Abstract
Background
Krüppel-like factor 4 (KLF4) is involved in many important cellular processes such as growth, development, differentiation, proliferation, and apoptosis. The purpose of this study was to determine the significance of KLF4 in both tumors and normal tissues of patients with colorectal cancer (CRC).
Methods
Between January 2003 and June 2005, 125 patients with CRC receiving treatment at the Yonsei Cancer Center were selected. We examined the mRNA level of the KLF4 gene in primary CRC specimens and matched normal colon tissues using real-time RT-PCR. Correlation of survival with clinicopathological parameters, including KLF4 level, was investigated with univariate and multivariate analyses.
Results
CRC tissue had a significantly lower KLF4 level when compared with matched normal tissues (KLF4 in tumors: 2007 ± 1531 copies/μl, KLF4 in normal tissues: 6586 ± 2834 copies/μl; P <0.0001). However, there was a correlation between the KLF4 level in tumors and normal tissues. Patients with a high KLF4 level in matched normal tissues were more likely than those with a low KLF4 level to develop recurrence and had poorer overall survival (P = 0.005). Therefore, the KLF4 level in the normal tissue of individuals was associated with prognosis of individuals.
Conclusions
Our data suggest that KLF4 mRNA expression level in normal tissues and tumors may be a useful prognostic marker in patients with CRC.
Keywords: KLF4, Prognostic marker, Colorectal cancer
Background
Worldwide, colorectal cancer (CRC) is the fourth most commonly diagnosed cancer in men and third in women. CRC is also the third most common cause of cancer-related death [1]. Despite advances in diagnosis and treatment of CRC, it remains a disease with high morbidity and mortality. Assessment of prognosis through tumor node metastasis (TNM) staging systems remains inappropriate due to the considerable diversity and heterogeneity among tissues of the same stage. Therefore, it is a necessary to establish novel prognostic markers for cancer recurrence and survival to improve treatment for individual patients.
Krüppel-like factors (KLFs) are a family of evolutionarily conserved mammalian zinc-finger transcription factors named for their homology with Krüppel, a Drosophila melanogaster protein [2]. It is now well established that KLFs are involved in many important cellular processes such as growth, development, differentiation, proliferation, and apoptosis [3-5]. KLF4 (also called gut-enriched KLF or GKLF) was one of the first KLF family members identified [6,7]. In addition to regulating many important physiological processes, KLF4 has been shown to play a role in pathologic conditions such as cancer and inflammation [8-13]. More recently, KLF4 was shown to play a crucial role in the reprogramming of somatic cells into induced pluripotent stem cells [10]. KLF4 is primarily expressed in postmitotic, terminally differentiated epithelial cells and is stimulated following p53-dependent DNA damage [14,15].
KLF4 expression is activated in breast cancer and oropharyngeal squamous cell carcinoma [16,17], whereas its expression in colorectal, gastric, and lung cancer is reduced relative to normal tissues [13,18-22]. Thus, the expression and role of KLF4 may vary according to cancer location. Several studies have shown that KLF4 mRNA and protein expression is reduced in human colorectal cancer [13,18,20,22]. Moreover, the level of KLF4 expression is significantly decreased in familiar and/or sporadic colonic adenomas and carcinomas when compared with normal colonic tissues [13]. Taken together, these findings suggest that downregulation of KLF4 protein expression in the colon may contribute to cellular hyperproliferation and malignant transformation. Until recently, the principal focus in cancer research has mostly been the malignant cell itself. However, we now know that tumor growth is not determined by malignant cells alone, but that the tumor microenvironment has a major impact on cancer growth, progression, and prognosis [23]. In this study, we examined the KLF4 mRNA level in colorectal tumors and normal tissues and evaluated the clinical significance of KLF4 expression on prognosis in CRC patients.
Methods
Tissue samples
A total of 125 paraffin-fixed colorectal cancer and matched normal tissue samples were obtained between January 2003 and June 2005 from the Yonsei Cancer Center, Severance Hospital (South Korea). All tumors and matched normal tissues were obtained from surgical specimens of patients with CRC. The matched normal tissues were at least 2 cm away from the edge of corresponding tumors. This study was approved by the Severance Hospital Institutional Review Board, which waived the requirement for informed consent.
Real-time reverse transcription-polymerase chain reaction analysis
Total RNA was isolated using the TRIzol™ method (Invitrogen, Carlsbad, California, United States), and first-strand cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase (Promega, Madison, Wisconsin, United States). Single-strand cDNA was used as a template for subsequent PCR. Two microliters of cDNA from each tissue were used for the real-time reverse transcription-polymerase chain reaction (RT-PCR) assay. Templates were amplified using QuantiTect SYBR Green PCR kit (QIAGEN, Valencia, California, United States). Primer (Proligo, Singapore, Singapore) sequences used in RT-PCR were: 5'-GGCAAAACCTACACAAAGAG-3' and 5'-GTAGTGCCTGGTCAGTTCAT-3'. PCR was initiated at 95°C for 15 minutes to activate the HotstarTaq DNA polymerase (QIAGEN, Hilden, Germany), and then amplified for 35 cycles at 95°C for 20 s, 50°C for 30 s, and 72°C for 45 s on a Rotor Gene 2072D real-time PCR machine (Corbett Research, New South Wales, Sydney, Australia). The amplified fluorescence signal in each specimen was measured at the late extension step of each cycle. To quantify each gene, 10-fold dilution of human genomic DNA was used (Promega).
Data analysis
All statistical analyses were performed using SPSS for Windows software, version 18.0 (SPSS Inc, Chicago, Illinois, United States). We used a paired t-test to compare the KLF4 level between tumors and matched normal tissues. The tKLF4 (KLF4 level in tumors) cutoff level was set to 2150 pg/dl (Area under the curve (AUC) 0.61, sensitivity 60%, specificity 73%), and the nKLF4 (KLF4 level in normal tissues) cutoff level was set to 7969 pg/dl (AUC 0.58, sensitivity 40%, specificity 65%) in a Receiver operating characteritsitc (ROC) curve (Figure 1). The patients were classified into two groups using the KLF4 cutoff levels. The χ2 test was applied to find the correlation between the KLF4 level and clinicopathologic parameters. A binary logistic model was used in the multivariate analysis. Overall survival (OS) was measured from the date of the CRC diagnosis to the date of death. Disease-free survival (DFS) was defined as the time between the diagnosis of CRC and recurrence of the disease. Survival plots were estimated using the Kaplan-Meier method, and differences in survival distributions were evaluated using the log-rank test. In multivariate analysis, Cox proportional hazards models were used to analyze the effect of specified risk factors on OS. P < 0.05 was considered to be statistically significant.
Figure 1.
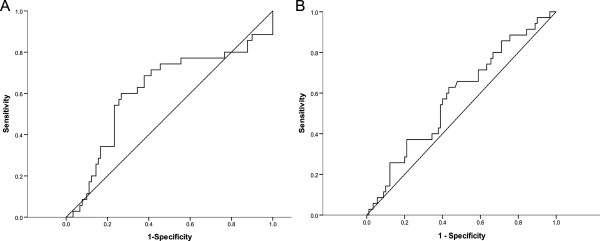
ROC curve. (A) ROC curve of tKLF4 (AUC 0.61). (B) ROC curve of nKLF4 (AUC 0.58).
Results
Patient characteristics
Patient characteristics are shown in Table 1. The median age of patients was 62 years (range 35 to 83 years), and 65.6% were male. On first diagnosis, 16.0% of the patients were stage I (n = 20), 24.8% were stage II (n = 31), 44.0% were stage III (n = 55), and 15.2% were stage IV (n = 19). The anatomic site of the tumors was defined as right colon (ascending colon cancer, transverse colon cancer), left colon (descending colon cancer, sigmoid colon cancer, rectosigmoid colon cancer), and rectum (rectal cancer). Tumors were distributed with 19.2% (n = 24) on the right colon, 36.0% (n = 45) on the left colon, and 44.8% (n = 56) on the rectum. A median of 20 lymph nodes (range 3 to 64) were taken and in only 12.8% (n = 16) of patients, less than 12 lymph nodes were taken. In 56.0% (n = 70) of patients, metastasis to the lymph nodes was shown. Curative resection was performed in 90.4% (n = 113) of patients, and palliative surgery was performed in 9.6% (n = 12) of patients. Among patients with stage IV (n = 19), 36.8% (n = 7) received curative resection (primary tumor resection and metastasectomy) and 63.2% (n = 12) received palliative surgery. The median follow-up period was 68.7 months (range 0.4 to 96.3 months), and the median survival of surviving patients was 94.8 months (range 72.8 to 102.6 months). Thirty-five patients died with a median survival of 29.8 months (range 3.0 to 75.1 months), and the overall five-year survival was 78.4% (98 out of 125).
Table 1.
Patient characteristics
| Number of patients (%) | |
|---|---|
| Total patients |
125 |
| Age in years (median) |
35-83 (62.0) |
| ≤60 |
50 (40.0) |
| ≥60 |
75 (60.0) |
| Sex |
|
| Male |
82 (65.6) |
| Female |
43 (34.4) |
| Positive nodes |
|
| absent |
55 (44.0) |
| present |
70 (56.0) |
| Stage |
|
| I |
20 (16.0) |
| II |
31 (24.8) |
| III |
55 (44.0) |
| IV |
19 (15.2) |
| Primary lesion |
|
| Right colon |
24 (19.2) |
| Left colon |
45 (36.0) |
| Rectum |
56(44.8) |
| Histology |
|
| Well differentiated |
16 (12.8) |
| Moderately differentiated |
100 (80.0) |
| Poorly differentiated |
5 (4.0) |
| Mucinous |
4 (3.2) |
| Blood/lymphatic vessel invasion |
|
| Absent |
109 (87.2) |
| Present |
16 (12.8) |
| Surgery |
|
| Curative |
113 (90.4) |
| Palliative | 12 (9.6) |
Factors associated with KLF4 expression in normal and tumor tissues
KLF4 mRNA expression levels were significantly decreased in CRC tissues compared with matched normal tissues (tKLF4: 2007 ± 1531 copies/μl, nKLF4: 6586 ± 2834 copies/μl) (P <0.0001) (Figure 2). However, there was a correlation between the nKLF4 and tKLF4 level (P <0.0001) (Figure 3). In multivariate and univariate analysis, tKLF4 was significantly associated with the tumor stage (univariate analysis: P = 0.031, multivariate analysis: P = 0.044) (Table 2). nKLF4 level had a significant relationship with age and anatomic distribution in a multivariate logistic model that took into account age, stage, gender, lymph nodes metastasis, and anatomic distribution (P = 0.024, P = 0.046: right and left colon, P = 0.027: right colon and rectum, respectively) (Table 3). No significant differences were observed among gender, differentiation, or lymph node status.
Figure 2.
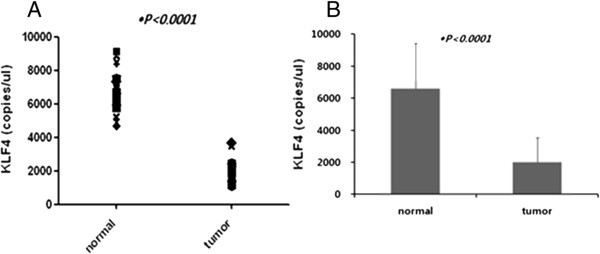
KLF4 expression in CRC and matched normal tissues. (A)KLF4 mRNA expression levels were significantly decreased in CRC tissues compared with matched normal tissues. (B) Bar graph of KLF4 mRNA expression levels.
Figure 3.
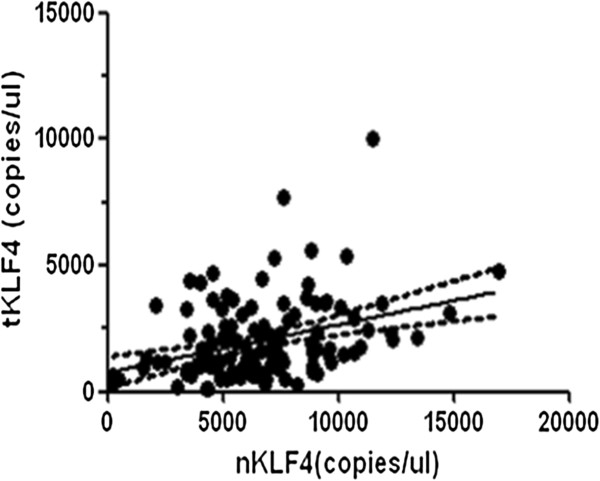
Correlation analysis of nKLF4 and tKLF4 expression. (r2 = 0.349, P <0.0001).
Table 2.
KLF4 expression in CRC and paired normal tissue
| Variable |
tKLF4 |
P value * |
nKLF4 |
P value * | ||
|---|---|---|---|---|---|---|
| <2150 | ≥2150 | <7969 | ≥7969 | |||
|
Age (years) | ||||||
| <60 |
36 (72.0%) |
14 (28.0%) |
|
42 (84.0%) |
8 (16.0%) |
|
| ≥60 |
44 (58.7%) |
31 (41.3%) |
0.128 |
51 (68.0%) |
24 (32.0%) |
0.045 |
|
Gender | ||||||
| Male |
49 (60.5%) |
32 (39.5%) |
|
59 (72.8%) |
22 (27.2%) |
|
| Female |
31 (70.5%) |
13 (29.5%) |
0.268 |
34 (77.3%) |
10 (22.7%) |
0.588 |
|
Stage |
|
|
|
|
|
|
| I-III |
72 (67.9%) |
34 (32.1%) |
|
79 (74.5%) |
27 (25.5%) |
|
| IV |
8 (42.1%) |
11 (57.9%) |
0.031 |
14 (73.7%) |
5 (26.3%) |
0.938 |
|
LN metastasis | ||||||
| + |
46 (64.8%) |
25 (35.2%) |
|
55 (77.5%) |
16 (22.5%) |
|
| − |
34 (63.0%) |
20 (37.0%) |
0.833 |
38 (70.4%) |
16 (29.6%) |
0.368 |
|
Anatomic distribution | ||||||
| Right |
14 (58.3%) |
10 (41.7%) |
|
22 (91.7%) |
2 (8.3%) |
|
| Left |
33 (73.3%) |
12 (26.7%) |
|
32 (71.1%) |
13 (28.9%) |
|
| Rectum | 33 (58.9%) | 23 (41.1%) | 0.264 | 39 (69.9%) | 17 (30.4%) | 0.096 |
*P value estimated by Chi-square χ2 test.
Table 3.
Multivariate analysis of nKLF4 expression
| Variable | Estimated coefficient | Estimated standard error | Wald X2 statistics | P value * | Estimated odds ratio | Confidence interval for odds ratio |
|---|---|---|---|---|---|---|
| Stage (I-III, IV) |
0.108 |
0.611 |
0.031 |
0.860 |
1.114 |
(0.336-3.685) |
| Age in years (<60, ≥60) |
1.087 |
0.482 |
5.076 |
0.024 |
2.965 |
(1.152-7.631) |
| Gender (M, F) |
−0.314 |
0.471 |
0.443 |
0.505 |
0.731 |
(0.290-1.840) |
| LN metastasis (+,−) |
0.649 |
0.459 |
1.994 |
0.158 |
1.913 |
(0.778-4.705) |
| Anatomic distribution (right, left colon) |
1.644 |
0.826 |
3.964 |
0.046 |
5.178 |
(1.026-26.135) |
| Anatomic distribution (right colon, rectum) | 1.801 | 0.815 | 4.880 | 0.027 | 6.054 | (1.225-29.916) |
*P value estimated by binary logistic model including all available covariates.
Implication of KLF4 on survival
Figure 4 shows that OS was significantly poorer in patients with a high expression level of both nKLF4 and tKLF4 (P = 0.036, P <0.0001, respectively). In multivariate analysis, a higher level of nKLF4 (nKLF4 ≥ 7696) was an independent risk factor for survival (P = 0.005). Other independent risk factors for survival were advanced stage, lymph node metastasis, and poor differentiation.
Figure 4.
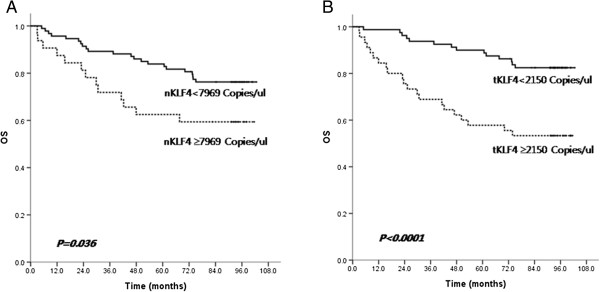
Kaplan-Meier overall survival (OS) curve of nKLF4 and tKLF4 expression in all patients. (A) OS was significantly poorer in patients with high expression level of both nKLF4 (P = 0.036). (B) OS was significantly poorer in patients with high expression level of both tKLF4 (P < 0.0001).
In patients without distant metastases (stage I-III), those with higher nKLF4 had decreased DFS and OS (P = 0.024, P = 0.007, respectively) (Figure 5). In multivariate models that included age, stage, lymph node status, and nKLF4, only nKLF4 was a significant predictor for DFS (P = 0.023) and OS (P = 0.011).
Figure 5.
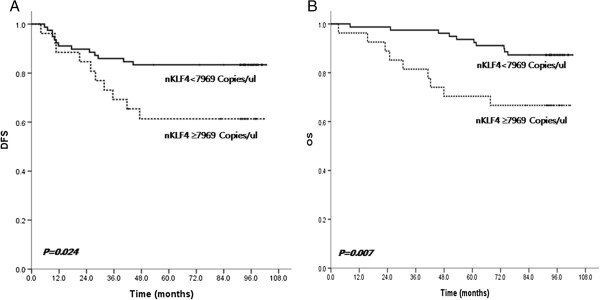
Kaplan-Meier disease-free survival (DFS) and overall survival (OS) curve of nKLF4 in stage I-III patients. (A) In patients without distant metastases (stage I-III), those with higher nKLF4 had decreased DFS (P = 0.024). (B) In patients without distant metastases (stage I-III), those with higher nKLF4 had decreased OS (P = 0.007).
Discussion
Diverse molecular alterations are associated with the initiation and progression of CRC. The zinc-finger transcription factor KLF4 is predominantly expressed in the epithelial cells of the gastrointestinal tract and is an important regulator of intestinal epithelial cell homeostasis and tumorigenesis [18,24,25]. In the few studies that have analyzed the role of KLF4 as a prognostic marker in CRC, a clear relationship has yet to emerge. A recent study showed that loss of KLF4 was an independent predictor of survival and disease recurrence, whereas another study found that loss of KLF4 expression was not associated with survival and disease recurrence [13,22].
In this study, we evaluated the level of KLF4 mRNA expression level using real time RT-PCR in tumors and matched normal tissues. Similar to previous studies, we found a lower expression level of KLF4 in tumors than in normal tissues. Additionally, we observed an elevated KLF4 level in tumors from CRC patients with high KLF4 levels in matched normal tissues. The patients with high KLF4 levels in normal tissues had shorter OS and DFS than those with low KLF4 levels. Therefore, high KLF4 in the normal tissue of individuals appears to be associated with poor prognosis of those individuals with CRC.
A previous study showed that CRC patients who lack KLF4 protein expression in tumors have a poor prognosis [22]. However in our study, patients with high KLF4 mRNA expression in tumors had a worse prognosis than those with low KLF4. A few possibilities exist that may explain this conflicting result. The first is that the different methods used to measure KLF4 expression may produce different results. We measured the level of KLF4 mRNA using real-time RT-PCR, whereas the level of KLF4 protein was detected by immunohistochemistry in the other study [22]. The difference in protein and mRNA expression levels suggests that KLF4 may be regulated at multiple levels. The second possibility is that CRC tissues contained not only tumor cells but also other cells such as surrounding stromal cells. In such a case, elevated KLF4 expression in surrounding stromal cells, despite low KLF4 expression in the tumor cell itself, would skew our analysis, leading us to incorrectly conclude that KLF4 expression in tumors is elevated. We are unaware of any other study that has analyzed the association between KLF4 expression in normal tissues and prognosis. Our study is the first to identify high KLF4 expression in normal tissues as an independent prognostic marker for poor survival and disease recurrence.
Recent studies have revealed that hypermethylation of the KLF4 5′-untranslated region and microRNA-146a (miR-146a), miR-145, or miR-143 expression are associated with altered KLF4 expression in human CRC tissue specimens [18,26,27]. Further studies investigating hypermethylation of the KLF4 5′-untranslated region and microRNA expression will be important to determine how KLF4 expression is regulated in normal and tumor tissues. The most striking aspect of our study is that KLF4 mRNA expression level in normal tissues is an independent predictor of OS and DFS. We identified a significant association between the KLF4 level in both tumor and normal tissue and survival.
Conclusions
Our data suggests that KLF4 mRNA expression level in both normal and tumor tissue is a potential prognostic marker in patients with CRC. Investigating how the microenvironment influences KLF4 expression and thus tumorigenesis, tumor progression, and prognosis, will be important in future studies.
Abbreviations
AUC: Area under curve; CRC: colorectal cancer; DFS: Disease-free survival; KLFs: Krüppel-like factors; nKLF4: KLF4 level in normal tissues; OS: Overall survival; ROC curve: Receiver operation characteristic curve; RT-PCR: Real-time reverse transcription-polymerase chain reaction; tKLF4: KLF4 level in tumors; TNM: tumor-node-metastasis.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HYL wrote the manuscript. NKK collected and provided the tissues. JBA, SYR, HCC and SJS have contributed part of the experiment, research design, and the data collection. KHP and TSK performed the experiments. SJS oversaw the design of the study, was involved in the critically revised the manuscript. All authors approved the final manuscript.
Contributor Information
Ha-young Lee, Email: hieveryone@hanmail.net.
Joong Bae Ahn, Email: Vvswm513@yuhs.ac.
Sun Young Rha, Email: rha7655@yuhs.ac.
Hyun Cheol Chung, Email: unchung8@yuhs.ac.
Kyu Hyun Park, Email: kyuhyun@yuhs.ac.
Tae Soo Kim, Email: taesoo@yuhs.ac.
Nam Kyu Kim, Email: namkyu@yuhs.ac.
Sang Joon Shin, Email: SSJ338@yuhs.ac.
Acknowledgments
This research was supported by the Public Welfare and Safety Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (grant number: 2010–0020841). We thank Eun sil Baek and Han na Park for data collection.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Preiss A, Rosenberg UB, Kienlin A, Seifert E, Jäckle H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313:27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- Ghaleb AM, Aggarwal G, Bialkowska AB, Nandan MO, Yang VW. Notch inhibits expression of the Kruppel-like factor 4 tumor suppressor in the intestinal epithelium. Mol Cancer Res. 2008;6:1920–1927. doi: 10.1158/1541-7786.MCR-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaleb AM, McConnell BB, Nandan MO, Katz JP, Kaestner KH, Yang VW. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 2007;67:7147–7154. doi: 10.1158/0008-5472.CAN-07-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le X, Yao J, Xie K. Loss of Kruppel-like factor 4 expression contributes to Sp1 overexpression and human gastric cancer development and progression. Clin Cancer Res. 2006;12:6395–6402. doi: 10.1158/1078-0432.CCR-06-1034. [DOI] [PubMed] [Google Scholar]
- Xu J, Lu B, Xu F, Gu H, Fang Y, Huang Q, Lai M. Dynamic down-regulation of Kruppel-like factor 4 in colorectal adenoma-carcinoma sequence. J Cancer Res Clin Oncol. 2008;134:891–898. doi: 10.1007/s00432-008-0353-y. [DOI] [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai SK, Yang MH, Chang SY, Chang YC, Li WY, Tsai TL, Wang YF, Chu PY, Hsieh SL. Persistent Kruppel-like factor 4 expression predicts progression and poor prognosis of head and neck squamous cell carcinoma. Cancer Sci. 2011;102:895–902. doi: 10.1111/j.1349-7006.2011.01859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Li J, Chen H, Fu J, Ray S, Huang S, Zheng H, Ai W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- Choi BJ, Cho YG, Song JW, Kim CJ, Kim SY, Nam SW, Yoo NJ, Lee JY, Park WS. Altered expression of the KLF4 in colorectal cancers. Pathol Res Pract. 2006;202:585–589. doi: 10.1016/j.prp.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Hu W, Hofstetter WL, Li H, Zhou Y, He Y, Pataer A, Wang L, Xie K, Swisher SG, Fang B. Putative tumor-suppressive function of Kruppel-like factor 4 in primary lung carcinoma. Clin Cancer Res. 2009;15:5688–5695. doi: 10.1158/1078-0432.CCR-09-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NV, Ghaleb AM, Nandan MO, Yang VW. Expression of the tumor suppressor Kruppel-like factor 4 as a prognostic predictor for colon cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:2631–2638. doi: 10.1158/1055-9965.EPI-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak HF, Weaver VM, Tlsty TD, Bergers G. Tumor microenvironment and progression. J Surg Oncol. 2011;103:468–474. doi: 10.1002/jso.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SG, Zheng B, Han M, Han M, Fang XM, Li HX, Miao SB, Su M, Han Y, Shi HJ, Wen JK. miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12:56–62. doi: 10.1038/embor.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]


