Abstract
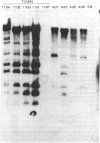
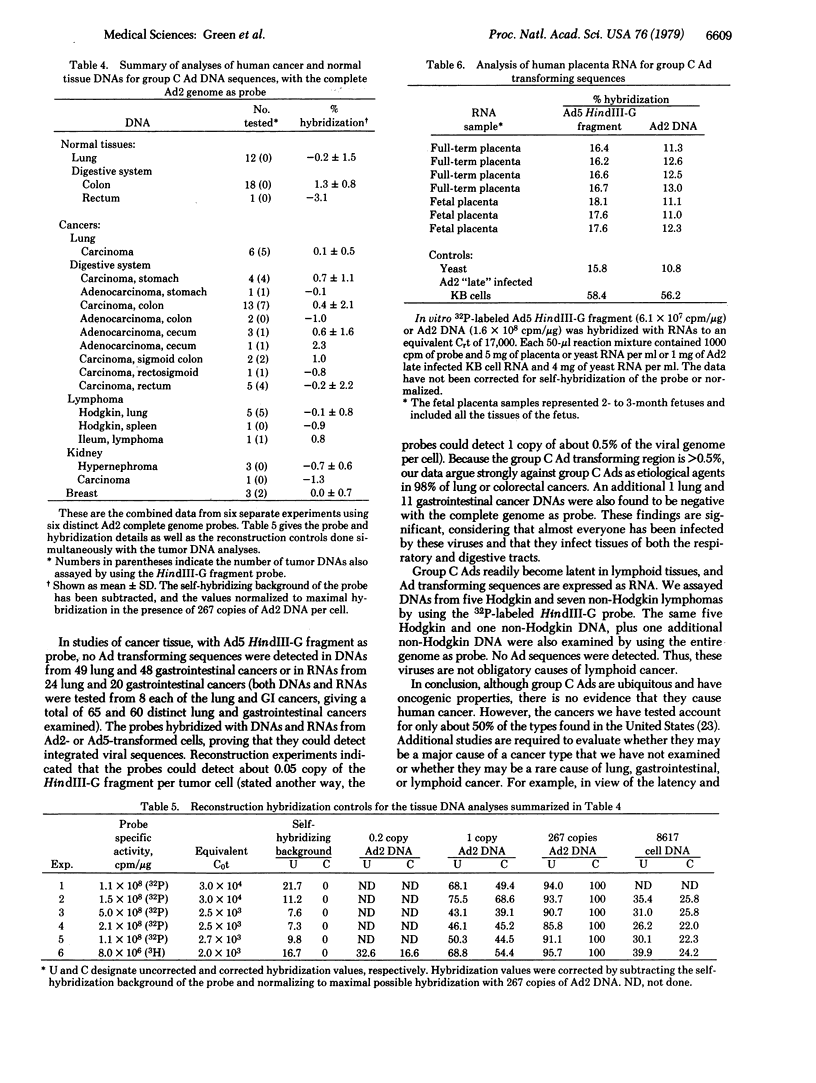
Group C human adenoviruses (Ads) of serotypes 1, 2, 5, and 6 infect most children and commonly cause latent infections of lymphoid tissues. Ads transform cells into a malignant-like phenotype; the oncogenic genetic information is in the left 8% of the viral genome, in the HindIII-G DNA fragment. We have investigated the molecular basis for group C Ad latent infections in human tonsils as well as whether these viruses are linked to human cancer. Tonsil or cancer DNAs and RNAs were assayed for Ad sequences by liquid-phase saturation-hybridization with in vitro-labeled Ad5 HindIII-G fragment. About 25% of the 52 tonsils analyzed contained DNA or RNA sequences specific to HindIII-G, indicating that Ad transforming sequences are expressed as RNA in tonsils. Southern blotting analysis of four tonsil DNAs revealed multiple copies of the complete Ad genome in a free state and provided evidence for an unusual form of the Ad genome, possibly Ad DNA integrated into cellular DNA. In assays of human cancers, no Ad sequences were detected in DNAs from 26 squamous cell carcinomas (Cas), 3 adenocarcinomas, 4 oat cell Cas, 5 stomach Cas, 5 small intestine Cas, 15 colon Cas, 6 rectum Cas, 5 Hodgkin and 6 non-Hodgkin lymphomas, and 2 breast Cas. Reconstruction experiments indicated that the HindIII-G probe could detect 1 copy per cell of 0.2-0.3% of the viral genome. No HindIII-G-specific sequences were detected in RNAs from 21 squamous cell Cas, 3 oat cell Cas, 2 stomach Cas, or 18 colon Cas. In six other experiments using the complete Ad2 genome as probe, no Ad sequences were found in DNAs from 6 lung Cas, 12 normal lung tissues, 33 gastrointestinal Cas, 19 normal gastrointestinal tissues, 6 Hodgkin lymphomas, 3 breast Cas, or 4 kidney Cas, at a sensitivity of about 1 copy per tumor cell of 5-10% of the Ad2 genome. All Ad-induced cancer cells should contain at least 1 copy of 1-6% of the viral genome, the minimal size of the transforming region, and probably should contain multiple copies of more of the genome. Therefore, our data are definitive evidence against group C Ads being the cause of the cancers tested, which represent about 50% of the cancer incidence in the United States. Of additional interest, we did not detect Ad2 sequences in RNAs from 7 human placentas, 12 normal lungs, or 19 normal gastrointestinal tissues (nor in 44 cancer or 23 tonsil RNAs). Thus, we did not confirm a recent report of the presence of Ad2 RNA in RNAs from human placentas; the possibility that a small population of cells in placenta expresses group C “related” sequences is not ruled out.
Keywords: human cancer, latent tonsil infections, molecular hybridization, Southern blot technique
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt C. D., Kim H. W., Jeffries B. C., Pyles G., Christmas E. E., Reid J. L., Chanock R. M., Parrott R. H. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. II. Variation in adenovirus infections by year and season. Am J Epidemiol. 1972 Mar;95(3):218–227. doi: 10.1093/oxfordjournals.aje.a121389. [DOI] [PubMed] [Google Scholar]
- Chinnadural G., Fujinaga K., Rho M. H., van der Eb A. J., Green M. Transcription of the transforming region of adenovirus type 5 in a rat cell line transformed by a small restriction fragment. J Virol. 1978 Dec;28(3):1011–1014. doi: 10.1128/jvi.28.3.1011-1014.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. P., Brandt C. D., Wassermann F. E., Hall C. E., Spigland I., Kogon A., Elveback L. R. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody response, efficiency of surveillance, patterns of infections, and relation to illness. Am J Epidemiol. 1969 Jan;89(1):25–50. doi: 10.1093/oxfordjournals.aje.a120913. [DOI] [PubMed] [Google Scholar]
- Fox J. P., Hall C. E., Cooney M. K. The Seattle Virus Watch. VII. Observations of adenovirus infections. Am J Epidemiol. 1977 Apr;105(4):362–386. doi: 10.1093/oxfordjournals.aje.a112394. [DOI] [PubMed] [Google Scholar]
- Gallimore P. H. Viral DNA in transformed cells. II. A study of the sequences of adenovirus 2 DNA IN NINE LINES OF TRANSFORMED RAT CELLS USING SPECIFIC FRAGMENTS OF THE VIRAL GENOME;. J Mol Biol. 1974 Oct 15;89(1):49–72. doi: 10.1016/0022-2836(74)90162-4. [DOI] [PubMed] [Google Scholar]
- Graham D. E. The isolation of high molecular weight DNA from whole organisms or large tissue masses. Anal Biochem. 1978 Apr;85(2):609–613. doi: 10.1016/0003-2697(78)90262-2. [DOI] [PubMed] [Google Scholar]
- Green M. R., Mackey J. K., Green M. Multiple copies of human adenovirus 12 genomes are integrated in virus-induced hamster tumors. J Virol. 1977 Apr;22(1):238–242. doi: 10.1128/jvi.22.1.238-242.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M., Mackey J. K., Wold W. S., Rigden P. Thirty-one human adenovirus serotypes (Ad1-Ad31) form five groups (A-E) based upon DNA genome homologies. Virology. 1979 Mar;93(2):481–492. doi: 10.1016/0042-6822(79)90251-4. [DOI] [PubMed] [Google Scholar]
- Green M., Wold W. S. Human adenoviruses: growth, purification, and transfection assay. Methods Enzymol. 1979;58:425–435. doi: 10.1016/s0076-6879(79)58157-9. [DOI] [PubMed] [Google Scholar]
- HUEBNER R. J., ROWE W. P., CHANOCK R. M. Newly recognized respiratory tract viruses. Annu Rev Microbiol. 1958;12:49–76. doi: 10.1146/annurev.mi.12.100158.000405. [DOI] [PubMed] [Google Scholar]
- Jones K. W., Kinross J., Maitland N., Norval M. Normal human tissues contain RNA and antigens related to infectious adenovirus type 2. Nature. 1979 Jan 25;277(5694):274–279. doi: 10.1038/277274a0. [DOI] [PubMed] [Google Scholar]
- Mackey J. K., Brackmann K. H., Green M. R., Green M. Preparation and characterization of highly radioactive in vitro labeled adenovirus DNA and DNA restriction fragments. Biochemistry. 1977 Oct 4;16(20):4478–4483. doi: 10.1021/bi00639a023. [DOI] [PubMed] [Google Scholar]
- Mackey J. K., Green M., Wold W. S., Rigden P. Analysis of human cancer DNA for DNA sequences of human adenovirus type 4. J Natl Cancer Inst. 1979 Jan;62(1):23–26. [PubMed] [Google Scholar]
- Mackey J. K., Rigden P. M., Green M. Do highly oncogenic group A human adenoviruses cause human cancer? Analysis of human tumors for adenovirus 12 transforming DNA sequences. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4657–4661. doi: 10.1073/pnas.73.12.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey J. K., Wold W. S., Rigden P., Green M. Transforming region of group A, B, and C adenoviruses: DNA homology studies with twenty-nine human adenovirus serotypes. J Virol. 1979 Mar;29(3):1056–1064. doi: 10.1128/jvi.29.3.1056-1064.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STROHL W. A., SCHLESINGER R. W. QUANTITATIVE STUDIES OF NATURAL AND EXPERIMENTAL ADENOVIRUS INFECTIONS OF HUMAN CELLS. II. PRIMARY CULTURES AND THE POSSIBLE ROLE OF ASYNCHRONOUS VIRAL MULTIPLICATION IN THE MAINTENANCE OF INFECTION. Virology. 1965 Jun;26:208–220. doi: 10.1016/0042-6822(65)90048-6. [DOI] [PubMed] [Google Scholar]
- Sohier R., Chardonnet Y., Prunieras M. Adenoviruses. Status of current knowledge. Prog Med Virol. 1965;7:253–325. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Van der Eb A. J., Houweling A. Transformation with specific fragments of adenovirus DNAs. II. Analysis of the viral DNA sequences present in cells transformed with a 7% fragment of adenovirus 5 DNA. Gene. 1977;2(3-4):133–146. doi: 10.1016/0378-1119(77)90013-0. [DOI] [PubMed] [Google Scholar]
- Van der Eb A. J., Mulder C., Graham F. L., Houweling A. Transformation with specific fragments of adenovirus DNAs. I. Isolation of specific fragments with transforming activity of adenovirus 2 and 5 DNA. Gene. 1977;2(3-4):115–132. doi: 10.1016/0378-1119(77)90012-9. [DOI] [PubMed] [Google Scholar]
- Wold W. S., Mackey J. K., Brackmann K. H., Takemori N., Rigden P., Green M. Analysis of human tumors and human malignant cell lines for BK virus-specific DNA sequences. Proc Natl Acad Sci U S A. 1978 Jan;75(1):454–458. doi: 10.1073/pnas.75.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W. S., Mackey J. K., Rigden P., Green M. Analysis of human cancer DNA's for DNA sequence of human adenovirus serotypes 3, 7, 11, 14, 16, and 21 in group B1. Cancer Res. 1979 Sep;39(9):3479–3484. [PubMed] [Google Scholar]