Casparian strip membrane proteins are potentially involved in the generation of plasma membrane domains and the modification of cell walls.
Abstract
CASPARIAN STRIP MEMBRANE DOMAIN PROTEINS (CASPs) are four-membrane-span proteins that mediate the deposition of Casparian strips in the endodermis by recruiting the lignin polymerization machinery. CASPs show high stability in their membrane domain, which presents all the hallmarks of a membrane scaffold. Here, we characterized the large family of CASP-like (CASPL) proteins. CASPLs were found in all major divisions of land plants as well as in green algae; homologs outside of the plant kingdom were identified as members of the MARVEL protein family. When ectopically expressed in the endodermis, most CASPLs were able to integrate the CASP membrane domain, which suggests that CASPLs share with CASPs the propensity to form transmembrane scaffolds. Extracellular loops are not necessary for generating the scaffold, since CASP1 was still able to localize correctly when either one of the extracellular loops was deleted. The CASP first extracellular loop was found conserved in euphyllophytes but absent in plants lacking Casparian strips, an observation that may contribute to the study of Casparian strip and root evolution. In Arabidopsis (Arabidopsis thaliana), CASPL showed specific expression in a variety of cell types, such as trichomes, abscission zone cells, peripheral root cap cells, and xylem pole pericycle cells.
Biological membranes are conceptually simple structures that may be generated in vitro according to simple physicochemical principles. In vivo, however, membranes are highly complex and host a plethora of proteins that mediate the transfer of molecules and communication across the membrane. Proteins may be trapped in membrane by their transmembrane domains, anchored by lipid tails, or attach to membrane-integral proteins. A further level of complexity is seen when membrane proteins are not equally distributed but occupy only a limited fraction of the available surface (i.e. when they are polarly localized or when they form small membrane subdomains in the micrometer range). The question of how membrane proteins are retained locally and prevented from diffusing freely is of high importance to cell biology. Polarly localized proteins may be retained in their respective domains by membrane fences; in such a situation, polarly localized proteins are mobile in their domains but cannot diffuse through tightly packed scaffold proteins forming a molecular fence within the membrane. Membrane fences delimiting polar domains have been described in different organisms. For example, diffusion between membrane compartments is prevented in budding yeast (Saccharomyces cerevisiae) at the level of the bud neck (Barral et al., 2000; Takizawa et al., 2000); in ciliated vertebrate cells, between ciliary and periciliary membranes (Hu et al., 2010); in epithelial cells, between apical and basolateral membranes (van Meer and Simons, 1986); in neurons, between axon and soma (Kobayashi et al., 1992; Winckler et al., 1999; Nakada et al., 2003); and in spermatozoa, at the level of the annulus (Myles et al., 1984; Nehme et al., 1993). The existence of membrane scaffolds that prevent free protein diffusion has also been described in bacteria (Baldi and Barral, 2012; Schlimpert et al., 2012). In plants, we have shown the existence of a strict membrane fence in the root endodermis, where a median domain splits the cell in two lateral halves occupied by different sets of proteins (Alassimone et al., 2010). The situation in the plant endodermis is analogous to the separation of animal epithelia into apical and basolateral domains; indeed, a parallel between epithelia and endodermal cells has been drawn, despite the different origin of multicellularity in plants and animals (Grebe, 2011).
The protein complexes responsible for the formation of membrane fences have been identified. Septins are a family of proteins able to oligomerize and form filaments (Saarikangas and Barral, 2011); their role in the formation of membrane fences has been demonstrated in several organisms and cellular situations, including the yeast bud neck (Barral et al., 2000; Takizawa et al., 2000), animal cilia (Hu et al., 2010), and mammalian spermatozoa (Ihara et al., 2005; Kissel et al., 2005; Kwitny et al., 2010). At the axonal initial segment of neurons, AnkyrinG is necessary to establish and maintain a membrane scaffold where different membrane proteins are immobilized and stabilized (Hedstrom et al., 2008; Sobotzik et al., 2009). In Caulobacter crescentus, the stalk protein Stp forms a complex that prevents diffusion between the cell body and stalk and between stalk compartments. Claudins and occludin are the main components of epithelial tight junctions (Furuse et al., 1993, 1998). Occludins are four-membrane-span proteins and belong to the MARVEL protein family (Sánchez-Pulido et al., 2002), as do Tricellulin and MARVELD3, which are also tight junction-associated proteins (Furuse et al., 1993; Ikenouchi et al., 2005; Steed et al., 2009).
In Arabidopsis (Arabidopsis thaliana), our group identified a family of proteins that form a membrane fence in the endodermis (Roppolo et al., 2011). These CASPARIAN STRIP MEMBRANE DOMAIN PROTEINS (CASP1 to CASP5) are four-transmembrane proteins that form a median domain referred to as the Casparian strip membrane domain (CSD). CASPs are initially targeted to the whole plasma membrane, then they are quickly removed from lateral plasma membranes and remain localized exclusively at the CSD; there, they show an extremely low turnover, although they are eventually removed (Roppolo et al., 2011). The membrane proteins NOD26-LIKE INTRINSIC PROTEIN5;1 and BORON TRANSPORTER1 are restricted from diffusing through the CSD and remain polarly localized in the outer and inner lateral membranes, respectively; a fluorescent lipophilic molecule, when integrated in the outer endodermal membrane, was blocked at the level of the CSD and could not diffuse into the inner membrane (Roppolo et al., 2011). Besides making a plasma membrane diffusion barrier, CASPs have an important role in directing the modification of the cell wall juxtaposing their membrane domain: by interacting with secreted peroxidases, they mediate the deposition of lignin and the building up of the Casparian strips (Roppolo et al., 2011; Naseer et al., 2012; Lee et al., 2013). The two CASP activities, making membrane scaffolds and directing a modification of the cell wall, can be uncoupled: indeed, (1) formation of the CASP domain is independent from the deposition of lignin, and (2) interaction between CASPs and peroxidases can take place outside the CSD when CASPs are ectopically expressed (Lee et al., 2013).
As CASPs are currently the only known proteins forming membrane fences in plants and because of their essential role in directing a local cell wall modification, we were interested in characterizing the repertoire of a large number of CASP-like (CASPL) proteins in the plant kingdom. Our aim was to provide the molecular basis for the discovery of additional membrane domains in plants and for the identification of proteins involved in local cell wall modifications. We extended our phylogenetic analysis outside of the plant kingdom and found conservation between CASPLs and the MARVEL protein family. Conserved residues are located in transmembrane domains, and we provide evidence suggesting that these domains are involved in CASP localization. We explored the potential use of the CASPL module in plants by investigating CASPL expression patterns and their ability to form membrane domains in the endodermis. Moreover, we related the appearance of the Casparian strips in the plant kingdom to the emergence of a CASP-specific signature that was not found in the genomes of plants lacking Casparian strips.
RESULTS
CASPLs Belong to the MARVEL Protein Family
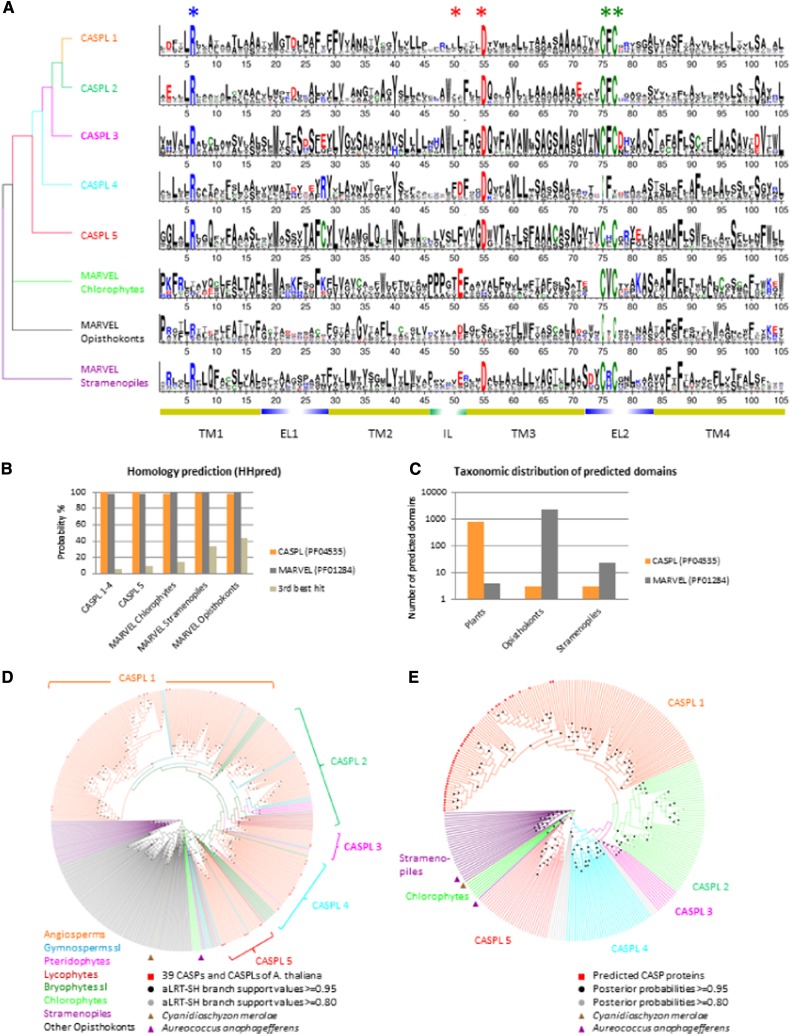
In an attempt to understand the evolutionary history of the CASPs, we analyzed sequenced plant genomes and EST databases for their repertoire of CASP homologs that we termed CASPLs. In all, we annotated over 350 proteins from more than 50 plant species. Arabidopsis CASPs (AtCASPs) have four predicted transmembrane domains, with cytoplasmic N and C termini, variable N terminus length, short C terminus, and short intracellular loop. Homologous plant proteins are conserved in the transmembrane domains, particularly the first (TM1) and the third (TM3): an Arg in TM1 and an Asp in TM3 are present in the vast majority of CASPLs (Fig. 1A; Supplemental Fig. S1). Six proteins with sequence similarity were identified in green algae: Chlorokybus atmophyticus (charophyte), Ostreococcus tauri, Ostreococcus lucimarinus, and Micromonas pusilla (chlorophyte), and Micromonas spp. strains RCC299 and NOUM17. Interestingly, Chlorokybus and Micromonas spp. CASPLs are annotated in UniProtKB as carrying a MARVEL-like domain (IPR021128). Proteins carrying a MARVEL domain show high similarity in their transmembrane domains, but not necessarily in their extracellular or intracellular exposed regions (Sánchez-Pulido et al., 2002). Conserved basic (Arg, His, and Lys) and acidic (Asp and Glu) amino acids are present in TM1 and TM3 of MARVELs from stramenopiles and fungi, a situation that very much resembles the conservation in transmembrane domains among CASPLs. CASPLs and MARVELs are predicted with high probability to be members of both families (Fig. 1B; Supplemental Table S1), indicating that CASPL and MARVEL domains are likely to be homologous. Also notable is the almost complementary taxonomic distribution of species with predicted CASPL (DUF588 and PF04535) or MARVEL (PF01284) domains in opisthokonts and plants (Fig. 1C; Supplemental Table S2), which led to the assumption that CASPLs could be the plant orthologs of the MARVEL family. Indeed, a reconstruction of the CASPL/MARVEL domain phylogeny places algae at the base of the plant clade when rooting with stramenopiles (Fig. 1D; Supplemental Fig. S2); however, three genes in key positions of the CASPL/MARVEL domain phylogeny challenge this hypothesis by taking no stable position in the obtained trees (Fig. 1, D and E; Supplemental Figs. S2–S4). Thus, an origin of CASPLs by speciation seems as likely as an origin by gene duplication ancestral to the divergence of plants and animals. Taken together, the CASPL/MARVEL family includes 1,792 homologs in the UniProtKB reference proteomes (545 CASPLs and 1,247 MARVELs; UniProt release 2014_04).
Figure 1.
Evidence for a common origin of CASPLs and MARVELs. A, Sequence similarity and five conserved residues. Sequence logos covering the four transmembrane domains and conserved adjacent regions of the CASPL/MARVEL domains have been constructed for the five plant groups, as well as for chlorophytes, stramenopiles, and opisthokonts. Conserved residues are marked by asterisks. Yellow bars below logos correspond to the predicted transmembrane domains of CASP1 from Arabidopsis, and the adjacent conserved regions of the assumed extracellular loops (EL1 and EL2) and of the intracellular loop (IL) are indicated. The predicted relationship of the five CASPL groups and MARVEL taxonomic groups is shown as a tree at the left of the sequence logos. The color code of the protein groups corresponds to that in E. B, Homology prediction. CASPL and MARVEL domains are predicted to belong to both domain families with a probability of at least 97.3% (Supplemental Table S1). C, Taxonomic range for domain predictions. A total of 99.5% of the CASPL domains are predicted in plant genes, and 99.9% of the MARVEL domains are predicted in genes from species of the animal lineage. D, Circular cladogram based on the maximum likelihood phylogeny of the CASPL and MARVEL domains. Genes from species in key positions of the CASPL/MARVEL domain phylogeny with no stable position in the tree are marked by triangles: two genes from the stramenopiles Aureococcus anophagefferens (UniProtKB nos. F0YFA3 and F0Y3N5) and one gene from the rhodophyte Cyanidioschyzon merolae (UniProtKB no. M1VAW8). Branch colors indicate taxonomic species groups. E, Circular cladogram of the CASPL phylogeny with MARVELs from stramenopiles and algae as an outgroup, based on a Bayesian inference analysis. The five CASPL subfamilies are marked by colors, and stramenopiles and chlorophytes are colored as in D; unclassified genes are gray. [See online article for color version of this figure.]
Based on stable clades of the inferred gene phylogenies, we classified the CASPL family into five groups, all of which contain homologs from bryophytes, lycophytes, or pteridophytes in addition to members from euphyllophytes such as conifers, dicotyledons, and monocotyledons (Fig. 1, D and E; Supplemental Figs. S3 and S4). Furthermore, subgroups were defined so that potential functionally related homologs can be easily identified between species (Supplemental Table S3). Only a minor portion of the predicted proteins could not be stably attributed to any group. Figure 1A illustrates the predicted relationship of the five CASPL groups and MARVEL taxonomic groups and visualizes conserved sequence positions. The two conserved charged residues as well as the two Cys residues are present in four of the five CASPL subfamilies and MARVEL stramenopiles; members of CASPL group 4 possess a short extracellular loop 2, which lacks the two Cys residues. Surprisingly, homologs from chlorophytes possess none of the conserved charged residues. Instead, we identify a Glu four positions upstream of the conserved acidic amino acid, which is found also in MARVEL stramenopiles, many opisthokonts, and CASPL group 4, suggesting that both acidic residues in TM3 are likely an ancestral characteristic of this family.
Extracellular Loops Are Dispensable for AtCASP1 Localization at the CSD
To begin understanding how CASPs get localized, we exploited conservation in the CASPL family to identify potential residues necessary for AtCASP1 localization (Fig. 2; Supplemental Fig. S5). We generated 14 AtCASP1-GFP variants, expressed them under the control of the AtCASP1 promoter, and compared their localization with a wild-type AtCASP1-mCherry (Vermeer et al., 2014). Besides the transmembrane domains, conservation in CASPLs is found in the second extracellular loop (EL2). EL1 is poorly conserved among CASPLs, even inside subgroups; however, AtCASPs present in their EL1 a stretch of nine residues that is highly conserved in all spermatophytes (see below). We decided to mutagenize specific residues in TM3 and EL2 and to delete either loop. When we mutagenized the MARVEL/CASPL conserved Asp residue in TM3 (AtCASP1D134H), we did not recover any lines in which fluorescence was visible, which suggests that this residue is essential for correct protein folding (Supplemental Table S4). In EL2, mutations of residues conserved only in the CASP subgroup (CASPL1A) did not affect the localization of AtCASP1 (A155S, H156D, N163D, and Q170E; Fig. 2); in contrast, mutations in residues shared among most CASPLs affected AtCASP1 localization to different extents. C168S, F174V, and C175S persisted longer than AtCASP1-mCherry at the lateral plasma membrane, although they started localizing at the CSD at the same time as the wild type; G158S localized normally at the lateral plasma membrane, but its localization at the CSD was strongly delayed and signal there was extremely low (Fig. 2; Supplemental Fig. S5). W164G showed the strongest effect, being initially excluded from the CSD and almost undetectable later (Fig. 2; Supplemental Fig. S5). Despite the fact that mutations in EL2 affected AtCASP1 localization, when the entire EL2 (Δ158:175) was deleted, AtCASP1 was still able to localize at the CSD, although its signal faded out faster than in the wild type (Fig. 2; Supplemental Fig. S5). Deletions of EL1 (Δ72:80, Δ73:79, and Δ74:78) did not affect localization at the CSD, although a longer persistence at the lateral membranes was observed and the exclusive enrichment at the CSD was delayed compared with AtCASP1-mCherry; the nine-amino acid deletion (Δ72:80) seemed less stable than the wild type at the CSD (Fig. 2; Supplemental Fig. S5). In summary, this analysis shows that AtCASP1 extracellular loops are dispensable for localization at the CSD; single-residue substitution in EL2 affects AtCASP1 localization, indicating that those residues contribute to, but are not essential for, localization at the CSD.
Figure 2.
Extracellular loops are dispensable for AtCASP1 localization at the CSD. Representative confocal images of AtCASP1-GFP mutant/wild type-mCherry crosses (green and red labels, respectively) are shown. Numbers indicate positions of the cell shown counted from the first elongating endodermal cell (the lower the number, the closer the cell is to the root apical meristem). White arrows point to the position of the CSD. The schematic summarizes the position of deletions and mutagenized residues that affect or do not affect localization (above and below, respectively). Mutations are annotated on a sequence logo of the CASPL1A subgroup to highlight the conservation of the mutagenized residues. Yellow bars indicate the positions of the extracellular loops. [See online article for color version of this figure.]
CASPs Differ from CASPLs in Their First Extracellular Loop
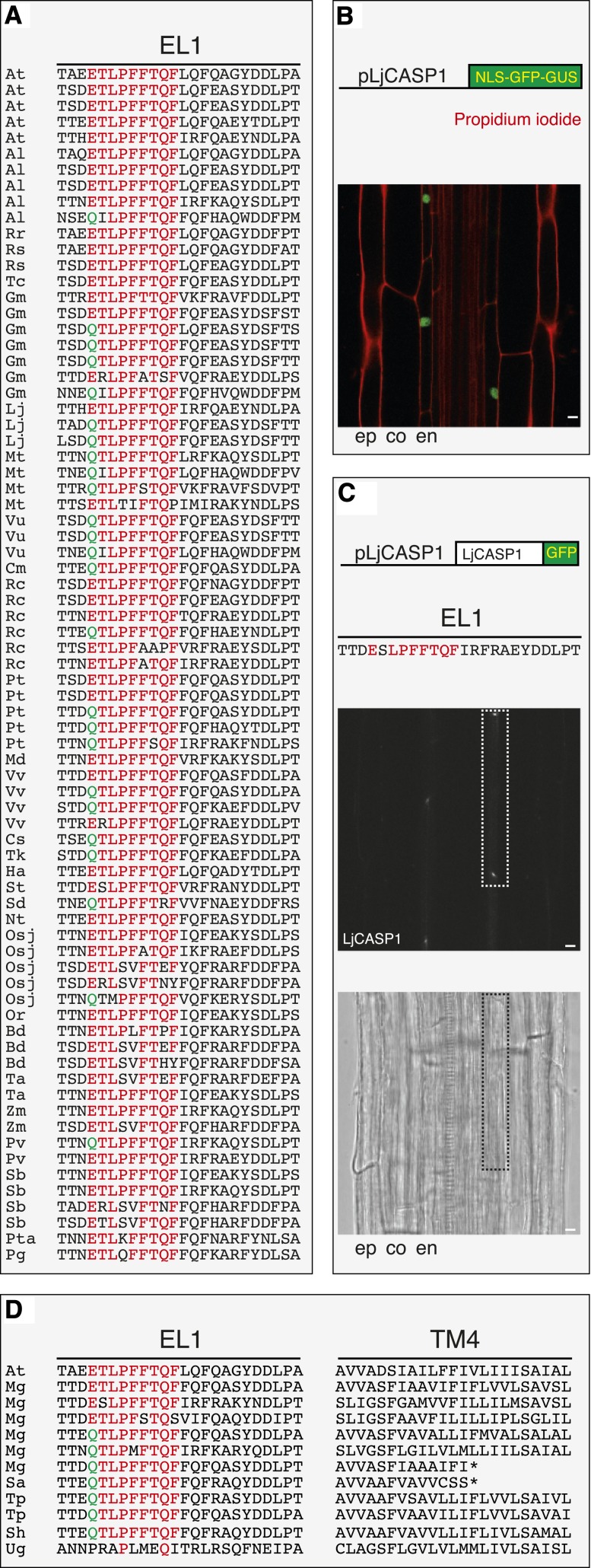
Although dispensable for the localization at the CSD, the conservation of EL1 in euphyllophytes suggests a conserved function of this stretch (Fig. 3A). To test if CASP homologs containing the EL1 stretch could be potential functional homologs of the AtCASPs, we cloned from Lotus japonicus a member of the family containing the nine-amino acid signature (ESLPFFTQF) and expressed it in Arabidopsis under the control of its own putative promoter. A 2-kb genomic fragment upstream of the translational start codon of this L. japonicus gene was sufficient to drive expression in the endodermis of Arabidopsis roots, showing the ease of predicting potential CASP functional homologs following identity in the first extracellular loop (Fig. 3B). An L. japonicus GFP translation fusion perfectly recapitulated the localization of the endogenous AtCASP1 at the CSD (Fig. 3C), and the reporter reflected temporally and spatially the expression of the AtCASP1 promoter. Therefore, these data suggest that the well-conserved EL1 serves some endodermis-specific function and that conservation extends to regulatory elements. CASP homologs with this nine-amino acid signature are absent in Physcomitrella patens and Selaginella moellendorffii, which have no roots or have roots of different evolutionary origin, respectively, while they are present in all roots of Casparian strip-bearing organisms (Raven and Edwards, 2001) for which we could extensively assess the genome (no genomes have been fully sequenced in the Moniliformopses). To extend further the correlation between the presence of the CASP EL1 signature and the appearance of Casparian strips, we analyzed the genomes of plants that have acquired parasitic behavior and show extensively modified root anatomy. The genus Striga contains obligate hemiparasites that do not have functional roots (Westwood et al., 2012); it belongs to the Orobanchaceae, a family of the Lamiales order containing plants that have evolved different parasitic behaviors (Westwood et al., 2010). Intriguingly, we could identify a single CASP homolog with a perfectly conserved EL1 signature in Striga asiatica; however, this protein encodes a premature stop codon that prevents the complete translation of the fourth transmembrane domain (Sa in Fig. 3D) and, therefore, is likely to be nonfunctional. In Striga hermontica, by contrast (Sh in Figure 3D), a potentially functional CASP gene can still be identified. Functional alleles are also found in another member of the Orobanchaceae, Triphysaria pusilla, which, however, is a facultative hemiparasite (Tp in Fig. 3D). The Striga asiatica truncated allele is also found in Mimulus guttatus, where we could identify five more presumably functional CASP homologs (Mg in Fig. 3D). The genus Mimulus belongs to the same order as the genus Striga (Lamiales) but shows normal root and nonparasitic lifestyle. In the Lamiales, a complete loss of the EL1 stretch happened in the carnivore plant Utricularia gibba, whose genome was recently sequenced (Ibarra-Laclette et al., 2013). In correlation with the lack of true roots in this carnivore plant species, Ibarra-Laclette et al. (2013) reported the presence of a single CASP homolog. We reassessed the U. gibba genome for CASPLs and compared it with the genome of M. guttatus. The U. gibba and M. guttatus genomes contain over 20 CASP homologs (Supplemental Table S5); in M. guttatus, six of them contain the EL1 signature, in three members perfectly conserved and in three showing a single-residue divergence (Fig. 3D). In contrast, the closest CASP homolog in U. gibba shows a clear divergence of the entire EL1: only two residues are identical to the AtCASP EL1 stretch (Ug in Fig. 3D). The unique absence of conservation of the EL1 stretch in U. gibba, P. patens, and S. moellendorffii correlates perfectly with an absence of Casparian strips in these species; in the Orobanchaceae with parasitic behavior, potential CASP pseudogenization has occurred in Striga asiatica. The above correlations support our hypothesis that the first extracellular loop bears residues necessary for the function of the CASPs in the deposition of the Casparian strips. For this reason, we named as CASP all the members of the family that carry the AtCASP EL1 signature (E/QTLPPFFTQF, with two amino acid substitutions accepted; Supplemental Table S3), designating them as potential functional homologs of the AtCASPs.
Figure 3.
The AtCASP first extracellular loop is conserved in euphyllophytes. A, EL1 of AtCASP1 is aligned to potential functional homologs. The nine-amino acid signature specific to AtCASPs and absent in AtCASPLs is highlighted in red. In the first position, E (red) and Q (green) are both found. Species abbreviations are as follows: At, Arabidopsis; Al, Arabidopsis lyrata; Rr, Raphanus raphanistrum; Rs, Raphanus sativus; Tc, Theobroma cacao; Gm, Glycine max; Lj, Lotus japonicus; Mt, Medicago truncatula; Vu, Vigna unguiculata; Cm, Cucumis melo; Rc, Ricinus communis; Pt, Populus trichocarpa; Md, Malus domestica; Vv, Vitis vinifera; Cs, Cynara scolymus; Tk, Taraxacum kok-saghyz; Ha, Helianthus annuus; St, Solanum tuberosum; Sd, Solanum demissum; Nt, Nicotiana tabacum; Osj, Oryza sativa subsp. japonica; Or, Oryza rufipogon; Bd, Brachypodium distachyon; Ta, Triticum aestivum; Zm, Zea mays; Pv, Panicum virgatum; Sb, Sorghum bicolor; Pta, Pinus taeda; and Pg, Picea glauca. B and C, Expression of an L. japonicus potential AtCASP functional homolog (LjCASP1) in Arabidopsis. The L. japonicus promoter drives expression exclusively in the endodermis (B), and LjCASP1-GFP localizes to the Casparian strip membrane domain (C). Images show confocal sections of a 5-d-old root. Rectangles in C highlight an endodermal cell; co, cortex; en, endodermis; and ep, epidermis. Bars = 10 μm. D, AtCASP1 EL1 and TM4 are aligned to their closest homologs from the Lamiales. Mimulus guttatus (Mg) and Striga asiatica (Sa) present an AtCASP homolog potentially nonfunctional due to a premature stop codon (asterisks) in TM4. The Utricularia gibba (Ug) closest AtCASP homolog does not show conservation in the EL1 signature. Striga hermontica (Sh), Triphysaria pusilla (Tp), and Mimulus guttatus encode for potential functional AtCASP homologs. See also Supplemental Table S5. [See online article for color version of this figure.]
AtCASPLs from All Groups Localize in the Endodermis as AtCASPs
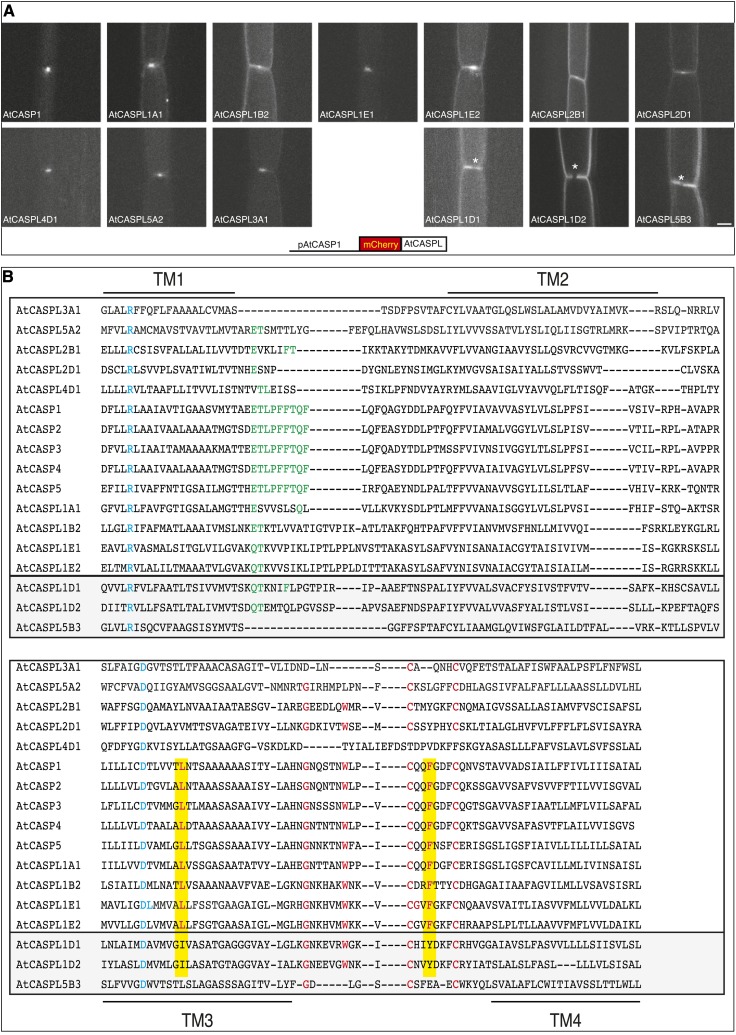
To see if membrane domain formation may be a general feature in the CASPL family, we expressed several AtCASPLs in the endodermis, where they are not endogenously expressed. We then evaluated their ability to localize at the plasma membrane and at the CSD. Twenty-two AtCASPLs and AtCASP1, AtCASP2, and AtCASP3 were expressed as N-terminal mCherry fusions under the control of the AtCASP1 promoter (Supplemental Table S6). As expected, mCherry-AtCASPs localized correctly (Fig. 4A). For nine AtCASPLs, we could not identify any lines expressing the transgene or the expression was extremely weak (Supplemental Table S6); when expression was detectable, mCherry signal was observed mainly in the vacuole, suggesting that these AtCASPLs were quickly degraded (data not shown). Twelve AtCASPLs were able to reach the plasma membrane, and nine of these showed a clear localization at the CSD (Fig. 4A). Interestingly, the CSD-localized AtCASPLs cover all CASPL groups, which suggests that the propensity to form membrane domains is broadly conserved within the family. As none of the tested CASPLs are endogenously expressed in the endodermis (Brady et al., 2007; Birnbaum et al., 2008), their localization at the CSD may be caused by homo-interaction, interaction with the endogenous AtCASPs, or interaction with other factors that mediate AtCASP localization and also recognize AtCASPLs. Because of the high divergence between CASPL groups, the comparison of the primary sequence of localized and nonlocalized CASPLs did not provide any useful information (Fig. 4B). For example AtCASPL3A1, AtCASPL5A2, and AtCASPL4D1 do not contain in the second extracellular loop the Trp-164 residue that, when mutated, prevents AtCASP1 localization, although they do localize at the CSD. When we limited the primary sequence analysis to AtCASPLs from group 1, we found two residues that differ between localized and nonlocalized AtCASPL1 (Fig. 4B): localized AtCASPL1s and AtCASPs share the AtCASP1 Leu-140 and Phe-171 residues in TM3 and EL2, respectively (positions 61 and 76 in Fig. 1A); AtCASPL1s excluded from the CSD but present at the lateral plasma membrane contain in the corresponding position an Ile (versus AtCASP1 Leu-140; one methyl group missing) and Tyr (versus AtCASP1 Phe-171; one extra hydroxyl group). These findings point to a predominant role of TM3 and EL2 in the localization of AtCASPs. In addition, the global analysis presented here shows that CASPLs with very divergent extracellular loops can have similar behaviors in the endodermis.
Figure 4.
AtCASPLs can localize at the CSD when expressed in the endodermis. A, Root confocal sections of 5-d-old seedlings expressing mCherry-AtCASPLs under the control of the AtCASP1 promoter. Expression is seen only in endodermal files, where AtCASPLs are retained or excluded from the CSD (asterisks mark exclusion). Bar = 5 µm. B, Alignment of AtCASPs and AtCASPLs that localize at the CSD (top rectangles) or not (bottom rectangles). Residues in blue are found in all CASPLs; residues in red are necessary for a correct localization of AtCASP1. The AtCASP first extracellular loop signature is highlighted in green. Yellow shading shows residues that are conserved in AtCASPL1s that localize at the CSD but absent in AtCASPL1s that do not localize at the CSD. Note that conservation in extracellular loops is not an absolute requirement for localization at the CSD. See also Supplemental Table S6. [See online article for color version of this figure.]
AtCASPLs Are Expressed in a Tissue-Specific Manner
AtCASPs are expressed exclusively in the endodermis, where Casparian strips are deposited. To see if other CASPLs have acquired tissue-specific function, we experimentally assessed the expression pattern of some AtCASPLs (Fig. 5).
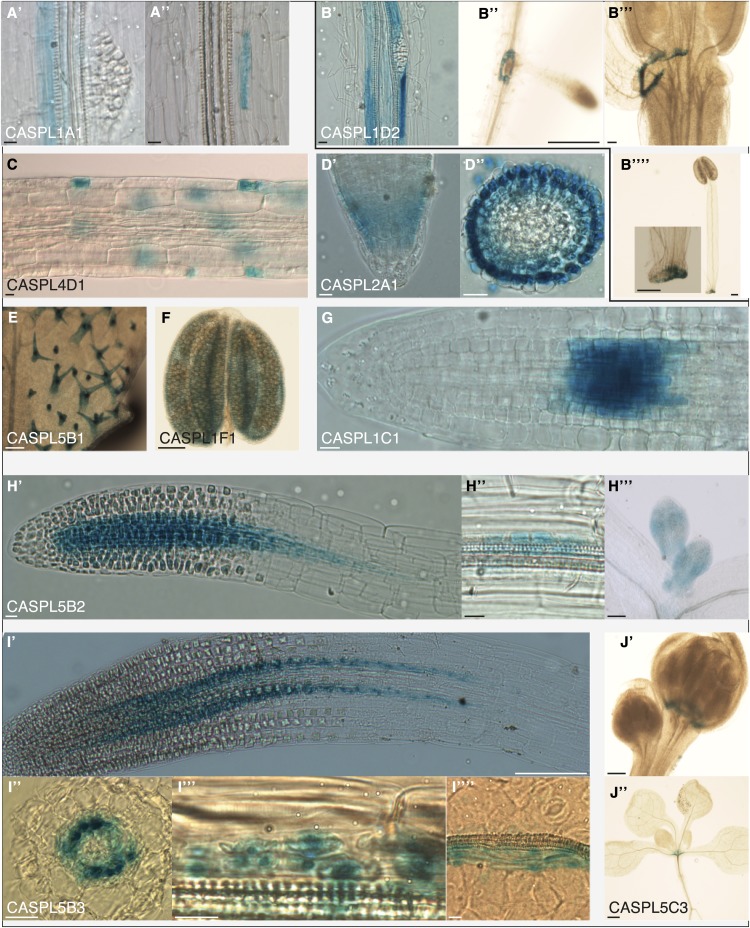
Figure 5.
AtCASPLs are expressed in different tissues. A, AtCASPL1A1 is expressed in the endodermis at late stages, initially in all cells (′), later only in isolated cells (′′); its expression is reduced in the endodermis overlaying lateral root primordia (′). B, AtCASPL1D2 is expressed in late endodermis, particularly in cells overlaying lateral root primordia (′); expression persists at the base of emerged lateral roots (′′); it is also expressed in the floral organ abscission zone (′′′) on the side of the shed organ (′′′′). C, AtCASPL4D1 is expressed in root epidermis from the maturation zone; its expression quickly declines. D, AtCASPL2A1 is expressed in the lateral root cap (′; D′′ shows a transverse section of the root tip); it is also expressed in leaves (data not shown). E, AtCASPL5B1 is expressed in immature and differentiated trichomes in leaves. F, AtCASPL1F1 is expressed in anthers. G, AtCASPL1C1 is expressed in the root maturation zone. H, AtCASPL5B2 is expressed in dividing cells in the root (′), lateral root primordia (′′), and leaves (′′′). I, AtCASPL5B3 is expressed in root xylem pole pericycle (′), lateral root primordia (′′′), and leaves (′′′′); I′′ shows a transverse section at the root tip. J, AtCASPL5C3 is expressed early in the floral organ abscission zone (′) and in the rosette vestigial abscission zone (′′). All images represent whole-mount stainings, except D′′ and I′′. Bars = 10 µm for A, B′, B′′′, C, D, H′, H′′, I′′, I′′′, and I′′′′′; 100 µm for B′′, B′′′′, E, F, H′′′, I′, and J′; and 500 µm for J′′. [See online article for color version of this figure.]
The closest CASP homolog, AtCASPL1A1, is expressed in the root endodermis at a late developmental stage, coinciding with the appearance of metaxylem vessels (Fig. 5A). This area of the root is situated at around 25 cells from the appearance of differentiated xylem vessel cells and Casparian strips. In 10-d-old roots, expression fades out in proximity to the hypocotyl, except in isolated endodermal cells (Fig. 5A′′). Expression is also reduced in endodermal cells overlaying lateral root primordia (Fig. 5A′). No expression was detected in leaves or in the hypocotyl of 4-d-old seedlings.
In the root, AtCASPL1D2 is expressed exclusively in the endodermis at a late developmental stage; no expression was detected in leaves or in the hypocotyl of 4-d-old seedlings. Interestingly, and in contrast to AtCASPL1A1, AtCASPL1D2 expression is strongly enhanced in endodermal cells overlaying lateral root primordia (Fig. 5B′); when lateral roots have emerged, its expression is seen in a collar of endodermal cells at the base of the emerged root (Fig. 5B′′). AtCASPL1D2 is also expressed in the floral organ abscission zone in cells that detach along with the shed organs (Fig. 5, B′′′ and B′′′′).
Apart from AtCASPL1D2, AtCASPL2A2 is reported to be expressed in the floral organ abscission zone (González-Carranza et al., 2012). AtCASPL5C3 is expressed in the floral organ abscission zone as well, but its early expression in floral buds precedes the activation of the abscission zone and the expression of most of the genes known to be involved in floral organ shedding (Fig. 5J).
AtCASPLs are also found expressed in epidermal tissues. AtCASPL4D1 is expressed in the root epidermis (Fig. 5C); its expression begins in the root maturation zone and quickly declines. Expression was not detected in leaves or in the hypocotyl. AtCASPL2A1 is expressed in the peripheral root cap; it is also expressed in leaf epidermis (Fig. 5D). In leaf, AtCASPL5B1 is expressed exclusively in hair cells, both in differentiated trichomes and immature cells (Fig. 5E).
Apart from the above-mentioned AtCASPLs in the abscission zone, we have also found in the flower AtCASPL1F1 expressed in the anther wall (Fig. 5F).
We identified three AtCASPLs expressed in the stele of the root. AtCASPL5B2 is expressed in 4-d-old seedlings exclusively in the root meristematic zone and in young leaves (Fig. 5, H′ and H′′′); in 10-d-old roots, expression is also detected in lateral root primordia (Fig. 5H′′) and in the stele in proximity to the hypocotyl (data not shown). In the strongest line, expression is detected in the stele all along the root. AtCASPL5B3 is expressed in the root maturation zone (Fig. 5I′) and in lateral root primordia (Fig. 5I′′′). Longitudinal view and transversal sections show expression in two parallel files that we interpret as xylem pole pericycle cells (Fig. 5, I′ and I′′); this is consistent with later expression in lateral root primordia. We also detected AtCASPL5B3 expression in leaves in files of cells parallel to the vasculature (Fig. 5I′′′′). AtCASPL1C1 is expressed in the root maturation zone; no expression was detected in leaves or hypocotyl (Fig. 5G). This expression pattern is consistent among different transgenic lines, but it is not seen in all root tips. In a 10-d-old plant, we could detect AtCASPL1C1 expression in five of 42 tips of emerged lateral roots, which may reflect a very transient expression in these organs.
DISCUSSION
CASPLs Are Related to MARVEL
Our analysis reveals that the plant family of CASPLs is related to the MARVEL protein family that had been experimentally described only in metazoans. Conservation is very limited, but strikingly specific, at the level of the transmembrane domains and in the overall tetraspan protein structure; a putative Cys bridge in the second extracellular loop is often present in both MARVELs and CASPLs. Besides the results presented here, an independent analysis classified CASPLs and MARVELs as members of clan CL0396 (Bateman et al., 2004). Only a handful of MARVELs have been characterized; although a general function cannot be ascribed to this group of proteins, some intriguing common features emerge upon comparison of the available functional data: (1) MARVELs are integral membrane proteins, found either at the plasma membrane or in vesicle membranes; (2) when overexpressed, they tend to form lamellae in the smooth endoplasmic reticulum; (3) they are associated with membrane fusion events or membrane apposition; and (4) they can occupy membrane subdomains. Among the characterized MARVELs are the tight junction-associated proteins Occludin, Tricellulin, and MARVELD3 (Furuse et al., 1993; Ikenouchi et al., 2005; Steed et al., 2009); Synaptophysin, the most abundant protein in synaptic vesicles (Jahn et al., 1985; Wiedenmann and Franke, 1985); and Singles Bar, necessary for myoblast membrane fusion in Drosophila melanogaster (Estrada et al., 2007). In a simple scenario, the last eukaryotic common ancestors possessed the MARVEL four-transmembrane domain functional skeleton: transmembrane domains have been conserved during evolution, while extracellular loops diverged in different eukaryotic divisions. The conservation in transmembrane domains may be necessary for the interactions of CASPL/MARVELs with each other within the membrane. Indeed, the site-directed mutagenesis of AtCASP1 and the gene-swap analysis presented here show that extracellular loops are dispensable for localization, pointing to a central role of the transmembrane regions (note that apart from the N terminus, which shows no conservation, intracellular regions are extremely short in the CASPL family). CASPLs, as well as MARVELs, may share the ability to undergo controlled polymerization of their transmembrane domains and thereby drive the formation of membrane scaffold microdomains in a variety of circumstances. Our hypothesis is supported by the few studies concerning the importance of the transmembrane domains of MARVELs: in Synaptophysin, loss of the acid residues in TM1 prevents protein accumulation (Leube, 1995); in Occludin, the transmembrane domain has been suggested to mediate dimerization (Yaffe et al., 2012); in MAL, the founder member of the family, transmembrane domain deletions prevent plasma membrane localization or the formation of a correct microdomain (Magal et al., 2009).
For their expression in typical animal structures (e.g. epithelia and synapses), MARVEL family members have been used to trace the origin of typical animal and typical multicellular proteins. When the genome of a sponge was released, Synaptogyrin, Synaptophysin, and Occludin were considered holozoa, metazoa, and vertebrate specific, respectively (Srivastava et al., 2010). Our phylogenetic analysis now demonstrates a much more ancient origin of the MARVEL family, predating multicellularity, although it does not challenge the idea that individual members may be associated with the appearance of specific structures.
CASPLs as Membrane Organizers and/or Cell Wall Modifiers
CASPs are the earliest known proteins localizing at the CSD and may be necessary for its formation. The ability of CASPLs from all groups to localize at the CSD when expressed in the endodermis supports the existence of a conserved CASPL module for membrane subdomain formation and prompts us to predict that many yet-undiscovered plasma membrane domains in other cell types are mediated by CASPLs. Our expression analysis shows that AtCASPLs are expressed in a highly tissue-specific manner. A straightforward speculation is that CASPLs and CASPs perform similar, but specialized, functions that would be (1) the generation of membrane scaffolds and/or (2) the recruitment of cell wall-modifying enzymes. Polar or local secondary cell wall modifications have been described in most tissues discussed above. For example, in anthers, the endothecium cell walls thicken laterally, forming bar-like structures that are enriched in cellulose and lignin (Dawson et al., 1999); polar secretion of cell wall-degrading enzyme is expected during abscission (Estornell et al., 2013). CASPLs may be involved in addressing different cell wall-modifying machineries in different tissues as well as in delimiting the region of the plasma membrane where such modifications must take place. CASPLs expressed in dividing cells may help establish cell polarity by ensuring the asymmetric distribution of plasma membrane proteins or contribute to callose deposition in plasmodesmata. Based on our results, we believe that CASPLs represent a promising new entry point to unravel membrane domain formation in plants and to uncover the mechanisms behind local modifications of plant cell walls.
Evolution of CASPs, Casparian Strips, and the Euphyllophyte Root
Roots have evolved independently in the Lycopodiopsida and the euphyllophytes, the two groups of the tracheophytes (Raven and Edwards, 2001). Casparian strips are a typical feature of the euphyllophyte root, both in spermatophytes and Moniliformopses. Casparian strip-like material has been sporadically reported in Lycopodiopsida; however, those structures should be considered at best analogous to Casparian strips, given the independent origin of roots in Lycopodiopsida and euphyllophytes. We report here the conservation of the CASP EL1 in all spermatophyte genomes we could extensively assess; presence/absence in Moniliformopses could not be assessed because of the absence of fully sequenced genomes in this group. Despite the lack of evidence in the Moniliformopses, we proposed the CASP first extracellular loop as a signature for Casparian strips. How the CASP first extracellular loop would mediate Casparian strip deposition is currently under investigation. Extracellular molecular players such as Peroxidase64 and Enhanced Suberin1 have recently been identified in the Casparian strips (Hosmani et al., 2013; Lee et al., 2013): CASP EL1 may function to restrict the action of those players in the area of the cell wall juxtaposing the CSD.
MATERIALS AND METHODS
Bioinformatics
Data Collection
Arabidopsis (Arabidopsis thaliana) CASP1, CASP2, CASP3, CASP4, and CASP5 protein sequences were used as templates to identify related plant protein sequences in the UniProtKB database with the BLAST tool available at www.uniprot.org. For organisms not present in UniProtKB, additional protein sequences have been deduced from nucleic acid sequences through BLAST tools available at http://blast.ncbi.nlm.nih.gov (e.g. Adiantum capillus-veneris, Chlorokybus atmophyticus, Cucumis melo, Cynara cardunculus, Helianthus annuus, Lactuca saligna, Malus domestica, Marchantia polymorpha, Mimulus guttatus, Nicotiana tabacum, Osmunda lancea, Panicum virgatum, Picea glauca, Pinus taeda, Raphanus raphanistrum, Raphanus sativus, Striga hermonthica, Taraxacum kok-saghyz, Theobroma cacao, Triphysaria pusilla, Vigna unguiculata, and Zea mays) and with local BLAST on nucleic sequence sets (e.g. Pteridium aquilinum and Ginkgo biloba). All sequences have been annotated following gold standards of UniProtKB/Swiss-Prot and are publicly available at http://www.uniprot.org/uniprot/?query=family:(Casparian+strip+membrane+proteins+(CASP)+family).
Homology Prediction
We collected 134 proteins with a predicted MARVEL domain from stramenopiles and opistokonts. A multiple sequence alignment (MSA) of CASPL- and MARVEL-domain containing proteins was calculated with MAFFT (version 7; option L-INS-I, gap opening penalty of 1.3, and gap offset value of 0; BLOSUM30; Katoh and Standley, 2014). The region homologous to the CASP domain was selected and realigned. The MSA was inspected and edited with JalViewLite (version 2.6.1; Waterhouse et al., 2009). From this alignment, we obtained subdata sets for plants (CASPL), algae, stramenopiles, and other MARVELs, which were used to scan the PfamA library version 27.0 (Finn et al., 2014) and PDB version 70 (November 30, 2013) databases (Berman et al., 2003) with the profile hidden Markov model-based homology recognition search method HHpred (v) (Hildebrand et al., 2009; Supplemental Table S1).
Phylogenetic Analysis
Two data models were selected from the MSA, taking into account scores calculated with GUIDANCE (version 1.3.1; Penn et al., 2010). In order to keep a maximum number of aligned positions, we constructed a data model under less stringent conditions by masking MSA residues with low confidence. Alignment positions with a high number of gap positions and masked positions were subsequently removed. The second data model was selected manually and consisted only of the four transmembrane domains and the conserved adjacent regions to optimize the analysis for deep nodes of the domain phylogeny. As an outgroup, we retained sequences from stramenopiles and algae; all other MARVEL sequences were removed from the alignment. Finally, we reduced the sequence redundancy to 90%. Gene phylogenies were estimated by maximum likelihood from both data models and by Bayesian inference analysis from the more stringently selected data model. The best fitting model of protein evolution according to ProtTest (version 2.4; Darriba et al., 2011) was determined for the CASPL data set to be JTT+I+G+F under the Akaike information criterion. The maximum likelihood phylogeny was reconstructed with PhyML (version 3.0; Guindon et al., 2009), modeling among-site rate heterogeneity using a discrete γ-distribution of eight categories and calculating nonparametric branch support by applying the approximate likelihood ratio test based on the Shimodaira-Hasegawa-like procedure (Anisimova and Gascuel, 2006). The Bayesian analysis was performed with MrBayes (version 3.2.0; Ronquist et al., 2012) under the same evolutionary model as described above, in addition allowing rate variation across the tree. Two independent runs of four Metropolis-coupled Markov chain Monte Carlo chains were run for 5 million generations, and the potential scale reduction factor approached a value of 1. A sample was taken every 1,000 generations from the MCMC chains after a burn-in phase of 1 million generations. Phylogenetic trees were colorized according to the taxonomic classification of the species and the predicted protein subfamilies using Archaeopteryx (version 0.957 beta; www.phylosoft.org/archaeopteryx).
Sequence Conservation Logos
Sequence logos were constructed from the most stringent data model using the WebLogo3 server (weblogo.threeplusone.com). For CASPL group 1, we reduced the sequence redundancy above 90%, as this group was of special interest in this study and hence included in addition homologs from species other than those selected for the analysis of the superfamily. No redundancies were removed from the CASPL groups 2 to 5 and the MARVEL groups from algae, stramenopiles, fungi, and other opisthokonts.
Sequence analyses were performed on the Vital-IT High Performance Computing Center (www.vital-it.ch), MAFFT (mafft.cbrc.jp/alignment/server/), and GUIDANCE (guidance.tau.ac.il).
Molecular Cloning, PCR Mutagenesis, and Transgenic Lines
Classical cloning was used to generate GUS reporter lines (pCASPL:NLS-GUS), Gateway cloning for the generation of pCASP1:CASP1-mCherry, pLjCASP1:LjCASP1-GFP, and pCASP1:CASP1mutant-GFP, and Cre-Lox-based recombination for the generation of swap lines (pCASP1:mCherry-CASPL). Transgenic lines were generated by floral dipping in the Arabidopsis Columbia-0 background. Point mutations in CASP1 CDS have been generated by site-directed mutagenesis. Additional details are available in Supplemental Table S4.
Staining and Microscopy
Confocal images were taken with a Leica SP/2 confocal microscope. Excitation and detection windows were set as follows: GFP 488 nm, 500 to 600 nm; mCherry 594 nm, 600 to 700 nm; GFP and mCherry 488 and 594 nm, 500 to 550 nm and 600 to 700 nm; GFP and propidium iodide 488 nm, 500 to 550 nm and 600 to 700 nm. Pinhole was set between 1 and 1.5 airy units; gain was adjusted to 600 to 700 V for GFP and propidium iodide and to 800 to 850 V for mCherry. Propidium iodide staining was performed by incubating seedlings in the dark for 10 min in a fresh solution of 15 μm (10 μg mL−1) propidium iodide and then rinsing in water before imaging. For confocal and root GUS staining, seeds were germinated on one-half-strength Murashige and Skoog medium and observed 5 d after imbibition (unless indicated differently in the main text); for flower GUS staining, plants were grown in an 18-h, 22°C/6-h, 18°C day/night cycle. For GUS staining, roots or flowers were stained in 10 mm EDTA, 0.1% (v/v) Triton X-100, 2 mm K4Fe(CN)6, 2 mm K3Fe(CN)6, 50 mm phosphate buffer, pH 7.2, and 4 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid. Incubation time was between 2 and 4 h at 37°C; samples were preincubated 1 h at −20°C in 90% (v/v) acetone, when this treatment increased staining efficiency. After staining, roots were cleared in 0.24 m HCl and 20% (v/v) methanol for 15 min at 57°C, then in 7% (w/v) NaOH and 60% (v/v) ethanol, and then rehydrated according to Malamy and Benfey (1997). For sections, stained samples were fixed in 4% (w/v) paraformaldehyde overnight at 4°C, washed in phosphate-buffered saline, dehydrated in an ethanol series (30%–100% [v/v]), treated with Histoclear (25%–100% [v/v]), included in Paraplast, sectioned with a microtome at 10 to 12 µm, dried overnight at 42°C, and mounted in Merckoglas.
UniProt accession numbers are provided in Supplemental Table S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of representative CASPL and MARVEL domains.
Supplemental Figure S2. Maximum likelihood phylogeny of the CASPL/MARVEL domain.
Supplemental Figure S4. Maximum likelihood phylogeny of the CASPL domain.
Supplemental Figure S5. Magnification of selected images presented in Figure 2.
Supplemental Table S1. Homology prediction with HHpred for CASPL groups and MARVELs.
Supplemental Table S5. CASPLs in Mimulus guttatus and Utricularia Gibba.
Supplemental Table S6. Localization of mCherry-AtCASPLs under the control of AtCASP1 promoter.
Supplemental Data Set S1. Data model 1: CASPL/MARVEL domains.
Supplemental Data Set S3. Maximum likelihood CASPL/MARVEL domain phylogeny (NHX format).
Supplemental Data Set S4. Bayesian inference CASPL phylogeny (NHX format).
Supplemental Data Set S5. Maximum likelihood CASPL phylogeny.
Supplementary Material
Acknowledgments
We thank Cris Kuhlemeier for hosting the group of D.R.
Glossary
- CSD
Casparian strip membrane domain
- MSA
multiple sequence alignment
Footnotes
This work was supported by the Swiss National Science Foundation (to D.R. and N.G.) and the European Research Council (to N.G.). The Swiss-Prot group (B.B., E.B., and I.X.) is part of the Swiss Institute of Bioinformatics, and its activities are supported by the Swiss Federal Government through the State Secretariat for Education, Research, and Innovation.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Alassimone J, Naseer S, Geldner N. (2010) A developmental framework for endodermal differentiation and polarity. Proc Natl Acad Sci USA 107: 5214–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Baldi S, Barral Y. (2012) Bacterial border fence. Cell 151: 1159–1160 [DOI] [PubMed] [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M. (2000) Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell 5: 841–851 [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, et al. (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H. (2003) Announcing the worldwide Protein Data Bank. Nat Struct Biol 10: 980. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2008) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson J, Sozen E, Vizir I, Vav Waeyenberge S, Wilson ZA, Mulligan BJ. (1999) Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol 144: 213–222 [Google Scholar]
- Estornell LH, Agustí J, Merelo P, Talón M, Tadeo FR. (2013) Elucidating mechanisms underlying organ abscission. Plant Sci 199-200: 48–60 [DOI] [PubMed] [Google Scholar]
- Estrada B, Maeland AD, Gisselbrecht SS, Bloor JW, Brown NH, Michelson AM. (2007) The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev Biol 307: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. (1998) Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol 141: 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. (1993) Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol 123: 1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Shahid AA, Zhang L, Liu Y, Ninsuwan U, Roberts JA. (2012) A novel approach to dissect the abscission process in Arabidopsis. Plant Physiol 160: 1342–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M. (2011) Unveiling the Casparian strip. Nature 473: 294–295 [DOI] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. (2009) Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537: 113–137 [DOI] [PubMed] [Google Scholar]
- Hedstrom KL, Ogawa Y, Rasband MN. (2008) AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J Cell Biol 183: 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand A, Remmert M, Biegert A, Söding J. (2009) Fast and accurate automatic structure prediction with HHpred. Proteins (Suppl 9) 77: 128–132 [DOI] [PubMed] [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE. (2013) Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci USA 110: 14498–14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E, Lyons E, Hernández-Guzmán G, Pérez-Torres CA, Carretero-Paulet L, Chang TH, Lan T, Welch AJ, Juárez MJA, Simpson J, et al. (2013) Architecture and evolution of a minute plant genome. Nature 498: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Kinoshita A, Yamada S, Tanaka H, Tanigaki A, Kitano A, Goto M, Okubo K, Nishiyama H, Ogawa O, et al. (2005) Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev Cell 8: 343–352 [DOI] [PubMed] [Google Scholar]
- Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S. (2005) Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 171: 939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Schiebler W, Ouimet C, Greengard P. (1985) A 38,000-dalton membrane protein (p38) present in synaptic vesicles. Proc Natl Acad Sci USA 82: 4137–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2014) MAFFT: iterative refinement and additional methods. Methods Mol Biol 1079: 131–146 [DOI] [PubMed] [Google Scholar]
- Kissel H, Georgescu MM, Larisch S, Manova K, Hunnicutt GR, Steller H. (2005) The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev Cell 8: 353–364 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Storrie B, Simons K, Dotti CG. (1992) A functional barrier to movement of lipids in polarized neurons. Nature 359: 647–650 [DOI] [PubMed] [Google Scholar]
- Kwitny S, Klaus AV, Hunnicutt GR. (2010) The annulus of the mouse sperm tail is required to establish a membrane diffusion barrier that is engaged during the late steps of spermiogenesis. Biol Reprod 82: 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N. (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Leube RE. (1995) The topogenic fate of the polytopic transmembrane proteins, synaptophysin and connexin, is determined by their membrane-spanning domains. J Cell Sci 108: 883–894 [DOI] [PubMed] [Google Scholar]
- Magal LG, Yaffe Y, Shepshelovich J, Aranda JF, de Marco MdC, Gaus K, Alonso MA, Hirschberg K. (2009) Clustering and lateral concentration of raft lipids by the MAL protein. Mol Biol Cell 20: 3751–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Myles DG, Primakoff P, Koppel DE. (1984) A localized surface protein of guinea pig sperm exhibits free diffusion in its domain. J Cell Biol 98: 1905–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada C, Ritchie K, Oba Y, Nakamura M, Hotta Y, Iino R, Kasai RS, Yamaguchi K, Fujiwara T, Kusumi A. (2003) Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat Cell Biol 5: 626–632 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme CL, Cesario MM, Myles DG, Koppel DE, Bartles JR. (1993) Breaching the diffusion barrier that compartmentalizes the transmembrane glycoprotein CE9 to the posterior-tail plasma membrane domain of the rat spermatozoon. J Cell Biol 120: 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Privman E, Landan G, Graur D, Pupko T. (2010) An alignment confidence score capturing robustness to guide tree uncertainty. Mol Biol Evol 27: 1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Edwards D. (2001) Roots: evolutionary origins and biogeochemical significance. J Exp Bot 52: 381–401 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Tendon VD, Pfister A, Alassimone J, Vermeer JEM, Yamazaki M, Stierhof YD, Beeckman T, Geldner N. (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473: 380–383 [DOI] [PubMed] [Google Scholar]
- Saarikangas J, Barral Y. (2011) The emerging functions of septins in metazoans. EMBO Rep 12: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. (2002) MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci 27: 599–601 [DOI] [PubMed] [Google Scholar]
- Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, et al. (2012) General protein diffusion barriers create compartments within bacterial cells. Cell 151: 1270–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotzik JM, Sie JM, Politi C, Del Turco D, Bennett V, Deller T, Schultz C. (2009) AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc Natl Acad Sci USA 106: 17564–17569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed E, Rodrigues NTL, Balda MS, Matter K. (2009) Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. (2000) Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290: 341–344 [DOI] [PubMed] [Google Scholar]
- van Meer G, Simons K. (1986) The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J 5: 1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JEM, von Wangenheim D, Barberon M, Lee Y, Stelzer EHK, Maizel A, Geldner N. (2014) A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183 [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, Claude W, Das M, Fernández-Aparicio M, Honaas A, Timko MP, Wafula EK, Wickett NJ, Yoder JI, Honaas LA. (2012) The Parasitic Plant Genome Project: new tools for understanding the biology of Orobanche and Striga. Weed Sci 60: 295–306 [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. (2010) The evolution of parasitism in plants. Trends Plant Sci 15: 227–235 [DOI] [PubMed] [Google Scholar]
- Wiedenmann B, Franke WW. (1985) Identification and localization of synaptophysin, an integral membrane glycoprotein of Mr 38,000 characteristic of presynaptic vesicles. Cell 41: 1017–1028 [DOI] [PubMed] [Google Scholar]
- Winckler B, Forscher P, Mellman I. (1999) A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature 397: 698–701 [DOI] [PubMed] [Google Scholar]
- Yaffe Y, Shepshelovitch J, Nevo-Yassaf I, Yeheskel A, Shmerling H, Kwiatek JM, Gaus K, Pasmanik-Chor M, Hirschberg K. (2012) The MARVEL transmembrane motif of occludin mediates oligomerization and targeting to the basolateral surface in epithelia. J Cell Sci 125: 3545–3556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.