Abstract
The p75 neurotrophin receptor (p75NTR) is normally expressed in cerebellar Purkinje cells throughout the lifespan. Children with autism spectrum behavior exhibit apparent cerebellar Purkinje cell loss. Cerebellar transcriptome changes seen in the murine prenatal valproate exposure model of autism include all of the proteins known to constitute the p75NTR interactome. p75NTR is a modulator of cytoplasmic and mitochondrial redox potential, and others have suggested that aberrant response to oxidant stress has a major role in the pathogenesis of autism. We have created Purkinje cell-selective p75NTR knockout mice that are the progeny of hemizygous Cre-Purkinje cell protein 2 C57Bl mice and p75NTR floxed C57Bl mice. These Cre-loxP mice exhibit complete knockout of p75NTR in ~50% of the cerebellar Purkinje cells. Relative to Cre-only mice and wild-type C57Bl mice, this results in a behavioral phenotype characterized by less allogrooming of (P<0.05; one-way analysis of variance) and socialization or fighting with (each P<0.05) other mice; less (1.2-fold) non-ambulatory exploration of their environment than wild-type (P<0.01) or Cre only (P<0.01) mice; and almost twofold more stereotyped jumping behavior than wild-type (P<0.05) or Cre (P<0.02) mice of the same strain. Wild-type mice have more complex dendritic arborization than Cre-loxP mice, with more neurites per unit area (P<0.025, Student's t-test), more perpendicular branches per unit area (P<0.025) and more short branches/long neurite (P<0.0005). Aberrant developmental regulation of expression of p75NTR in cerebellar Purkinje cells may contribute to the pathogenesis of autism.
Introduction
Autism is a neurodevelopmental condition characterized by sparse and inappropriate social interaction; inability to properly ‘read' non-verbal cues and social context; and stereotypic, repetitive behaviors.1 Its prevalence has been estimated to be 1 in 80 children2 and it occurs alone and in association with a large number of defined neurodevelopmental disorders.1 Aberrancy of cerebellar Purkinje cell connectivity, expression of brain-derived neurotrophic factor (BDNF) and handling of inflammation and oxidative stress have been implicated in the manifestations of autism.3, 4, 5, 6 Candidate molecules for involvement in the pathogenesis of autism should best demonstrate developmental regulation and relevance to these processes and functions.
p75NTR is ubiquitously expressed in the embryonic brain. Its expression is increasingly anatomically restricted as the brain matures. In the adult brain, it persists in the cerebellum, hippocampus and basal forebrain. p75NTR is one of two receptors for which BDNF is a ligand. p75NTR is expressed by Purkinje cells and has been associated with modulation of cell survival and intracellular redox state. In fact, we and others have shown that p75NTR exhibits many of the characteristics one would predict are exhibited by molecular mediators of autism (see Table 1).7, 8, 9, 10, 11, 12, 13 We now demonstrate that knockout of the p75 neurotrophin receptor (p75NTR) from cerebellar Purkinje cells of mice results in autism-like stereotypies, diminished social interaction, decreased interest in novel environments and decreased complexity of the dendritic arborization of cerebellar Purkinje cells.
Table 1. Alignment of characteristics of putative molecular mediators of autism and known characteristics of p75NTR.
| Putative molecular mediators of autism | p75NTR |
|---|---|
| Developmentally regulated brain expression or function with an inflection point at ~2 years in the human1 | Developmentally regulated brain expression with an inflection point at ~2 years in human7,13 |
| Differential expression in cerebellar Purkinje cells, cholinergic basal forebrain, hippocampal pyramidal cells relative to rest of the brain3,14 | Differential expression in cerebellar Purkinje cells, cholinergic basal forebrain, hippocampal pyramidal cells relative to rest of the brain8,13 |
| Modulated by oxidative stress and inflammation5,6 | Induces enhanced GSSG → GSH cycling in oxidative conditions9,10 |
| Involved in BDNF activity4 | BDNF is one of its cognate ligands7 |
| Expression and/or activity (itself and/or downstream effectors) altered in known models of autism15 | All molecular species known and published to be part of the p75NTR ‘interactome' exhibit altered (and most, upregulated) expression in the prenatal valproate model of autism16 |
Abbreviations:
BDNF, brain-derived neurotrophic factor; p75NTR, p75 neurotrophin receptor.
Materials and methods
Cerebellar Purkinje cell-specific p75NTR knockout mice
Mice of a C57Bl6J background with loxP sites flanking all p75NTR spliced isoforms (exons 4–6) were obtained from Vesa Kaartinen, PhD (University of Michigan).17 Mice of a C57Bl6 background with a hemizygous Cre gene driven by the Purkinje cell protein-2 promoter (Pcp-2) and previously shown to express Cre selectively in cerebellar Purkinje cells were obtained from Jackson Laboratories (Bar Harbor, ME, USA; strain B6.129-Tg(Pcp2-cre)2Mpin/J; stock number 004146). These mice were developed as follows: the transgene vector was electroporated into R1 ES cells and positive clones were injected into C57BL/6J blastocysts. Resulting chimeras were crossed with C57BL/6NCrl. The strain has been backcrossed for a total of six generations to C57BL/6J. Cre is variably turned on in post-natal life and half of the progeny resulting from mating of these hemizygous mice with floxed mice exhibit knockout of the floxed gene in ~ 50% of their Purkinje cells. We used this hemizygous targeted Cre mouse because the Pcp-2 promoter-linked homozygous Cre mouse dies during embryonic life (http://jaxmice.jax.org/strain/004146.html, accessed 16 December 2013). The Pcp-2 Cre mice were cross-bred with the p75NTR-floxed mice. The genomic DNA of the resulting progeny was extracted from tail snips by using the Wizard SV Genomic Purification System Kit (Promega, Madison, WI, USA, no. A2360). Genotyping was performed with PCR using an Eppendorf Master Cycler. The protocol for detection of Cre was obtained from Jackson Laboratories (http://jaxmice.jax.org/protocolsdb/f?p=116:2:2606126352577965::NO:2:P2_MASTER_PROTOCOL_ID,P2_JRS_CODE:288,010536; accessed 16 December 2013); the protocol for detection of p75NTR was obtained from Dr Kaartinen.17 Primers used were as follows: Cre, 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and 5′-GTGAAACAGCATTGCTGTCACTT-3′ P75-C, 5′-TGCAGAAATCATCGACCCTTCCC-3′ P75-D: 5′-TCCTCACCCCGTTCTTTCCCC-3′. Littermates with the range of genotypes were used, and a total of only three litters were used in these studies of 15 mice. Control (non-Cre, non-loxP) mice were of C57BL/6J background, as recommended by Jackson Laboratories (http://jaxmice.jax.org/strain/004146.html, accessed 16 December 2013).
Mice were housed three to four per cage in the University of Rochester Medical Center Animal Facility, under routine, 12-h light–dark cycle, and allowed access to food and water ad libitum.
Brain immunohistochemistry
Mice were killed by pentobarbital injection, followed by saline/4% paraformaldehyde perfusion. Brain sectioning was cryostatically performed with stereotactic coordinates noted in all cases. Cerebellar Purkinje cells were identified and their dendritic networks visualized by immunohistochemical staining with anti-calbindin antibodies (Monoclonal Anti-Calbindin-D-28K Clone CB-955, mouse IgG, 1:1000; Sigma, St Louis, MO, USA). Staining for p75NTR was performed using anti-p75 antibodies (Anti-Nerve Growth Factor Receptor Antibody, p75, AB1554, rabbit IgG, 1:1000; Millipore, Billerica, MA, USA). Secondary antibodies used included Alexa Fluor 488, A11008 goat anti-rabbit and Alexa Fluor 594, A11005 goat anti-mouse, 1:1000 (Life Technologies, Carlsbad, CA, USA). Quantitative analysis of stained images was performed as described below and in the legends for Figures 1 and 2. Statistical analysis of differences between groups was performed using Student's t-test and considering P-values maximally 0.05 to be indicative of statistical significance.
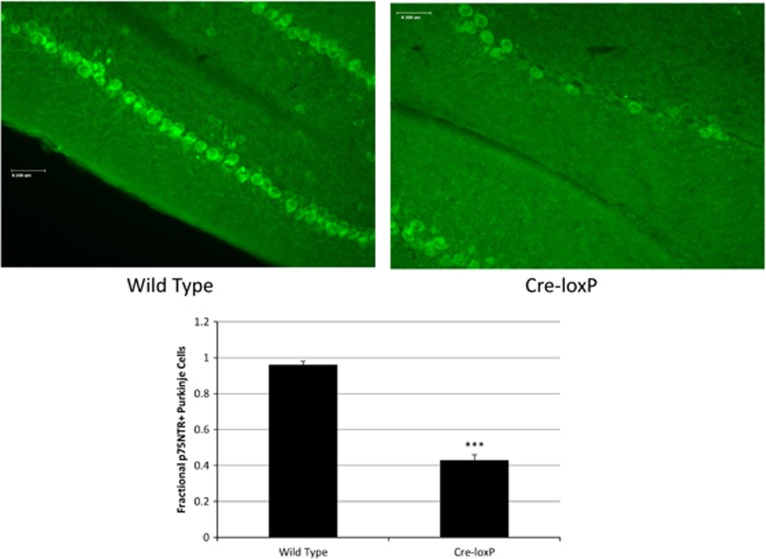
Figure 1.
Representative cerebellar sections from wild-type and Cre-loxP mouse, respectively, stained with antibodies to p75NTR ( × 20; scale bar=0.5 μm). The graph depicts the fraction of calbindin-positive (that is, Purkinje) cells that are p75NTR-positive in each of ‘n' 1 μm2 areas along the folium for each mouse genotype (n=82 areas for wild-type mice; n=59 areas for Cre-loxP mice; error bar=s.e.m.; ***P<0.0005 relative to wild type).
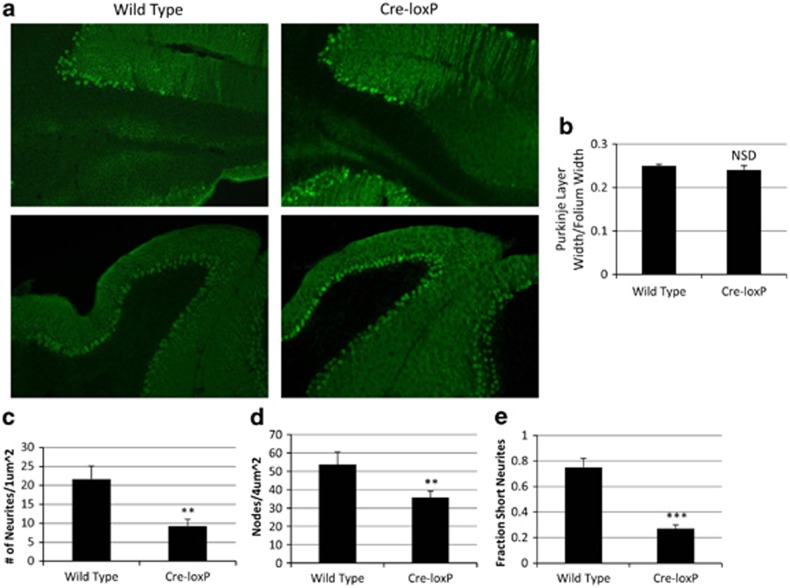
Figure 2.
(a) Representative areas of a calbindin-stained sagittal section of cerebellum ( × 10; Bregma-6.84) from one wild-type and one Cre-loxP mouse, respectively, of three each examined. (b) Fraction of folium width occupied by Purkinje cell layer in wild-type versus Cre-loxP mice. (n=3 for each genotype; error bar=s.e.m.; NSD, wild type and Cre-loxP do not differ significantly from one another). (c, d) Number of neurites per μm2 area (c); visualized at × 20; n=3 for each genotype; error bar=s.e.m.; **P<0.025 relative to wild type and number of dot-like ‘nodes' per 4 μm2 area (d); visualized at × 10; n=12 for each genotype; error bar=s.e.m.; **P<0.025 relative to wild type. (e) Number of short neurite branches off of long neurites/total number of neurites (that is, fraction of neurites that are short branches off of longitudinal neurites; n=3 for each genotype; visualized at × 20; error bar=s.e.m.; ***P<0.0005 relative to wild type).
Quantitative microanatomy of Purkinje cells
The cerebellum from each of the three Cre-loxP mice and the cerebellum from each of three wild-type mice were cryostatically sectioned. Sections from Cre-loxP mice and wild-type mice, respectively, with the same stereotactic coordinates were stained with anti-calbindin antibodies as noted above, visualized at × 10 and × 20 magnification, and compared by manual counting by an observer blind to the genotype of the animal of its origin of the number of longitudinal dendrites; shorter branches off of longitudinal dendrites; and dot-like ‘nodes' representing dendrite branches perpendicular to the plane of section, per standardized square (4 μm2 for × 10 sections; 1 μm2 for × 20 sections). The standardized square was moved along the folia in approximately one Purkinje cell diameter increments and within-square counts were performed for each position of the square. Positions for which the width of a given folium was smaller than a side of the square were excluded from the determination.
Studies of murine behavior
Motor and behavioral studies were performed in the University of Rochester Medical Center Behavioral Sciences Facility Core (URMC Behavioral Sciences Facility Core; http://www.urmc.rochester.edu/environmental-health-sciences/integrative-health-sciences-core/behavioral-sciences-facility-core/animal-facilities.aspx#Behaviors; accessed 7 November 2013).
Spontaneous locomotor activity
In brief, spontaneous locomotor activity was measured by the Open Field Test in photobeam chambers equipped with a transparent acrylic arena with a 48-channel infrared source, detector and controller (Med Associates). Data acquired from the Open Field Test included total jump counts, total jump time, average ambulatory velocity, number and duration of ambulatory episodes, ambulatory distance, total ambulatory time, total stereotypic time, total stereotypic counts, total resting time, total vertical (rearing) counts and total vertical time. Ambulatory counts are defined as number of episodes in which three successive horizontal photobeams were broken; vertical counts are defined by breaks of the z axis photobeam (located 5.25 cm above the arena floor) and stereotypy by beam breaks within a small defined area within the larger field. The Open Field Test session was videotaped and scored by an observer blinded to the experimental conditions for time spent in and number of episodes of stereotypies, respectively.
Social behavior
The mouse to be tested (‘resident') was first habituated to the Plexiglas chamber in which social dyadic interaction with a novel (‘stranger') mouse was to be assessed; testing occurred 24 h after this habituation session, at which time the resident mouse was placed in the arena and a novel mouse was introduced. The subsequent behavior was videotaped. An observer blinded to all experimental conditions scored each videotape for total number of occurrences per 5 min epoch of each of several interactive, operationally defined behaviors initiated by the resident mouse, including social investigation (sniffing), allogrooming, pursuit, sideways offensive posture, submissive posture, defensive upright boxing, side-by-side activity and escape, respectively.
Statistical analysis of differences between groups was performed using one-way analyses of variance with Statview version 5.0 (SAS Institute, Cary, NC, USA); P<0.05 was considered statistically significant.
Results
Expression of p75NTR in cerebellar Purkinje cells of Purkinje cell Cre-p75NTR loxP (Cre-loxP) mice
We have studied expression of p75NTR in Cre-loxP mice. These mice are the result of cross-breeding of the floxed p75NTR C57Bl/6J mouse developed by Bogenmann et al.17 with the hemizygous Cre-Purkinje cell protein-2 C57Bl/6J mouse (Jackson Laboratories). (Note that the homozygous Purkinje cell protein-2-driven Cre C57Bl/6J mouse dies in embryonic life (JAX Mice Database, http://jaxmice.jax.org/strain/010536.html, accessed 23 August 2013). Figure 1 demonstrates that, in wild-type C57Bl/6J mice, 100% of the calbindin-positive cells in 82 1-μm2 areas counted were p75NTR-positive; in contrast, in Cre-loxP mice, ~50% of calbindin-positive cells in 59 1- μm2 areas counted were p75NTR-positive.
Purkinje cell layer cytoarchitecture in Cre-loxP mice
There is no significant difference between wild-type and Cre-loxP mice in the relative thickness of the cerebellar Purkinje cell layer (Figures 2a and b). However, as Figure 2a shows, the cerebellar folia of wild-type mice have a more densely packed, homogeneous appearance than those of Cre-loxP mice. In addition, the numbers of calbindin-positive longitudinal dendrites (Figure 2c) and dot-like nodes, representing dendritic branches perpendicular to the plane of section (Figure 2d) per unit area are greater in wild-type mice than in Cre-loxP mice. Furthermore, the number of transverse short branches (within the plane of section) per longitudinal dendrite is greater in wild-type mice than in Cre-loxP mice (Figure 2e). This is consistent with the finding of others in hippocampal neurons that expression of p75NTR enhances the density of the dendritic network.18
Behavioral phenotype of Cre-loxP mice
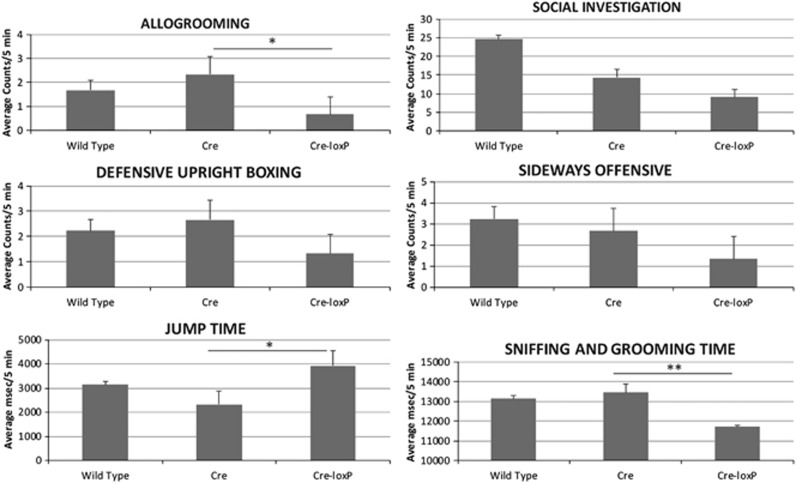
We have studied the behavior of wild-type, Purkinje cell Cre only (Cre) and Cre-loxP mice. Figure 3 shows that 6-week-old male Cre-loxP mice demonstrate less allogrooming of (P<0.05; one-way analysis of variance) and socialization or fighting (each P<0.05) with other mice. They also demonstrate less (1.2-fold) non-ambulatory exploration of their environment (for example, sniffing, rearing on hind legs) than wild-type (P<0.01) or Cre only (P<0.01) mice. Finally, they exhibit almost twofold more stereotyped jumping behavior than wild-type (P<0.05) or Cre (P<0.02) mice of the same strain. Ambulatory time, velocity/distance traversed and gait were similar for all three mouse genotypes (data not shown).
Figure 3.
Behavioral testing of wild-type (C57Bl6; n=9), Cre only (CrePCP2; n=3) and Cre-loxP (CrePCP2P75FL/FL; n=3) mice. Behavioral testing was performed as detailed in the Materials and Methods section, performance epochs were captured on videotape and the analysis was completed from photobeam break recordings and from the videotapes scored by an observer blinded to the genotypes of the mice. Ordinate labels for allogrooming, social investigation, defensive upright boxing and sideways offensive denote counts per 5-min epochs. Ordinate labels for jump time, vertical time and sniffing and grooming time denote ms spent in the behavior per 5-min epochs. *P<0.05; **P<0.01 (error bar=s.e.m.).
Discussion
Neuronal p75NTR expression in the human central nervous system is widespread during early embryonic life. It becomes successively more and more restricted and is limited to a few loci by the age of 2 years. The normal time at which the abundance and localization of p75NTR expression change coincides with the time of loss of language function and onset of noticeably aberrant social interactions in children with autism.7,8,13 By the age of 2 years in human, its expression is limited to the hippocampus, cholinergic basal forebrain and cerebellum (particularly the external granular and the Purkinje cell layers).8,13,19 All of these regions have been implicated in autism. Recent studies of others have implicated the cerebellum, and particularly abnormalities of the cerebellar vermis, in autistic behaviors.3,20,21 Patients with autism of many etiologies have lower cerebellar vermis size and Purkinje cell number than non-autistic matched control patients. Knockout of the tuberous sclerosis-associated protein, tuberin, in animal models results in Purkinje cell apoptosis and loss, a finding that is of particular interest, given the robust association of tuberous sclerosis with autistic behaviors.14,22
The role of BDNF and p75NTR in prenatal cerebellar foliation has been studied in the p75NTR exon III deletion mutant mouse;23 however, our studies8 and those of others24 have demonstrated expression of p75NTR alternate splice variants, many of which retain biological activity, in this model, making it difficult to know what these ‘knockout' studies mean for the function of p75NTR signaling in the cerebellum during embryogenesis. The present studies employ a true p75NTR conditional knockout mouse17 cross-bred with a Purkinje cell protein-2-linked hemizygous Cre mouse to create knockout of p75NTR in 50% of cerebellar Purkinje cells. They demonstrate altered dendritic arborization of the cerebellar Purkinje cells in these focal knockout mice.
In our comparison of the p75NTR proteomic ‘interactome' with the set of genes the expression of which is altered in the murine valproate autism model16 and with genes associated with human autism (The Wall Lab, Autworks, http://autworks.hms.harvard.edu/, accessed 30 August 2013), we have demonstrated many points of overlap. Most notably, 20 of the 39 genes in the p75NTR interactome are highly correlated with genes implicated in human autism and 18 of them are themselves implicated in neurobehavioral and/or neurodegenerative disorders. In addition, the genes with most different gene expression in the hippocampi of young autistic versus young normal brains fall into four functional categories: (a) genes related to apoptosis; (b) genes related to cell cycle and differentiation; (c) genes related to lipid metabolism and intracellular shuttling; and (d) genes related to the cytoskeleton and extracellular matrix.25 We have similarly reported that changed gene expression in p75NTR-positive versus p75NTR-negative PC12 cells demonstrates the same functional groups, including genes responsible for apoptosis induction and prevention; genes that code for mediators of inflammation that also have a role in inflammatory and neuroblastic cell cycling and differentiation; genes that code for the enzymes involved in de novo cholesterol synthesis; and genes that code for NCAMs.8,13
Mutations of the NGFB gene have been described that impair the processing of proNGF to NGF and the binding of both proNGF and NGF to p75NTR.26 These mutations lead to the absence of pain perception. Autism is hypothesized to include among its primary features anomalous interpretation of sensory signals—touch, sound and vision.27 Furthermore, loss of p75NTR leads to changes in hippocampal cholinergic signaling, which may be involved in regulation of stress-enabled hippocampal long-term depression and in modulating behaviors related to stress and anxiety28—a common feature of autism spectrum disorders.1
Several studies have shown increased lipid peroxidation and levels of oxidized glutathione and decreased levels of cellular antioxidant enzymes and proteins in the brains of autistic subjects. Markers of inflammation, mitochondrial and immune dysfunction, and altered metabolism of homocysteine and methionine, all evidence of oxidant stress, have also been associated with autism.5,6 We have reported that p75NTR is a cytoplasmic antioxidant and mitochondrial oxidant in neural crest cell lines.9, 10, 11 We have defined its antioxidant signaling pathway to involve neurotrophin binding to p75NTR; upregulation of the cholesterol biosynthetic pathway; palmitoylation of p75NTR; phosphorylation of p75NTR; cleavage of p75NTR by α- and γ-secretase to its intracellular fragment, p75ICD; translocation of p75ICD to the nucleus; and phosphorylation of Akt.10,11 Its mitochondrial oxidant pathway involves phosphorylation of JNK and ‘leakiness' of Complex II to superoxide.11
BDNF-p75NTR signaling is required for migration of cerebellar neurons during development.23 It is therefore perhaps not surprising that aberrations in BDNF regulation and signaling have been implicated in autism. Furthermore, induction of expression of p75NTR in neural crest cells induces expression of CHOP-10, an effector in the BDNF signaling pathway in neurons. We have shown CHOP-10 to be variably pro- or anti-apoptotic depending on its stoichiometric relationship with the related bZIP heterodimeric factors, cEBP-β and ATF-4, and whether or not expression of Tribbles-3, an inhibitor of Akt1, is induced.29 Interestingly, cEBP-β has previously been identified as an environmentally responsive gene in a region that has been found to harbor autism-associated mutations.15,30
In recent genome-wide association studies, genes for cell adhesion molecules such as the cadherins and neuroligin fulfill criteria for autism spectrum disorder-associated genes.15 Our studies of genes that are coordinately regulated with p75NTR demonstrated the relationship between expression of p75NTR and expression of the neural cell adhesion molecules, NCAM and L1.13 In addition, a high-density single nucleotide polymorphism study implicated chromosome 17q21 and the CACNA1G gene in autism.30 The NGFR gene, which codes for p75NTR, is located at chromosome 17q21–22, and may be a viable candidate gene, as well.
One prevalent theory of the pathogenesis of autism relates to a deficiency of plasticity of axonal sprouting and synaptic connectivity.31 p75NTR has been demonstrated to modulate dendrite complexity, the balance of excitatory and inhibitory synapses, and functional and structural plasticity in the cerebellum and hippocampus.18
These parallels between the functional mechanisms that underlie p75NTR ontogeny and signaling and the clinical and neuroanatomic characteristics of autism led us to hypothesize that knockout of p75NTR in cerebellar Purkinje cells would result in behaviors and neuroanatomic features that resemble those seen in autistic individuals. Our studies demonstrate that, indeed, mice with cerebellar Purkinje cell p75NTR knockout exhibit stereotypic motor automatisms, decreased social interaction with stranger mice, decreased exploration of novel environments and diminished complexity of Purkinje cell dendritic arborization, characteristics also reported in individuals with autism.
p75NTR functions both as a co-receptor with Trk receptors and an independently signaling receptor.7 Its co-receptor function enhances the affinity of TrkA for NGF and TrkB for BDNF,32 both relationships that may have relevance to autism.4,33 Its independent receptor function is thought to involve binding of NGF, BDNF or NT-3,7 cleavage of the holoreceptor to its intracellular fragment,34 and translocation of the intracellular fragment to the nucleus,35 where it acts as a transcription factor to upregulate expression of, among other things, cell matrix proteins13 and enzymes responsible for the de novo synthesis of cholesterol.8 The ‘naked' p75NTR includes a ‘death domain' and has been shown to have apoptosis-inducing properties,36 whereas the NGF-bound p75NTR is variably pro- or anti-apoptotic12,37 and pro- or anti-oxidant,9,11 depending on the cellular milieu in which it is expressed. The latter paradox most likely relates to the finding that signaling through p75NTR proceeds through a branch point that can lead to activation of apotosis-inducing JNK activation or apoptosis-suppressing NF-κB and IAP activation, respectively.38 The present findings and their relationship to the known transcriptional and signaling effectors of p75NTR will fuel identification and pharmacological targeting of druggable contributors to autism.
Acknowledgments
We are grateful to Vesa Kaartinen for providing the p75NTR-floxed founder mice for these studies and to Robert H Schor and Simeng Wang for assistance with preparation of the figures. We also thank Christopher Stodgell for pointing out the relationship between the p75NTR interactome and the valproate model transcriptome. These studies were funded by the William H. Eilinger Endowment of the Golisano Children's Hospital at the University of Rochester Medical Center and by a Strong Children's Research Center Pilot Grant. NFS and DC-S have been funded by the NIH.
The authors declare no conflicts of interest.
References
- Rapin I. Autistic children: diagnosis and clinical features. Pediatrics. 1991;87:751–760. [PubMed] [Google Scholar]
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2008 Principal Investigators Prevalence of Autism Spectrum Disorders—Autism and Developmental Disabilities Monitoring Network, 14 Sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Sheikh AM, Malik M, Wen G, Chauhan A, Chauhan V, Gong CX, et al. BDNF-Akt-Bcl2 antiapoptotic signaling pathway is compromised in the brain of autistic subjects. J Neurosci Res. 2010;88:2641–2647. doi: 10.1002/jnr.22416. [DOI] [PubMed] [Google Scholar]
- Ghanizadeh A, Akhondzadeh S, Hormozi M, Makarem A, Abotorabi-Zarchi M, Firoozabadi A. Glutathione-related factors and oxidative stress in autism, a review. Curr Med Chem. 2012;19:4000–4005. doi: 10.2174/092986712802002572. [DOI] [PubMed] [Google Scholar]
- Khansari PS, Sperlagh B. Inflammation in neurological and psychiatric diseases. Inflammopharmacol. 2012;20:103–107. doi: 10.1007/s10787-012-0124-x. [DOI] [PubMed] [Google Scholar]
- Schor NF. p75NTR in development and disease. Prog Neurobiol. 2005;77:201–214. doi: 10.1016/j.pneurobio.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Korade Z, Mi Z, Portugal C, Schor NF. Expression and p75 neurotrophin receptor dependence of cholesterol synthetic enzymes in adult mouse brain. Neurobiol Aging. 2007;28:1522–1531. doi: 10.1016/j.neurobiolaging.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Nylander KD, Mirnics ZK, Portugal C, Yan C, Zaccaro C, et al. The intracellular domain of p75NTR as a determinant of cellular reducing potential and response to oxidant stress. Aging Cell. 2005;4:187–196. doi: 10.1111/j.1474-9726.2005.00160.x. [DOI] [PubMed] [Google Scholar]
- Mi Z, Rogers DA, Korade Mirnics Z, Schor NF. p75NTR-dependent modulation of cellular handling of reactive oxygen species. J Neurochem. 2009;110:296–306. doi: 10.1111/j.1471-4159.2009.06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan V, Ashton J, Schor NF. p75NTR: an enhancer of fenretinide toxicity in neuroblastoma. Cancer Chemother Pharmacol. 2013;71:777–787. doi: 10.1007/s00280-013-2071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Mirnics ZK, Portugal CF, Liang Y, Nylander KD, Rudzinski M, et al. Cholesterol biosynthesis and the pro-apoptotic effects of the p75 nerve growth factor receptor in PC12 pheochromocytoma cells. Mol Brain Res. 2005;139:225–234. doi: 10.1016/j.molbrainres.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Mirnics ZK, Yan C, Portugal C, Kim TW, Saragovi HU, Sisodia SS, et al. P75 neurotrophin receptor regulates expression of neural cell adhesion molecule 1. Neurobiol Dis. 2005;2:969–985. doi: 10.1016/j.nbd.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt TL, Stodgell CJ, Rodier PM. The teratology of autism. Int J Dev Neurosci. 2005;23:189–199. doi: 10.1016/j.ijdevneu.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Bogenmann E, Thomas PS, Li Q, Kim J, Yang LT, Pierchala B, et al. Generation of mice with a conditional allele for the p75(NTR) neurotrophin receptor gene. Genesis. 2011;49:862–869. doi: 10.1002/dvg.20747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M, Holz A, Dechant G, Barde YA, Bonhoeffer T, Korte M. The p75 neurotrophin receptor negatively modulates dendrite complexity and spine density in hippocampal neurons. J Neurosci. 2005;25:9989–9999. doi: 10.1523/JNEUROSCI.2492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes M, Eberhart CG, Collins R, Tihan T. Expression of p75NTR in fetal brain and medulloblastomas: evidence of a precursor cell marker and its persistence in neoplasia. J Neurooncol. 2009;92:193–201. doi: 10.1007/s11060-008-9755-6. [DOI] [PubMed] [Google Scholar]
- Bolduc ME, du Plessis AJ, Sullivan N, Guizard N, Zhang X, Robertson RL, et al. Regional cerebellar volumes predict functional outcome in children with cerebellar malformations. Cerebellum. 2012;11:531–542. doi: 10.1007/s12311-011-0312-z. [DOI] [PubMed] [Google Scholar]
- O'Halloran CJ, Kinsella GJ, Storey E. The cerebellum and neuropsychological functioning: A critical review. J Clin Exp Neuropsychol. 2011;1:1–22. doi: 10.1080/13803395.2011.614599. [DOI] [PubMed] [Google Scholar]
- Reith RM, McKenna J, Wu H, Hashmi SS, Cho SH, Dash PK, et al. Loss of the tuberous sclerosis complex protein tuberin causes Purkinje cell degeneration. Neurobiol Dis. 2011;43:113–122. doi: 10.1016/j.nbd.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AR, Berry EM, Segal RA. Regional expression of p75NTR contributes to neurotrophin regulation of cerebellar patterning. Mol Cell Neurosci. 2003;22:1–13. doi: 10.1016/s1044-7431(02)00015-5. [DOI] [PubMed] [Google Scholar]
- Dhanoa NK, Krol KM, Jahed A, Crutcher KA, Kawaja MD. Null mutations for exon III and exon IV of the p75 neurotrophin receptor gene enhance sympathetic sprouting in response to elevated levels of nerve growth factor in transgenic mice. Exp Neurol. 2006;198:416–426. doi: 10.1016/j.expneurol.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Chow ML, Pramparo T, Winn ME, Barnes CC, Li HR, Weiss L, et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLOS Genet. 2012;8:e1002592. doi: 10.1371/journal.pgen.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capsoni S, Covaceuszach S, Marinelli S, Ceci M, Bernardo A, Minghetti L, et al. Taking pain out of NGF: a ‘painless' NGF mutant, linked to hereditary sensory autonomic neuropathy type V, with full neurotrophic activity. PLoS ONE. 2011;6:e17321. doi: 10.1371/journal.pone.0017321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JJ, Fagiolini M. Autism: a ‘critical period' disorder. Neural Plast. 2011;2011:921680. doi: 10.1155/2011/921680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag N, Baxter MG, Berger-Sweeney JE. Efficacy of a murine-p75-saporin immunotoxin for selective lesions of basal forebrain cholinergic neurons in mice. Neurosci Lett. 2009;452:247–251. doi: 10.1016/j.neulet.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Halterman MW, Gill M, DeJesus C, Ogihara M, Schor NF, Federoff HJ. The ER stress response factor, chop-10, protects against hypoxia-induced neuronal death. J Biol Chem. 2010;285:21329–21340. doi: 10.1074/jbc.M109.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom SP, Stone JL, Ten Bosch JR, Merriman B, Cantor RM, Geschwind DH, et al. High-density SNP association study of the 17q21 chromosomal region linked to autism identifies CACNA1G as a novel candidate gene. Mol Psychiatr. 2010;15:996–1005. doi: 10.1038/mp.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR, Russo JP, Yang S, Roohi J, Blaxill M, Kahler SG, et al. Autism and environmental genomics. Neurotoxicology. 2006;27:671–684. doi: 10.1016/j.neuro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Barker PA, Murphy RA. The nerve growth factor receptor: a multicomponent system that mediates the actions of the neurotrophin family of proteins. Mol Cell Biochem. 1992;110:1–15. doi: 10.1007/BF02385000. [DOI] [PubMed] [Google Scholar]
- Dinçel N, Ünalp A, Kutlu A, Öztürk A, Uran N, Ulusoy S. Serum nerve growth factor levels in autistic children in Turkish population: a preliminary study. Indian J Med Res. 2013;138:900–903. [PMC free article] [PubMed] [Google Scholar]
- Ceni C, Kommaddi RP, Thomas R, Vereker E, Liu X, McPherson PS, et al. The p75NTR intracellular domain generated by neurotrophin-induced receptor cleavage potentiates Trk signaling. J Cell Sci. 2010;123:2299–2307. doi: 10.1242/jcs.062612. [DOI] [PubMed] [Google Scholar]
- Frade JM. Nuclear translocation of the p75 neurotrophin receptor cytoplasmic domain in response to neurotrophin binding. J Neurosci. 2005;25:1407–1411. doi: 10.1523/JNEUROSCI.3798-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabizadeh S, Ye X, Sperandio S, Wang JJ, Ellerby HM, Ellerby LM, et al. Neurotrophin dependence domain: a domain required for the mediation of apoptosis by the p75 neurotrophin receptor. J Mol Neurosci. 2000;15:215–229. doi: 10.1385/JMN:15:3:215. [DOI] [PubMed] [Google Scholar]
- Cortazzo MH, Kassis ES, Sproul KA, Schor NF. Nerve growth factor (NGF)-mediated protection of neural crest cells from antimitotic agent-induced apoptosis: the role of the low-affinity NGF receptor. J Neurosci. 1996;16:3895–3899. doi: 10.1523/JNEUROSCI.16-12-03895.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zeng J, Cen L, Chen Y, Wang X, Yao G, et al. Multiple roles of the p75 neurotrophin receptor in the nervous system. J Int Med Res. 2009;37:281–288. doi: 10.1177/147323000903700201. [DOI] [PubMed] [Google Scholar]