A Ca2+-dependent protein kinase not only phosphorylates a substrate but also acts as a scaffold that promotes the interaction between a phosphorylation product and its binding protein.
Abstract
A molecular mechanism to ensure signaling specificity is a scaffold. REPRESSION OF SHOOT GROWTH (RSG) is a tobacco (Nicotiana tabacum) transcription factor that is involved in gibberellin feedback regulation. The 14-3-3 proteins negatively regulate RSG by sequestering it in the cytoplasm in response to gibberellins. The N. tabacum Ca2+-dependent protein kinase NtCDPK1 was identified as an RSG kinase that promotes 14-3-3 binding of RSG by phosphorylation of RSG. CDPKs are unique sensor responders of Ca2+ that are only found in plants and some protozoans. Here, we report a scaffolding function of CDPK. 14-3-3 proteins bound to NtCDPK1 by a new mode. Autophosphorylation of NtCDPK1 was necessary for the formation of the binding between NtCDPK1 and 14-3-3 but not for its maintenance. NtCDPK1 formed a heterotrimer with RSG and 14-3-3. Furthermore, we found that NtCDPK1 transfers 14-3-3 to RSG after phosphorylation of RSG and that RSG dissociates from NtCDPK1 as a complex with 14-3-3. These results suggest that NtCDPK1 is an interesting scaffolding kinase that increases the specificity and efficiency of signaling by coupling catalysis with scaffolding on the same protein.
A basic property of living systems is the ability to respond to extracellular signals by intracellular signaling, which leads to changes in gene expression and cellular activities. Signaling pathways are complex networks of biochemical reactions, such as protein-protein interactions and various covalent modifications. Intensive studies have revealed the principal components of signaling, including kinases, transcription factors, and adaptors. They are frequently shared by functionally diverse distinct signaling pathways, providing a potential for unwanted cross talk. Thus, a central question of signaling in recent years is how the system of signal transduction is able to specifically process external and internal stimuli. A molecular mechanism to ensure signaling specificity is a molecular scaffold. Scaffold proteins function as wiring elements, which direct the flow of signaling information by tethering signal components into complexes and localizing them to specific sites in the cell. Scaffolds are thought both to enhance interactions between the correct signaling components so that the signals can be relayed with precision, speed, and efficiency and to insulate them from interactions with competing proteins (Good et al., 2011).
REPRESSION OF SHOOT GROWTH (RSG) is a tobacco (Nicotiana tabacum) transcriptional activator with a basic Leu zipper domain. RSG regulates the transcription of GA biosynthetic genes (Fukazawa et al., 2000). RSG plays a role in the homeostasis of GAs through direct binding to the promoter of NtGA20ox1 encoding a GA 20-oxidase (Fukazawa et al., 2010). The 14-3-3 proteins D5, D31, and D75 were identified as RSG-binding proteins and exhibited higher similarity to ω-, ω-, and μ-types of 14-3-3 proteins in Arabidopsis (Arabidopsis thaliana), respectively (Igarashi et al., 2001). The function of RSG is negatively regulated by 14-3-3 signaling proteins, which form a highly conserved family of homodimeric and heterodimeric proteins in eukaryotes (for review, see van Hemert et al., 2001; Tzivion and Avruch, 2002). The dimeric 14-3-3 proteins bind to phosphorylated motifs containing phosphoserine residues of RSXpSXP and RXY/FXpSXP (where pS indicates a critical phosphoserine) in their target proteins (Yaffe et al., 1997). Through these binding reactions, the 14-3-3 proteins appear to act as molecular scaffolds or chaperones. The biological roles of 14-3-3 proteins have been demonstrated in signal transduction, development, and cell cycle control by the promotion of protein-protein interaction, subcellular targeting, and protection of phosphorylation sites from phosphatases (Bridges and Moorhead, 2005). The 14-3-3 proteins can also act as allosteric cofactors modulating the catalytic activity of their binding partners. The 14-3-3 proteins bind to RSG depending on the RSG phosphorylation of Ser-114 and thereby sequester RSG in the cytoplasm so that it is unable to regulate its target genes in the nucleus (Igarashi et al., 2001; Ishida et al., 2004). We found that GA levels regulate the intracellular localization of RSG; that is, RSG is translocated into the nucleus in response to a reduction in GA levels, and GA treatment could reverse this nuclear accumulation (Ishida et al., 2004). The GA-dependent cytoplasmic migration of RSG requires 14-3-3 binding and Ser/Thr kinase activity (Ishida et al., 2004). We identified a Ca2+-dependent protein kinase, N. tabacum (Nt)CDPK1, as an RSG kinase that promotes 14-3-3 binding of RSG by phosphorylation of Ser-114 in RSG (Ishida et al., 2008). NtCDPK1 interacts with RSG in a Ca2+-dependent manner in vivo and in vitro and specifically phosphorylates Ser-114 in RSG in response to GAs. NtCDPK1 decodes the Ca2+ signal produced by GAs and regulates the intracellular localization of RSG.
CDPKs are unique Ca2+ decoders that are found only in plants and some protozoans. They are sensor responders that have both a Ca2+-sensing function and kinase activity within one protein. Among Ca2+-binding sensory proteins in plants, CDPKs are thought to play central roles in Ca2+ signaling because protein kinase C and conventional Ca2+/calmodulin-dependent protein kinase (CaMK), which represent the two major types of Ca2+-regulated kinases in animal systems, are missing from Arabidopsis (Hrabak et al., 2003). There are 34 genes encoding CDPKs in Arabidopsis (Arabidopsis Genome Initiative, 2000) and 29 genes in rice (Oryza sativa; Asano et al., 2005). CDPKs have been shown to play important roles in various physiological processes, including plant growth and development and abiotic and biotic stress responses in plants (Harper et al., 1991; Suen and Choi, 1991; Cheng et al., 2002; Harper and Harmon, 2005; Takahashi and Ito, 2011). CDPKs are Ser/Thr protein kinases that are composed of a variable N-terminal domain, a catalytic domain, a junction domain containing an autoinhibitory segment, and a calmodulin-like domain (Harper et al., 2004). Although amino acid sequences of a catalytic domain, a junction domain, and a calmodulin-like domain are highly conserved among CDPK isoforms, the variable N-terminal domain of CDPKs is diversified not only in amino acid sequence but also in length, ranging from 25 to 180 amino acids in Arabidopsis. Little was known about the functions of the variable N-terminal domain of CDPKs; however, our study on NtCDPK1 has shown that the variable N-terminal domain is involved in substrate recognition (Ito et al., 2010). The catalytic domain exhibits the classic bilobed architecture, with the N-terminal β-lobe and the C-terminal helical lobe sandwiching the catalytic active site (Wernimont et al., 2011). Recently, the structures of parasitic CDPK in both autoinhibited and activated (i.e. Ca2+-bound) conformations have been solved (Wernimont et al., 2010, 2011). Those authors showed that all the regions downstream of the catalytic domain (i.e. the junction domain and the calmodulin-like domain) work together for activation, and so they termed the entire C-terminal region the CDPK activation domain. Ca2+ binding triggers the translocation of the entire CDPK activation domain to a new position roughly 135° clockwise from the substrate-binding site. This movement makes the important catalytic residue available to interact with ATP, allows the activation loop to assume the active orientation, and removes occlusion of the autoinhibitory segment from the substrate-binding state. Because residues involved in making the conformational change are mostly conserved between parasitic CDPKs and plant CDPKs, the overall mechanism of the activation of CDPKs may be similar (Wernimont et al., 2011). Autophosphorylation is a key event in the activation of many protein kinases (Giese et al., 1998; Lochhead et al., 2005). For example, cAMP-dependent protein kinase A is activated by phosphorylation at Thr-197 in the activation loop (Johnson et al., 1996). The equivalent position in CDPKs is Asp or Glu that mimics a phosphoactivated state, suggesting that CDPKs exhibit full activities without phosphorylation in the activation loop. Although phosphorylation has been observed in many CDPKs, a physiological role of phosphorylation has not been demonstrated (Takahashi and Ito, 2011).
The molecular basis by which commonly used signaling components are able to elicit stimulus-specific responses in cells has become an important topic in recent years. Both 14-3-3 and CDPK form large protein families in plants and are involved in diverse physiological processes, including development, hormone signaling, and stress responses (Gökirmak et al., 2010; Takahashi and Ito, 2011). How they can preserve the specificity of a signal in order to ultimately elicit distinct, specific responses is still poorly understood. In this study, we found a new function for CDPK. NtCDPK1 interacts with 14-3-3 in an autophosphorylation- and Ca2+-dependent manner. 14-3-3 is transferred from NtCDPK1 to the immediate product, phosphorylated RSG. Our results suggest that NtCDPK1 not only phosphorylates RSG but also functions as a scaffold that promotes the interaction between 14-3-3 and RSG, which ensures the fidelity of signaling.
RESULTS
14-3-3 Interacts with Autophosphorylated NtCDPK1 in the Presence of Ca2+
14-3-3 proteins generally bind to phosphorylated consensus motifs, RSXpSXP and RXY/FXpSXP, in target proteins (Bridges and Moorhead, 2005). 14-3-3 has been reported to interact with an Arabidopsis CDPK, AtCPK1 (Camoni et al., 1998). AtCPK1 was predicted to interact with 14-3-3 by the conserved 14-3-3-binding motif in the variable N-terminal domain of AtCPK1 in an autophosphorylation-dependent manner (Camoni et al., 1998). AtCPK1 and NtCDPK1 belong to subgroups I and II of the CDPK family, respectively (Boudsocq and Sheen, 2013). Although the variable N-terminal domains of NtCDPK1 showed low similarity to that of AtCPK1 at the amino acid sequence level (Ito et al., 2010) and the typical 14-3-3-binding motif was not found in NtCDPK1, we examined whether NtCDPK1 interacts with 14-3-3. NtCDPK1 has been reported to be autophosphorylated in a Ca2+-dependent manner (Yoon et al., 1999; Supplemental Fig. S1). Glutathione S-transferase (GST)-NtCDPK1 was autophosphorylated and absorbed into glutathione beads. The immobilized GST-NtCDPK1 was incubated with His-tagged 14-3-3 (His-14-3-3), followed by affinity chromatography. Nonphosphorylated GST-NtCDPK1 interacted slightly with His-14-3-3 in the presence of Ca2+; however, autophosphorylation of GST-NtCDPK1 significantly increased its binding affinity to His-14-3-3 (Fig. 1A). Because no conserved consensus 14-3-3-binding motif was found in NtCDPK1, 14-3-3 might interact with NtCDPK1 through a previously unidentified 14-3-3-binding sequence.
Figure 1.
14-3-3 interacts with NtCDPK1. A, 14-3-3 interacts with NtCDPK1 in an autophosphorylation- and Ca2+-dependent manner. GST and nonphosphorylated or autophosphorylated GST-NtCDPK1 were immobilized on glutathione beads and incubated with His-14-3-3 in the presence of Ca2+ or EGTA (EG). NtCDPK1-bound proteins were subjected to SDS-PAGE, followed by immunoblot analysis with anti-14-3-3 antibody for the detection of His-14-3-3. GST fusion proteins were visualized by Coomassie Brilliant Blue (CBB) staining. The values below the top gel indicate the relative strength levels of signals after standardization using the signals of Coomassie Brilliant Blue staining of GST or GST-NtCDPK1 as a loading control. The value of GST as a negative control was set to 1. This experiment was repeated three times with similar results. B, Statistical analysis of the data shown in A. The bar graph represents means and sd (n = 3). A two-way ANOVA was used to separate the effects of Ca2+, autophosphorylation, and their combination. Ca2+ main effect (P < 0.05), autophosphorylation main effect (P < 0.05), and Ca2+ × autophosphorylation interaction effect (P < 0.05) were detected. Different letters above the bars indicate significant differences among the treatments (P < 0.001, Tukey’s honestly significant difference test). C, BiFC analysis reveals in vivo interaction between NtCDPK1 and 14-3-3. BiFC constructs were delivered into leaf cells of tobacco by particle bombardment. After 24 h, the cells were visualized by epifluorescence microscopy. Coexpression of YFPN-14-3-3 and NtCDPK1-YFPC (top) and coexpression of YFPN-14-3-3 and YFPC (bottom) are shown. Reconstituted YFP fluorescence (left), RFP fluorescence as a control for transfection efficiency (center), and bright-field images (right) are shown. This experiment was repeated three times with similar results. Bars = 50 µm. D, Coimmunoprecipitation (Co-IP) assay also reveals the in vivo interaction between NtCDPK1 and 14-3-3. Leaf cell extracts from a transgenic plant overexpressing NtCDPK1-GFP were immunoprecipitated with anti-NtCDPK1 or anti-His antibody. Anti-His antibody was used as a negative control for immunoprecipitation (IP). Coprecipitated 14-3-3 with NtCDPK1-GFP was detected by immunoblot analysis with anti-14-3-3 antibody. NtCDPK1-GFP was detected by anti-NtCDPK1 antibody. [See online article for color version of this figure.]
NtCDPK1 interacts with its substrate RSG in a Ca2+-dependent manner (Ishida et al., 2008); therefore, we examined whether NtCDPK1 also interacts with 14-3-3 in a Ca2+-dependent manner. Autophosphorylated GST-NtCDPK1 interacts with His-14-3-3 in the presence of Ca2+ but not EGTA (Fig. 1A), showing that 14-3-3 interacts with NtCDPK1 in a Ca2+-dependent manner even if NtCDPK1 is autophosphorylated. These results suggested that the interaction between 14-3-3 and NtCDPK1 requires not only the autophosphorylation of NtCDPK1 but also the Ca2+-induced conformational change of NtCDPK1. Statistical analysis also demonstrated that both Ca2+ and autophosphorylation are required for 14-3-3 binding of NtCDPK1 (Fig. 1B).
A bimolecular fluorescence complementation (BiFC) assay (Hu et al., 2002) was used to examine the interaction between NtCDPK1 and 14-3-3 in plant cells. 14-3-3 was translationally fused to the N-terminal portion of yellow fluorescent protein (YFPN), which generated YFPN-14-3-3. For the other partner, NtCDPK1 was translationally fused to the C-terminal portion of yellow fluorescent protein (YFPC), which generated the NtCDPK1-YFPC fusion protein. The corresponding constructs were codelivered into leaf cells of tobacco by particle bombardment. The reconstituted YFP signal, caused by the interaction between YFPN-14-3-3 and NtCDPK1-YFPC, was observed in leaf epidermal cells (Fig. 1C, top row). Control experiments in which YFPN-14-3-3 was coexpressed with unfused YFPC did not show any fluorescence (Fig. 1C, bottom row). These results indicated that NtCDPK1 interacts with 14-3-3 in living cells.
Additionally, we confirmed the interaction between NtCDPK1 and 14-3-3 by coimmunoprecipitation assay. To this end, transgenic tobacco expressing NtCDPK1-GFP was used (Ishida et al., 2008). NtCDPK1-GFP protein was immunoprecipitated with anti-NtCDPK1 antibodies, and NtCDPK1-bound endogenous 14-3-3 proteins were immunoblotted with anti-14-3-3 antibodies. Anti-His antibodies were used as a negative control for immunoprecipitation. As shown in Figure 1D, 14-3-3 proteins were coimmunoprecipitated with NtCDPK1. These results confirmed that NtCDPK1 interacts with 14-3-3 in plant cells.
14-3-3 Binds to the Catalytic Domain of NtCDPK1
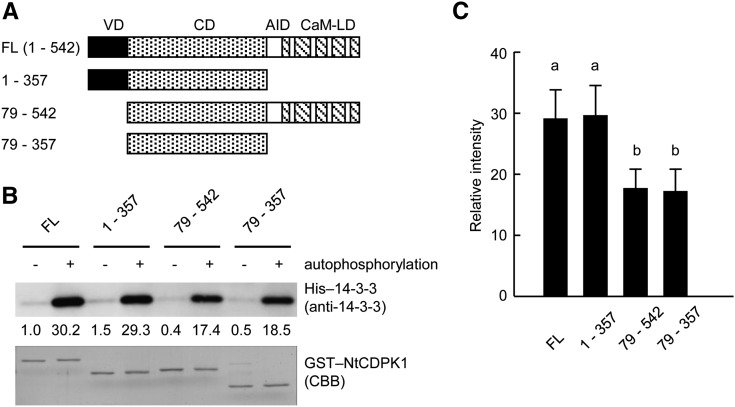
To determine which region of NtCDPK1 is important for the interaction with 14-3-3, we performed an in vitro pull-down assay using various deletion mutants of NtCDPK1 (Fig. 2A). Deletion mutants were autophosphorylated and used for an in vitro pull-down assay. As shown in Figure 2, B and C, deletion of the autoinhibitory domain and the calmodulin-like domain (1–357) did not affect 14-3-3 binding. Deletion of the variable N-terminal domain (79–542) slightly affected 14-3-3 binding. The binding ability of the catalytic domain (79–357) to 14-3-3 was comparable to that of the deletion mutant of the variable N-terminal domain (79–542). These results indicated that 14-3-3 binds to the catalytic domain of NtCDPK1 in an autophosphorylation-dependent manner and that the variable N-terminal domain of NtCDPK1 contributes slightly to the 14-3-3 binding.
Figure 2.
14-3-3 interacts with the catalytic domain of NtCDPK1. A, Schematic diagram of NtCDPK1 constructs used in the pull-down assay. The numbers indicate the position of each amino acid residue. VD, Variable N-terminal domain; CD, catalytic domain; AID, autoinhibitory domain; CaM-LD, calmodulin-like domain. B, The catalytic domain of NtCDPK1 is sufficient for binding to 14-3-3. Nonphosphorylated or various autophosphorylated GST-NtCDPK1s were immobilized on glutathione beads and incubated with His-14-3-3 in the presence of Ca2+. NtCDPK1-bound proteins were subjected to SDS-PAGE, followed by immunoblot analysis with anti-14-3-3 antibody for the detection of His-14-3-3. GST-NtCDPK1s were visualized by Coomassie Brilliant Blue (CBB) staining. The values below the top gel indicate the relative strength levels of signals after standardization using the signals of Coomassie Brilliant Blue staining of GST-NtCDPK1s as a loading control. The value of nonphosphorylated GST-NtCDPK1 (full length [FL]) as a negative control was set to 1. This experiment was repeated three times with similar results. C, Statistical analysis of the data shown in B. The bar graph represents means and sd (n = 3). To determine significant differences of 14-3-3 binding affinity among GST-NtCDPK1s, the values of autophosphorylated GST-NtCDPK1s were used. Different letters above the bars indicate significant differences of 14-3-3 binding affinity among GST-NtCDPK1s (P < 0.001, one-way ANOVA with Tukey’s honestly significant difference test).
NtCDPK1 Forms a Heterotrimer with RSG and 14-3-3
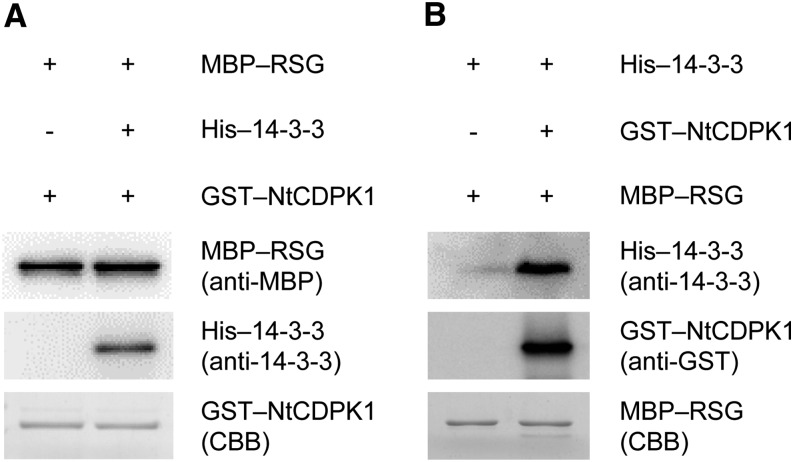
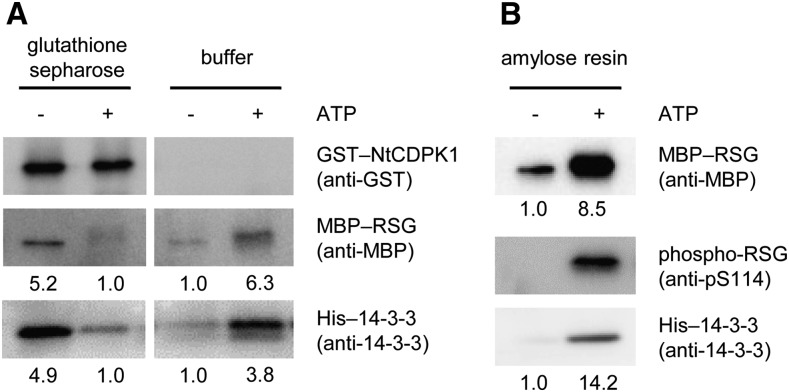
We previously found that RSG binds to the variable N-terminal domain of NtCDPK1 (Ito et al., 2010). In this study, we found that 14-3-3 bound to the catalytic domain of NtCDPK1 (Fig. 2). Therefore, we hypothesized that NtCDPK1 might form a heterotrimer with RSG and 14-3-3. To examine this hypothesis, GST-NtCDPK1 was autophosphorylated and absorbed into glutathione beads. The immobilized GST-NtCDPK1 was incubated with His-14-3-3. Removal of ATP is required for the detection of binding between NtCDPK1 and RSG, because RSG appears to dissociate from NtCDPK1 after RSG is phosphorylated by NtCDPK1. Glutathione beads into which GST-NtCDPK1 and His-14-3-3 had been absorbed were washed with a pull-down buffer to remove ATP and incubated with maltose-binding protein (MBP)-RSG, followed by affinity chromatography. As shown in Figure 3A, binding of His-14-3-3 to GST-NtCDPK1 did not affect the interaction between MBP-RSG and GST-NtCDPK1, suggesting that NtCDPK1 can form a heterotrimer with RSG and 14-3-3. However, we cannot rule out the possibility that the result was merely the sum of independent interactions between GST-NtCDPK1 and His-14-3-3 and between GST-NtCDPK1 and MBP-RSG. To solve this issue, we performed an alternative pull-down assay based on the fact that nonphosphorylated RSG does not directly interact with 14-3-3 (Ishida et al., 2004). Nonphosphorylated MBP-RSG is absorbed into amylose resin and incubated with autophosphorylated GST-NtCDPK1 and His-14-3-3. The assay showed that His-14-3-3 associates with nonphosphorylated MBP-RSG in the presence of autophosphorylated GST-NtCDPK1 but not in its absence (Fig. 3B), showing that nonphosphorylated MBP-RSG interacts indirectly with His-14-3-3 via autophosphorylated GST-NtCDPK1. These results suggested that NtCDPK1 forms a heterotrimer with RSG and 14-3-3.
Figure 3.
NtCDPK1 forms a heterotrimer with RSG and 14-3-3. A, RSG binds to the complex of NtCDPK1 with 14-3-3. GST-NtCDPK1 was autophosphorylated and absorbed into glutathione beads. The immobilized GST-NtCDPK1 was incubated with His-14-3-3. Glutathione beads into which GST-NtCDPK1 and His-14-3-3 had been absorbed were washed with a pull-down buffer to remove ATP and incubated with MBP-RSG. NtCDPK1-bound proteins were subjected to SDS-PAGE, followed by immunoblot analysis with anti-14-3-3 and anti-MBP antibodies for the detection of His-14-3-3 and MBP-RSG, respectively. GST-NtCDPK1 was visualized by Coomassie Brilliant Blue (CBB) staining. This experiment was repeated three times with similar results. B, 14-3-3 interacts with nonphosphorylated RSG via autophosphorylated NtCDPK1. Nonphosphorylated MBP-RSG was absorbed into amylose resin and incubated with autophosphorylated GST-NtCDPK1 and His-14-3-3. RSG-bound proteins were subjected to SDS-PAGE, followed by immunoblot analysis with anti-GST and anti-14-3-3 antibodies for the detection of GST-NtCDPK1 and His-14-3-3, respectively. MBP-RSG was visualized by Coomassie Brilliant Blue staining. This experiment was repeated three times with similar results. In Supplemental Figure S2, MBP-RSG and His-14-3-3 were shown not to bind directly to glutathione beads and amylose resin, respectively.
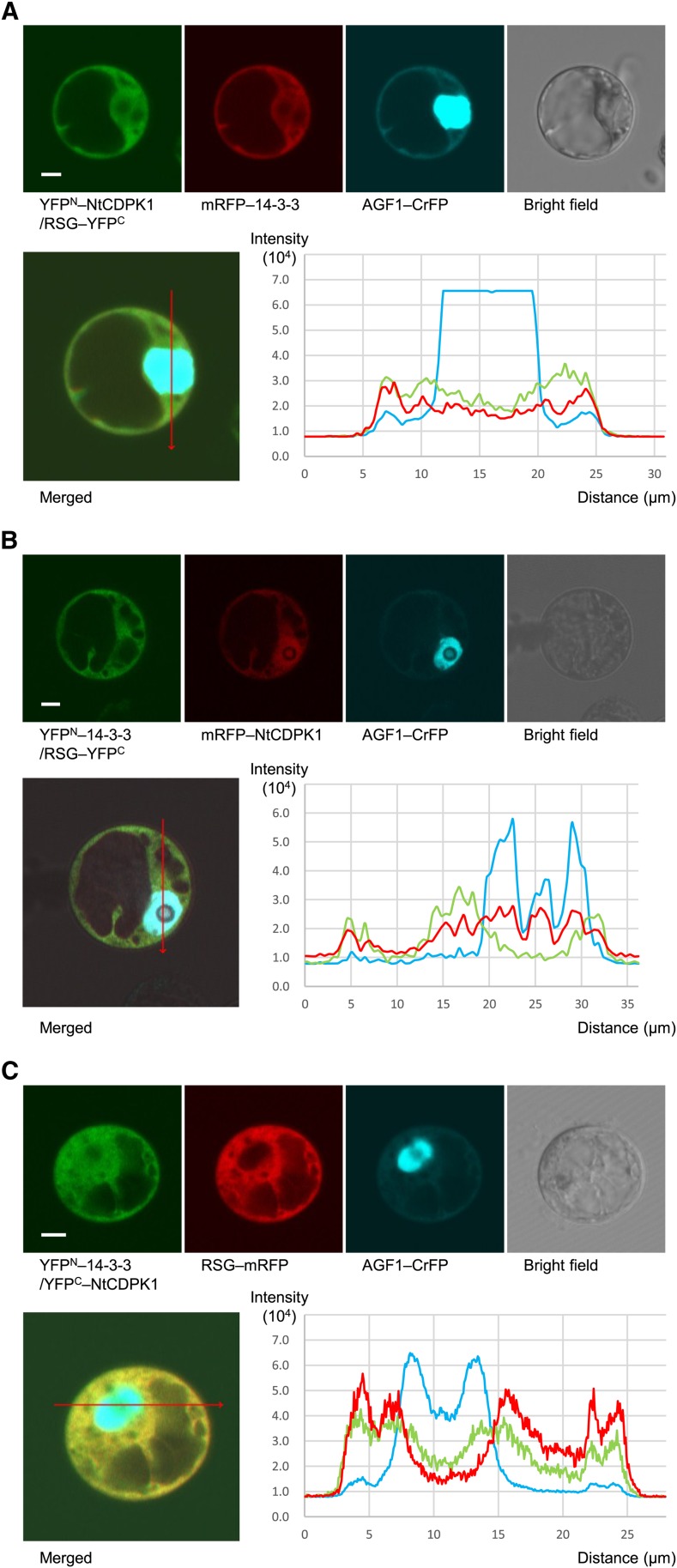
Since NtCDPK1 formed a heterotrimer with RSG and 14-3-3 in vitro, we examined the intracellular colocalization of NtCDPK1, RSG, and 14-3-3. We cotransfected constructs expressing YFPN-NtCDPK1, RSG-YFPC, monomeric red fluorescent protein (mRFP)-14-3-3, and AT-hook protein of GA feedback regulation1 (AGF1)-cerulean fluorescent protein (CrFP) into Arabidopsis cultured cell line T87. AGF1-CrFP was used as a control for nuclear localization (Matsushita et al., 2007). After 24 h, the cells were visualized by confocal laser scanning microscopy and fluorescence intensities were quantified (Fig. 4A). The reconstituted YFP signal indicated the interaction between NtCDPK1 and RSG. Fluorescence intensities revealed the overlapping patterns of YFP and mRFP signals in cytoplasm. This result suggested that 14-3-3 colocalizes with the NtCDPK1-RSG complex in cytoplasm. Similarly, Figure 4, B and C, suggested that NtCDPK1 colocalizes with the 14-3-3-RSG complex in cytoplasm and that RSG colocalizes with the 14-3-3-NtCDPK1 complex in cytoplasm, respectively. Taking these results all together, NtCDPK1 appears to coexist with RSG and 14-3-3, and NtCDPK1 could form a ternary complex with RSG and 14-3-3 in living cells.
Figure 4.
NtCDPK1 largely colocalizes with RSG and 14-3-3. Constructs expressing fluorescent protein fusion proteins were cotransfected into Arabidopsis T87 protoplasts. AGF1-CrFP was used as a control for nuclear localization. After 24 h, the cells were visualized by confocal laser scanning microscopy. Fluorescence intensities were quantified by ZEN 2009 software (Carl Zeiss). Distance in the graph of fluorescence intensity corresponds to the position of the arrow (from nock to head) in the merged image. This experiment was repeated three times with similar results. Bars = 5 µm. A, 14-3-3 colocalizes with the NtCDPK1-RSG complex in the cytoplasm. Shown are the reconstituted YFP fluorescence resulting from the interaction between YFPN-NtCDPK1 and RSG-YFPC (top left), the mRFP fluorescence of mRFP-14-3-3 (top second from left), the CrFP fluorescence of AGF1-CrFP (top second from right), the bright-field image (top right), the merged image (bottom left), and the quantified fluorescence intensities (bottom right). Pinhole settings are 2.78 airy units (AU) for YFP, 2.43 AU for mRFP, and 2.86 AU for CrFP. B, NtCDPK1 colocalizes with the 14-3-3-RSG complex in the cytoplasm. Shown are the reconstituted YFP fluorescence resulting from the interaction between YFPN-14-3-3 and RSG-YFPC (top left), the mRFP fluorescence of mRFP-NtCDPK1 (top second from left), the CrFP fluorescence of AGF1-CrFP (top second from right), the bright-field image (top right), the merged image (bottom left), and the quantified fluorescence intensities (bottom right). Pinhole settings are 1 AU for YFP, 1 AU for mRFP, and 1 AU for CrFP. C, RSG colocalizes with the 14-3-3-NtCDPK1 complex in the cytoplasm. Shown are the reconstituted YFP fluorescence resulting from the interaction between YFPN-14-3-3 and YFPC-NtCDPK1 (top left), the mRFP fluorescence of RSG-mRFP (top second from left), the CrFP fluorescence of AGF1-CrFP (top second from right), the bright-field image (top right), the merged image (bottom left), and the quantified fluorescence intensities (bottom right). Pinhole settings are 1.48 AU for YFP, 1.43 AU for mRFP, and 1.25 AU for CrFP. [See online article for color version of this figure.]
14-3-3 Has No Effect on the Kinase Activity of NtCDPK1
In human cells, 14-3-3 is phosphorylated by c-Jun N-terminal kinase (JNK) (Tsuruta et al., 2004). Bax is one of the target proteins of 14-3-3. Phosphorylation of 14-3-3s by JNK results in the dissociation of 14-3-3 from Bax. We showed that 14-3-3 binds to the catalytic domain of NtCDPK1 (Fig. 2), suggesting that 14-3-3 could be a substrate of NtCDPK1 as well as RSG. To examine this possibility, we performed an in vitro kinase assay. NtCDPK1 did not phosphorylate 14-3-3 (Supplemental Fig. S3). This result suggested that 14-3-3 is not the substrate of NtCDPK1, despite its binding to the catalytic domain of NtCDPK1.
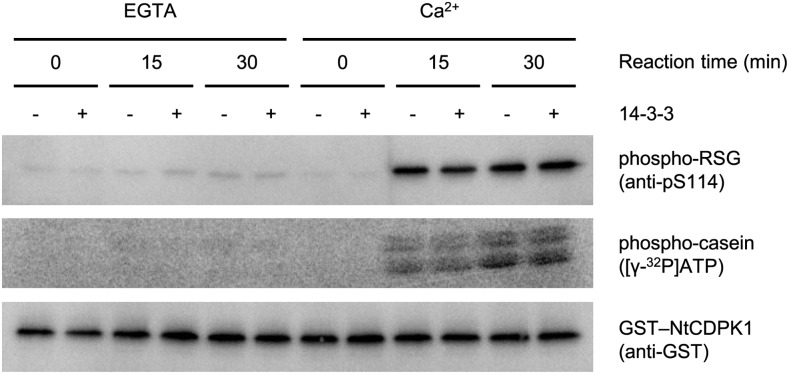
Bridges and Moorhead (2005) reported that 14-3-3 affects the enzyme activity of several proteins. We investigated whether the binding of 14-3-3 to NtCDPK1 affects the catalytic activity of NtCDPK1. We performed an in vitro kinase assay using full-length RSG as a substrate and antibodies that specifically recognize phosphor-Ser-114 of RSG (Ishida et al., 2004). The kinase activity of GST-NtCDPK1 with His-14-3-3 was comparable to that of GST-NtCDPK1 without His-14-3-3 (Fig. 5). This result indicated that 14-3-3 binding to NtCDPK1 does not affect the catalytic activity of NtCDPK1.
Figure 5.
14-3-3 has no effect on the kinase activity of NtCDPK1. Autophosphorylated GST-NtCDPK1 was immobilized on glutathione beads and incubated with or without His-14-3-3 in the presence of Ca2+. Glutathione beads into which GST-NtCDPK1 and His-14-3-3 had been absorbed were washed with a pull-down buffer to remove ATP and His-14-3-3 that was not bound to NtCDPK1. An in vitro phosphorylation assay was performed by incubating MBP-RSG and glutathione beads into which GST-NtCDPK1 and His-14-3-3 had been absorbed in a reaction mixture for the indicated periods of time in the presence of Ca2+ or EGTA. Aliquots of the reactions were subjected to SDS-PAGE, followed by immunoblot analysis with anti-GST and anti-pS114 (which specifically recognizes phosphorylated Ser-114 of RSG) antibodies. The enzymatic activity of recombinant NtCDPK1 was also confirmed by an in vitro phosphorylation assay on the general substrate casein using [γ-32P]ATP. This experiment was repeated three times with similar results.
The kinase domain of NtCDPK1 is up to 75% similar to that among mammalian Ca2+/CaMKII at the amino acid sequence level (Takahashi and Ito, 2011). CaMKII undergoes intersubunit autophosphorylation at Thr-287 (or Thr-286; specific numbering is isoform dependent), resulting in Ca2+/calmodulin-independent activity (Hudmon and Schulman, 2002). Thr-287 lies within the autoinhibitory segment of CaMKII, and autophosphorylation at Thr-287 produces Ca2+-autonomous activity by preventing reassociation of the catalytic domain and the autoinhibitory segment. Although the autoinhibitory segment of CDPKs shows similarity to that of CaMKII, Thr/Ser is missing at the corresponding position in CDPKs, suggesting the lack of a Ca2+-independent active state by autophosphorylation in the autoinhibitory segment of CDPKs. In fact, autophosphorylated NtCDPK1 did not phosphorylate RSG in the absence of Ca2+ (Supplemental Fig. S4).
A Ca2+-induced large conformational change of CDPK removes occlusion of the autoinhibitory segment from the substrate-binding domain (Wernimont et al., 2010, 2011). We supposed that the 14-3-3 binding to the catalytic domain of NtCDPK1 could confer the Ca2+-independent kinase activity by inhibiting reassociation of the catalytic domain with the autoinhibitory segment. As a prerequisite for this assumption, 14-3-3 must continue to interact with NtCDPK1 after the removal of Ca2+. The pull-down buffer containing Ca2+ was replaced with a new pull-down buffer containing EGTA after His-14-3-3 was bound to GST-NtCDPK1. The interaction of 14-3-3 with NtCDPK1 was maintained in the absence of Ca2+ (Supplemental Fig. S5), suggesting that Ca2+ is necessary for the formation of the binding between NtCDPK1 and 14-3-3 but not for its maintenance. Next, we examined the kinase activity of 14-3-3-bound NtCDPK1 in the presence of EGTA. The kinase assay showed that a complex of GST-NtCDPK1 with 14-3-3 does not phosphorylate MBP-RSG in the absence of Ca2+ (Fig. 5). These results suggested that 14-3-3 binding to NtCDPK1 does not affect the kinase activity and the dependency on Ca2+.
14-3-3 Binds to NtCDPK1 by a New Mode
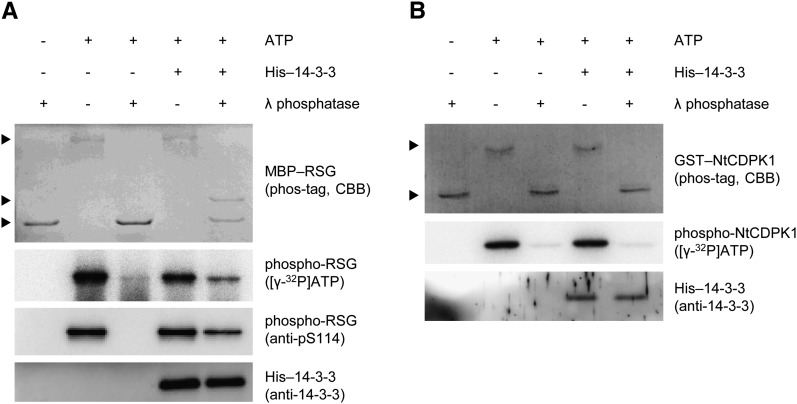
One of the physiological functions of 14-3-3 is to bind and protect phosphate groups of the target proteins against phosphatases (Bridges and Moorhead, 2005). To examine whether 14-3-3 protects phosphate groups of RSG or NtCDPK1, we performed an in vitro dephosphorylation assay. To reveal the phosphorylation state of proteins, we used phosphate-binding tag (Phos-tag) SDS-PAGE, which visualizes phosphorylated proteins as slower migration bands (Kinoshita et al., 2006). We initially examined whether 14-3-3 protects phosphate groups of RSG against λ-phosphatase. 14-3-3 binds to RSG through a conserved phosphorylated motif in RSG (Ishida et al., 2004). MBP-RSG was phosphorylated by GST-NtCDPK1 and was subjected to Phos-tag SDS-PAGE, showing that several amino acids of RSG are phosphorylated by NtCDPK1 (Fig. 6A). Treatment with λ-phosphatase could reverse the mobility shifts of phosphorylated MBP-RSG that did not bind to His-14-3-3, showing that phosphate groups of RSG are completely removed by λ-phosphatase when 14-3-3 does not bind to RSG. On the other hand, when MBP-RSG that bound to His-14-3-3 was treated with λ-phosphatase, one phosphorylated form of MBP-RSG was protected from λ-phosphatase. This result suggested that 14-3-3 directly binds to phosphor-Ser-114 of RSG (a phosphorylation site in RSG).
Figure 6.
The 14-3-3 binding mode of NtCDPK1 differs from that of RSG. A, The phosphorylation of Ser-114 in RSG is protected from λ-phosphatase by 14-3-3. MBP-RSG was phosphorylated by GST-NtCDPK1 with or without ATP. MBP-RSG was immobilized on amylose resin and incubated with or without His-14-3-3. Amylose resin was washed and treated with or without λ-phosphatase. The phosphorylation state of MBP-RSG was examined using Phos-tag SDS-PAGE and [γ-32P]ATP. Phos-tag SDS-PAGE was visualized by Coomassie Brilliant Blue (CBB) staining. The phosphorylation of Ser-114 in RSG was detected by immunoblot analysis using anti-pS114 antibody, which specifically recognizes phosphorylated Ser-114 of RSG. Top, middle, and bottom arrowheads represent multiply phosphorylated, Ser-114 phosphorylated, and dephosphorylated MBP-RSG, respectively. RSG-bound His-14-3-3 was detected by immunoblot analysis with anti-14-3-3 antibody. This experiment was repeated three times with similar results. B, NtCDPK1 is dephosphorylated by λ-phosphatase even if 14-3-3 is bound to NtCDPK1. GST-NtCDPK1 was autophosphorylated with or without ATP, immobilized on glutathione beads, and incubated with or without His-14-3-3. The phosphorylation state of GST-NtCDPK1 was examined using Phos-tag SDS-PAGE and [γ-32P]ATP. Top and bottom arrowheads represent autophosphorylated and dephosphorylated GST-NtCDPK1, respectively. NtCDPK1-bound His-14-3-3 was detected by immunoblot analysis with anti-14-3-3 antibody. This experiment was repeated three times with similar results.
Next, we examined whether 14-3-3 protects a phosphate group of NtCDPK1 against λ-phosphatase as well as that of RSG. Treatment with λ-phosphatase revealed that GST-NtCDPK1 was already phosphorylated in Escherichia coli; however, this phosphorylation did not affect the 14-3-3 binding (Supplemental Fig. S6). GST-NtCDPK1 required an autophosphorylation reaction in vitro for 14-3-3 binding (Fig. 1A). When 14-3-3-bound GST-NtCDPK1 was treated with λ-phosphatase, GST-NtCDPK1 was completely dephosphorylated, although His-14-3-3 maintained the binding to GST-NtCDPK1 (Fig. 6B). This result indicated that 14-3-3 does not directly bind to a phosphate group of NtCDPK1 and suggested that the autophosphorylation of NtCDPK1 is necessary for the formation of the binding between NtCDPK1 and 14-3-3 but not for its maintenance. The effect of the autophosphorylation of NtCDPK1 on 14-3-3 binding is similar to that of Ca2+ (Supplemental Fig. S5). In general, 14-3-3 directly binds to the phosphorylated motif in target proteins, including RSG; however, a consensus 14-3-3-binding motif was not found in NtCDPK1. Taken together, our results suggested that 14-3-3 binding to NtCDPK1 represents a new mode.
Phosphorylation of RSG Decreased Affinity for NtCDPK1
Phosphorylated proteins appear to rapidly dissociate from kinases, which enables kinases to efficiently phosphorylate substrates. Although RSG forms a complex with NtCDPK1, RSG should dissociate from NtCDPK1 after RSG is phosphorylated by NtCDPK1. Therefore, NtCDPK1 was predicted to have a higher affinity for nonphosphorylated RSG than for phosphorylated RSG. To confirm this, we performed an in vitro pull-down assay. Autophosphorylated GST-NtCDPK1 was absorbed into glutathione beads and incubated with nonphosphorylated or phosphorylated MBP-RSG, followed by affinity chromatography. The pull-down assay showed that autophosphorylated NtCDPK1 interacts with nonphosphorylated RSG more strongly than with phosphorylated RSG (Supplemental Fig. S7). These results suggested that phosphorylation of RSG decreases the affinity for NtCDPK1.
14-3-3 Shows Higher Affinity for RSG Than for NtCDPK1
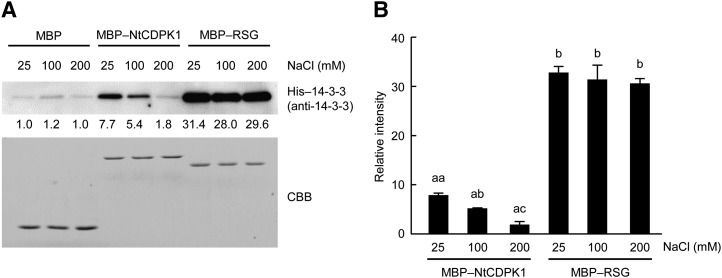
Because 14-3-3 binds to NtCDPK1 and RSG by different modes, we compared the binding affinities of 14-3-3 for phosphorylated RSG and autophosphorylated NtCDPK1. To examine this, autophosphorylated MBP-NtCDPK1 and phosphorylated MBP-RSG were absorbed into glutathione beads and amylose resin, respectively. The immobilized proteins were incubated with His-14-3-3. A pull-down assay showed that His-14-3-3 interacts with phosphorylated MBP-RSG more efficiently than with autophosphorylated MBP-NtCDPK1 (Fig. 7). Furthermore, higher concentrations of NaCl inhibited the binding of His-14-3-3 to MBP-NtCDPK1, whereas the interaction between His-14-3-3 and MBP-RSG was not affected by the concentrations of NaCl. These results suggested that the binding affinity of 14-3-3 for RSG is higher than that for NtCDPK1.
Figure 7.
14-3-3 shows higher affinity for RSG than for NtCDPK1. A, Autophosphorylated MBP-NtCDPK1 and phosphorylated MBP-RSG were immobilized on amylose resin and incubated with His-14-3-3. NtCDPK1- or RSG-bound proteins were subjected to SDS-PAGE, followed by immunoblot analysis with anti-14-3-3 antibody for the detection of His-14-3-3. MBP-NtCDPK1 and MBP-RSG were visualized by Coomassie Brilliant Blue (CBB) staining. The values below the top gel indicate the relative strength levels of signals after standardization using the signals of Coomassie Brilliant Blue staining of recombinant proteins as a loading control. The value of MBP at 25 mm NaCl was set to 1. This experiment was repeated three times with similar results. B, Statistical analysis of the data shown in A. The bar graph represents means and sd (n = 3). A significant difference of 14-3-3 binding affinity was detected between MBP-NtCDPK1 and MBP-RSG (P < 0.001, Tukey’s honestly significant difference test). Significant differences of 14-3-3 binding affinity of MBP-NtCDPK1 among the different concentrations of NaCl were determined using Tukey’s honestly significant difference test. Different letters above the bars (aa, ab, and ac) indicate significant differences among the different concentrations of NaCl (P < 0.01). No significant difference of 14-3-3 binding affinity of MBP-RSG was detected among the different concentrations of NaCl (P > 0.05).
14-3-3 Is Transferred from NtCDPK1 to RSG
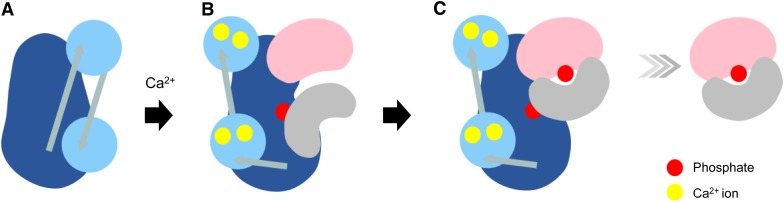
Although the binding of 14-3-3 to NtCDPK1 is precisely regulated by phosphorylation, the functional importance of the interaction between NtCDPK1 and 14-3-3 remained unclear. We observed in vivo interactions both between NtCDPK1 and 14-3-3 and between NtCDPK1 and RSG (Fig. 1; Ito et al., 2010) as well as a heterotrimer of NtCDPK1, RSG, and 14-3-3 in vitro (Fig. 3). Furthermore, the binding affinity of 14-3-3 for phosphorylated RSG was higher than that for autophosphorylated NtCDPK1 (Fig. 7). Based on these results, we hypothesized that 14-3-3 is transferred from NtCDPK1 to RSG after phosphorylation of RSG and that RSG dissociates from NtCDPK1 as a complex with 14-3-3. To examine this, a heterotrimer of GST-NtCDPK1, His-14-3-3, and MBP-RSG was formed on glutathione beads as described above (Fig. 3). When the pull-down buffer was replaced with the phosphorylation buffer containing ATP, MBP-RSG and His-14-3-3 dissociated from GST-NtCDPK1 (Fig. 8A). On the other hand, without ATP, dissociation of MBP-RSG and His-14-3-3 from NtCDPK1 was only a fraction. This result indicated that 14-3-3 is dissociated from NtCDPK1 together with RSG after phosphorylation of RSG by NtCDPK1. Next, to examine whether MBP-RSG and His-14-3-3 form a complex in the eluate from GST-NtCDPK1, we performed a successive pull-down assay using amylose resin. Immunoblot assays showed that His-14-3-3 bound to MBP-RSG in the eluate and that Ser-114 of RSG was phosphorylated (Fig. 8B), indicating that phosphorylated RSG dissociates from NtCDPK1 as a complex with 14-3-3. Collectively, our results suggested that 14-3-3 is transferred from NtCDPK1 to RSG. NtCDPK1 not only phosphorylates the substrate RSG but also may play a role as a scaffold that promotes binding between phosphorylated RSG and 14-3-3 (Fig. 9).
Figure 8.
14-3-3 is transferred from NtCDPK1 to RSG. A, RSG and 14-3-3 dissociate from NtCDPK1 after RSG is phosphorylated by NtCDPK1. GST-NtCDPK1 was heterotrimerized with RSG and 14-3-3 as described above. The pull-down buffer was replaced with a phosphorylation buffer with or without ATP. NtCDPK1-bound proteins and the phosphorylation buffer containing proteins were subjected to SDS-PAGE, followed by immunoblot analysis with anti-GST, anti-MBP, and anti-14-3-3 antibodies for the detection of GST-NtCDPK1, MBP-RSG, and His-14-3-3, respectively. The values below the middle and bottom gels indicate the relative strength levels of signals. The values of MBP-RSG and His-14-3-3 in the resin with ATP were set to 1. This experiment was repeated three times with similar results. B, MBP-RSG and His-14-3-3 dissociate from NtCDPK1 as a complex. The eluate from GST-NtCDPK1 was incubated with amylose resin. Amylose resin-bound proteins were subjected to SDS-PAGE, followed by immunoblot analysis with anti-MBP and anti-14-3-3 antibodies for the detection of MBP-RSG and His-14-3-3, respectively. Phosphorylation of Ser-114 in RSG was detected by anti-pS114 antibody. The values below the top and bottom gels indicate the relative strength levels of signals. The value of His-14-3-3 without ATP was set to 1. This experiment was repeated three times with similar results. The statistical analysis of these data is shown in Supplemental Figure S8.
Figure 9.
Model of 14-3-3 transfer from NtCDPK1 to RSG. A, In the absence of Ca2+, the catalytic activity of NtCDPK1 is inhibited by the autoinhibitory domain. RSG and 14-3-3 cannot access NtCDPK1. B, In the presence of Ca2+, the autoinhibitory domain is dissociated from the catalytic domain, and NtCDPK1 is autophosphorylated. RSG and 14-3-3 interact with the variable N-terminal domain and the catalytic domain of NtCDPK1, respectively. C, RSG is phosphorylated by NtCDPK1. 14-3-3 is transferred from NtCDPK1 to RSG. RSG and 14-3-3 are dissociated from NtCDPK1 as a complex. The catalytic domain of NtCDPK1 is shown in dark blue, the calmodulin-like domain of NtCDPK1 in light blue, RSG in pink, and 14-3-3 in gray. [See online article for color version of this figure.]
DISCUSSION
Many signal transduction events of eukaryotes are orchestrated by specific interactions of proteins mediated through phosphopeptide-binding motifs. Although several phosphospecific binding domains are now known, 14-3-3s were the first proteins recognized to specifically bind a phosphoserine or phosphothreonine motif (Muslin et al., 1996). In this study, we found a new binding mode of 14-3-3. Previous studies uncovered that binding to a target in the amphipathic groove of a 14-3-3 dimer often requires a phosphorylated consensus motif (mode I, RSXpSXP; mode II, RXY/FXpSXP; Yaffe et al., 1997) and can have a range of variation. An alternative binding motif (mode III, [pS/pT]X1-2-COOH) was found in the C terminus of several proteins, including H+-ATPase (Svennelid et al., 1999) and arylalkylamine N-acetyltransferase (Ganguly et al., 2005). Although it has been documented that 14-3-3s also can bind unphosphorylated targets (Petosa et al., 1998; Masters et al., 1999; Seimiya et al., 2000; Taoka et al., 2011), studies of the 14-3-3 interactome demonstrated that nonphosphorylated targets constitute only a small population of the total number of 14-3-3 interactors (Meek et al., 2004; Pozuelo-Rubio et al., 2004).
We showed that NtCDPK1 required an autophosphorylation reaction for 14-3-3 binding (Fig. 1A); however, 14-3-3 binding did not protect the phosphorylation of NtCDPK1 from λ-phosphatase (Fig. 6B). This suggested that the autophosphorylation reaction of NtCDPK1 triggered the binding of 14-3-3 to NtCDPK1, but 14-3-3 did not directly bind to phosphate groups of NtCDPK1. This notion is also supported by the observation that NtCDPK1 slightly bound to 14-3-3 without autophosphorylation in the presence of Ca2+ (Fig. 1A). The autophosphorylation could expose a hidden 14-3-3-binding site or create a new one by induction of a conformational change in NtCDPK1. Otherwise, the conformational change necessary for 14-3-3 binding could be induced by Ca2+ and ATP but not by the phosphate group in NtCDPK1 generated by autophosphorylation. In any case, 14-3-3 binds to NtCDPK1 through a previously unknown binding mode. Furthermore, the binding between NtCDPK1 and 14-3-3 was evidently weaker than that between RSG and 14-3-3 (Fig. 7). The property of 14-3-3 to readily dissociate from NtCDPK1 might be critical to the physiological function of NtCDPK1 as a scaffold.
Low-affinity 14-3-3 binding is also important in the functional regulation of H+-ATPase. The fungal phytotoxin fusicoccin, a diterpene glucoside, is a well-known activator of H+-ATPase and has been shown to bind to the preformed H+-ATPase/14-3-3 complex, thereby stabilizing the enzyme in its activated state (Oecking et al., 1997). Although the 14-3-3 binding to the C-terminal motif of H+-ATPase, mode III, is relatively weak, the affinity for the motif is increased 100-fold when fusicoccin is present (Würtele et al., 2003). Fusicoccin causes a constitutive activation of H+-ATPase, which results in the constant opening of the stomatal aperture and the subsequent wilting of plants. Thus, the weak binding of 14-3-3 in this case is indispensable for the normal physiological regulation of H+-ATPase. Comprehensive studies using affinity approaches revealed various 14-3-3 binding partners (Jin et al., 2004); however, the physiological significance of the weak binding of 14-3-3 could be underestimated.
A molecular mechanism to ensure the fidelity and specificity of information flow in a cell is a scaffold protein that interacts with multiple proteins of a pathway and links them into a signaling circuit. Scaffolds are thought both to enhance interactions between the correct signaling proteins and to insulate them from interactions with competing proteins (Good et al., 2011). One of the best-studied examples of a signaling scaffold is the Sterile5 (Ste5) protein of yeasts (Saccharomyces cerevisiae), which plays an essential role in signal transmission through the mating pathway (Bhattacharyya et al., 2006). The yeast mitogen-activated protein kinase kinase kinase (MAPKKK) Ste11 is a component of three distinct MAPK cascades that are involved in three different biological processes: mating, invasive growth, and high-osmolarity responses. Upon stimulation of a mating signal, a scaffold Ste5 forms a complex with Ste11, Ste7 (MAPKK), and Fusion3 (Fus3, MAPK) to prevent cross talk with other MAPK cascades (Choi et al., 1994). Furthermore, scaffold Ste5 plays a substrate-specific cocatalytic role for Fus3 phosphorylation by Ste7 (Good et al., 2009).
In addition to the typical scaffolds, a few kinases serve as scaffolds facilitating the subcellular localization of target proteins or the assembly of a complex in a catalytic activity-independent manner. Mammalian kinase p21-activated protein kinases (PAKs) are regulators of actin reorganization and cell migration. Although PAKs were originally identified as Ser/Thr protein kinases, some aspects of the PAK regulation of cytoskeletal reorganization and cell motility do not require its kinase activity. PAK serves as a scaffold protein that facilitates 3-phosphoinositide-dependent protein kinase1 (PDK1)-mediated activation (phosphorylation) of Ser/Thr protein kinase and the recruitment of Akt to the membrane by a kinase-independent mechanism (Higuchi et al., 2008). Dual-specificity Tyr-(Y)-phosphorylation-regulated kinase2 (DYRK2) is an evolutionally conserved animal protein kinase that regulates cell cycle progression and development (Lochhead et al., 2005; Varjosalo et al., 2008). DYRK2 functions as a scaffold for an E3 ubiquitin ligase complex, although the kinase activity of DYRK2 is dispensable for its ability to promote complex formation (Maddika and Chen, 2009). Another remarkable example is CaMKII in neurons. Following synaptic excitation, the proteasome redistributes rapidly to spines from the dendritic shaft and becomes more proteolytically active. CaMKII acts as a scaffold for the translocation of the proteasome independent of its kinase activity (Bingol et al., 2010). These studies demonstrated the existence of previously unrecognized functions of kinases: they facilitate signaling by recruiting proteins other than substrates to a signaling complex or to an organelle in a kinase-independent manner.
In this study, we showed that CDPK, which is only found in plants and some protozoans, may function as a scaffold that places 14-3-3 in proximity to phosphorylated RSG. Phylogenetic analyses have proposed that the CDPK gene family arose through the fusion of a CaMK and a calmodulin (Harper et al., 1991; Cheng et al., 2002). Scaffolding function is an interesting common feature of these two evolutionally related kinases. However, there are significant differences. Although known scaffolding functions of kinases are catalytic activity independent, the binding of 14-3-3 to NtCDPK1 requires the autophosphorylation of NtCDPK1 (Fig. 1). Another important property of NtCDPK1 as a scaffold is that NtCDPK1 transfers 14-3-3 to the immediate product of the own enzymatic reaction (i.e. phosphorylated RSG); however, other kinases with scaffolding functions, including CaMKII, promote the interaction of proteins other than substrates for kinase activity. NtCDPK1 may enhance the local concentration of 14-3-3 and orient 14-3-3 to allow for the specific and rapid interaction with phosphorylated RSG by restricting the conformational freedom, which protects the phosphoserine of RSG from phosphatase activity in order to maintain precision in signaling. Weak 14-3-3 binding to NtCDPK1 may help the efficient transfer of 14-3-3. Thus, NtCDPK1 is an interesting scaffolding kinase that increases the specificity and efficiency of signaling by coupling catalysis (i.e. phosphorylation of RSG) with scaffolding (i.e. transfer of 14-3-3 to phosphorylated RSG) on the same protein.
The 14-3-3s are a family of ancient signaling proteins that have been found in all eukaryotic organisms examined. The Arabidopsis 14-3-3 family consists of 13 members. Because the primary structures of 14-3-3 isoforms are highly conserved, it was considered unlikely that they would have binding and functional specificities. However, recent studies showed that 14-3-3 isoforms have distinct binding and functional specificities (Wilker et al., 2005, 2007; Taoka et al., 2011). If NtCDPK1 bound to only a limited number of 14-3-3 isoforms, the specificity would be maintained in the binding between RSG and 14-3-3, which could be important for the functional regulation of RSG.
The in vivo interactions between NtCDPK1 and 14-3-3 and between NtCDPK1 and RSG (Fig. 1; Ishida et al., 2008; Ito et al., 2010), together with in vitro experiments, suggest a scaffold function of NtCDPK1 in plant cells. To examine the in vivo role of the scaffolding function of NtCDPK1 on GA signaling, the mutant version of NtCDPK1 that lacks 14-3-3 binding but retains kinase activity is required. However, 14-3-3 binding to NtCDPK1 is autophosphorylation dependent and its binding site is located in the catalytic domain of NtCDPK1, making it difficult to discriminate between the residues that are necessary for the autophosphorylation reaction and those directly involved in 14-3-3 binding. A comprehensive mutational analysis of the catalytic domain of NtCDPK1 would generate a proper mutant to aid understanding of the physiological significance of the scaffolding function of NtCDPK1. CDPKs comprise a large protein family in the plant kingdom, and several CDPKs bind to 14-3-3 (Camoni et al., 1998; Moorhead et al., 1999). Furthermore, it has been demonstrated that basic-hydrophobic (ϕ)-X-basic-X-X-Ser/Thr-X-X-X-ϕ-basic is an optimal target sequence of CDPKs (Hernández Sebastià et al., 2004; Harper and Harmon, 2005) that is compatible with the 14-3-3-binding motifs modes I and II (Yaffe et al., 1997). Therefore, the function as a scaffold that transfers 14-3-3 to the phosphorylated product could be found in other CDPKs. Although signal transduction networks of eukaryotes are complicated, they often share core signaling components yet maintain specificity. Extensive analysis of the scaffolding function of CDPKs that play diverse physiological roles in plant signaling may help to reveal how cells coordinate multiple paralogous signaling systems to maintain the information flow while preventing unwanted cross talk.
MATERIALS AND METHODS
Preparation of Recombinant Proteins
GST-NtCDPK1, MBP-NtCDPK1, and MBP-RSG were expressed in Escherichia coli containing pGEX 4T-1-NtCDPK1, pMALc2-NtCDPK1, and pMALc2-RSG, respectively, and purified by Glutathione-Sepharose 4B (GE Healthcare) and amylose resin (New England Biolabs; Ito et al., 2010). In order to construct His-14-3-3, the complementary DNA fragments encoding 14-3-3 were amplified by PCR with specific primers (Supplemental Table S1) and cloned into pET16b (Novagen). His-14-3-3 was expressed in E. coli and purified by COSMOGEL His-Accept (Nacalai Tesque).
Autophosphorylation and Dephosphorylation Reactions
Purified recombinant GST-NtCDPK1 was autophosphorylated in a reaction mixture containing 20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 0.5 mm CaCl2, 0.1% (v/v) Triton X-100, and 0.05% (v/v) β-mercaptoethanol with 1 mm ATP (cold assay) or 1 mm ATP supplemented with [γ-32P]ATP (5,000 µCi mmol−1, 10–20 µCi per reaction; hot assay) at 30°C for 30 min. GST-NtCDPK1 was dephosphorylated by λ-protein phosphatase at 30°C for 30 min according to the manufacturer’s manual (New England Biolabs). MBP-RSG was phosphorylated by GST-NtCDPK1 in the reaction mixture at 30°C for 30 min.
In Vitro Pull-Down Assays
GST-NtCDPK1 (2 μg) and His-14-3-3 (3 μg) or GST-NtCDPK1 (2 μg) and MBP-RSG (3 μg) were incubated with 20 µL of Glutathione-Sepharose 4B (GE Healthcare) in 400 µL of a binding buffer containing 25 mm MOPS-NaOH, pH 7, 25 mm NaCl, 0.05% (v/v) β-mercaptoethanol, 0.1% (v/v) Triton X-100, 0.1 mm phenylmethylsulfonyl fluoride (PMSF), and 0.5 mm CaCl2 or 2 mm EGTA at 4°C for 30 min and washed with the binding buffer. MBP-RSG (2 μg) and His-14-3-3 (3 μg) or MBP-NtCDPK1 (2 μg) and His-14-3-3 (3 μg) were incubated with 20 µL of amylose resin (New England Biolabs) in 400 µL of a binding buffer. NaCl concentrations were changed from 25 to 200 mm in the experiment examining the binding affinity of 14-3-3 to RSG or NtCDPK1. Proteins binding to the beads were resolved by SDS-PAGE (Tris/Gly buffer) and visualized by Coomassie Brilliant Blue staining or immunoblot analysis as described below. The intensities of Coomassie Brilliant Blue-stained bands were measured using ImageJ software (version 1.46).
Coimmunoprecipitation Assay
Transgenic tobacco (Nicotiana tabacum) expressing NtCDPK1-GFP (Ishida et al., 2008) was used for the immunoprecipitation assay. A total cell extract of young tobacco leaves was prepared as follows: leaves were disrupted in liquid nitrogen by grinding with a mortar and pestle and then extracted in 3 volumes of an extraction buffer (40 mm MOPS-NaOH, pH 6.5, 25 mm NaCl, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 0.05% [v/v] β-mercaptoethanol, 1 mm PMSF, 0.05% [w/v] bovine serum albumin, a protease inhibitor cocktail [Roche], and a phosphatase inhibitor cocktail [Sigma-Aldrich]). The extract was centrifuged, and the supernatant was used for immunoprecipitation. Aliquots of each protein sample were immunoprecipitated with anti-NtCDPK1 (Ishida et al., 2008) or anti-His (Novagen) antibody for 3 h at 4°C, and then Protein G-conjugated beads (GE Healthcare) were added to each reaction. Immunoprecipitates were washed three times at 4°C with wash buffer (40 mm MOPS-NaOH, pH 7.5, 25 mm NaCl, 0.1% [v/v] Triton X-100, 0.05% [v/v] β-mercaptoethanol, and 0.1 mm PMSF), and coprecipitated proteins were subjected to immunoblot analysis.
Immunoblot Analysis
Aliquots of each protein sample were resolved by SDS-PAGE (Tris/Gly buffer), transferred onto an Immobilon-P transfer membrane (Millipore), and reacted with antiserum raised against the phosphorylated Ser-114 of RSG (antipS114; Ishida et al., 2004), anti-NtCDPK1 (Ishida et al., 2008), anti-14-3-3 (Igarashi et al., 2001), anti-GST (GE Healthcare), or anti-MBP (MBL), followed by the horseradish peroxidase-conjugated secondary antibody. Chemiluminescence was detected (Immobilon Western Chemiluminescent HRP Substrate; Millipore) and quantified using a CCD camera imaging system: LAS-1000 plus with LAS-1000 Lite software (Fuji Film) or ImageQuant LAS 4000 with ImageQuant TL software (GE Healthcare).
BiFC Analysis
For BiFC analysis, the DNA sequences coding for the N- and C-terminal fragments of YFP (YFPN, residues 1–155; YFPC, residues 156–239) were amplified by PCR with the pEYFP-Nuc vector (Clontech) as a template and the primers shown in Supplemental Table S1 and cloned into pJ4 (Igarashi et al., 2001), which was designed to express inserted genes under the control of the 35S promoter with the 5′-leader sequence (Ω) of tobacco mosaic virus RNA as a translational enhancer. We generated constructs expressing 14-3-3 N-terminally fused with YFPN and NtCDPK1 C-terminally fused with YFPC, which were named pJ4/YFPN-14-3-3 and pJ4/NtCDPK1-YFPC, respectively. As a negative control, pJ4/YFPC (stop codon +) was used. The primers used to generate these constructs are shown in Supplemental Table S1. For the expression of a set of YFP fusion proteins in tobacco, 4 μg of each plasmid DNA was transfected. As a control for transfection efficiency, 2 µg of a pJ4mRFP vector harboring mRFP complementary DNA driven by a cauliflower mosaic virus 35S promoter was cotransfected. Transfection was performed according to the protocol described previously (Ito et al., 2010). YFP and RFP fluorescence signals were detected with an epifluorescence microscope (ECLIPSE Ni-E; Nikon) using the YFP filter cube (500/20-nm excitation, 535/30-nm emission) and the RFP filter cube (540/25-nm excitation, 605/55-nm emission), respectively.
In Vitro Heterotrimerization Assay
Autophosphorylated GST-NtCDPK1 (2 μg) was incubated with Glutathione-Sepharose 4B (GE Healthcare) and His-14-3-3 (3 μg) in a pull-down binding buffer as described above at 4°C for 30 min. Glutathione beads into which GST-NtCDPK1 and His-14-3-3 had been absorbed were washed with a pull-down binding buffer to remove ATP and incubated with MBP-RSG (3 μg), followed by affinity chromatography. An alternative assay was performed as follows. Nonphosphorylated MBP-RSG (2 μg) was absorbed into amylose resin (New England Biolabs) and incubated with His-14-3-3 (3 μg) in the presence or absence of autophosphorylated GST-NtCDPK1 (3 μg). Proteins bound to the beads were resolved by SDS-PAGE (Tris/Gly buffer) and visualized by Coomassie Brilliant Blue staining or immunoblot analysis as described above.
Subcellular Colocalization Analysis
We generated constructs expressing YFPN-NtCDPK1, RSG-YFPC, mRFP-14-3-3, mRFP-NtCDPK1, YFPC-NtCDPK1, and RSG-mRFP, which were named pJ4/YFPN-NtCDPK1, pJ4/RSG-YFPC, pJ4/mRFP-14-3-3, pJ4/mRFP-NtCDPK1, pJ4/YFPC-NtCDPK1, and pJ4/RSG-mRFP, respectively. CrFP was generated by modifying enhanced cyan fluorescent protein (Zacharias et al., 2002; Rizzo et al., 2004). The plasmid construct expressing AGF1-CrFP, pJ4/AGF1-CrFP, was used as a control for nuclear localization, since AGF1 has been reported to localize in the nucleus (Matsushita et al., 2007). The primers used for generating these constructs are shown in Supplemental Table S1. These constructs were cotransfected into Arabidopsis (Arabidopsis thaliana) T87 protoplasts according to Satoh et al. (2004). CrFP fluorescence was excited at 458 nm with an argon-ion laser (reflected by the beam splitter HFT 458/514; Carl Zeiss), and emission was recorded through a 475- to 525-nm infrared band-pass filter. YFP fluorescence was excited at 488 nm with an argon-ion laser (reflected by the beam splitter HFT 405/488/543), and emission was recorded through a 505- to 530-nm infrared band-pass filter. mRFP fluorescence was excited at 543 nm with a helium-neon laser (reflected by the beam splitter HFT 405/488/543), and emission was recorded through a 560- to 615-nm infrared band-pass filter. Fluorescence signals were detected with the LSM 5 Pascal confocal microscope (Carl Zeiss). Colocalization was analyzed by ZEN 2009 software (Carl Zeiss).
In Vitro Kinase Assay
The catalytic activities of GST-NtCDPK1 (0.4 ng µL−1) were assayed in the reaction mixture as described for autophosphorylation reactions with 0.02 mg mL−1 MBP-RSG as a substrate with 1 mm ATP in the presence or absence of His-14-3-3 (4 ng µL−1) at 30°C for 10 to 30 min. After the reactions were subjected to SDS-PAGE (Tris/Gly buffer), the phosphorylation of RSG was detected by immunoblot analysis or with a bioimaging analyzer (BAS 1800II; Fuji Film).
Phosphatase Protection Assay
Autophosphorylated GST-NtCDPK1 was absorbed into glutathione beads with or without His-14-3-3 in a pull-down binding buffer as described above at 4°C for 30 min. Glutathione beads into which GST-NtCDPK1 and His-14-3-3 had been absorbed were washed with the binding buffer to remove ATP and incubated with 400 units of λ-protein phosphatase (New England Biolabs) in a pull-down binding buffer with 10 mm MgCl2 at 4°C for 30 min. Phosphorylated MBP-RSG was absorbed into amylose resin. Phosphatase treatment of MBP-RSG was performed in the same manner as for GST-NtCDPK1. Aliquots of the reactions were subjected to SDS-PAGE (Tris/Gly buffer), followed by immunoblot analysis with anti-14-3-3 antibody for the detection of 14-3-3 binding. Phosphorylation status was examined using a bioimaging analyzer (BAS 1800II; Fuji Film) or Phos-tag SDS-PAGE (Kinoshita and Kinoshita-Kikuta, 2011).
14-3-3 Transfer Analysis
After the heterotrimer of GST-NtCDPK1, His-14-3-3, and MBP-RSG was formed on glutathione beads at 4°C, the complex on beads was incubated in the reaction mixture as described above with or without 1 mm ATP at 4°C for 60 min. Eluate from GST-NtCDPK1 was subjected to a successive pull-down assay using amylose resin in a pull-down binding buffer at 4°C for 30 min. Proteins binding to glutathione beads or amylose resin and proteins in the phosphorylation reaction buffer were resolved by SDS-PAGE (Tris/Gly buffer), followed by immunoblot analysis for the detection of the proteins and the phosphorylation of Ser-114 in RSG.
Statistical Analysis
Statistical analysis was performed using R software (version 3.1.0). Significant differences between two groups were evaluated using Student’s t test. Comparisons among three or more groups were performed using one-way ANOVA with Tukey’s honestly significant difference test. The effects of Ca2+ and autophosphorylation on the 14-3-3 binding affinity of NtCDPK1 were assessed using two-way ANOVA.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB040471 (RSG), AB071967 (14-3-3, D31), and AF072908 (NtCDPK1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. NtCDPK1 is autophosphorylated in the presence of Ca2+.
Supplemental Figure S2. MBP-RSG and His-14-3-3 do not bind directly to glutathione beads and amylose resin, respectively.
Supplemental Figure S3. NtCDPK1 does not phosphorylate 14-3-3.
Supplemental Figure S4. Autophosphorylated NtCDPK1 does not phosphorylate RSG in the absence of Ca2+.
Supplemental Figure S5. 14-3-3 maintains the binding to NtCDPK1 in the absence of Ca2+.
Supplemental Figure S6. The phosphorylation of NtCDPK1 in E. coli does not affect the 14-3-3 binding ability of NtCDPK1.
Supplemental Figure S7. NtCDPK1 interacts with nonphosphorylated RSG rather than phosphorylated RSG.
Supplemental Figure S8. Statistical analysis for Figure 8.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Yuri Abe for technical assistance and Makoto Kusaba for helpful discussions.
Glossary
- BiFC
bimolecular fluorescence complementation
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
This work was supported by the Japan Society for the Promotion of Science (grant no. 23657038 to Y.T.) and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant no. 24118004 to Y.T.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S. (2005) Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46: 356–366 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Reményi A, Good MC, Bashor CJ, Falick AM, Lim WA. (2006) The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311: 822–826 [DOI] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. (2010) Autophosphorylated CaMKIIα acts as a scaffold to recruit proteasomes to dendritic spines. Cell 140: 567–578 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Sheen J. (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Moorhead GBG. (2005) 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 2005: re10. [DOI] [PubMed] [Google Scholar]
- Camoni L, Harper JF, Palmgren MG. (1998) 14-3-3 proteins activate a plant calcium-dependent protein kinase (CDPK). FEBS Lett 430: 381–384 [DOI] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA. (1994) Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78: 499–512 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y. (2010) The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J 62: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y. (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Weller JL, Ho A, Chemineau P, Malpaux B, Klein DC. (2005) Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci USA 102: 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese KP, Fedorov NB, Filipkowski RK, Silva AJ. (1998) Autophosphorylation at Thr286 of the α calcium-calmodulin kinase II in LTP and learning. Science 279: 870–873 [DOI] [PubMed] [Google Scholar]
- Gökirmak T, Paul AL, Ferl RJ. (2010) Plant phosphopeptide-binding proteins as signaling mediators. Curr Opin Plant Biol 13: 527–532 [DOI] [PubMed] [Google Scholar]
- Good M, Tang G, Singleton J, Reményi A, Lim WA. (2009) The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332: 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. (2004) Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Harper JF, Harmon A. (2005) Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol 6: 555–566 [DOI] [PubMed] [Google Scholar]
- Harper JF, Sussman MR, Schaller GE, Putnam-Evans C, Charbonneau H, Harmon AC. (1991) A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 252: 951–954 [DOI] [PubMed] [Google Scholar]
- Hernández Sebastià C, Hardin SC, Clouse SD, Kieber JJ, Huber SC. (2004) Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch Biochem Biophys 428: 81–91 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Onishi K, Kikuchi C, Gotoh Y. (2008) Scaffolding function of PAK in the PDK1-Akt pathway. Nat Cell Biol 10: 1356–1364 [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Hudmon A, Schulman H. (2002) Neuronal Ca2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu Rev Biochem 71: 473–510 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y. (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y. (2004) Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 16: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Yuasa T, Nakata M, Takahashi Y. (2008) A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. (2010) Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell 22: 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O’Donnell P, Taylor P, Taylor L, et al. (2004) Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol 14: 1436–1450 [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ. (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E. (2011) Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11: 319–323 [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5: 749–757 [DOI] [PubMed] [Google Scholar]
- Lochhead PA, Sibbet G, Morrice N, Cleghon V. (2005) Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121: 925–936 [DOI] [PubMed] [Google Scholar]
- Maddika S, Chen J. (2009) Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol 11: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters SC, Pederson KJ, Zhang L, Barbieri JT, Fu H. (1999) Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry 38: 5216–5221 [DOI] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y. (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek SEM, Lane WS, Piwnica-Worms H. (2004) Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem 279: 32046–32054 [DOI] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Cotelle V, Harthill J, Morrice N, Meek S, Deiting U, Stitt M, Scarabel M, Aitken A, et al. (1999) Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84: 889–897 [DOI] [PubMed] [Google Scholar]
- Oecking C, Piotrowski M, Hagemeier J, Hagemann K. (1997) Topology and target interaction of the fusicoccin-binding 14-3-3 homologs of Commelina communis. Plant J 12: 441–453 [Google Scholar]
- Petosa C, Masters SC, Bankston LA, Pohl J, Wang B, Fu H, Liddington RC. (1998) 14-3-3ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem 273: 16305–16310 [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BHC, Wood NT, Campbell DG, Morrice N, Mackintosh C. (2004) 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J 379: 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. (2004) An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 22: 445–449 [DOI] [PubMed] [Google Scholar]
- Satoh R, Fujita Y, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. (2004) A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol 45: 309–317 [DOI] [PubMed] [Google Scholar]
- Seimiya H, Sawada H, Muramatsu Y, Shimizu M, Ohko K, Yamane K, Tsuruo T. (2000) Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J 19: 2652–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen KL, Choi JH. (1991) Isolation and sequence analysis of a cDNA clone for a carrot calcium-dependent protein kinase: homology to calcium/calmodulin-dependent protein kinases and to calmodulin. Plant Mol Biol 17: 581–590 [DOI] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. (1999) Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Ito T (2011) Structure and function of CDPK: a sensor responder of calcium. In S Luan, ed, Coding and Decoding of Calcium Signals in Plants. Springer, Berlin, pp 129–146 [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Tsuruta F, Sunayama J, Mori Y, Hattori S, Shimizu S, Tsujimoto Y, Yoshioka K, Masuyama N, Gotoh Y. (2004) JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J 23: 1889–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Avruch J. (2002) 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem 277: 3061–3064 [DOI] [PubMed] [Google Scholar]
- van Hemert MJ, Steensma HY, van Heusden GP. (2001) 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. BioEssays 23: 936–946 [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Björklund M, Cheng F, Syvänen H, Kivioja T, Kilpinen S, Sun Z, Kallioniemi O, Stunnenberg HG, He WW, et al. (2008) Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell 133: 537–548 [DOI] [PubMed] [Google Scholar]
- Wernimont AK, Amani M, Qiu W, Pizarro JC, Artz JD, Lin YH, Lew J, Hutchinson A, Hui R. (2011) Structures of parasitic CDPK domains point to a common mechanism of activation. Proteins 79: 803–820 [DOI] [PubMed] [Google Scholar]
- Wernimont AK, Artz JD, Finerty P, Jr, Lin YH, Amani M, Allali-Hassani A, Senisterra G, Vedadi M, Tempel W, Mackenzie F, et al. (2010) Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol 17: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, Grant RA, Artim SC, Yaffe MB. (2005) A structural basis for 14-3-3σ functional specificity. J Biol Chem 280: 18891–18898 [DOI] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MATM, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, et al. (2007) 14-3-3σ controls mitotic translation to facilitate cytokinesis. Nature 446: 329–332 [DOI] [PubMed] [Google Scholar]
- Würtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C. (2003) Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J 22: 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91: 961–971 [DOI] [PubMed] [Google Scholar]
- Yoon GM, Cho HS, Ha HJ, Liu JR, Lee HS. (1999) Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol 39: 991–1001 [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296: 913–916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.