Abstract
Telomeres are repetitive sequences at the ends of chromosomes protected by DNA binding proteins of the shelterin complex that form capping structures. Through the interaction of shelterin complex-associated proteins, telomere length maintenance is regulated. Recently, the newly identified embryonic stem cell marker, Zinc finger and SCAN domain-containing 4 gene (Zscan4), was shown to be a telomere-associated protein, co-localizing to the shelterin complex. Furthermore, it was shown to play an essential role in genomic stability by regulating telomere elongation. Although it is known that Zscan4 regulates TRF2, POT1b, and Rap1 expression in embryonic stem cells, the relationship and the exact mechanism of action for ZSscan4-mediated telomere maintenance in cancer cells is unknown. In this study, we investigated Zscan4 expression and interactions with Rap1 in telomerase positive (HeLa, MCF7) and ALT pathway (SaOS2, U2OS) cancer cells. Through western, pulldown, siRNA, and overexpression assays we demonstrate, for the first time, that Zscan4 directly associates with Rap1 (physical association protein). Furthermore, by generating truncated versions of Zscan4, we identified its zinc finger domain as the Rap1 binding site. Using bimolecular fluorescence complementation, we further validate this functional interaction in human cancer cells. Our results indicate that Zscan4 functions as a mediator of telomere length through its direct interaction with Rap1, possibly regulating shelterin complex-controlled telomere elongation in both telomerase positive and alternative lengthening of telomere pathways. This direct interaction between Zscan4 and Rap1 may explain how Zscan4 rapidly increases telomere length, yielding important information about the role of these proteins in telomere biology.
Keywords: Zscan4, Rap1, cancer, telomeres, shelterin complex
Introduction
A telomere is a repetitive sequence of DNA that protects the ends of linear chromosomes from deterioration and repair activity.1,2 Mammalian telomeres consist of repetitive TTAGGG sequences that are crucial to formation of the capping structures, which are bound by telomere-binding factors called shelterin.3-5 The shelterin complex is a six subunit complex composed of directly binding proteins TRF1, TRF2, and POT16 and their associated proteins Rap1, TPP1, and TIN2.7 The importance of the shelterin complex in protecting telomere ends from chromosomal stability and in regulating telomere maintenance mechanisms has been previously shown.8,9
There are two types of telomere maintenance mechanisms. The default mechanism is through telomerase activation.10,11 Telomerase is a cellular DNA polymerase that lengthens telomeres by adding specific DNA sequence repeats to the ends of telomeres.7,12 The second mechanism identified is termed alternative lengthening of telomeres (ALT).13-17 Although the mechanism behind the ALT pathway is not well understood in mammalian tumors, it has been shown that telomere shortening in a telomerase-independent immortalized mammalian cell line induces immortalization by initiating homologous recombination (HR)18 between telomere sister chromatids, or telomere sister chromatid exchange (T-SCE).11,19 HR is one of the major pathways to repair double-strand breaks (DSBs).20 The functional Mre11-Rad50-Nbs1 (MRN) complex, which re-distributes to the sites of DSBs, is required for ataxia telangiectasia mutated (ATM), which is an initiator of DNA damage responses.21,22 The MRN complex is involved in all three pathways; classical or canonical nonhomologous end joining (C-NHEJ), homologous recombination repair (HRR), and alternative nonhomologous end joining (A-NHEJ).23 HR events give rise to SCE while T-SCE occurred more often in cells with ALT activity than in normal cells and telomerase positive cells.13
The shelterin complex has been identified as a mediator in regulating telomere elongation.6 It has been suggested that shelterin has a dual function as the core of its telomere length regulation. Based on different telomeric protein associations within the complex, shelterin can either promote or inhibit telomere elongation.7 These shelterin complex and telomere-associated factor exchanges are important in mediating telomere elongation, but many functions remain undefined. For instance, Repressor Activator Protein 1 (Rap1) plays an important role in transcriptional activation and repression recombination, although it does not directly bind to telomeric DNA.24 In addition, the C-terminal RCT domain of Rap1 is directly tethered to telomeres by binding the RBM domain of TRF2, initiating formation of heterochromatin-like telomere structures.25 This evidence suggests that the role of Rap1 in the human cell probably goes beyond the control of telomere length and is potentially a new regulator of telomere length.26
Recently, the newly identified embryonic stem cell marker Zinc finger and SCAN domain containing 4 gene (Zscan4), which has a key function in genomic stability by regulating telomere elongation, might also have a fundamental role in the mechanism controlling telomere length regulation.2,27 The role of Zscan4 in stem cell research suggests that it can replace the oncogenic reprogramming factor in somatic cells, resulting in induced pluripotent stem (iPS) cells.2,27 Another function of Zscan4 was found to promote telomere elongation during reprogramming. However, it is not associated with increased telomerase activity.2,27 Overexpression of Zscan4 rescues cell proliferation, causes rapid telomere extension, and suppresses TRF2, POT1b, and Rap1 which, in turn, inhibits T-SCE.2 Other functions of Zscan4 are control of T-SCE (strongly related to the ALT pathway) and an increase in the DNA repair protein SPO11.2,27
While current studies focus on Zscan4 as an ES cell-specific transcription factor for regulating ESC pluripotency,2,27 here, we sought to investigate Zscan4 expression in cancer cell lines, identify proteins members within the shelterin complex with which it associated, and whether these associations mirrored that of ESC. Given the current understanding of Zscan4 from ESC, we hypothesized that Zscan4 functions as a mediator of telomere length through its interaction with proteins in the shelterin complex and may control the function of the shelterin complex-controlled telomere elongation in cancer cell lines. Furthermore, this work was driven by the desire to discover how Zscan4 functions in elongating telomere length. By investigating the interactions between Zscan4 and certain shelterin complex members, we discovered that Zscan4 directly binds Rap1. Our results suggest that Zscan4 has the same function in cancer cells as ESC, directly interacts with the shelterin complex member Rap1, regulating telomere elongation. These studies highlight the importance of investigating the relationship between Zscan4 and the shelterin complex in cancer cells to elucidate the role of Zscan4 in elongating telomeres.
Results
Zscan4 expression in cancer cells
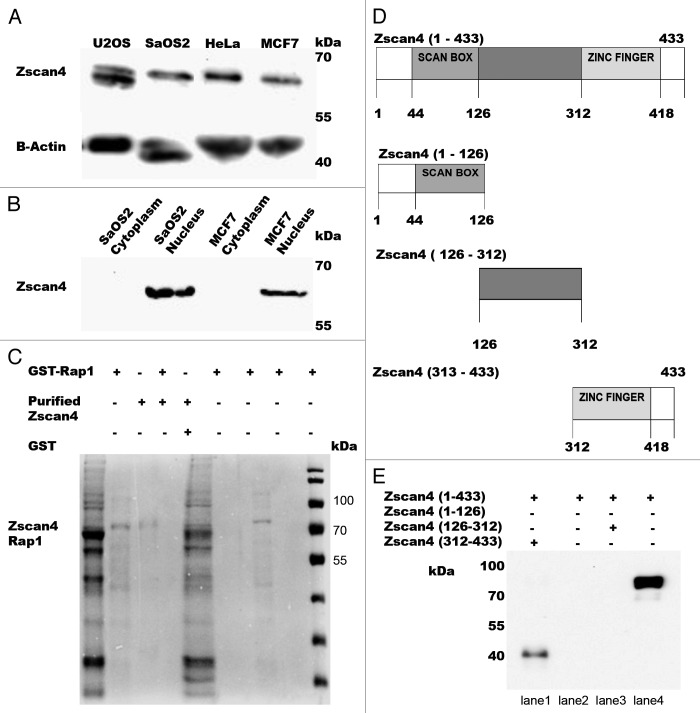
Zscan4 is transiently expressed specifically at the zygotic genome activation (ZGA) stage of embryogenesis and highly expressed exclusively in late 2-cell embryonic stem cells.2,27,28 Although, Zscan4 expression is low in human tissues, Zscan4 was shown to be highly expressed during inflammation.29 However, Zscan4 expression in cancer cells is not currently known. We hypothesized that Zscan4 would be expressed in cancer cells due to their requirement for telomere maintenance to maintain their immortality. Furthermore, we hypothesized that Zscan4 would directly interact with one of more members of the shelterin complex. Finally, we were interested in whether or not Zscan4 expression-interaction varied with telomerase activity. To that end, U2OS, SaOS2, HeLa, and MCF7 cancer cell lysates were separated by SDS-PAGE and then analyzed by western with anti-Zscan4 antibody. As predicted, Zscan4 was expressed all in cancer cells, although the expression levels of Zscan4 varied slightly different between cancer cell lines (Fig. 1A). MCF7 and SaOS2 cells were analyzed as they represented two different but important tissue lineages (epithelial/telomerase + and mesenchymal/telomerase −, respectively). Asynchronous cancer cell lysates from SaOS2 and MCF7 were fractionated into cytoplasmic and nuclear fractions. Zscan4 was detected in the nuclear fractions of MCF7 and SaOS2 by western (Fig. 1B). This data demonstrated for the first time that Zscan4 is also expressed in cancer cells with different types of telomere maintenance, and that the telomere-related functions of Zscan4 were not associated with telomerase activity.2,27
Figure 1. The expression levels of Zscan4 in the cancer cell lines analyzed and pulldown assay results between Rap1 and Zscan4. (A) Western results for total cell lysates in both telomerase positive and telomerase negative cancer cell lines using anti-Zscan4 antibody. (B) Zscan4 expression from nuclear and cytoplasmic fractions for SaOS2 and MCF7 cells. β-actin was used as the loading control. (C) The pulldown assay interaction between Rap1 and Zscan4 was analyzed by Coomassie detection (shown in lane 3). The GST negative control showed no signal with purified Zscan4 (lane 4). After GST-Rap1 was immobilized to the beads, eluents from the extensive washing (first and fifth wash) were loaded (shown in lanes 5 and 6). These results demonstrate that unbound GST-Rap1 was clearly removed. Weak signal and no signal were detected in the eluents from the first and fifth washings after purified Zscan4 was incubated with immobilized GST-Rap1 (shown in the lanes 7 and 8). This demonstrated that most of the purified Zscan4 was bound to GST-Rap1 and confirmed a direct interaction exists between Rap1 and Zscan4. GST-Rap1 and purified Zscan4 (shown in lanes 1 and 2) were used as positive controls. (D) Schematic representation of recombinant GST-fused Zscan4 truncations. To identify the minimal domain(s) responsible for the Zscan4-hRap1 interaction, full-length Zscan4 and Zscan4 fragments containing residues (1–126 N-terminal with SCAN BOX-Zscan4, 126–312 middle domain-Zscan4, and 312–433 Zscan4 with Zinc Fingers) were produced as a set of GST–Zscan4 plasmid constructs. (E) The pulldown assays were performed with full-length Zscan4/truncation of Zscan4 and Rap1. Captured proteins were analyzed by western with anti-Rap1 antibody. Signal was detected in the full-length of GST- Zscan4 (Zscan4 1–433) and GST-Zscan4 fragment containing residues 312–433 (Zscan4 312–433). N-terminal of Zscan4 (Zscan4 1–126) and middle domain of Zscan4 (126–312) were negative for signal. This data demonstrated that the C-terminal position of Zscan4 (312–433) was the specific binding site for Rap1.
Zscan4 directly interacts with Rap1 in vitro
Previous studies in ESC demonstrated Zscan4 co-localization with shelterin member foci.2,27,30 Thus, Zscan4 has been predicted to have a similar function in telomere elongation in cancer cells. Based on these findings, we hypothesized that Zscan4 interacts with one of the components in the shelterin complex to control the length of telomeres in cancer cells. In order to examine whether Zscan4 and shelterin components interact under physiological conditions, initially, we performed direct Zscan4 pulldown assays with purified TRF2 and POT1 because the expression levels of these two proteins was shown to be inversely related to overexpression of Zscan4 in ESC.2 Preliminary pulldown results showed no interaction with Zscan4/TRF2 or POT1 (results not shown). The next candidate protein we investigated was Rap1 because it is also the last shelterin complex protein that showed changes in expression levels correlating with overexpression of Zscan4 in ESC.2 To investigate a possible relationship, purified Zscan4 protein was used as the prey protein in the pulldown assay. The TNT® Transcription/Translation method was performed to purify GST-Rap1, which was then immobilized to the GST-beads. Purified Zscan4 protein was incubated with immobilized GST-Rap1 beads and detected by Coomassie staining (Fig. 1C, lane 3). This data indicated that revealed Rap1 as a novel Zscan4 interacting protein.
The Zinc finger domain of Zscan4 is the sight of Rap1 binding
Even though the shelterin complex member, Rap1, was identified as a Zscan4 binding protein, nothing is known about whether this interaction has any role in telomere regulation. To begin to address this question, we first needed to identify the specific domain involved with Zscan4 binding. The SCAN binding domain of Zscan4 had been mapped to amino acids (aa) 1–126 in the N-terminal. The middle domain was (aa) 126–312 and the zinc finger domains of Zscan4 comprise (aa) 312–433 (Fig. 1D). A pulldown assay was performed with truncations of Zscan4 to identify the domain involved in the Zscan4/Rap1 interaction. The results for Zscan4/Rap1 domain binding showed that purified Rap1 bound to the C-terminal of Zscan4 (312–433) (Fig. 1E, lane 1). Pulldown assay interaction with the middle domain of Zscan4/Rap1 and the N-terminal of Zscan4/Rap1 showed no signal (Fig. 1E, lanes 2 and 3). Full-length Zscan4 (1–433) directly interacts with Rap1 (Fig. 1E, lane 4) and this data was confirmed in the purified pulldown assay (GST-Rap1 and purified Zscan4) (Fig. 1B) using a different detection method and prey/bait proteins. Through these results, we demonstrate that the C-terminal of Zscan4, containing the 4 zinc finger motifs, plays a key role in binding Rap1.
BiFC results show an interaction between Zscan4 and Rap1 in cancer cells
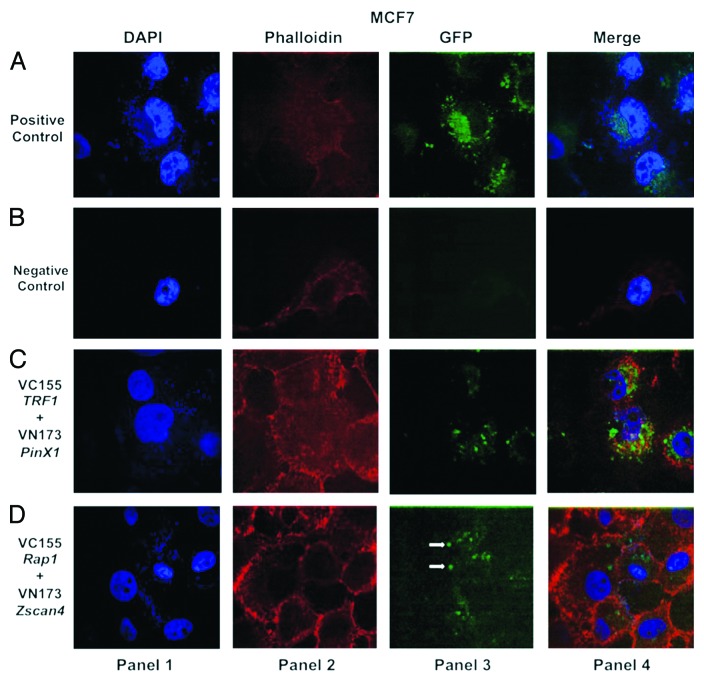
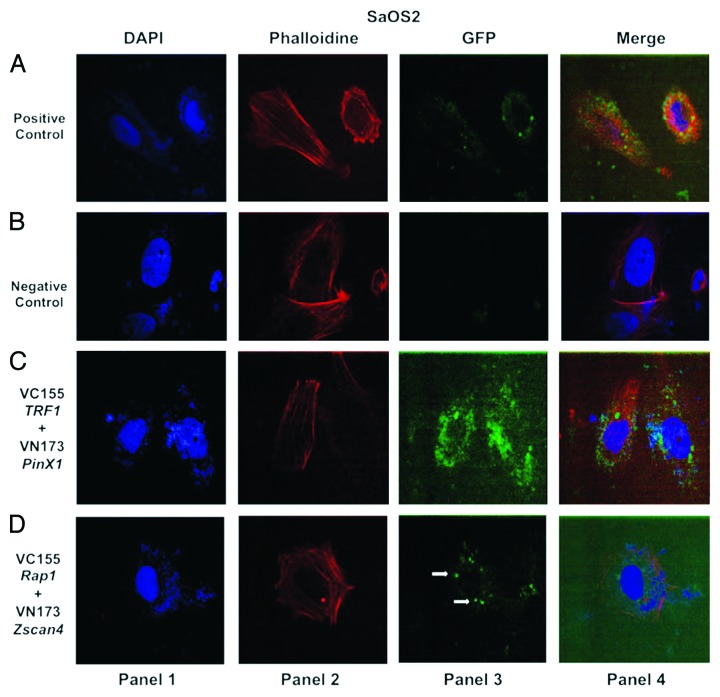
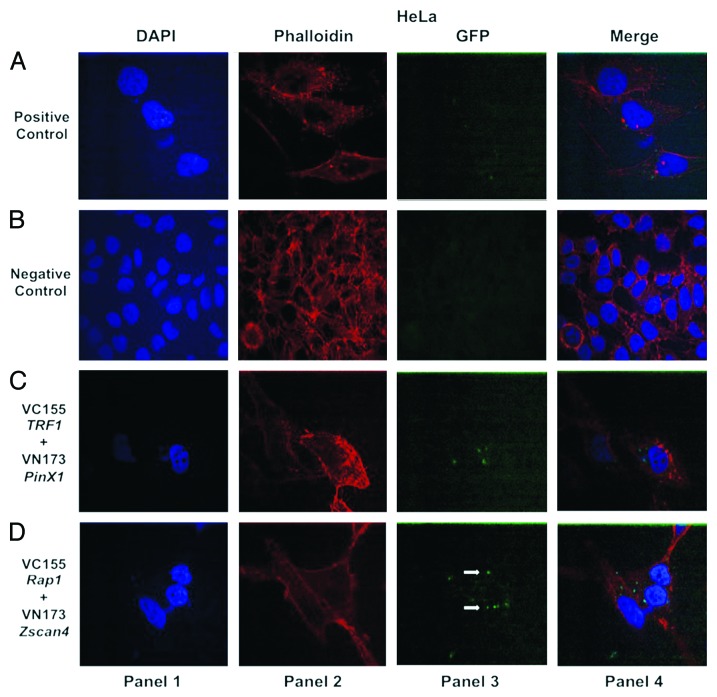
In the previous pulldown assay experiment, we observed that Zscan4 and Rap1 interacted directly. Although Zscan4 does directly interact with Rap1, it was important to verify whether the data generated from the pulldown assay were biologically relevant in living cells. Because protein-protein interactions are essential for coordinating many cellular processes and mediating signal-transduction pathways,31 bimolecular fluorescence complementation (BiFC) was employed to determine the subcellular localization and visualization of Zscan4 and Rap1 as well as gain an idea of their proximity in living cells.32-34 GFP has used as a positive control for transfection efficiency (Figs. 2A, 3A, and 4A, panel 3) and two empty vector fragments were used as negative controls for BiFC in the cancer cell lines (Figs. 2B, 3B, and 4B, panel 3). The interaction between PinX1 and TRF1 was used as a positive control for the pulldown assay. This was decided because PinX1 has been shown to be a direct interacting TRF1-associated protein.35 Results showed that pBiFC-VN173 PinX1 and pBiFC-VC155 TRF1 formed BiFC complexes in the cancer cell lines (Figs. 2C, 3C, and 4C, panel 3). These controls indicated that the BiFC system was working correctly to allow subsequent visualization of the protein interactions. BiFC fluorescent signals between Zscan4 and Rap1 were detected in the cancer cells tested (Figs. 2–4, panel 3). Thus, direct and functionally relevant interactions of Zscan4 and Rap1 were verified through both molecular and cellular biology techniques in the different cancer cell types.
Figure 2. Confocal images of bimolecular fluorescence complementation (BiFC) results in MCF7 cancer cells. DNA was labeled with DAPI (blue, shown in panel 1). Actin filaments were labeled with a fluorescent phalloidin (red, shown in panel 2). Panel 3 is the BiFC signal generated by interaction of the GFP fluorophore components based on proximity. The last panel (panel 4) represents the merged images. (A) GFP signal (green) (shown in the panel 3) was used as a positive control for transfection efficiency in MCF7 cancer cells. The corresponding overlaid images are shown in the last panel. (B) Two vector fragments (pBiFC-VC-155 and pBiFC-VN-173) (shown in panel 3) were used as negative control for BiFC in MCF7 cancer cell line. (C) Two vector fragments (pBiFC-VC-155-TRF1 and pBiFC-VN-173-PinX1) were combined to produce a bimolecular fluorescent complex (green, shown in panel 3). (D) Visualization of Zscan4 and Rap1 complex in MCF7 cancer cells using BiFC analysis (green, shown in panel 3). The images (shown in panel 3) are representative of BiFC analysis for the interaction between Zscan4 and Rap1. Images were generated under an Olympus APON 60X TIRF NA 1.4 objective.
Figure 3. Confocal images of bimolecular fluorescence complementation (BiFC) results in SaOS2 cancer cells. BiFC analysis was performed in SaOS2 cancer cells as described in Figure 2 and reflects the same pattern observed for MCF7 cells. The interaction between Zscan4 and Rap1 (shown in panel 3 of Fig. 4D) occurs in SaOS2 cancer cell line as well. Cells were visualized using the same conditions as described in Figure 2.
Figure 4. Confocal images of bimolecular fluorescence complementation (BiFC) results in HeLa cancer cells. The BiFC performed in HeLa cancer cells reflects the same patterns observed for MCF7 cells (Fig. 2) and SaOS2 cells (Fig. 3). The interaction between Zscan4 and Rap1 (shown in panel 3 of [D]) demonstrates the same relationship in HeLa cancer cells. Cells were visualized using the same conditions as described in Figure 2.
Zscan4 expression in cancer cells is dependent on Rap1
Next, we were interested in determining whether Zscan4 expression was dependent upon Rap1 expression. Therefore, we analyzed Zscan4 expression changes when Rap1 was either knocked down (siRNA) or overexpressed in the cancer cells. To that end, Rap1 constructs with an HA tag at the N-terminus (Table 1) was introduced into cancer cells. Western analysis revealed that cancer cells transfected with Rap1 siRNA expressed significantly less Zscan4 (Fig. 5A, lane 2). In contrast, greater Zscan4 expression was observed when Rap1 was overexpressed (Fig. 5A, lane 3). Previous reports have shown that overexpression of Zscan4 decreased the expression level of some shelterin proteins (TRF2, Rap1, and POT1) related to the inhibition of telomere sister chromatid exchange (T-SCE) in embryonic stem cells.2 However, in our study, overexpression of Rap1 increased the expression level of Zscan4 in cancer cell lines. This suggested that Zscan4 rapidly increased and bound to endogenous Rap1 to disrupt its function as a negative regulator of telomere length. Thus, there may be a dosage effect such that, the amount of Zscan4 binding to endogenous Rap1 acts as a switch to positively regulate telomere biology in cancer cells. In addition, we observed that inhibition of Rap1 expression downregulated the expression of Zscan4 (Fig. 5A, lane 1). This suggests that telomere elongation of Zscan4 might be the result of its binding activity with Rap1. Thus, Zscan4 might be a potential key protein in controlling the shelterin complex. However, more study is needed in this area.
Table 1. List of vectors used in this project.
| Name | Application |
|---|---|
| pGEX-4T-1-ZSCAN4 (1–433) | Bacterial protein expression vector |
| pGEX-4T-1-ZSCAN4 (1–126) | Bacterial protein expression vector |
| pGEX-4T-1-ZSCAN4 (126–312) | Bacterial protein expression vector |
| pGEX-4T-1-ZSCAN4 (312–433) | Bacterial protein expression vector |
| pT7CFE1-HA-TRF1 | Cell free protein expression vector |
| pCMV-HA-RAP1 | Eukaryotic protein expression vector |
| BiFC VN173-ZSCAN4 | Eukaryotic protein expression vector |
| BiFC VN173-PinX1 | Eukaryotic protein expression vector |
| BiFC VN155-TRF1 | Eukaryotic protein expression vector |
| pGEX-4T-1-TRF1 | Bacterial protein expression vector |
| pANT7-cGST-TRF1 | Eukaryotic protein expression vector |
| pANT7-cGST-RAP1 | Eukaryotic protein expression vector |
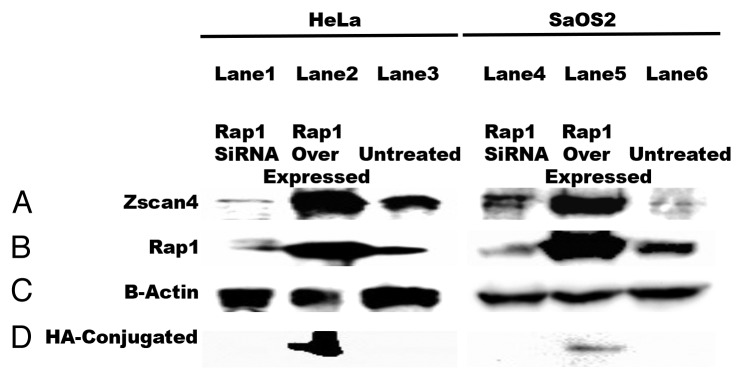
Figure 5. Zscan4 expression in cancer cells is dependent on Rap1. Either transfecting with Rap1 siRNA knockdown or overexpressing Rap1 with the addition of Rap1-HA modulated Zscan4 expression in cancer cells. (A) When Rap1 siRNA knockdown was performed, Zscan4 levels decreased. In contrast, Zscan4 levels increased when Rap1-HA (overexpression) was transfected. (B) Transfection efficiency was confirmed by western blot with anti-Rap1antibody. Cancer cells transfected with Rap1 siRNA had no detectable Rap1 signal, but cancer cell expression in cultures transfected with the Rap1-HA had significantly upregulated Zscan4 expression compared with untreated. (C) β-actin was used as the loading control. (D) HA signal was only detected in the overexpressed Rap1-HA lysates.
Discussion
While the field of telomere biology is an active research area, a complete understanding of the regulation of telomere function has not yet been achieved. Studies on the newly identified ESC marker, Zinc finger and SCAN domain containing 4 gene (Zscan4), demonstrated that endogenous Zscan4 rapidly increases telomere length corresponding with the high frequency of T-SCE in ESC, and changes the expression levels of DNA repair proteins.2,27 These results suggest that Zscan4 is involved in telomere length maintenance in the ALT pathway, because this telomerase negative pathway maintains telomeres by initiating DNA repair systems such as HR between T-SCE.11,18,36 However, in our study, the expression levels of Zscan4 were detected in cancer cells with two different mechanism of telomere maintenance, telomerase positive and negative. While current studies focus on Zscan4 as an ES cell-specific transcription factor for regulating ESC pluripotency,2,27 Zscan4’s functional role in binding telomeres, activating T-SCE and regulating telomere elongation for genomic stability in ES cells2,27 was the basis for this research.
Our results showed that Zscan4 directly binds to the shelterin complex member Rap1. This is the first report of a direct (physical) interaction and visualization between Rap1 and Zscan4 in telomerase-positive and -negative cancer cells. To gain a better understanding of this relationship between Zscan4 and Rap1, we addressed whether Rap1 directly interacts with Zscan4 and how this functions in the telomere biology of cancer cells. The BRCT and MYB domain of Rap1 are required domains for a negative regulation of telomere length.24,26 However, mutations in the BRCT and MYB domains in the N-terminal of Rap1 induced telomere elongation and these two domains are sites predicted to be present in a telomere elongating protein.24,26 Using truncated versions of Zscan4, our results identified the specific domain of Zscan4 that is able to bind to Rap1. Thus, we predict that the binding domain of Zscan4 functions as a mediator of the BRCT and MYB domain of Rap1 to regulate the function of the shelterin complex-controlled telomere elongation in cancer cell lines.
Using Rap1 overexpression and knockdown techniques, we observed that endogenous Rap1 significantly increased the Zscan4 expression levels and knockdown of Rap1 decreased the Zscan4 levels in cancer cells. Parallel changes in expression levels occurred in both genes. Thus, we posit that abundant Rap1 elevates Zscan4 expression levels in order to block the dominant negative function of overexpressed Rap1 and rapidly elongate telomere length in cancer cells. Results from Rap1 knockdown experiments suggest a potential dosage effect in which a small quantity of Zscan4 expression in cancer cells may be sufficient to sustain the uncapping of the telomeric termini and activate DNA repair proteins to accumulate DSB. As a result, Zscan4 may corrupt normal DNA repair function by increasing SPO1127 to accumulate a large amount of DSB and controlling TRF2 to limit ATM,37-39 which in turn modulates SPO11,40,41 finally driving cancer progression. Thus, the binding of Zscan4 with Rap1 may be required for disrupting telomere protection or the dissociation of the t-loop to control telomere length in telomere biology of cancer cells.
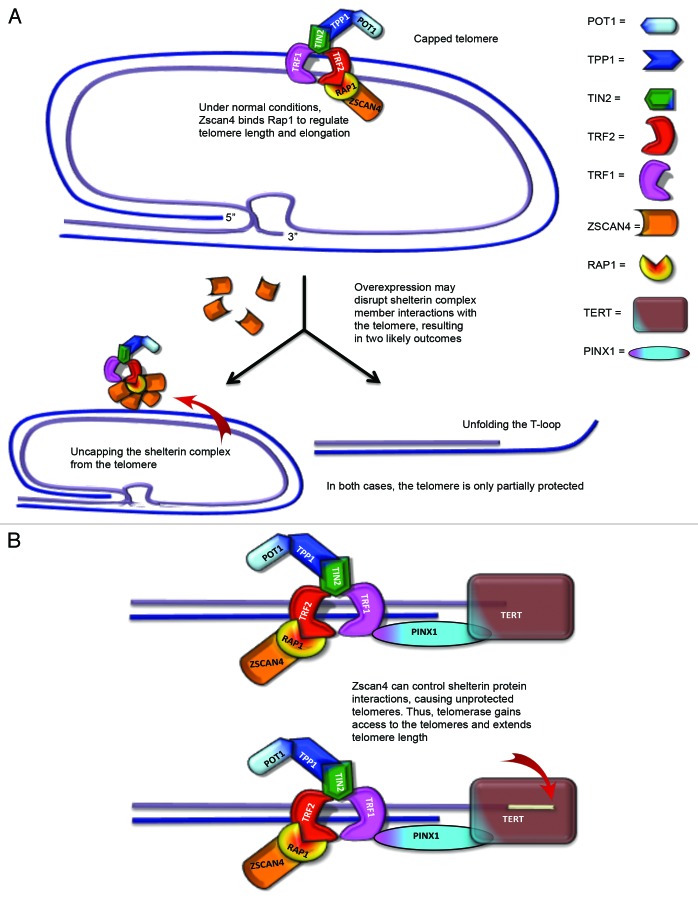
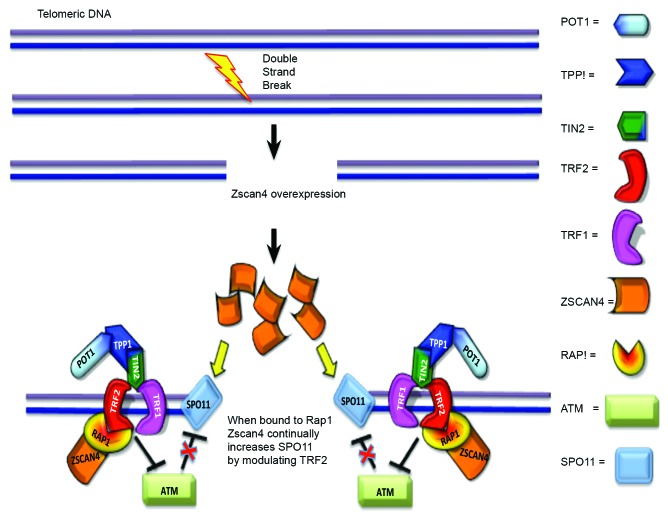
In summary, in telomerase positive cancer cells, one way that Zscan4 may maintain telomere length is by recruiting Rap1 and redirects its participation in the DNA repair system. TRF2/Rap1 effects may lose the function of protecting chromosome ends when Zscan4 binds to Rap1. Thus, Zscan4/Rap1 may bind to TRF2 in the shelterin complex of telomeric DNA and the loss of TRF2/Rap1 in the telomere results in an open state (uncapped telomere end). A model is proposed for this possibility is included in Figure 6. In telomerase negative cancer cells, one possibility to be considered is that Zscan4/Rap1 recruits TRF2 in DNA repair regions of non-telomeric DNA and telomeric DNA. Zscan4 continually increases SPO11 by controlling TRF238,42 when Zscan4 is bound to Rap1. Alternatively, abundant TRF2, which is released from the disrupted shelterin complex by uncapped telomeric ends or through binding of Zscan4/Rap1 to TRF2, might inhibit ATM control of SPO11. Thus, there may be insufficient TRF2 available to prevent the telomere damage response for DSB formation or DNA repair because the binding of Zscan4/Rap1 to TRF2 may cause the loss of t-loop protection and control TRF2 function.43 In other words, it is possible that Zscan4, when associated with the Rap1, switches Rap1 function from a negative regulator of telomere length to a positive regulator of telomere length in both telomerase positive and negative cancer cells. A proposed model for this explanation is included in Figure 7. To confirm this possible “Jekyll and Hyde” behavior of Rap1 induced by Zscan4 association will require further study in the context of telomere biology as well as DNA damage repair. Another possibility is that Zscan4 may compete for the BRTC or MYB domain-binding site with other unknown proteins that promote telomere length shortening in association with Rap1. Further study is needed to clarify whether these proposed interactions occur.
Figure 6. Proposed model of Zscan4–Rap1 interactions in telomere regulation in telomerase-positive cells. (A) TRF2/Rap1 may lose their function of protecting chromosome ends when Zscan4 binds to Rap1. Thus, Zscan4/Rap1 may bind to TRF2 in the shelterin complex of telomeric DNA. (B) The loss of TRF2/Rap1 in the telomere results in an open state (uncapped telomere end) allowing telomerase access to the 3′ overhang.
Figure 7. Proposed model of Zscan4–Rap1 interactions in telomere regulation in ALT-pathway cells. When DSBs occur, Zscan4/Rap1 may recruit TRF2 in DNA repair regions of non-telomeric DNA and telomeric DNA. Zscan4 continually increases SPO11 by controlling TRF2 when Zscan4 is bound to Rap1. Alternatively, abundant TRF2, (released from the disrupted shelterin complex through binding of Zscan4/Rap1 to TRF2), might inhibit ATM control of SPO11.
In conclusion, our study demonstrates that Zscan4 is expressed in telomerase positive and ALT pathway cancer cells, and interacts directly with Rap1 in the shelterin complex to regulate telomere elongation regardless of telomerase status. Thus, our results suggest a similar function for Zscan4 in cancer cells to that in ESC. This may make it an attractive target for future anticancer treatment strategies.
Materials and Methods
Cell culture
The following human cancer cell lines: HeLa (cervical), MCF7 (breast), U2OS, and SaOS2 (both derived from osteosarcomas [OS]) were purchased from the American Tissue and Culture Collection (ATCC). HeLa, U2OS, and MCF7 cancer cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 IU/mL and 100 μg/mL, respectively) under 5% CO2, in humid conditions, at 37 °C. SaOS2 cells were cultured in RPMI medium supplemented with 10% FBS, 2 mM l-glutamine, and 0.01% penicillin/streptomycin under humidity in 5% CO2 at 37 °C.
Cell extract and SDS-PAGE
The human cancer cells and transfected cells were washed with ice-cold phosphate buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (25 mM TRIS-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) (Thermo Fisher Scientific, Inc.) with protease inhibitor cocktail (G‐Biosciences, Inc.) added. Each sample was separated by SDS-PAGE (PAGE), and transferred to a nitrocellulose membrane. Membranes were incubated in a Zscan4 rabbit anti-human polyclonal (C-terminus) antibody (LSBio) (1:4000, overnight at 4 °C) or Rap1 rabbit anti-human monoclonal antibody (Cell Signaling) (1:4,000, overnight at 4 °C). Protein detection was performed using Super-Signal West Femto Chemiluminescent Substrate (Pierce) and the membrane was exposed using the FluorChem E system (ProteinSimple) optimized for analysis of chemiluminescent western blots, or detection with X-ray film. Nuclear and cytoplasmic extract in cancer cells were isolated and collected using the nuclear extraction kit (ActiveMotif) according to the manufacturer’s instructions.
Generating recombinant proteins
To generate N-terminal GST-Zscan4 fusion proteins, the Zscan4 gene was amplified from Zscan4 (NM_152677) purified human protein (OriGene) by standard polymerase chain reaction (PCR). PCR amplification was performed using Platinum pfx DNA polymerase (Invitrogen). The PCR product was digested with EcoRI and NotI, and then ligated into the pGEX-4T-1 vector (GE Healthcare Life Science). The pANT7-cGST-Rap1 was purchased from DNASU Plasmid Repository (DNASU) and the expression vector of the truncation of Zscan4 (GST-Zscan4 1–126, Zscan4 126–312, and Zscan4 312–433) and Rap1 were generated with primers, listed in Table 1.
Cell-free protein expression (in vitro translation)
Cell-free protein synthesis was performed using the TNT® T7 Quick Coupled system (Promega). The pANT7-cGST-Rap1 (DNASU) was expressed using the TNT® T7 Quick Coupled system according to the manufacturer’s instructions (Promega). For analyzing the results of translation, the membrane was visualized by western blot using standard protocols previously described in Lee and Gollahon.44
Recombinant fusion protein purification and quantitation
Recombinant pGEX-4T-1 vectors were transformed into E. coli BL21 (DE3). Isopropyl-β-D-1-thiogalactopyranoside (IPTG)-induced cells were harvested and pellets were resuspended in pulldown binding buffer containing 20 mM TRIS-HCl, pH 7.5, 100 mM NaCl, 0.5% NP-40, 50 mM EDTA, 5 mM DTT, and protease inhibitors. Cells were disrupted by sonication with a 3 mm probe in an Ultrasonic Processor (Cole Parmer). In order to obtain purified Zscan4 protein, the IPTG-induced-GST-Zscan4 cell supernatant was transferred to washed glutathione agarose beads, then wash three times with thrombin cleavage buffer (20 mM TRIS-HCl pH 8.5, 100 mM NaCl, 0.3 mM CaCl2, 1 mM DTT, and 0.1% Tween X-100). Thrombin (1 unit/mg protein) was added in 2 volumes with the thrombin cleavage buffer. Cleavage effectiveness was analyzed by SDS-PAGE. Thrombin was removed using a Thrombin Cleavage Capture kit according to manufacturer’s instructions (Novagen). The eluted sample was analyzed by SDS-PAGE to estimate the yield, purity and extent of digestion.
Cell free GST-pulldown assay system
The bait protein (pANT7-cGST-Rap1) generated using cell-free protein expression as described above, was incubated in pre-washed glutathione agarose beads. pANT7-cGST-Rap1 samples bound to glutathione beads were washed three times with binding buffer. The purified Zscan4 and truncated Zscan4, collected as described above, were incubated with the bead-GST-Rap1 sample. Immunoprecipitated protein complexes were collected for further immunoblot analysis against Zscan4.
Plasmid construction for bimolecular fluorescence complementation (BiFC)
Plasmid pairs pBiFC-VN173 and pBiFC-VC155 were purchased from Addgene. The Zscan4 gene was amplified from Zscan4 (NM_152677) purified human protein (OriGene) using a standard PCR protocol followed by enzyme restriction. The PCR product of Zscan4 amplification was cloned into a pBiFC-VC173 vector (Addgene). The PCR product was digested with KpnI and EcoRI, and then ligated into pBiFC-VN173 (Addgene). The recombinant construct was confirmed as described above. The same method was used to construct the following pBiFC fusion proteins: pBiFC–PinX1–VN173, pBiFC–TRF1–VC155 and pBiFC–Rap1–VC155. Primer sequences are listed in Table 2.
Table 2. List of primer sequences used in this project.
| Name | Sequence (5′ to 3′) | Application |
|---|---|---|
| ZSCAN4 (1–433)-GST forward (EcoRI) | TTGGGGGAAT TCATGGCTTT AGATCTAAGA ACC | pGEX-4T-1 |
| ZSCAN4 (1–433)-GST reverse (NotI) | TTGGGGGCGG CCGCTTAGGA AGCTTCTGGT GTGGA | pGEX-4T-1 |
| ZSCAN4 (1–126)-GST forward (EcoRI) | TTGGGGGAAT TCATGGCTTT AGATCTAAGA ACC | pGEX-4T-1 |
| ZSCAN4 (1–126)-GST reverse (NotI) | GGGCCCGCGG CCGCGCTGTC ATCAGTCAGG TCTTCTAA | pGEX-4T-1 |
| ZSCAN4 (126–312)-GST forward (EcoRI) | GGGCCCGAAT TCATGATAAA TCCACCTGCC TTAGTC | pGEX-4T-1 |
| ZSCAN4 (126–312)-GST reverse (NotI) | GGGCCCGCGG CCGCGTATGA TTTTTGGACT CCATGTAA | pGEX-4T-1 |
| ZSCAN4 (312–433)-GST forward (EcoRI) | GGGCCCGAAT TCATGAAATG TGAAGAATGC CCCAAG | pGEX-4T-1 |
| ZSCAN4 (312–433)-GST reverse (NotI) | TTGGGGGCGG CCGCTTAGGA AGCTTCTGGT GTGGA | pGEX-4T-1 |
| RAP1- pCMV forward (EcoRI) | GGGCCCGAAT TCACATGGCG GGAGGCGATG GAT | pCMV-HA vector |
| RAP1-pCMV reverse (KpnI) | GGCCCGGTAC CTTATTTCTT TCGAAATTCA ATCC | pCMV-HA vector |
| ZSCAN4- BiFC forward (EcoRI) | GGGCCCGAAT TCGGCTTTA GATCTAAGAA CC | BiFC VC 155 vector |
| ZSCAN4-BiFC reverse (KpnI) | TTGGGGGGTA CCAAGGAAGC TTCTGGTGTG GA | BiFC VC 155 vector |
| TRF1-BiFC forward (EcoRI) | TTTCCCGAAT TCTGGCTACG CCCCTGGTGG CG | BiFC VC 155 vector |
| TRF1-BiFC reverse (KpnI) | AACCCCGGTA CCCAAAGGTC TAGAACTGTC | BiFC VC 155 vector |
| PinX1-BiFC forward (EcoRI) | TTTCCCGAAT TCGTCTATGC TGGCTGAACG T | BiFC VC 155 vector |
| PinX1-BiFC reverse (KpnI) | TTTCCCGGTA CCAATTTGGA ATCTTTCTTC TT | BiFC VC 155 vector |
| RAP1-BiFC forward (EcoRI) | TTTCCCGAAT TCGGCGGAGG CGATGGATTT G | BiFC VC 155 vector |
| RAP1-BiFC reverse (KpnI) | TTTCCCGGTA CCAATTTCTT TCGAAATTCA AT | BiFC VC 155 vector |
Transfections for BiFC analysis
Cancer cells were transfected with the plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Each cancer cell line was transfected with BiFC VN173-Zscan4 and BiFC VN155-Rap1, BiFC VN173-PinX1, and BiFC VN155-TRF1, control BiFC vectors, GFP (Bio-Rad), and SiRNA. Post-transfection the cells proteins cells were fixed, washed with PBS, and permeabilized. DNA was counterstained with DAPI containing 5% Phalloidin (Invitrogen). The cells were washed and visualized using an Olympus IX2-DSU (Disk Scanning Unit) confocal microscopy (Hamamatsu Photonics) under an Olympus APON 60X TIRF NA 1.4 objective. Slidebook 5.5 software by 3i (Intelligent Imaging Innovations) was used for image capture and processing.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank TTU Department of Biological Sciences for their support, TTU Association of Biologist for their Grants-in-Aid, the TTU Imaging Center for access the fluorescence microscopes, Dr Boyd Butler for help and access to the Spinning Disk LSCM as well as training to generate the BiFC images for analysis. We would also like to thank the Biology and Biotechnology Core Facilities for access to specialized, high-end instrumentation.
Glossary
Abbreviations:
- A-NHEJ
alternative nonhomologous end joining
- ALT
alternative lengthening of telomeres
- ATM
ataxia telangiectasia mutated
- BiFC
bimolecular fluorescence complementation
- BRCT
BRCA1 C-terminus
- C-NHEJ
canonical nonhomologous end joining
- DSB
double–strand breaks
- ESC
embryonic stem cell
- GFP
green fluorescent protein
- HR
homologous recombination
- HRR
homologous recombination repair
- iPS
induced pluripotent stem
- MEFs
mouse embryonic fibroblasts
- MRN
mre11-rad50-nbs1
- MYB
myeloblastosis
- POT1
protection of telomeres 1
- Rap1
repressor activator protein 1
- SCE
sister chromatid exchange
- T-SCE
telomere sister chromatid exchange
- TIN2
TRF1- interacting nuclear protein 1
- TPP1
tripeptidyl peptidase 1
- TRF1
telomere repeat binding factor 1
- TRF2
telomere repeat binding factor 2
- ZGA
zygotic genome activation
- Zscan4
zinc finger and SCAN domain-containing 4
References
- 1.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, et al. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thanasoula M, Escandell JM, Martinez P, Badie S, Muñoz P, Blasco MA, Tarsounas M. p53 prevents entry into mitosis with uncapped telomeres. Curr Biol. 2010;20:521–6. doi: 10.1016/j.cub.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85:6622–6. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci U S A. 1989;86:7049–53. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 7.Diotti R, Loayza D. Shelterin complex and associated factors at human telomeres. Nucleus. 2011;2:119–35. doi: 10.4161/nucl.2.2.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–52. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou XZ, Huang P, Shi R, Lee TH, Lu G, Zhang Z, Bronson R, Lu KP. The telomerase inhibitor PinX1 is a major haploinsufficient tumor suppressor essential for chromosome stability in mice. J Clin Invest. 2011;121:1266–82. doi: 10.1172/JCI43452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuo T, Shimose S, Kubo T, Fujimori J, Yasunaga Y, Ochi M. Telomeres and telomerase in sarcomas. Anticancer Res. 2009;29:3833–6. [PubMed] [Google Scholar]
- 11.Wu XM, Tang WR, Luo Y. [ALT--alternative lengthening of telomere] Yi Chuan. 2009;31:1185–91. [PubMed] [Google Scholar]
- 12.Zvereva MI, Shcherbakova DM, Dontsova OA. Telomerase: structure, functions, and activity regulation. Biochemistry (Mosc) 2010;75:1563–83. doi: 10.1134/S0006297910130055. [DOI] [PubMed] [Google Scholar]
- 13.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Xiao BK, Liu JP, Chen SM, Tao ZZ. Alternative lengthening of telomeres in hTERT-inhibited laryngeal cancer cells. Cancer Sci. 2010;101:1769–76. doi: 10.1111/j.1349-7006.2010.01611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg G, Sehic D, Länsberg JK, Øra I, Frigyesi A, Castel V, Navarro S, Piqueras M, Martinsson T, Noguera R, et al. Alternative lengthening of telomeres--an enhanced chromosomal instability in aggressive non-MYCN amplified and telomere elongated neuroblastomas. Genes Chromosomes Cancer. 2011;50:250–62. doi: 10.1002/gcc.20850. [DOI] [PubMed] [Google Scholar]
- 16.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–60. doi: 10.1016/0092-8674(93)90234-H. [DOI] [PubMed] [Google Scholar]
- 17.Lundblad V. Telomere maintenance without telomerase. Oncogene. 2002;21:522–31. doi: 10.1038/sj.onc.1205079. [DOI] [PubMed] [Google Scholar]
- 18.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–5. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 19.Brault ME, Autexier C. Telomeric recombination induced by dysfunctional telomeres. Mol Biol Cell. 2011;22:179–88. doi: 10.1091/mbc.E10-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conrad S, Künzel J, Löbrich M. Sister chromatid exchanges occur in G2-irradiated cells. Cell Cycle. 2011;10:222–8. doi: 10.4161/cc.10.2.14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, et al. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–79. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen J, Cerosaletti K, Schultz KJ, Wright JA, Concannon P. NBN phosphorylation regulates the accumulation of MRN and ATM at sites of DNA double-strand breaks. Oncogene. 2013;32:4448–56. doi: 10.1038/onc.2012.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–24. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 24.O’Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–91. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol. 2011;18:213–21. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell. 2003;14:5060–8. doi: 10.1091/mbc.E03-06-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–63. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–50. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko SB, Azuma S, Yokoyama Y, Yamamoto A, Kyokane K, Niida S, Ishiguro H, Ko MS. Inflammation increases cells expressing ZSCAN4 and progenitor cell markers in the adult pancreas. Am J Physiol Gastrointest Liver Physiol. 2013;304:G1103–16. doi: 10.1152/ajpgi.00299.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shyu YJ, Suarez CD, Hu CD. Visualization of AP-1 NF-kappaB ternary complexes in living cells by using a BiFC-based FRET. Proc Natl Acad Sci U S A. 2008;105:151–6. doi: 10.1073/pnas.0705181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1:1278–86. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–87. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–86. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soohoo CY, Shi R, Lee TH, Huang P, Lu KP, Zhou XZ. Telomerase inhibitor PinX1 provides a link between TRF1 and telomerase to prevent telomere elongation. J Biol Chem. 2011;286:3894–906. doi: 10.1074/jbc.M110.180174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bechter OE, Zou Y, Shay JW, Wright WE. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 2003;4:1138–43. doi: 10.1038/sj.embor.7400027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–5. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao Z, Seluanov A, Jiang Y, Gorbunova V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A. 2007;104:13068–73. doi: 10.1073/pnas.0702410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huda N, Tanaka H, Mendonca MS, Gilley D. DNA damage-induced phosphorylation of TRF2 is required for the fast pathway of DNA double-strand break repair. Mol Cell Biol. 2009;29:3597–604. doi: 10.1128/MCB.00944-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–40. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inagaki A, Schoenmakers S, Baarends WM. DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics. 2010;5:255–66. doi: 10.4161/epi.5.4.11518. [DOI] [PubMed] [Google Scholar]
- 42.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–51. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 44.Lee J, Gollahon L. Mitotic perturbations induced by Nek2 overexpression require interaction with TRF1 in breast cancer cells. Cell Cycle. 2013;12:3599–614. doi: 10.4161/cc.26589. [DOI] [PMC free article] [PubMed] [Google Scholar]