Abstract
Purpose
This study was to analyze the spectrum and frequency of rhodopsin gene (RHO) mutations in Chinese patients with retinitis pigmentosa (RP).
Methods
Patients were given physical examinations, and blood samples were collected for DNA extraction. The RHO mutations were screened with direct sequencing.
Results
Eight heterozygous nucleotide changes were detected in eight of 300 probands with RP, including six novel mutations and two known mutations. p.R21C, p.C110S, p.G182V, p.C187G, c.409–426delGTGGTGGTGTGTAAGCCC, and p.P347L were found in six autosomal dominant families. p.T92I and p.Y178C were found in two isolated cases.
Conclusions
The results reveal the spectrum and frequency of RHO mutations in Chinese patients with different forms of RP and demonstrate that RHO mutations account for a high proportion of autosomal dominant RP (adRP) cases.
Introduction
Retinitis pigmentosa (RP) is an inherited, degenerative eye disease that causes severe vision impairment and often blindness. RP has a prevalence of about 1:3,000–1:7,000 and is the most frequent form of inherited retinal degeneration, without apparent ethnic or racial distinctions [1-3]. RP is a clinically and genetically heterogeneous disease. Clinically, the age of onset and the rate of progression are different. Symptoms usually emerge as night blindness and progress to legal blindness. Genetically, many different mutations in different genes cause similar consequences, yet other mutations in the same gene may cause different diseases [4].
There are dominant, recessive, and X-linked forms of inheritance. Some cases are sporadic and lack a family history of the disease. RP may occur alone or as part of a more complex syndrome [2]. Since the first disease-causing gene (RHO; gene ID: 6010, OMIM: 180380) was discovered in 1990, more than 60 genes that cause RP have been reported (RetNet) [5]. Most genes were only a small proportion of cases. The RHO gene accounts for approximately 25% of autosomal dominant RP (adRP) in Americans [6,7]. The rate of RHO mutations in Koreans, Japanese, and Chinese is lower than in Americans [8-10].
In this study, we had collected 300 probands with RP in an outpatient setting. In the follow-up genetic study, we identified eight mutations in the RHO gene that is responsible for the disease.
Methods
Patients and clinical data
300 probands enrolled in this study were come maily from Hebei Province, Henan province and Shanxi province. Probands include 171 males and 129 females. Patients come to our hospital from ages 5 to 72 years. After probands were diagnosed RP, their family history inquiry were collected in detail and their family members were recruited further. The probands enrolled in this study were from Hebei Province, China. Clinical examination, peripheral blood collection, and DNA extraction were performed at the Department of Ophthalmology, Hebei Ophthalmic Hospital. Peripheral blood was drawn after register and preserved at -80 °C Ultra-low temperature freezer prior to use. Genomic DNA was isolated with DNA extraction kit (DP332, Tiangen, Beijing, China). Informed consent was obtained from all participants in accordance with the Declaration of Helsinki, the Institutional Review Board, and Ethics Committee of Hebei Province. Clinical data for the subjects were ascertained with detailed ocular examinations. In addition, physical examinations were performed to exclude systemic diseases.
In the mutation analysis, coding exons of RHO were amplified with PCR using a set pair of primers [8]. The PCR products were sequenced on an ABI3730 Automated Sequencer (PE Biosystems, Foster City, CA). Briefly, PCR amplification conditions were: Reaction Mixture Set Up (50 μl); 2 μl (40 ng/ml) of DNA, 1.5 μl (10 pM) of each exon primer, 20 μl of water, and 25 μl of PCR mix (2× EasyTaq PCR SuperMix, TransGen, Beijing, China) in a final reaction volume of 50 μl. Thermal cycling conditions were: an initial denaturation step at 95 °C for 5 min, 36 cycles (30 s at 95 °C, 30 s at 58 °C, and 30 s at 72 °C) and a final 10 min 72 °C extension. The PCR products were sequenced on an ABI3730 automated sequencer (PE Biosystems, Foster City, CA).
Results
Clinical findings
All probands showed typical RP disease (Table 1). The 300 probands included 40 with adRP, 38 with autosomal recessive RP (arRP), and 222 sporadic cases. The physical examinations excluded systemic disorders in all patients.
Table 1. The clinical features of probands from the eight families.
| Proband/Family | Age/Sex | Mutation | Onset of blindness | BCVA | Fundusa | ERG |
|---|---|---|---|---|---|---|
| III:1/Family 12 |
24 y/F |
p.R21C |
16 y |
0.25/0.25 |
No |
No rod responses |
| II:4/Family 245 |
39 y/F |
p.T92I |
Early Childhood |
0.10/0.10 |
YES |
No rod responses |
| III:1/Family 30 |
43 y/M |
p.C110S |
Early Childhood |
0.12/0.12 |
YES |
Data not available |
| II:7/Family 13 |
42 y/F |
p.G182V |
Early Childhood |
0.15/0.12 |
YES |
No rod responses |
| II:5/Family 22 |
48 y/F |
p.C187G |
Early Childhood |
0.30/0.30 |
YES |
No rod responses |
| III:3/Family 7 |
42 y/F |
c.409-426del |
25 y |
0.15/0.12 |
YES |
No rod responses |
| II:5/Family 171 |
39 y/M |
p.Y178C |
Early Childhood |
0.20/0.30 |
YES |
No rod responses |
| III:1/Family 21 | 27 y/M | p.P347L | Early Childhood | 0.40/0.30 | YES | No rod responses |
Abbreviations: BCVA, best corrected visual acuity; ERG, electroretinogram; a,bone spicule pigment deposits.
Mutation analysis
Sequence analysis of the coding region of RHO was performed in the probands. Eight heterozygous nucleotide changes were detected in eight of the 300 probands with RP (Figure 1, Figure 2). p.R21C, p.C110S, p.G182V, p.C187G, c.409–426delGTGGTGGTGTGTAAGCCC, and p.P347L were found in six autosomal dominant families. p.T92I and p.Y178C were found in two isolated cases. The eight mutations include six novel mutations (p.R21C, p.T92I, p.C110S, p.G182V, p.C187G, and c.409–426delGTGGTGGTGTGTAAGCCC) and two known mutations (p.Y178C and p.P347L). Of the eight mutations, seven were missense changes, and one was a small deletion (Figure 3). Missense mutations were predicted to be pathogenic with SIFT and Polyphen-2 prediction software (Table 2). These mutations were not found in 100 control individuals from the same ethnic background. The mutation frequencies were 15% (6/40) for adRP, 0.9% (2/222) for sporadic RP, and 2.7% (8/300) in total. No mutation was detected in families with arRP.
Figure 1.
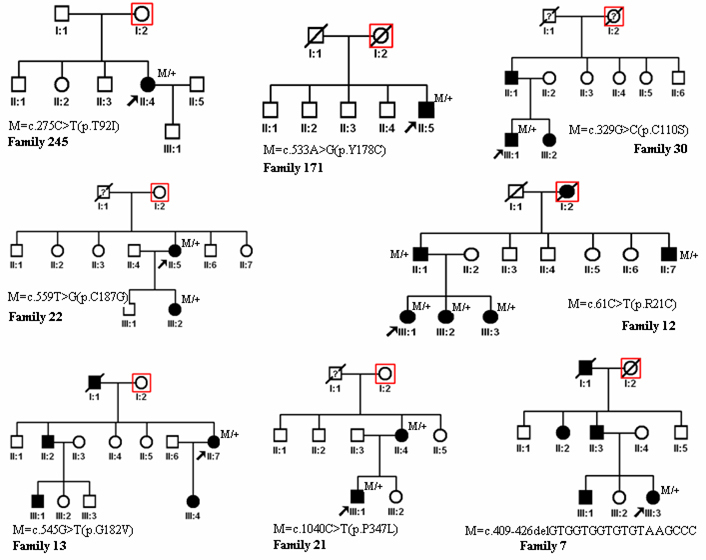
Pedigree of the families. Squares indicate men, circles indicate women, slashed symbols indicate deceased individuals, solid symbols indicate the affected, and open symbols indicate the unaffected. Red squares are extra squares when we use the Genogram software of cyrillic.
Figure 2.
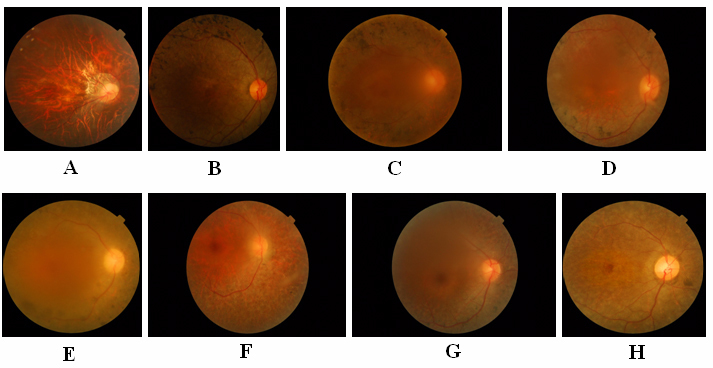
Fundus photographs of probands (in Figure 1). A: III:1 in family 12. B: II:4 in family 245. C: III:1 in family 30. D: II:5 in family 171. E: II:7 in family 13. F: II:5 in family 22. G: III:1 in family 21. H: III:3 in family 7.
Figure 3.
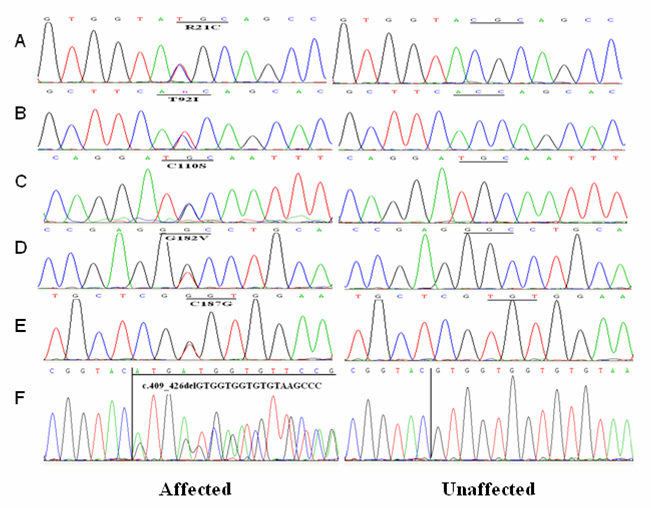
DNA sequences of RHO in patient and control individuals.
Table 2. Prediction of the functional impact of missense mutations.
| Mutation | SIFT | Polyphen-2 (HumDiv) | Polyphen-2 (HumVar) |
|---|---|---|---|
| R21C |
0.01 |
Probably damaging |
Probably damaging |
| T92I |
0.07 |
Possibly damaging |
Possibly damaging |
| C110S |
0 |
Probably damaging |
Probably damaging |
| G182V |
0 |
Probably damaging |
Probably damaging |
| C187G |
0 |
Probably damaging |
Probably damaging |
| Y178C |
0 |
Probably damaging |
Probably damaging |
| P347L | 0 | Probably damaging | Probably damaging |
Discussion
In this study, 300 families with RP in an outpatient setting in the past 2 years were collected consecutively. In the follow-up genetic study, we identified eight mutations in the RHO gene. The results show that RHO mutations accounted for a high proportion (15%, 6/40) in Chinese patients with adRP. RHO mutations in arRP and sporadic RP are not common.
Up to now, more than 160 mutations of RHO have been found in different racial and ethnic populations (HGMD). Studies show that mutations in this gene are responsible for approximately 25% of adRP in American patients. The P23H mutation occurs most frequently in American patients but has not been reported in patients with RP in other ethnic populations [6,10]. P347L and Y178C mutations have been reported in several ethnic populations [6,8-13].
The proportion of RHO mutations in Asia is lower than in the United States and Europe. The proportion of RHO mutations in Chinese patients was 2.0–5.6%, 2.0% in Koreans, 2.0% in Japanese, and 2.0% in Indians [8-10,14].
In conclusion, our study provides useful material for evaluating the proportion of RHO mutations in Chinese patients with RP. We revealed the spectrum and frequency of RHO mutations in Chinese patients with RP. RHO mutations were found in 2.7% of the Chinese patients.
Acknowledgments
Dr. Minglian Zhang (zhangminglian001@126.com) and Dr. Jialiang Zhao (zhaojialiang1234@aliyun.com) are co-corresponding authors.
References
- 1.Wissinger B. Kohl Susanne, Langenbeck U. Genetics in ophthalmology [M]. Berlin: Karger, 2003:109–110. [Google Scholar]
- 2.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002;233:1–34. doi: 10.1046/j.1395-3907.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- 4.van Soest S, Westerveld A, de Jong PT, Bleeker-Wagemakers EM, Bergen AA. Retinitis pigmentosa: defined from a molecular point of view. Surv Ophthalmol. 1999;43:321–34. doi: 10.1016/s0039-6257(98)00046-0. [DOI] [PubMed] [Google Scholar]
- 5.Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, Berson EL. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–6. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- 6.Dryja TP, McGee TL, Hahn LB, Cowley GS, Olsson JE, Reichel E, Sandberg MA, Berson EL. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med. 1990;323:1302–7. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 7.Sohocki MM, Daiger SP, Bowne SJ, Rodriquez JA, Northrup H, Heckenlively JR, Birch DG, Mintz-Hittner H, Ruiz RS, Lewis RA, Saperstein DA, Sullivan LS. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001;17:42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim KJ, Kim C, Bok J, Kim KS, Lee EJ, Park SP, Chung H, Han BG, Kim HL, Kimm K, Yu HG, Lee JY. Spectrum of rhodopsin mutations in Korean patients with retinitis pigmentosa. Mol Vis. 2011;17:844–53. [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Xiao X, Wang P, Guo X, Zhang Q. Mutation spectrum and frequency of the RHO gene in 248 Chinese families with retinitis pigmentosa. Biochem Biophys Res Commun. 2010;401:42–7. doi: 10.1016/j.bbrc.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, Garcia CA, Ruiz RS, Blanton SH, Northrup H, Gire AI, Seaman R, Duzkale H, Spellicy CJ, Zhu J, Shankar SP, Daiger SP. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–64. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XL, Liu M, Meng XH, Fu WL, Yin ZQ, Huang JF, Zhang X. Mutational analysis of the rhodopsin gene in Chinese ADRP families by conformation sensitive gel electrophoresis. Life Sci. 2006;78:1494–8. doi: 10.1016/j.lfs.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Audo I, Manes G, Mohand-Saïd S, Friedrich A, Lancelot ME, Antonio A, Moskova-Doumanova V, Poch O, Zanlonghi X, Hamel CP, Sahel JA, Bhattacharya SS, Zeitz C. Spectrum of rhodopsin mutations in French autosomal dominant rod-cone dystrophy patients. Invest Ophthalmol Vis Sci. 2010;51:3687–700. doi: 10.1167/iovs.09-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrar GJ, Kenna P, Redmond R, Shiels D, Mcwilliam P, Humphries MM, Sharp EM, Jordan S, Kumarsingh R, Humphries P. Autosomal Dominant Retinitis-Pigmentosa - A Mutation in Codon 178 of the Rhodopsin Gene in Two Families of Celtic Origin. Genomics. 1991;11:1170–1. [PubMed] [Google Scholar]
- 14.Gandra M, Anandula V, Authiappan V, Sundaramurthy S, Raman R, Bhattacharya S, Govindasamy K. Retinitis pigmentosa: mutation analysis of RHO, PRPF31, RP1, and IMPDH1 genes in patients from India. Mol Vis. 2008;14:1105–13. [PMC free article] [PubMed] [Google Scholar]


