (Heil and Baldwin, 2002). Evidently, BIK1 is necessary for normal plant growth (Veronese et al., 2006) and seed production (Supplemental Table S1). High levels of SA may be a major causal factor for the aberrant development and reduced growth of bik1 because SA depletion by sid2 and nahG largely restored the wild-type stature of bik1 plants (Fig. 5; Supplemental Fig. S1B). Furthermore, the defect in SA accumulation in pad4 could be responsible for the near wild-type plant form and leaf shape of the bik1 pad4 double mutant (Fig. 8; Supplemental Fig. S1D). Many lesion-mimic mutants display altered plant morphology due to production of elevated levels of SA and its constitutive interaction with other pathways (Lorrain et al., 2003). Therefore, it is very likely that BIK1 regulates normal plant growth in part by controlling SA levels. Conversely, bik1 ein2-1 and bik1 ein3-1 double mutants suffered the same growth suppression and aberrant development as the bik1 single mutant and did not show any phenotypic recovery (Fig. 7; Supplemental Fig. S1C). Therefore, despite the essential role of ET in plant development, it is unlikely that the elevated ET level contributed to the bik1 growth abnormality.
Notably, although BIK1 enhanced susceptibility to aphids, its presence did not block induction of effective aphid resistance genes but reduced their basal expression (Fig. 8). Perhaps, without BIK1, the penalty in general plant fitness imposed by maintaining a defense system in a no-pest environment outweighs an immediately available defense when plants are facing aphid attack. In addition to plant development, BIK1 confers resistance to necrotrophic pathogens (Veronese et al., 2006) and is involved in activation of PAMP-triggered signaling pathways (Lu et al., 2010). This study showcased the cross talk among signaling pathways involved in plant development and defense against insects versus pathogens.
In contrast to our results showing that BIK1 negatively regulated resistance to a phloem sap feeder and had no effect on a chewing insect, studies on the BIK1 homolog in tomato, TPK1b, indicate that TPK1b positively regulates plant resistance against herbivory of tobacco hornworm, also a chewing insect (Abuqamar et al., 2008). Because TPK1b rescues the phenotype of the Arabidopsis bik1 mutant, i.e. restoring its resistance to Botrytis, TPK1b and BIK1 are thought to perform similar functions in their respective species. The differential, even opposing, functions exhibited by BIK1 and TPK1 suggests that the involvement of BIK1 in plant defense against insects could be shaped by specific insects through their distinct feeding styles and unique interactions with their host plants formed over the long history of coevolution.
Our study has drawn an important link between ROS production/cell death and plant resistance to aphids. However, uncoupling cell death from insect resistance has also been reported in studies with Medicago truncatula (Klingler et al., 2009). In these studies, it is clearly demonstrated that HR lesions are not required for resistance to the pea aphid (Acyrthosiphon pisum). In plant-pathogen interactions where the HR is often considered a major form of resistance, it has been shown that the Arabidopsis defense, no death mutant exhibits enhanced resistance against pathogen infection in the virtual absence of HR cell death (Yu et al., 1998). Further investigation is needed to establish whether the hypersensitivity is the basis for aphid resistance in bik1 plants. It also remains to be elucidated whether HR lesions directly cause plant defense or if they are the consequence of defensive biochemical reactions activated by aphids.
MATERIALS AND METHODS
Plant Growth and Aphid Rearing
Arabidopsis (Arabidopsis thaliana) was grown in LP5 potting medium (Sun Gro Horticulture) in environmental chambers at 23°C (day)/21°C (night), 65% relative humidity (RH), and 12-h light/12-h dark photoperiod with a photosynthetic photon flux density of 85 µmol m–2 s–1. For plant damage evaluation, histochemical assays, and aphid no-choice tests, 4- to 5-week-old plants were used. For plant gene expression analyses and hormone measurements, as well as for aphid choice tests, 3- to 4-week-old plants were used.
Phloem sap-feeding green peach aphids (Myzus persicae) a tobacco (Nicotiana tabacum)-adapted red lineage (kind gift from Dr. Georg Jander, Boyce Thompson Institute for Plant Research, Cornell University) were cultured on cabbage (Brassica oleracea) and maintained in an environmental chamber at 21°C, 65% RH, and 12-h light/12-h dark photoperiod (63 µmol m–2 s–1). All insect treatments and bioassays were performed in this chamber.
Arabidopsis Lines
The previously reported Arabidopsis lines, wild-type ecotype Columbia-0, and mutants fls2 (SALK_141277), fls2 (SALK_062054), efr, bak1-3, bak1-4, bik1, sid2, nahG, bik1 sid2, bik1 nahG, ein2-1, ein3-1, pad4, bik1 pad4, and the bik1 complementation line bik1+BIK1 used in this study (Jirage et al., 1999; Veronese et al., 2006; Lu et al., 2010; Laluk et al., 2011; Lin et al., 2013) were kindly provided by Dr. Tesfaye Mengiste (Purdue University) or obtained from the Arabidopsis Biological Resource Center (Ohio State University). To generate bik1 ein2-1 and bik1 ein3-1 double mutants, we crossed bik1 with ein2-1 and ein3-1 using bik1 as the female parental line. The F2 seeds were germinated in the dark on Murashige and Skoog agar medium containing 50 µm 1-aminocyclopropane-1-carboxylic acid. The seedlings that lacked a triple response were selected and transferred to soil. The presence of ein2-1 and ein3-1 was confirmed by the derived cleaved amplified polymorphic sequence method as previous described, with modification (Nandi et al., 2003; Binder et al., 2007; Bouchez et al., 2007; Chen et al., 2009). For ein2-1 genotyping, a 195-bp fragment flanking the point mutation was amplified by PCR, followed by purification and AflII restriction digestion. AflII cut the mutant sequence into 160- and 35-bp fragments but left the wild-type sequence intact. For ein3-1, the 222-bp PCR product remained intact in the mutant sequence but was cut by HaeIII into 190- and 32-bp fragments in the wild-type sequence. DNA fragments were resolved on 2% (w/v) agarose gel. For bik1 genotyping, a procedure developed previously was followed (Lu et al., 2010). Primer sequences are provided in Supplemental Table S2.
Insect Bioassays
Aphid no-choice and choice tests were performed to assess the antibiotic and antixenotic resistance of different Arabidopsis genotypes. For the no-choice tests, six age-synchronized second-instar nymphs (within 24 h) were placed on 4-week-old plants. The total aphid population (adult and nymph) on each plant was counted 7 d after infestation. Each genotype had at least 10 replicates. For the choice tests, 35 adults were released at an equal distance between two plants of different genotypes. The number of adult aphids settled on each plant was recorded 6 and 24 h after releasing. At least 10 pairs of plants were used in each comparison. All experiments were repeated at least three times, and a representative data set was presented.
To obtain the average adult aphid body weight, adult aphids were transferred to wild-type or bik1 plants and removed 24 h later to produce age-synchronized progenies. Ten days later, the new generations of adults reared on Arabidopsis genotypes were collected and were weighed as six groups of 10 aphids each.
Eggs of fall armyworm (Spodoptera frugiperda), purchased from Benzon Research, were incubated in a growth chamber (27°C and 65% RH). Newly hatched larvae were transferred to 4-week-old wild-type or bik1 plants. Plants were replaced once a week to ensure sufficient food supply. Larvae reared on Arabidopsis genotypes were weighed after feeding for 16 or 22 d. At least 30 larvae were measured for each genotype.
Ninhydrin Staining and Quantification of Aphid Honeydew
Honeydew production served as an indicator of insect feeding activity. To determine honeydew secretion, Whatman filter papers, protected by a plastic membrane to avoid absorbance of water from soil, were placed under Arabidopsis plants of various genotypes infested by 30 adult aphids. These filter papers were collected 1, 2, and 3 d after aphid infestation, soaked in 0.1% (w/v) ninhydrin in acetone, and dried in a 65°C oven for 30 min. Honeydew stained by ninhydrin was shown as purple spots (Kim and Jander, 2007).
To quantify the honeydew stains, the filter papers were cut into pieces and stains were extracted into 1 mL of 90% (v/v) methanol for 1 h at 4°C with continuous agitation. After centrifugation at 6,000g for 1 min, the absorbance of the supernatant was measured at 500 nm (Nisbet et al., 1994). Methanol (90%) served as a blank.
Plant Damage and Histochemical Assays
Four- to five-week-old Arabidopsis plants were infested with adult aphids, taking into consideration the variation of the rosette size of each genotype. Accordingly, 48 aphids were placed on the wild type, fls2, efr, bak1-3, bak1-4, bik1+BIK1, sid2, nahG, ein2-1, ein3-1, and pad4 (sizes comparable to the wild type), 12 on bik1, bik1 ein2-1, and bik1 ein3-1 (one-quarter the size of the wild type), and 24 on bik1 sid2, bik1 nahG, and bik1 pad4 (one-half the size of the wild type). Plants were examined daily to identify symptoms of yellowing and lesion formation. Digital images were taken of representative leaves at 6 d post aphid infestation. Leaves obtained in the same manner were subjected to histochemical assay (see below). For every experiment, eight plants or more of each genotype were used. All experiments were repeated at least three times.
To visualize H2O2 accumulation, DAB staining was performed. Leaves at 6 d post infestation, as well as control leaves, were collected and vacuum infiltrated with DAB solution (1 mg mL–1 of DAB in pH 3.5 water) in a six-well titer plate. After an overnight incubation in the same solution in darkness, the leaves were destained in 95% (v/v) ethanol until they turned clear. Images were then captured with a digital camera.
To determine local and systemic ROS accumulation, aphids were placed in clear plastic cups (4-cm diameter, 4-cm height) with mesh cloth replacing the bottoms for ventilation. Twenty insects were used for the wild type and 10 for bik1. The cage was fitted around the leaf petiole between the cap and the cup and sealed with cotton to avoid wounding as well as aphid escape, restricting the aphids onto one 4-week-old Arabidopsis leaf for the desired time (Kim and Jander, 2007). Caged leaves without aphids served as controls. After treatments, the cages were removed, and leaves were excised for DAB staining.
Trypan blue staining was performed to visualize cell death. Trypan blue was dissolved in lactophenol solution (phenol:lactic acid:glycerol:water [1:1:1:1]) at a concentration of 0.125 mg mL–1. Leaves prepared as above were boiled in this staining solution for 1 min. After cooling, leaf samples were destained in 95% (v/v) ethanol and photographed with an Olympus SZX2-ILLK microscope.
The accumulation of autofluorescent compounds and deposition of callose are features of HR lesions (Hunt et al., 1997). Lesions on Arabidopsis leaves were examined 6 d after aphid infestation using the Olympus microscope under bright-field or UV excitation with a GFP filter. Images of lesions and autofluorescence emitted from the same lesion sites were recorded (Stewart et al., 2009).
Aniline blue staining (Clay et al., 2009) was performed to detect callose deposition. Arabidopsis leaves were fixed in buffer containing 10% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol at 37°C overnight. Slightly translucent leaves were then washed in 95% ethanol several times until clear, rinsed twice in water, and then stained for 4 h or longer in the dark with 0.01% (w/v) aniline blue in 150 mm K2HPO4 (pH 9.5). Callose deposits were visualized with an Olympus IX-81 microscope at 10× magnification under UV illumination with a broadband DAPI filter set.
JA, SA, and ABA Measurements
For SA, JA, and ABA measurements, 3-week-old plants were infested with aphids (30 per plant). Two days later, treated or control plants were ground to a fine powder in liquid nitrogen. For each sample replicate, ground tissue (60 mg) and a mixture of stable isotope-labeled hormones including 10 ng of 2H4-SA, 3.8 ng of 13C2-JA, and 1 ng of 2H6-ABA were added to a 5-mL glass tube with 500 µL of methanol at 55°C and extracted by vortexing three times during a 10-min incubation. The tissue was reextracted with 500 µL of methanol and then once with 500 µL of 80% ethanol warmed to 55°C, centrifuging and pooling the cleared supernatants after each extraction. The pooled extracts were dried, and the residue was resuspended in 800 µL of chloroform and partitioned against 1 mL of water adjusted to pH 9.0 with NH4OH. The aqueous fraction was recovered, adjusted to pH 5.0 with acetic acid, and partitioned against 1 mL of ethyl acetate. The organic fraction was transferred to a Reactivial, dried, and then methylated with ethereal diazomethane. Samples were then analyzed on an Agilent 7890A/7693A/5975C XL GC-MS equipped with a 0.25-mm × 30-m DB-5MS column (0.25-μm film) using pulsed splitless injection. Helium was used as the carrier gas at 0.75 mL min–1. The inlet was maintained at 250°C, and the oven was ramped from 45°C (2.25-min initial hold) to 250°C at 40°C per minute, held at 250°C for 3 min, and then ramped to 290°C at 40°C per minute. The ion source temperature was maintained at 230°C, and the quadrupole was heated to 150°C. The ion source was operated in electron impact mode, and both scan and selected ion data were acquired. Two ions were monitored for each hormone, and the larger fragment was used for peak area quantification (SA: 120, 124, 152, and 156 mass-to-charge ratio [m/z]; JA: 193, 195, 224, and 226 m/z; ABA: 162, 166, 190, and 194 m/z).
ET Measurement and 1-MCP Treatment
Three-week-old Arabidopsis plants were infested with aphids (30 per plant) for 2 d. Shoots were excised, weighed, and kept in 10-mL syringes with three-way stopcocks to seal them. One hour later, 1 mL of headspace gas was injected into a Photovac 10SPlus gas chromatograph. At least six individual plants were averaged for each treatment. Each experiment was repeated at least three times. ET was quantified by integration of peak area, relative to an authentic standard (Finlayson et al., 2007).
1-MCP gas was generated by dissolving a solid formulation of a proprietary 1-MCP α-cyclodextrin complex (AgroFresh) in 0.1 n NaOH in a flask fitted with a septum. The mass of the 1-MCP α-cyclodextrin complex used was calculated to produce 1000 μL L−1 1-MCP gas in the headspace of the flask. An aliquot of the concentrated 1-MCP gas was then injected into a desiccator to give a final calculated concentration of 1 ppm. Plants in the desiccator thus were subjected to 1-MCP treatment. After 1-h exposure to 1-MCP, plants were brought to a normal environmental atmosphere. This procedure was repeated every 12 h for 5 d to maintain the effect of 1-MCP, followed by aphid choice tests. Control plants were handled in the same manner without 1-MCP gas.
Quantitative RT-PCR
Plant samples were harvested, frozen, and ground in liquid nitrogen to a fine powder. Total RNA was extracted with TRIzol Reagent (Invitrogen) and then treated with RNase-Free DNase (Qiagen). Equal amounts of RNA (2 µg) were used to synthesize cDNA with random hexamer primers and SuperScript II Reverse Transcriptase (Invitrogen). Quantitative reverse transcription (RT)-PCR reactions were performed using SYBR Green Master Mix (BioRad) according to the manufacturer’s protocol. Primers were designed using PerlPrimer software, and their quality was examined using Primer-BLAST (National Center for Biotechnology Information). Primer sequences are provided in Supplemental Table S2. Arabidopsis UBIQUITIN10 (AT4G05320) served as an internal control for data normalization. Quantitative RT-PCR was run on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Controls using untranscribed RNA confirmed that there was no genomic DNA contamination. Dissociation curve analyses were applied to check amplification specificity. The mean fold change in gene expression was calculated as described previously (Zhu-Salzman et al., 2003).
Statistical Analysis
SPSS 16.0 software was used for analyses of all data. The no-choice tests of aphid performance among genotypes were analyzed by one-way ANOVA. Tukey’s multiple range test analysis was used for pairwise comparisons of the difference between treatments for mean separation (P < 0.05). The χ2 test was applied to the aphid choice tests (P < 0.05).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BIK1 (AT2G39660), FLS2 (AT5G46330), EFR (AT5G20480), BAK1 (AT4G33430), ERF1 (AT3G23240), PDF1.2 (AT5G44420), PR1 (AT2G14610), MYC2 (AT1G32640), SID2 (AT1G74710), EIN2 (AT5G03280), EIN3 (AT3G20770), PAD4 (AT3G52430), and SAG13 (AT2G29350).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mature shoot phenotypes of various Arabidopsis genotypes used in the study.
Supplemental Figure S2. The effect of ein3-1 mutation on bik1-mediated resistance against aphids.
Supplemental Figure S3. Loss of BIK1 function did not confer Arabidopsis resistance to fall armyworm.
Supplemental Table S1. Loss of BIK1 function negatively affects growth and reproduction traits in Arabidopsis.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Tesfaye Mengiste (Purdue University) for kindly sharing the mutant plant seeds, the Arabidopsis Biological Resource Center for the transfer DNA insertion and point mutation lines, Dr. Hisashi Koiwa (Department of Horticulture, Texas A&M University) for thoughtful discussions, Dr. Xiuren Zhang (Department of Biochemistry, Texas A&M University) for critical evaluation of the manuscript, and Dr. Martin Dickman (Institute for Plant Genomics and Biotechnology, Texas A&M University) for use of the Olympus microscope.
Glossary
- HR
hypersensitive response
- ROS
reactive oxygen species
- SA
salicylic acid
- JA
jasmonic acid
- ET
ethylene
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- PAMP
pathogen-associated molecular pattern
- MAMP
microbe-associated molecular pattern
- RLK
receptor-like kinase
- cDNA
complementary DNA
- DAB
3,3′-diaminobenzidine
- 1-MCP
1-methylcyclopropene
- RH
relative humidity
- m/z
mass-to-charge ratio
- RT
reverse transcription
Footnotes
This work was supported in part by the U.S. Department of Agriculture Agriculture and Food Research Initiative (grant no. 2014–67013–21781).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abuqamar S, Chai MF, Luo H, Song F, Mengiste T. (2008) Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20: 1964–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P, Winge P, Kusnierczyk A, Tran DH, Bones AM. (2013) Molecular signatures in Arabidopsis thaliana in response to insect attack and bacterial infection. PLoS ONE 8: e58987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi JL, Felton GW. (1995) Foliar oxidative stress and insect herbivory: primary compounds, secondary metabolites, and reactive oxygen species as components of induced resistance. J Chem Ecol 21: 1511–1530 [DOI] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. (2007) The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bos JIB, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA. (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez O, Huard C, Lorrain S, Roby D, Balagué C. (2007) Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1. Plant Physiol 145: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Delauré SL, De Bolle MF, Cammue BP. (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn JR, Martin GB. (2005) Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J 44: 139–154 [DOI] [PubMed] [Google Scholar]
- Cooper WR, Goggin FL. (2005) Effects of jasmonate-induced defenses in tomato on the potato aphid, Macrosiphum euphorbiae. Entomol Exp Appl 115: 107–115 [Google Scholar]
- Couldridge C, Newbury HJ, Ford-Lloyd B, Bale J, Pritchard J. (2007) Exploring plant responses to aphid feeding using a full Arabidopsis microarray reveals a small number of genes with significantly altered expression. Bull Entomol Res 97: 523–532 [DOI] [PubMed] [Google Scholar]
- De Vos M, Jander G. (2009) Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana. Plant Cell Environ 32: 1548–1560 [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Dong HP, Peng J, Bao Z, Meng X, Bonasera JM, Chen G, Beer SV, Dong H. (2004) Downstream divergence of the ethylene signaling pathway for harpin-stimulated Arabidopsis growth and insect defense. Plant Physiol 136: 3628–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Turner JG. (2002) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant Microbe Interact 15: 1025–1030 [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Hays DB, Morgan PW. (2007) phyB-1 sorghum maintains responsiveness to simulated shade, irradiance and red light: far-red light. Plant Cell Environ 30: 952–962 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Girousse C, Moulia B, Silk W, Bonnemain JL. (2005) Aphid infestation causes different changes in carbon and nitrogen allocation in alfalfa stems as well as different inhibitions of longitudinal and radial expansion. Plant Physiol 137: 1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin FL. (2007) Plant-aphid interactions: molecular and ecological perspectives. Curr Opin Plant Biol 10: 399–408 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Heil M, Baldwin IT. (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7: 61–67 [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, Woltering EJ. (2003) Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant-specific regulators. BioEssays 25: 47–57 [DOI] [PubMed] [Google Scholar]
- Hunt MD, Delaney TP, Dietrich RA, Weymann KB, Dangl JL, Ryals JA. (1997) Salicylate-independent lesion formation in Arabidopsis lsd mutants. Mol Plant Microbe Interact 10: 531–536 [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloshian I. (2004) Gene-for-gene disease resistance: bridging insect pest and pathogen defense. J Chem Ecol 30: 2419–2438 [DOI] [PubMed] [Google Scholar]
- Kerchev PI, Karpińska B, Morris JA, Hussain A, Verrall SR, Hedley PE, Fenton B, Foyer CH, Hancock RD. (2013) Vitamin C and the abscisic acid-insensitive 4 transcription factor are important determinants of aphid resistance in Arabidopsis. Antioxid Redox Signal 18: 2091–2105 [DOI] [PubMed] [Google Scholar]
- Kettles GJ, Drurey C, Schoonbeek HJ, Maule AJ, Hogenhout SA. (2013) Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol 198: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Jander G. (2007) Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J 49: 1008–1019 [DOI] [PubMed] [Google Scholar]
- Klingler JP, Nair RM, Edwards OR, Singh KB. (2009) A single gene, AIN, in Medicago truncatula mediates a hypersensitive response to both bluegreen aphid and pea aphid, but confers resistance only to bluegreen aphid. J Exp Bot 60: 4115–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ. (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuśnierczyk A, Winge P, Jørstad TS, Troczyńska J, Rossiter JT, Bones AM. (2008) Towards global understanding of plant defence against aphids—timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ 31: 1097–1115 [DOI] [PubMed] [Google Scholar]
- Kusnierczyk A, Winge P, Midelfart H, Armbruster WS, Rossiter JT, Bones AM. (2007) Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicae and oligophagous Brevicoryne brassicae. J Exp Bot 58: 2537–2552 [DOI] [PubMed] [Google Scholar]
- Laluk K, Luo H, Chai M, Dhawan R, Lai Z, Mengiste T. (2011) Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 23: 2831–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L. (2013) Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci USA 110: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Williams CE, Nemacheck JA, Wang H, Subramanyam S, Zheng C, Chen MS. (2010) Reactive oxygen species are involved in plant defense against a gall midge. Plant Physiol 152: 985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM. (2013) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 110: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balagué C, Roby D. (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Louis J, Gobbato E, Mondal HA, Feys BJ, Parker JE, Shah J. (2012) Discrimination of Arabidopsis PAD4 activities in defense against green peach aphid and pathogens. Plant Physiol 158: 1860–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J. (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Mantelin S, Bhattarai KK, Kaloshian I. (2009) Ethylene contributes to potato aphid susceptibility in a compatible tomato host. New Phytol 183: 444–456 [DOI] [PubMed] [Google Scholar]
- Mewis I, Appel HM, Hom A, Raina R, Schultz JC. (2005) Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol 138: 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohase L, van der Westhuizen AJ. (2002) Salicylic acid is involved in resistance responses in the Russian wheat aphid-wheat interaction. J Plant Physiol 159: 585–590 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Moran PJ, Cheng Y, Cassell JL, Thompson GA. (2002) Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch Insect Biochem Physiol 51: 182–203 [DOI] [PubMed] [Google Scholar]
- Moran PJ, Thompson GA. (2001) Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol 125: 1074–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A, Kachroo P, Fukushige H, Hildebrand DF, Klessig DF, Shah J. (2003) Ethylene and jasmonic acid signaling affect the NPR1-independent expression of defense genes without impacting resistance to Pseudomonas syringae and Peronospora parasitica in the Arabidopsis ssi1 mutant. Mol Plant Microbe Interact 16: 588–599 [DOI] [PubMed] [Google Scholar]
- Nisbet AJ, Woodford JAT, Strang RHC. (1994) Quantifying aphid feeding on non-radioactive food sources. Entomol Exp Appl 72: 85–89 [Google Scholar]
- Nombela G, Williamson VM, Muñiz M. (2003) The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant Microbe Interact 16: 645–649 [DOI] [PubMed] [Google Scholar]
- Pegadaraju V, Knepper C, Reese J, Shah J. (2005) Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol 139: 1927–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju V, Louis J, Singh V, Reese JC, Bautor J, Feys BJ, Cook G, Parker JE, Shah J. (2007) Phloem-based resistance to green peach aphid is controlled by Arabidopsis PHYTOALEXIN DEFICIENT4 without its signaling partner ENHANCED DISEASE SUSCEPTIBILITY1. Plant J 52: 332–341 [DOI] [PubMed] [Google Scholar]
- Prince DC, Drurey C, Zipfel C, Hogenhout SA. (2014) The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol 164: 2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM. (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95: 9750–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Hodge S, Ismail N, Mansfield JW, Feys BJ, Prospéri JM, Huguet T, Ben C, Gentzbittel L, Powell G. (2009) The RAP1 gene confers effective, race-specific resistance to the pea aphid in Medicago truncatula independent of the hypersensitive reaction. Mol Plant Microbe Interact 22: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL. (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57: 755–766 [DOI] [PubMed] [Google Scholar]
- Tjallingii WF. (2006) Salivary secretions by aphids interacting with proteins of phloem wound responses. J Exp Bot 57: 739–745 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. (2006) Reactive oxygen species in plant cell death. Plant Physiol 141: 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, et al. (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol 16: 1365–1369 [DOI] [PubMed] [Google Scholar]
- Walling LL. (2008) Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol 146: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, van Bel AJE. (2007) Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA 104: 10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, van Bel AJE. (2006) Physical and chemical interactions between aphids and plants. J Exp Bot 57: 729–737 [DOI] [PubMed] [Google Scholar]
- Williams B, Kabbage M, Kim HJ, Britt R, Dickman MB. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog 7: e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IC, Parker J, Bent AF. (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95: 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhou G, Qi J, Ren N, Cheng J, Erb M, Mao B, Lou Y. (2009) Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J 60: 638–648 [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K, Koiwa H, Salzman RA, Shade RE, Ahn JE. (2003) Cowpea bruchid Callosobruchus maculatus uses a three-component strategy to overcome a plant defensive cysteine protease inhibitor. Insect Mol Biol 12: 135–145 [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K, Salzman RA, Ahn JE, Koiwa H. (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.