Abstract
CD81 is a ubiquitously expressed member of the tetraspanin family. It forms large molecular platforms, so-called tetraspanin webs that play physiological roles in a variety of cellular functions and are involved in viral and parasite infections. We have investigated which part of the CD81 molecule is required for the formation of domains in the cell membranes of T-cells and hepatocytes. Surprisingly, we find that large CD81 platforms assemble via the short extracellular δ-domain, independent from a strong primary partner binding and from weak interactions mediated by palmitoylation. The δ-domain is also essential for the platforms to function during viral entry. We propose that, instead of stable binary interactions, CD81 interactions via the small δ-domain, possibly involving a dimerization step, play the key role in organizing CD81 into large tetraspanin webs and controlling its function.
Introduction
Tetraspanins constitute a family of small, conserved four transmembrane spanning proteins with 33 members in humans and mice (1). They form extended networks of molecular interactions with other surface molecules called tetraspanin webs or tetraspanin-enriched microdomains (TEMs or TERMs) by means of their strong tendency to associate laterally with one another, with integrins, members of the Ig superfamily and/or signaling receptors (2–5). TEMs play a role in an astonishing diverse range of cellular processes, including cell proliferation, adhesion, spreading and migration, signal-transduction, endocytic trafficking, cell-cell fusion, and host-pathogen interactions (5,6). Due to their involvement in malignancy, the immune system, and infectious diseases, they emerged as potential targets in therapeutic strategies (7).
One of the most studied tetraspanins is CD81 (Cluster of Differentiation 81), also known as 26 kDa cell surface protein or tetraspanin 28. Initially, functions of CD81 were discovered in the immune system including B- and T-cell activation, as target of an antiproliferative antibody (8). Meanwhile other roles in many different cell types have been documented, including brain development (9), retinal pigment epithelium development (10), and fertility (11) (for review see (1,12)).
Apart from its biological functions, CD81 is clinically significant acting as an entry factor for infections with the hepatitis C virus (HCV) (13), the malaria Plasmodium parasite (14), and Listeria monocytogenes (15). Moreover, TEMs provide entry and/or exit platforms for human papilloma virus (HPV) and human immunodeficiency virus (16–18) and other pathogens (19,20).
It is still under debate how CD81 or tetraspanins in general form tetraspanin webs. Complexes found in immunoprecipitates after lysing cells with detergents of different strength have led to a hierarchical classification of interactions involved in tetraspanin web formation. Interactions between tetraspanins and their nontetraspanin partners that are resistant to Triton X-100 and digitonin are referred to as primary complexes (7,21). Under these conditions tetraspanin-tetraspanin interactions are disrupted but can be maintained using milder detergents such as Brij 97 or CHAPS. Interactions preserved only under mild solubilization conditions are thought to be weaker, less defined second-level interactions (1,22) that presumably lead to the association of primary complexes forming a large tetraspanin network. Interactions are further stabilized by protein palmitoylation (7,23,24). For instance, palmitoylation of the tetraspanin CD81 promotes its association with the tetraspanin CD9 and one of its major primary binding partners EWI-2 from the Ig superfamily (25,26). Accordingly, a palmitoylation deficient mutant of the tetraspanin CD151 associates less with CD81 and CD82 (27). Still, it is debated what mechanism drives the coalescence of such a variety of different complexes/molecules into large tetraspanin webs (5).
To date, the complete three-dimensional structure has been predicted for only one tetraspanin, CD81 (28). As in all tetraspanins, a small and a large extracellular loop (LEL, further subdivided into the predominantly helical domains α, β, γ, δ, and ε) are each flanked by transmembrane regions (TMs), TM1/TM2 and TM3/TM4, respectively. The TMs are closely packed and in continuity of TM3 and TM4 two extracellular antiparallel alpha-helices (α and ε) form the central stalk of the LEL conferring a compact rod-shaped structure to the molecule, with only the short δ-domain exposed at the side of the extracellular terminus. The short intracellular segments harbor palmitoylation sites, two at each of the N- and C-terminal tails and two at the linker between TM2 and TM3. The LEL is structurally subdivided into a conserved and a nonconserved section, the so-called constant (α, β, and ε) and variable region (γ and δ), respectively (28).
Different conformations of the LEL are observed in hexagonal and monoclinic crystals and in solution suggesting that conformational flexibility of this region plays a key role in tetraspanin web assembly and function (29–31). This is in line with the findings that the variable region is crucial for tetraspanin protein-protein interactions and presumably defines the diverse tetraspanin classes (3,32,33).
Here, we have examined how the lateral organization of CD81 in the plasma membrane depends on CD81 protein domains and identified a small part of the variable region, the δ-domain, to organize CD81 into tetraspanin platforms, a mechanism supported but in principle independent from primary complexes.
Materials and Methods
Cloning
From a cDNA library we obtained by polymerase chain reaction (PCR) the sequences for CD81 (NM_004356.3), EWI-2 (NM_052868.4), and CD9 (NM_001769.3), which were subcloned using the pGEM-T easy vector system (catalogue No. A1360, Promega, Madison, WI). All constructs were expressed using the expression vector pEGFP-C1 (catalogue No. 6084-1, clontech, Mountain View, CA) backbone, lacking EGFP. Via fusion PCR, CD81 was fused directly to the N-terminus of monomeric-enhanced GFP (carrying the mutation A207K, which corresponds to A206K described in (34)) with a stop codon and inserted via the NheI/KpnI sites. Substitution and deletion constructs of CD81 were produced by fusion PCR and inserted using a C-terminal Kpn2I restriction site within the CD81 sequence, via the NheI/Kpn2I sites. In cases the modification was downstream of the Kpn2I site, PCR was performed over the entire CD81-mGFP construct and the NheI/KpnI sites were used. The following constructs were produced according to this procedure. C/A, substitution of the juxtamembrane cysteines by alanines in the positions 6, 9, 80, 89, 227, and 228; Δαβ lacking aa 115–155; Δγ, lacking aa 156–174; Δγδ. lacking aa156–190; Δδ, lacking aa 176–186. Following the same procedure, a CD81-mRFP construct was cloned fusing C-terminally to CD81 mRFP ((35); carrying two silent mutations for introducing a restriction site) with a stop codon.
The C-terminus of EWI-2 or CD9 was fused directly to the N-terminus of a C-terminally myc-tagged mRFP (myc sequence; GAACAAAAACTTATTTCTGAAGAAGATCTG followed by a stop codon) via PCR and the EWI-2-mRFP-myc or CD9-mRFP-myc sequence was inserted into pEGFP-C1 via the AgeI/KpnI sites (for EWI-2) or NheI/KpnI (for CD9).
Cell culture
Jurkat E6.1 cells and HepG2 cells were cultured essentially as described previously (36). For transfections, 60 μg plasmid (in double transfections 30 μg plasmid of each construct) was added per electroporation cuvette. Electroporation was performed with a Gene pulser Xcell electroporation system (Bio-Rad, Hercules, CA). Jurkat T cells and HepG2 cells were used for experiments 2 days and 1 day after transfection, respectively.
Antibodies
Mouse monoclonal antibodies were used for human CD81 (catalogue No. 16-0819, eBioscience, San Diego, CA) and GFP (clone 3E6, catalogue No. A11120, Invitrogen, Carlsbad, CA). For staining of pseudovirions (PsVs) we used the polyclonal antibody K75 raised against the L1 protein of the PsVs (16). As secondary antibodies, we used AlexaFluor594-labeled donkey antimouse IgG (H+L) (catalogue No. A21203, Invitrogen), AlexaFluor546-labeled donkey antirabbit IgG (catalogue No. A10040, Invitrogen), or AlexaFluor488 donkey antimouse IgG (H+L) (catalogue No. A21202, Invitrogen).
Epifluorescence microscopy
For imaging, we used an Olympus IX81 microscope equipped with a 60 × 1.49 NA Apochromat objective applying a 1.6× magnifying lens (Olympus, Tokyo, Japan) and an EMCCD camera (16 × 16 μm2 pixel size, ImagEM C9100-13, Hamamatsu Photonics, Hamamatsu, Japan) with a 2× magnifying lens. For illumination, we used a 488 laser (LAS/488/20, Olympus) or a 150 W Xenon lamp integrated into the MT20E-fluorescence illumination system (Olympus) in combination with the following filter sets: CMR-U-MTIR-488-HC (Olympus) and F36-500 DAPI HC, F36-525 EGFP HC, and F36-503 Tritc HC (AHF Analysentechnik, Tuebingen, Germany). All images are shown applying a linear lookup table at arbitrary scalings.
Confocal microscopy
For confocal microscopy we used an Olympus FluorView 1000 microscope as described previously (37). Fluorescence recovery after photobleaching measurements were essentially performed and analyzed as described ((37); pixel size was adjusted to 207 nm; for recording the 488 nm laser line and for bleaching the 488 and 405 nm laser lines were used). Otherwise, pixel size was adjusted to 103 nm and DAPI (blue), GFP (green), and Alexa546 (red) were excited at 405, 488, and 543 nm, respectively. Fixed cells were mounted on microscopy slides with 15 μl mounting medium with antifade (catalogue No. P10144, Invitrogen) and cured for 24 h at room temperature (RT). Transfected cells were identified in the green channel, followed by taking at different axial positions optical sections in the blue, green, red, and bright-field channels. For live cell imaging at RT in Ringer solution (130 mM NaCl, 4 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 48 mM D(+)glucose, 10 mM HEPES, pH 7.4) areas were selected in the bright-field mode, followed by taking a bright-field and fluorescence images.
Pearson correlation coefficient
The Pearson correlation coefficient (PCC) quantifies the degree of colocalization between two images (38). In principle, its value ranges from 1 (perfect overlap) through 0 (no relation) to −1 (perfectly anticorrelating images). Using the program ImageJ we employed the plugin Align slice (Gabriel Landini, University of Birmingham) for manual alignment of images from different channels allowing corrections of lateral shifts in the x and y directions that occasionally occurred during imaging. The PCC was calculated for the aligned images using the plugin Colocalization_Indices (Kouichi Nakamura, Kyoto University).
Autocorrelation analysis
An image region defined by a region of interest (ROI) was duplicated yielding the reference image. Using the ImageJ plugin Align slice, the ROI was repeatedly shifted by 1 pixel in one direction subsequently duplicating the respective region. The PCCs between the reference and the duplicates from the shifted images were then calculated using the Colocalization_Indices plugin and the values (starting with 1) were plotted against the pixel shift. For analysis of stimulated emission depletion (STED) micrographs, we also used the GDSC stack correlation analyzer plugin and always increased Pixel intensity by one to avoid zero values that cannot be processed. For each independent experiment, values from different cell membranes were averaged and a curve was fitted (y = 1 – (a × x)/(b + x)) yielding the 50% decay value. For STED microscopy, correlation was performed on three different ROIs for each membrane sheet; the curves were averaged and fitted using a simple exponential decay function, yielding the 50% decay value for the respective membrane.
Immunostaining for CD81
Membrane sheets were generated from transfected cells in ice-cold sonication buffer (120 mM potassium glutamate, 20 mM potassium acetate, 10 mM EGTA, 20 mM HEPES, pH 7.2) as described previously (37) and fixed directly with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, pH 7.4) for 30 min at RT. PFA was quenched for 30 min at RT with 50 mM NH4Cl in PBS, followed by 3 washes of 10 min with PBS. Membrane sheets were incubated with the first antibody (diluted 1:100 in 3% bovine serum albumin (BSA)-PBS) for 1 h at RT followed by 3 PBS washing steps of 10 min. Incubation with the secondary antibody (AlexaFluor594 labeled donkey anti-mouse diluted 1:200 in 3% BSA-PBS) at RT lasted 1 h for HepG2 membrane sheets and overnight for Jurkat T cell membrane sheets. Afterward, the coverslips were washed 3 times with PBS for 10 min and imaged in PBS containing 1-(4-tri-methyl-ammonium-phenyl)-6-phenyl-1,3,5-hexatriene p-toluene-sulfonate (TMA-DPH, catalogue No. T-204, Invitrogen) for visualization of membranes. In the green channel, exposure times for recording of Jurkat and HepG2 membrane sheets were 100 ms and 1 s, respectively. In the red channel, recording times were 100 and 500 ms for Jurkat and HepG2 membrane sheets, respectively. Background corrected intensities were determined for the green and red channels of individual membrane sheets using ImageJ. The PCC between the images from the red and the green channels was determined as described previously. Corrections for lateral shifts were performed manually using the Align slice plugin referring to at least 4 to 5 different structures apparent in both channels or to 100 nm Tetraspeck beads (TetraSpeck Microspheres, catalogue No.T-7279, Invitrogen) for Jurkat and HepG2 membrane sheets, respectively.
STED microscopy sample preparation and imaging
Membrane sheets were generated from HepG2 cells transfected and fixed as described previously. After quenching for 20–30 min with 50 mM NH4Cl in PBS, membranes were washed three times with PBS and blocked for 1 h at RT in 3% BSA-PBS. Membranes were then incubated with anti-GFP antibody, diluted 1:100 in 1% BSA-PBS overnight at 4°C. After 4 washes with PBS, incubation with the secondary antibody (Alexa488 labeled-donkey antimouse), diluted 1:100 in 1% BSA-PBS, was performed for 2 h at RT. After 3 washes with PBS, coverslips were mounted on microscopy slides and embedded as described for confocal microscopy samples. Samples were sealed with clear nail polish and stored at 4°C.
For imaging, we used a TCS-SP8 gated-STED microscope (Leica, Mannheim, Germany) at the Light Microscopy Facility (LMF) at the German Centre for Neurodegenerative Diseases (DZNE) in Bonn. First, confocal images were taken into the red channel to detect EWI-2-RFP and into the green channel to measure CD81-GFP intensity. STED images were recorded at 200 Hz, 40% STED beam intensity, gating between 1 and 6.5 ns and Pixel sizes were close to 20 nm. Analyzed STED micrographs were composed of 6 averaged frames.
Coclustering assay
From cotransfected cells, expressing either CD81-RFP and CD81-GFP or a GFP-labeled CD81 variant, EWI-2-RFP or CD9-RFP and GFP-labeled CD81 or CD81 variants membrane sheets were generated, fixed, quenched, and washed as described previously. Imaging was performed in PBS containing TMA-DPH, after adding 100 nm Tetraspeck beads for the correction of lateral shifts that occasionally occurred between filter changes. Green, red (which tended to yield less clustered patterns), and blue channels were recorded in the epifluorescence mode of the microscope. Images were analyzed using the program ImageJ. First, an image stack including all three channels was created and, if necessary, corrected for lateral shifts (referring to Tetraspeck beads) using the Align Slice plugin of ImageJ. A ROI was then placed on the basal membrane for generating duplicates from the green and red channels, which were used for calculating the PCC using the Colocalization Indices plugin of ImageJ. In experiments involving EWI-2-RFP, we also performed autocorrelation analysis on the green channel.
Total internal reflection fluorescence (TIRF) microscopy
Jurkat cells were cultured in suspension for 2 days after transfection and harvested through 3 min of centrifugation at 1000 rpm in an Eppendorf (Hamburg, Germany) centrifuge 5810. The cell pellet was resuspended in Ringer solution and aliquots were plated into 6-well plates at a concentration of ∼1.5 × 106 cells/well, containing poly-L-lysine coated glass coverslips with a diameter of 25 mm. Cells adhered in the cell incubator for 20 min and were imaged within the following 20 min. Imaging was performed in the TIRF mode of the microscope, which selectively excites fluorescence in a thin layer spanning only a few 100 nm at the glass-water interface, including the basal membrane of the adhered cells. Movies were recorded at 2 Hz for ∼15 s. Images were analyzed using the program ImageJ, calculating the PCC between two successive frames separated by a time interval of 0.5 s. Values obtained from one movie were averaged. For each independent experiment the values obtained from all movies were averaged.
Pseudovirion-induced endocytosis
Jurkat cells were transfected with either CD81-GFP or CD81-Δδ-GFP and cultured for 2 days in suspension. 106 cells were pelleted for 3 min at 1000 rpm in an Eppendorf centrifuge 5810, resuspended in 5 ml prewarmed medium, and transferred into a 25 cm2 flask. Sulforhodamine101 (catalogue No. s7635, Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 20 μM and, where required, 2 μl of PsVs were added (corresponding to 0.42 μg total protein or 0.2 μg of the major capsid protein L1 of HPV; PsVs were prepared as previously described (39)). The mixture was incubated for 10 min in the cell incubator with gentle agitation from time to time to avoid cell sedimentation. Cells were then centrifuged for 3 min at 800 rpm (using an Allegra X-15R centrifuge from Beckman Coulter, Brea, CA), the pellet was resuspended in 1 ml prewarmed Ringer solution and 0.5 ml aliquots each were directly plated onto poly-L-lysine coated glass coverslips. The cells were adhered for 20 min in the cell incubator and directly fixed for 30 min with 4% PFA in PBS after membrane sheet generation. To correct for potential lateral shifts that occur occasionally during imaging, the samples were supplemented with 100 nm Tetraspeck beads which can be observed in all channels. Images were then recorded using epifluorescence microscopy in the green, red, and blue channels visualizing the CD81/CD81-Δδ distribution, endosomes or the plasma membrane, respectively. For analysis, we manually determined the number of endosomes (as visualized by the fluid phase marker in the red channel) per membrane sheet colocalizing with a CD81/CD81-Δδ cluster in the green channel.
In a second set of experiments, 5⋅105 transfected cells were incubated in the absence of Sulforhodamine in 1 ml prewarmed medium adding 5 μl of PsVs. The suspension was incubated for 1 h at 37°C in a rotating Eppendorf tube. Cells were adhered and fixed as previously mentioned before they were permeabilized for 2 min in 0.2% Triton X-100 in PBS. After a PBS wash, cells were blocked for 30 min with 1% BSA-PBS. Incubation with the first antibody K75, diluted 1:1000 in 1% BSA-PBS, was then performed for 1 h at 37°C. After one wash in PBS and another wash in 1% BSA-PBS, the secondary antibody was applied (Alexa546 donkey anti-rabbit IgG, diluted 1:300) in 1% BSA-PBS containing Hoechst dye (diluted 1:10,000) for 1 h at 37°C. After two washes in PBS, cells were embedded for confocal microscopy as described above.
Results
To investigate CD81 membrane organization, we used unroofed cells (i.e., plasma membrane sheets). For this detergent-free preparation, a 100 ms ultrasound pulse generates essentially pure basal plasma membranes through mechanical shearing forces (40). Because the membranes remain attached to the glass coverslip, they constitute flat objects on which the distribution of plasmalemmal components can be imaged by epifluorescence microscopy with high sensitivity. Previous work showed that the morphology of the inner membrane leaflet is retained on immediately fixed membrane sheets (41) and that diffusion of a membrane protein in freshly prepared membranes is the same as in intact cells (42). This suggests that plasma membrane sheets are a suitable and convenient preparation for studying selectively proteins expressed on the cell surface.
First, we examined how the distribution of CD81 in the cell membrane depends on its concentration. We expressed GFP-labeled CD81 (for C-terminal tagging of CD81 by GFP see also (43)) in cells endogenously lacking (HepG2 cells) or expressing CD81 (Jurkat T cells). Membrane sheets were fixed immediately and also immunostained for CD81. In the green channel, we recorded overexpressed CD81-GFP and in the red channel immunostaining of overexpressed CD81 and, in the case of Jurkat T cells, also endogenous CD81. Over the entire concentration range CD81 was concentrated in spotty structures, though the cluster pattern was potentially not completely resolved due to diffraction-limited imaging. Moreover, as shown in Fig. S1, in the Supporting Material: i), HepG2 cells lack, as expected, endogenous CD81; ii), in Jurkat T cells overexpression in most membranes hardly doubled total CD81; and iii), overexpressed and endogenous CD81 do not form distinct patterns.
For a more detailed analysis we turned to superresolution light microscopy on HepG2 cells, in which the clustering of expressed CD81 can be studied in the absence of endogenous CD81. At lower concentrations up to densities of 40 clusters per μm2, cluster density was linearly related to CD81 concentration: ∼twofold more clusters formed upon doubling the concentration (see inset in Fig. 1 C). Upon a further increase in CD81, cluster density rose less, almost certainly because individual clusters can no longer be resolved properly (even at superresolution) and their number is underestimated (Fig. 1; see membrane sheets #3 and #4 at STED microscopy resolution). From this data, we conclude that more CD81 molecules primarily form more clusters rather than increasing their size or causing a more uniform distribution of molecules.
Figure 1.
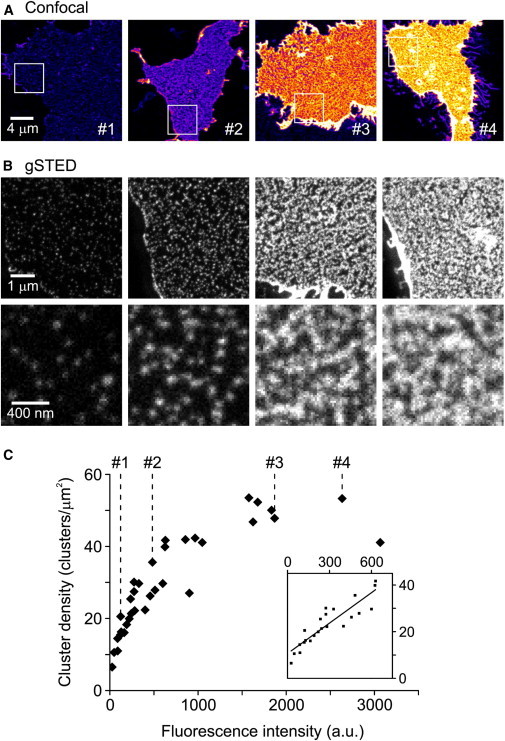
Elevation of CD81 levels generates more clusters. CD81-GFP was expressed in HepG2 cells and for gated STED imaging of membrane sheets, and the signal was enhanced by immunostaining for GFP. (A) Confocal micrographs (to minimize bleaching) for quantification of CD81 expression levels. Shown are four membrane sheets (see labels #1 to #4) from cells with varying CD81 expression levels applying the fire lookup table at the same scaling. Immunostaining intensity was quantified and taken as readout for relative CD81 concentration. (B) Upper panel, sections from larger STED micrographs, corresponding to the white squares in A. On each original STED micrograph three ROIs (one shown in the lower panel) were selected for manual cluster counting; averaging all three values yielded the cluster density obtained from the respective membrane sheet. Images are shown at the same scaling. (C) Cluster density plotted against CD81 concentration. Labels #1 to #4 mark intensity values obtained for the respective membrane sheets shown in (A). The inset illustrates the linear relationship between cluster density and CD81 concentration in the lower concentration range. Membrane sheets were collected from three independent experiments (8–13 membrane sheets per experiment). To see this figure in color, go online.
Next, we aimed for identifying the CD81 protein domain(s) or feature(s) necessary for targeting the molecule into CD81 microdomains. In Jurkat T cells, we overexpressed CD81-RFP together with CD81-GFP or GFP-fused to a CD81 variant and analyzed whether the red and green constructs are enriched in the same domains (Fig. 2). For quantification of the similarity between the green and the red channels we calculated the PCC, which is independent from offsets and absolute intensities in the two channels, weighting the departure from the mean (38).
Figure 2.
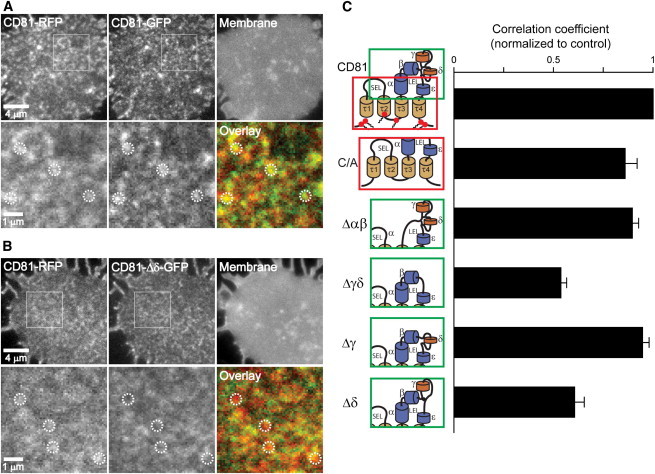
The δ-domain of the LEL is required for targeting of the CD81 molecule into CD81-enriched domains. Membrane sheets from Jurkat T cells co-overexpressing CD81-RFP along with CD81-GFP (A) or CD81-Δδ-GFP (B). Images were recorded in the red (RFP), green (GFP), and blue (TMA-DPH, a general membrane stain) channels. RFP- and GFP-labeled CD81 are enriched in the same domains, whereas deletion of the δ-domain strongly decreases the overlap between full length CD81-RFP and CD81-Δδ-GFP (for comparison see dotted circles marking identical locations). (C) PCC between CD81-RFP and CD81-GFP or a GFP-labeled CD81 variant. Pictograms show the respective GFP-labeled constructs whose lateral distribution was compared to CD81-RFP; please note that the pictograms are schemes not reflecting the real size and orientation of the domains; for the relation between the pictogram and the predicted 3D structure see Fig. S3, which also illustrates the size of the deleted molecule portions on a realistic scale. From top: CD81; C/A, with all cysteine palmitoylation sites substituted by alanines; Δαβ, Δγδ, Δγ, and Δδ, deletion of the respective domains of the LEL (large extracellular loop). SEL, small extracellular loop; red solid circles with acyl chains indicate cysteine palmitoylation sites. τ1–τ4 indicate transmembrane regions. Values are given as means ± SE (n = 3–10 independent experiments; for each experiment values from 5 to 22 membrane sheets were averaged and normalized to control (CD81-GFP)). To see this figure in color, go online.
As suggested by a high PCC (on average 0.6; Fig. S2), CD81-RFP and CD81-GFP were located in the same domains, indicating that the targeting mechanism did not discriminate between the differently labeled constructs (Fig. 2 A). When all six palmitoylation sites were replaced by alanines, coenrichment decreased only slightly (Fig. 2 C), suggesting that palmitoylation plays some yet not the major role. We then deleted almost half of the LEL, including the α- and β-helical domains, expected to have a strong impact on the structural orientation of the remaining helices (see Fig. S3 for illustration). However, despite such dramatic modification of the molecule it was still capable of forming CD81 domains (Fig. 2 C), indicating that the previously described dimerization via the αβ-interface (29,30,44) is not a prerequisite for domain targeting. The variable region within the LEL plays the major role in specific interactions. In fact, deletion of the variable region and its two flanking cysteines (aa 156–190; (12,29); also see Fig. S3 for illustration of the deleted domains) strongly decreased domain targeting (Fig. 2 C). The variable region contains two alpha-helical domains separated by a disulfide bridge, referred to as γ-domain (158–174, not including the flanking cysteines) and δ-domain (176–189) (29). A construct lacking the γ-domain (Δ156–174) essentially behaved like wild-type CD81, whereas deletion of the δ-domain (Δ176–186) diminished domain targeting to a level indistinguishably from that seen for the construct lacking the entire variable region (Fig. 2, B and C). This effect was not caused by incomparable cell surface expression levels of CD81-GFP and CD81-Δδ-GFP (Fig. S4), and we also verified by confocal microscopy that CD81-Δδ-GFP was not retained more strongly in intracellular structures (Fig. S5). As an alternative approach to validate the result from our microscopic assay, we performed immunoprecipitation experiments after cell solubilization with CHAPS. CD81-GFP pulled down endogenous CD81, as expected, whereas CD81-Δδ-GFP was less efficient (Fig. S6). This indicates that CD81-Δδ-GFP and CD81 are still interacting, presumably via their α/β-domains, albeit in steady state the number of complexes is reduced as CD81-Δδ-GFP is less enriched in CD81 microdomains (Fig. 2) due to diminished binding either to other CD81 molecules (forming dimers by a mechanism that in this case would be independent from the described α/β-interface dimerization) or to other TEM components. Alternatively, the capability to form dimers is reduced and dimer formation is a prerequisite for TEM targeting.
The δ-domain-dependent CD81 targeting was also observed in HepG2 cells (Fig. S7). Furthermore, CD81-Δδ was not uniformly distributed but formed separate clusters in both cell types. From these data we conclude that correct targeting of CD81 into CD81 domains requires the δ-domain, whereas other regions of the LEL and palmitoylation play a less prominent role, at least when analyzing the static distribution of molecules in the membrane.
Large tetraspanin webs or TEMs are circular in shape and relatively stable in position (45,46). In case CD81 domains represent large tetraspanin webs or TEMs, they should be less dynamic than putative non-TEM domains formed by CD81-Δδ. For clarification, we performed fluorescence recovery after photobleaching measurements in living cells (Fig. S8). For CD81, we determined an apparent lateral diffusion coefficient of 0.03 μm2/s. Compared to previously published values in polarized and nonpolarized HepG2 cells, in which 0.07 and 0.11 μm2/s have been determined (47), respectively, the lower value is expected as in Jurkat T cells CD81 forms larger clusters (see below). Consistent with this view, we find a strongly increased diffusion coefficient of CD81-Δδ (Fig. S8). To study the stability of the domains we further examined CD81 and CD81-Δδ cluster dynamics using TIRF microscopy. As shown in Fig. 3, most CD81 clusters were stable over seconds, whereas CD81-Δδ clusters translocated and/or disassembled. When the entire variable region was deleted, dynamics of platforms did not increase further, and deletion of the γ-domain alone showed a cluster pattern comparable to wild-type CD81 (see Fig. 3 E). The δ-domain-dependent effect on stability correlates well with the results from targeting analysis, suggesting that CD81 is organized in larger and stable molecular platforms only in the presence of the δ-domain.
Figure 3.
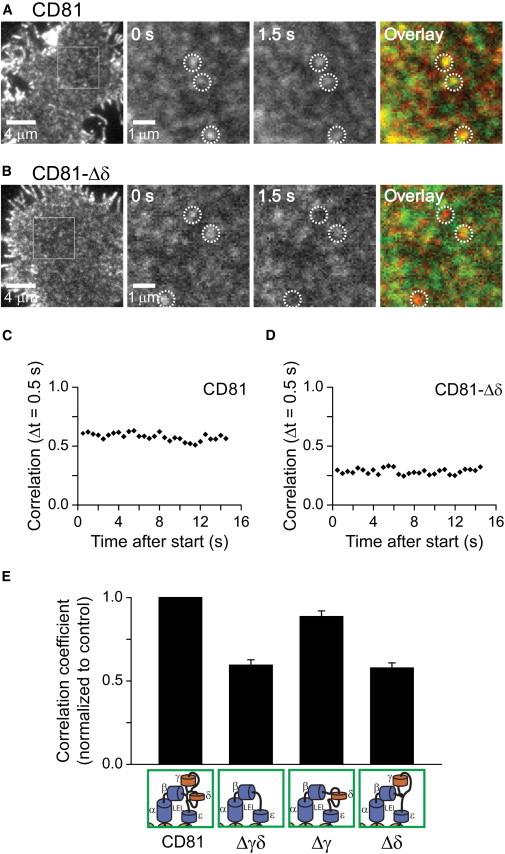
Dynamics of CD81 and CD81 lacking the γ- and/or δ-domain. Live Jurkat cells expressing (A) GFP-labeled CD81 or (B) CD81-Δδ imaged at 2 Hz in Ringer solution at RT using total internal reflection fluorescence microscopy. From left to right, overview, magnified views from two recordings separated by 1.5 s and overlay. Yellow color indicates areas in which the distribution pattern remains unchanged. In contrast to CD81-Δδ, CD81 domains regularly remain largely unchanged between two or more frames. Dotted circles indicate identical regions. (C and D) CD81 and CD81-Δδ, for comparative illustration of the lateral dynamics, over the entire movie the PCC between two successive images was determined, essentially representing repetitive measurements between two successive frames over time. The individual PCC values were then plotted against the time the second image of the correlated pair of images was recorded (plots are derived from the cells shown in A and B). (E) Averaged traces from cells expressing GFP-labeled CD81, CD81-Δγδ, CD81-Δγ, and CD81-Δδ were generated and the average PCC over all time points was calculated. For one experiment and condition 5–8 cells were averaged and normalized to control. Values are given as means ± SE (n = 4). To see this figure in color, go online.
The primary binding partner EWI-2 forms stable Triton X-100 resistant complexes with CD81 (25,48) via interactions involving from EWI-2 the short cytoplasmic tail and the glycine-zipper motif in the TM, and from CD81 the TM3 and TM4, with some contributions of the extracellular domains (49). To clarify whether CD81 and CD81-Δδ clusters differ in their capability to incorporate primary binding partners, we elevated the concentration of EWI-2. As shown in Fig. 4, in Jurkat T cells EWI-2 and CD81 coenriched in the same domains, indicating the formation of primary complexes (Fig. 4, A and C). As for domain targeting of CD81 (Fig. 2) and domain dynamics (Fig. 3), also for the coenrichment of CD81 and EWI-2 only the δ-domain was required (Fig. 4 C).
Figure 4.
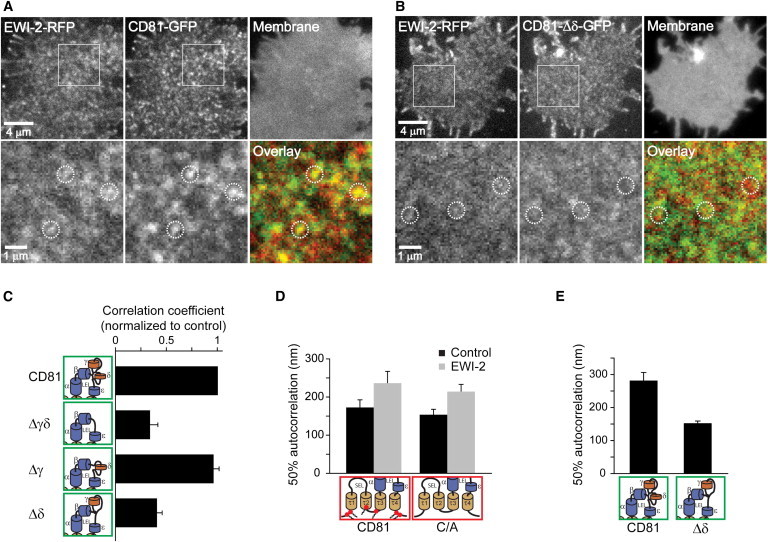
For large TEMs the δ-domain is required. (A and B) Membrane sheets from Jurkat cells co-overexpressing the CD81 interaction partner EWI-2 (RFP-labeled) together with GFP-labeled CD81 (A) or CD81-Δδ (B). Lower panels show magnified views and overlays from the marked regions in the upper panels. Upon overexpression of CD81-Δδ-GFP, the EWI-2-RFP signal tended to show a less clustered pattern. Dotted circles indicate identical pixel locations. (C) Strong overlap between EWI-2 and the clusters in the green channel required the CD81 δ-domain. Overlap was quantified by the PCC. Values are given as means ± SE (n = 3–5 independent experiments and for each experiment values from 5 to 20 membrane sheets were averaged and normalized to the respective control). (D) Cells expressing CD81-GFP or CD81-C/A-GFP without (black bars) or with (gray bars) cotransfected EWI-2-RFP. Autocorrelation analysis (see Fig. S9 for method evaluation) of the green channel shows that EWI-2 increases the size of both CD81 and the palmitoylation deficient mutant. Values are given as means ± SE (n = 3 independent experiments and for each experiment values from 7 to 8 membrane sheets were averaged). (E) Coexpression of EWI-2-RFP and CD81-GFP or CD81-Δδ-GFP. CD81 forms about threefold larger domains (referring to area and assuming circular shape) when compared to CD81-Δδ domains. Values are given as means ± SE (n = 3 independent experiments and for each experiments values from 7 to 11 membrane sheets were averaged). Please note that the condition CD81-GFP/EWI-2-RFP was performed in D as well as in E, yielding a smaller effect in D, which we explain that here we had 38% of very low EWI-2 expressing cells compared to 22% in E. To see this figure in color, go online.
Next, we tested association of CD81 with another tetraspanin, CD9. Compared to CD81, overlap with CD9 was diminished (0.35; Fig. S2) but also dependent on the presence of the δ-domain (Fig. 5). Considering that the overlap between CD81 and EWI-2 (0.53; Fig. S2) was higher than that between CD81 and CD9, the strength of interactions found by immunoprecipitation after cell lysis (primary and secondary interactions, see above) is reflected in the degree of overlap found in the microscopic assay.
Figure 5.
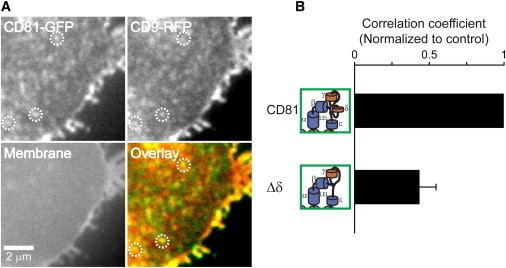
Colocalization of CD81 and CD9. (A) Membrane sheet from a Jurkat T cells co-overexpressing CD81-GFP and CD9-RFP. The upper panel shows the fluorescent protein-tagged constructs, the lower panel a TMA-DPH membrane staining and an overlay. (B) Overlap was analyzed as described in Fig. 2. Values are given as means ± SE (n = 3 independent experiments; for each experiment values from 13 to 20 membrane sheets were averaged and the mean of CD81-Δδ was normalized to control). To see this figure in color, go online.
EWI-2 also increased the platform size strongly (Fig. 4, D and E), an effect that was not dependent on palmitoylation of CD81 (Fig. 4 D). Although a diameter of 560 nm for CD81 domains (Fig. 4 E; correction for the blurring effect would reduce this value only by ∼10%) is clearly above the resolution limit, the size of CD81-Δδ clusters is possibly limited by diffraction and may represent an upper estimate. However, we can safely conclude that EWI-2 increases the cluster area size at least by a factor of 3 (Fig. 4 E). In contrast, CD81-Δδ clusters overlapped less with EWI-2 and remained close to diffraction-limited size (Fig. 4 E). These findings show that CD81 and EWI-2 interact with each other and that, as a consequence, the increase in primary complexes leads to larger tetraspanin platforms. This is in line with a recent report showing that EWI-2wint (EWI-2 without its N-terminus, a slightly shorter cleavage product from EWI-2 (50)) decreases CD81 mobility in the cell membrane and increases the fraction of confined molecules (51). The observation that CD81-Δδ clusters failed to incorporate EWI-2 might be explained by the putatively diminished stabilizing effect of the large extracellular loop on the interaction with EWI-2, though interactions still should be mediated via the TMs of both CD81 and EWI-2 (49). In contrast to Jurkat T cells, elevation of EWI-2 in HepG2 cells did not produce larger CD81 domains (Fig. S10). This suggests that an increase of primary complexes promotes platform growth (in Jurkat T cells) but per se is not sufficient for the coalescence of primary complexes into webs.
CD81 acts as a coreceptor during the entry of a variety of pathogens into different cell types. Hence, we tested whether the function of the δ-domain, or in other words the formation of large CD81 platforms, is also relevant for pathogen induced endocytosis. Using pseudovirions (PsVs) from HPV type 16, it has been shown that TEMs are involved in HPV endocytosis and infection (16,52) and we tested whether our CD81-GFP platforms are also capable of mediating PsV uptake. As shown in Fig. 6, PsVs readily triggered the formation of membrane sheet-associated endosomes that overlap with CD81 platforms, whereas in the absence of the δ-domain hardly any PsV-triggered endocytosis was detected. At sites of endocytosis, we frequently observed patching of CD81, but not of CD81-Δδ. Such CD81 accumulation sites, overlapping with immunostained PsV particles, were also observed on intact cells with confocal microscopy (Fig. S11). These observations are similar to a recent study showing that a hepatitis C viral particle engaged CD81-GFP before internalization (53).
Figure 6.
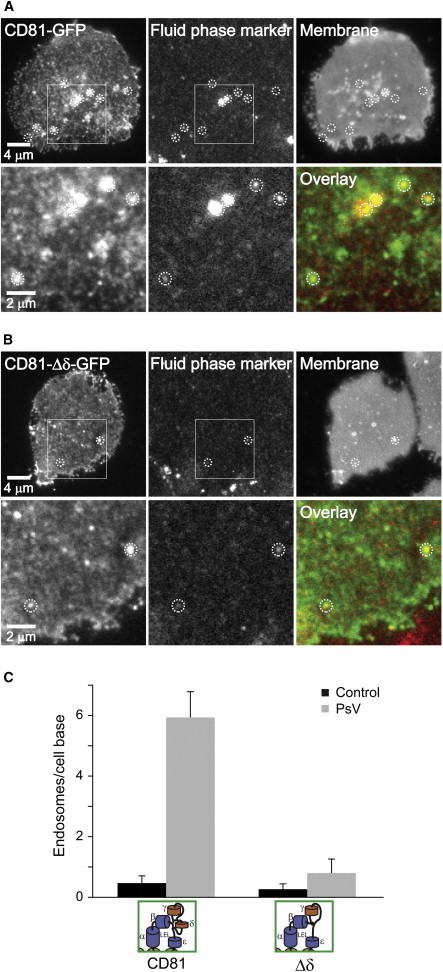
Virus-triggered endocytosis requires the CD81 δ-domain. (A–C) Jukat cells expressing either CD81-GFP (A) or CD81-Δδ-GFP (B) were incubated in the presence of a fluid phase marker (20 μM sulforhodamine) for 10 min at 37°C with (A and B; PsV) or without (control) HPV pseudovirions. Cells were then adsorbed onto coverslips at 37°C for 20 min, sonicated for the production of membrane sheets, fixed, washed, and images were taken in the green channel (CD81-GFP or CD81-Δδ-GFP), red channel (shows internalized fluid phase marker; red spots indicate sealed organelles associated with the plasma membrane), and blue channel (membrane, staining by the lipophilic dye TMA-DPH). As shown in (A), pseudovirions changed the pattern of CD81 domain distribution, leading to local accumulation of domains that often overlap with an endosome. At the site of endosome formation, the accumulation of plasma membrane was also detected in the TMA-DPH channel (see bright spots; dotted circles indicate identical pixel locations). (B) The pattern of CD81-Δδ-GFP is unaffected by PsVs and less endocytic organelles are observed. Squares in the upper panels indicate regions magnified and overlaid in the lower panels. (C) Quantification of endosomes per membrane sheet. Values are given as mean ± SE (n = 3 independent experiments; for one experiment 5 membrane sheets were analyzed). To see this figure in color, go online.
This indicates that CD81-Δδ clusters are incapable of acting as sites at which endocytosis of PsV is triggered and that the δ-domain is essential for CD81 integration into platforms functioning in pathogen entry.
Discussion
CD81-enriched membrane domains
Our data show that over a wide concentration range CD81 is enriched in domains. Though we cannot exclude that some CD81-enriched domains contain exclusively CD81 or CD81 together with non-TEM components, several arguments indicate that in particular the large molecular CD81 platforms represent bona fide TEMs. First, the primary partner EWI-2 is recruited into the CD81 domains increasing their size. Second, the tetraspanin CD9, known to be coenriched with CD81 in TEMs, also overlaps with CD81 domains. Third, CD81 domains are platforms at which PsV-triggered endocytosis occurs. Fourth, the lateral mobility of CD81 domains is much lower when compared to non-TEM CD81-Δδ clusters that do not overlap with EWI-2.
All these findings have been obtained with fluorescent protein-labeled CD81 and are in line with data from previous reports on immunoprecipitation experiments, indirectly indicating that fluorescent protein tagging does not seriously alter the behavior of CD81. This view is also supported by our immunoprecipitation experiment verifying that CD81-GFP still binds to endogenous CD81. In addition, we find CD81-GFP to be coenriched with endogenous CD81 (for earlier reports using FP tagged CD81 also see (43,53)).
Role of CD81 TEMs in pathogen entry and targeting strategies
A number of studies have documented that the CD81 molecule plays a key role as an entry factor for pathogens (19,20). The mechanisms apparently vary as in the case of HCV, it is questionable whether association of CD81 with TEMs is truly essential for the early steps of HCV entry (51,54), whereas for other pathogens, such as Plasmodium, association with TEMs is a prerequisite (55). As demonstrated for HPV, virus/TEM association mediates endocytosis during which tetraspanins and viral particles are cointernalized into endosomes (52,56). Our study shows that HPV pseudovirions trigger endocytosis of CD81 molecules organized into platforms and that the formation of functional platforms requires the δ-domain.
One strategy for interfering with pathogen entry employs antibodies directed against CD81/the large extracellular loop of CD81 (14,16,57). Our findings suggest that it would be sufficient to interfere with a much smaller binding interface (the δ-domain is composed of only 11 aa), possibly using an aptamer that would be much cheaper to produce and capable of inhibiting proper CD81 platform formation, thereby preventing viral entry.
Mechanisms of CD81 web formation and stabilization
Tetraspanins participate in different categories of interactions (5) that have been classified according to their resistance to detergents. The direct, first-level tetraspanin-partner interactions (22) survive harsh solubilization conditions and are supposed to be the most stable ones, whereas tetraspanin-tetraspanin contacts are second-level interactions and are only preserved using milder detergents (1,22). In some cases, the classification level 1 has also been used to refer to direct interactions (also including direct tetraspanin-tetraspanin interactions that are disrupted by strong detergents), whereas level 2 refers to secondary interactions via palmitate residues (3). In any case, the tetraspanin-partner pair model confers the interaction between CD81 and EWI-2 a pivotal role in the organization of TEMs. In addition, TEMs and microdomains in general are assumed to be the result of a developing network of interactions, suggesting that in a snapshot of the TEM, more or less every molecule has established direct contact to at least one other molecule and in a sense a TEM constitutes one large supramolecular complex.
In two cell types, we found that the δ-domain is required for CD81 segregation into CD81 rich clusters. Such clusters, but not those formed by CD81-Δδ, are capable of growing larger in the presence of EWI-2, but only in Jurkat T cells. The data confirm that the CD81/EWI-2 pair interaction plays a central role, albeit the primary complex per se is not sufficient. Additional interactions are required for the development of large TEMs, putatively involving also a CD81 dimerization step, as suggested by the decreased association of CD81 with CD81-Δδ in the immunoprecipitation experiments.
In the absence of weak hydrophobic interactions, for instance upon deletion of the α- and β-helices (required for CD81 dimerization via a hydrophobic interface) or upon removal of palmitoylation sites, only small effects are observed. However, as we largely analyzed the static distribution of molecules in the plasma membrane, we cannot exclude that other regions of the LEL or palmitoylation play a more prominent role upstream of trafficking, e.g., in the endoplasmic reticulum or the Golgi-complex or that the mean residence time of CD81 in TEMs is reduced in their absence. Regarding palmitoylation, as pointed out by (1), palmitate moieties are dispensable for interactions between tetraspanins aside from stabilizing them, in line with the observation that depalmitoylation of CD9 slightly increases the diffusion coefficient by ∼20% and mildly reduces the fraction of confined molecules by ∼10% (46).
In any case, we conclude that the δ-domain is essential for formation of CD81 clusters (as well as for colocalization of CD9 and CD81 (see Fig. 5) and EWI-2 and CD81 (see Fig. 4)), which serve as a starting point for the development of large TEMs, and thereby plays an essential role in regulating tetraspanin web dynamics.
Structural comparison of tetraspanins revealed that the variable domain of the LEL, inserted within the conserved domain and containing only a few conserved residues, differs between tetraspanins in length and secondary structure (58). If the δ-domains in other tetraspanins were found to be equally essential for the formation of large TEMs (with other binding partners), it would be tempting to speculate that this region defines in which biological process the respective tetraspanin acts via its ability to generate large TEMs. This view is in line with previous studies showing that CD9, carrying the LEL of CD81, is capable of acting like CD81 in viral entry (59).
Assuming a sequence of interactions, by which a supramolecular TEM is generated, is an explanation complying with the previous expectation about how most microdomains in general, or in this case, TEMs do form. However, as will be discussed below, some findings may point to an alternative explanation based on cluster phases.
Cluster phases as alternative explanation?
For many years, the enrichment and sorting of proteins in microdomains has been a major biological issue, yet it remains poorly understood. Originally, the phenomenon was explained by, e.g., picket fences capturing proteins in diffusion compartments (60) or by enrichment of certain proteins in lipid rafts generated through clustering of sphingolipids and cholesterol in the outer membrane (61,62). However, such mechanisms cannot explain the high degree of micropatterning observed, for instance, when two isoforms of a protein segregate into separate domains despite their high structural similarity (63–65). These observations, including findings made in this study, indicate that a more fundamental explanation, based on gain in free energy, plays a major role in micropatterning mechanisms, similar to the transition during condensed phase formation in solution.
Cluster phases arise from the competition between moderate short-range attractions—a few times the thermal energy kBT—and longer range repulsion—in the range of, or lower than the thermal energy (66). Accordingly, small modulations of the attractive forces lead to the segregation (or sorting) of different proteins into specialized, distinct clusters, because the subsequent cost in entropy is counterbalanced by the gain of stabilizing interactions (66). Fig. S12 illustrates and explains the model in greater detail. In contrast to the classical image outlined previously, cluster phases rely on a dynamical point of view. For example, the interaction strength of the small δ-domain can be only moderate, because it involves few residue-residue interactions. Such moderate attractions are unlikely to be stable on long timescales, but metastable, and thus to break through thermal agitation, explaining why in cluster phases most molecules are free to diffuse inside clusters and even, from time to time, to escape clusters and to diffuse freely in the membrane before being captured again by another cluster.
In this view, δ-domains are involved in interactions of higher energetic affinity between CD81 and its partners (see Fig. S12), in addition to less specific forces. The low level of colocalization observed between CD81-Δδ and EWI-2 (Fig. 4 C) is then the result of a weakened interaction between CD81 and EWI-2, in line with previous reports that the LEL is supporting (but not essential) for CD81-EWI-2 interactions (49). Pushing this hypothesis further, modulation of the small δ-domains between different tetraspanins would promote their targeting to different cluster types to perform specific biological functions. Conversely, in the absence of δ-domains, a different pattern of preferential interactions energies (as a result of interacting with other partners) generates different clusters (in our study, they are more unstable and not able to grow). The colocalization of CD81 and CD9 appears to follow the same logic. Apart from the findings in this study, other examples document that small modifications of a protein domain cause missorting or even declustering see, e.g. (67), which cannot be explained exclusively by weakening specifically one direct primary interaction, because that should decrease only the number of binary complexes but not their capability to coalesce into clusters.
The cluster phase model theoretically predicts (68) that increasing the concentration of a cluster component only marginally changes cluster size but significantly increases the number of clusters. In agreement with this prediction, we find more and apparently not larger clusters upon CD81 increase (see Fig. 1), a typical cluster phase behavior also observed previously, e.g., in (69). By contrast, the cluster phase model predicts growth of clusters when increasing the attractive forces, e.g., here, by overexpression of EWI-2 or by soluble interaction partners on the surface of viruses.
These arguments support the proposition that CD81 and its partners form cluster phases driven by free-energy gain, related either to weak interactions (e.g., hydrophobic surfaces or palmitoylation) or to moderate, more specific interactions involving the δ-domain. Presently, this scenario can be considered just as a conceptual framework to be explored in the future to assess its relevance with respect to the classical one.
Acknowledgments
The authors thank Dr. Fedor Berditchevski for critical comments on the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB704 to T.L.) and the Johannes Gutenberg University of Mainz (intern-funding program to L.F.).
Supporting Material
References
- 1.Charrin S., le Naour F., Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 2009;420:133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 2.Rubinstein E., Le Naour F., Boucheix C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur. J. Immunol. 1996;26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 3.Hemler M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 4.Berditchevski F., Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 5.Berditchevski F., Rubinstein E. Springer; NY: 2013. Tetraspanins Proteins and Cell Regulation. [Google Scholar]
- 6.Zhang X.A., Huang C. Tetraspanins and cell membrane tubular structures. Cell. Mol. Life Sci. 2012;69:2843–2852. doi: 10.1007/s00018-012-0954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemler M.E. Targeting of tetraspanin proteins—potential benefits and strategies. Nat. Rev. Drug Discov. 2008;7:747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oren R., Takahashi S., Levy S. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 1990;10:4007–4015. doi: 10.1128/mcb.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geisert E.E., Jr., Williams R.W., Levy S. Increased brain size and glial cell number in CD81-null mice. J. Comp. Neurol. 2002;453:22–32. doi: 10.1002/cne.10364. [DOI] [PubMed] [Google Scholar]
- 10.Pan Y., Geisert D.F., Geisert E.E. The effects of a CD81 null mutation on retinal pigment epithelium in mice. Neurochem. Res. 2011;36:569–573. doi: 10.1007/s11064-010-0268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubinstein E., Ziyyat A., Boucheix C. Reduced fertility of female mice lacking CD81. Dev. Biol. 2006;290:351–358. doi: 10.1016/j.ydbio.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 12.Levy S., Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 13.Pileri P., Uematsu Y., Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 14.Silvie O., Rubinstein E., Mazier D. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 2003;9:93–96. doi: 10.1038/nm808. [DOI] [PubMed] [Google Scholar]
- 15.Tham T.N., Gouin E., Pizarro-Cerda J. Tetraspanin CD81 is required for Listeria monocytogenes invasion. Infect. Immun. 2010;78:204–209. doi: 10.1128/IAI.00661-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spoden G., Freitag K., Florin L. Clathrin- and caveolin-independent entry of human papillomavirus type 16—involvement of tetraspanin-enriched microdomains (TEMs) PLoS ONE. 2008;3:e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nydegger S., Khurana S., Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krementsov D.N., Rassam P., Thali M. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic. 2010;11:1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Spriel A.B., Figdor C.G. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 2010;12:106–112. doi: 10.1016/j.micinf.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Monk P.N., Partridge L.J. Tetraspanins: gateways for infection. Infect. Disord. Drug Targets. 2012;12:4–17. doi: 10.2174/187152612798994957. [DOI] [PubMed] [Google Scholar]
- 21.Boucheix C., Rubinstein E. Tetraspanins. Cell. Mol. Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yáñez-Mó M., Barreiro O., Sánchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19:434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Yang X., Claas C., Hemler M.E. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell. 2002;13:767–781. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charrin S., Manié S., Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 2002;516:139–144. doi: 10.1016/s0014-5793(02)02522-x. [DOI] [PubMed] [Google Scholar]
- 25.Stipp C.S., Kolesnikova T.V., Hemler M.E. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 2001;276:40545–40554. doi: 10.1074/jbc.M107338200. [DOI] [PubMed] [Google Scholar]
- 26.Delandre C., Penabaz T.R., Clem R.J. Mutation of juxtamembrane cysteines in the tetraspanin CD81 affects palmitoylation and alters interaction with other proteins at the cell surface. Exp. Cell Res. 2009;315:1953–1963. doi: 10.1016/j.yexcr.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berditchevski F., Odintsova E., Gilbert E. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 2002;277:36991–37000. doi: 10.1074/jbc.M205265200. [DOI] [PubMed] [Google Scholar]
- 28.Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 2006;90:212–227. doi: 10.1529/biophysj.105.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitadokoro K., Bordo D., Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitadokoro K., Ponassi M., Bolognesi M. Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop. Biol. Chem. 2002;383:1447–1452. doi: 10.1515/BC.2002.164. [DOI] [PubMed] [Google Scholar]
- 31.Rajesh S., Sridhar P., Overduin M. Structural basis of ligand interactions of the large extracellular domain of tetraspanin CD81. J. Virol. 2012;86:9606–9616. doi: 10.1128/JVI.00559-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stipp C.S., Kolesnikova T.V., Hemler M.E. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 2003;28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 33.DeSalle R., Mares R., Garcia-España A. Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol. Phylogenet. Evol. 2010;56:486–491. doi: 10.1016/j.ympev.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Zacharias D.A., Violin J.D., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 35.Campbell R.E., Tour O., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauria I., van Üüm J., Lang T. GLTP mediated non-vesicular GM1 transport between native membranes. PLoS ONE. 2013;8:e59871. doi: 10.1371/journal.pone.0059871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zilly F.E., Halemani N.D., Lang T. Ca2+ induces clustering of membrane proteins in the plasma membrane via electrostatic interactions. EMBO J. 2011;30:1209–1220. doi: 10.1038/emboj.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adler J., Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A. 2010;77:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 39.Spoden G.A., Besold K., Florin L. Polyethylenimine is a strong inhibitor of human papillomavirus and cytomegalovirus infection. Antimicrob. Agents Chemother. 2012;56:75–82. doi: 10.1128/AAC.05147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avery J., Ellis D.J., Jahn R. A cell-free system for regulated exocytosis in PC12 cells. J. Cell Biol. 2000;148:317–324. doi: 10.1083/jcb.148.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heuser J. Effects of cytoplasmic acidification on clathrin lattice morphology. J. Cell Biol. 1989;108:401–411. doi: 10.1083/jcb.108.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sieber J.J., Willig K.I., Lang T. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 43.Mittelbrunn M., Yáñez-Mó M., Sánchez-Madrid F. Cutting edge: dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APC. J. Immunol. 2002;169:6691–6695. doi: 10.4049/jimmunol.169.12.6691. [DOI] [PubMed] [Google Scholar]
- 44.Drummer H.E., Wilson K.A., Poumbourios P. Determinants of CD81 dimerization and interaction with hepatitis C virus glycoprotein E2. Biochem. Biophys. Res. Commun. 2005;328:251–257. doi: 10.1016/j.bbrc.2004.12.160. [DOI] [PubMed] [Google Scholar]
- 45.Barreiro O., Zamai M., Sánchez-Madrid F. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J. Cell Biol. 2008;183:527–542. doi: 10.1083/jcb.200805076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espenel C., Margeat E., Milhiet P.E. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J. Cell Biol. 2008;182:765–776. doi: 10.1083/jcb.200803010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris H.J., Clerte C., McKeating J.A. Hepatoma polarization limits CD81 and hepatitis C virus dynamics. Cell. Microbiol. 2013;15:430–445. doi: 10.1111/cmi.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charrin S., Le Naour F., Rubinstein E. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 2003;373:409–421. doi: 10.1042/BJ20030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montpellier C., Tews B.A., Cocquerel L. Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J. Biol. Chem. 2011;286:13954–13965. doi: 10.1074/jbc.M111.220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocha-Perugini V., Montpellier C., Cocquerel L. The CD81 partner EWI-2wint inhibits hepatitis C virus entry. PLoS ONE. 2008;3:e1866. doi: 10.1371/journal.pone.0001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potel J., Rassam P., Cocquerel L. EWI-2wint promotes CD81 clustering that abrogates Hepatitis C Virus entry. Cell. Microbiol. 2013;15:1234–1252. doi: 10.1111/cmi.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheffer K.D., Gawlitza A., Florin L. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J. Virol. 2013;87:3435–3446. doi: 10.1128/JVI.02906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coller K.E., Berger K.L., Randall G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 2009;5:e1000702. doi: 10.1371/journal.ppat.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rocha-Perugini V., Lavie M., Cocquerel L. The association of CD81 with tetraspanin-enriched microdomains is not essential for Hepatitis C virus entry. BMC Microbiol. 2009;9:111. doi: 10.1186/1471-2180-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silvie O., Charrin S., Rubinstein E. Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J. Cell Sci. 2006;119:1992–2002. doi: 10.1242/jcs.02911. [DOI] [PubMed] [Google Scholar]
- 56.Scheffer K.D., Popa-Wagner R., Florin L. Isolation and characterization of pathogen-bearing endosomes enable analysis of endosomal escape and identification of new cellular cofactors of infection. Methods Mol. Biol. 2013;1064:101–113. doi: 10.1007/978-1-62703-601-6_7. [DOI] [PubMed] [Google Scholar]
- 57.Meuleman P., Hesselgesser J., Leroux-Roels G. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48:1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 58.Seigneuret M., Delaguillaumie A., Conjeaud H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 2001;276:40055–40064. doi: 10.1074/jbc.M105557200. [DOI] [PubMed] [Google Scholar]
- 59.Zhang J., Randall G., McKeating J.A. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kusumi A., Nakada C., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y., Yang B., Jacobson K. Transient confinement zones: a type of lipid raft? Lipids. 2004;39:1115–1119. doi: 10.1007/s11745-004-1337-9. [DOI] [PubMed] [Google Scholar]
- 62.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 63.Uhles S., Moede T., Leibiger I.B. Isoform-specific insulin receptor signaling involves different plasma membrane domains. J. Cell Biol. 2003;163:1327–1337. doi: 10.1083/jcb.200306093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kai M., Sakane F., Kanoh H. Lipid phosphate phosphatases 1 and 3 are localized in distinct lipid rafts. J. Biochem. 2006;140:677–686. doi: 10.1093/jb/mvj195. [DOI] [PubMed] [Google Scholar]
- 65.Low S.H., Vasanji A., Weimbs T. Syntaxins 3 and 4 are concentrated in separate clusters on the plasma membrane before the establishment of cell polarity. Mol. Biol. Cell. 2006;17:977–989. doi: 10.1091/mbc.E05-05-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meilhac N., Destainville N. Clusters of proteins in biomembranes: insights into the roles of interaction potential shapes and of protein diversity. J. Phys. Chem. B. 2011;115:7190–7199. doi: 10.1021/jp1099865. [DOI] [PubMed] [Google Scholar]
- 67.Schreiber A., Fischer S., Lang T. The amyloid precursor protein forms plasmalemmal clusters via its pathogenic amyloid-β domain. Biophys. J. 2012;102:1411–1417. doi: 10.1016/j.bpj.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Destainville N., Foret L. Thermodynamics of nanocluster phases: a unifying theory. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;77:051403. doi: 10.1103/PhysRevE.77.051403. [DOI] [PubMed] [Google Scholar]
- 69.Sieber J.J., Willig K.I., Lang T. The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane. Biophys. J. 2006;90:2843–2851. doi: 10.1529/biophysj.105.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flint M., Maidens C., McKeating J.A. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poger D., Mark A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: a comparison with experiment. J. Chem. Theory Comput. 2010;6:325–336. doi: 10.1021/ct900487a. [DOI] [PubMed] [Google Scholar]
- 72.Schmidt T.H., Kandt C. LAMBADA and inflateGRO2: efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J. Chem. Inf. Model. 2012;52:2657–2669. doi: 10.1021/ci3000453. [DOI] [PubMed] [Google Scholar]
- 73.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics Modell. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 74.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins: Struct. Funct. Bioinf. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 75.Weitz S., Destainville N. Attractive asymmetric inclusions in elastic membranes under tension: cluster phases and membrane invaginations. Soft Matter. 2013;9:7801. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


