Abstract
Molecular machines fueled by NTP play pivotal roles in a wide range of cellular activities. One common feature among NTP-driven molecular machines is that NTP binding is a major force-generating step among the elementary reaction steps comprising NTP hydrolysis. To understand the mechanism in detail,in this study, we conducted a single-molecule rotation assay of the ATP-driven rotary motor protein F1-ATPase using uridine triphosphate (UTP) and a base-free nucleotide (ribose triphosphate) to investigate the impact of a pyrimidine base or base depletion on kinetics and force generation. Although the binding rates of UTP and ribose triphosphate were 103 and 106 times, respectively, slower than that of ATP, they supported rotation, generating torque comparable to that generated by ATP. Affinity change of F1 to UTP coupled with rotation was determined, and the results again were comparable to those for ATP, suggesting that F1 exerts torque upon the affinity change to UTP via rotation similar to ATP-driven rotation. Thus, the adenine-ring significantly enhances the binding rate, although it is not directly involved in force generation. Taking into account the findings from another study on F1 with mutated phosphate-binding residues, it was proposed that progressive bond formation between the phosphate region and catalytic residues is responsible for the rotation-coupled change in affinity.
Introduction
Chemical energy liberated from the hydrolysis of ATP is a common chemical fuel for biomolecular machines that change their own conformation to exert force on substrates or partner proteins upon ATP hydrolysis. Many ATP-driven molecular machines share highly conserved structural features, such as the p-loop (1,2) (also termed Walker motif A) and are therefore thought to also share their functional principle. One common feature among p-loop ATP-driven molecular machines is that among the elementary reaction steps comprising ATP hydrolysis, ATP binding, together with the release of inorganic phosphate (Pi), is a major force-generating step, whereas chemical cleavage of the phosphate ester bond of ATP does not comparably contribute to force generation (3). Many ATP-driven molecular machines show a large conformational difference between the nucleotide-free and nucleotide-bound states (4,5), suggesting that a large conformational change occurs upon ATP binding. Thus, the ATP-binding step is crucial for the force-generation mechanism of biomolecular machines. However, several fundamental questions regarding the molecular mechanism of binding-induced force generation remain. One basic question involves the role of the chemical moieties composing NTP, i.e., the base, ribose, and phosphate, in force generation. In this study, we addressed this issue by using ATP analogs either with a pyrimidine base or without a base structure in a single-molecule rotation assay of F1-ATPase, which is one of the best-characterized molecular motors.
F1 is the water-soluble part of F0F1-ATP synthase and acts as an ATP-driven rotary motor when isolated from the membrane-embedded part of ATP synthase, F0, which is also rotary-molecular-motor-fueled by proton-motive force (pmf) across the membrane (6–8). In the F0F1-ATP synthase complex, F1 and F0 are connected to intertransmit torque. Under physiological conditions where pmf is sufficiently large, F0 exerts larger torque than F1, leading to reverse rotation in F1. As a result, the reverse reaction of ATP hydrolysis, ATP synthesis, is catalyzed by F1 (9,10). The catalysis of F0F1-ATP synthase is reversible: when pmf decreases, F1 reversibly rotates F0, hydrolyzing ATP to form pmf.
The minimum complex of F1 as a rotary motor is the α3β3γ subcomplex, hereafter referred to as F1 for simplicity. F1 is composed of the α3β3 stator ring and the central γ subunit (4,11,12). Upon ATP hydrolysis, the α3β3 ring rotates the γ subunit in the counterclockwise direction as viewed from the membrane side (from the top in Fig. 1 a) (13–17). The catalytic reaction centers are located on each αβ interface (4). The crystal structure of F1 shows that most directly interacting residues reside on the β subunit, whereas the α subunit has one of the most catalytically critical arginine residues (18–20). Each catalytic site performs ATP hydrolysis in a highly cooperative manner to induce the unidirectional rotation of the γ subunit (21).
Figure 1.
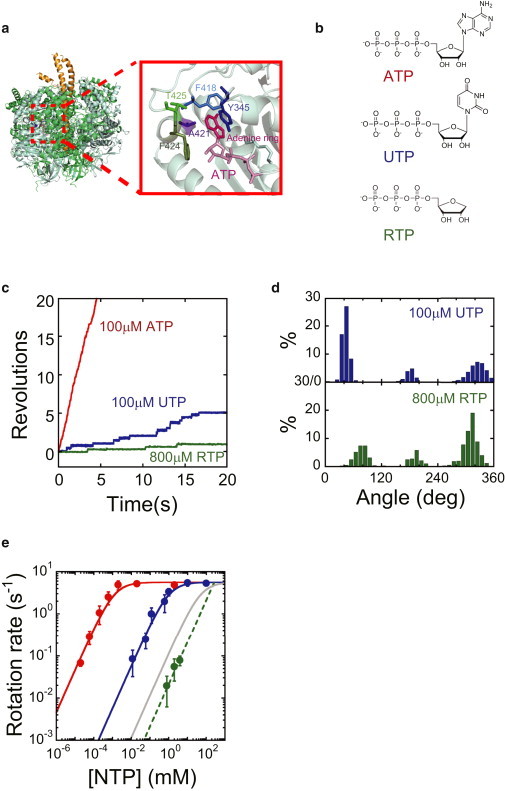
Rotary motion of F1 in the presence of UTP or RTP. (a) Side view of the crystal structure of F1 (PDB code 1BMF): interface between αDP and βDP. The α, β, and γ subunits are shown in green, light green, and yellow, respectively. (b) Structural formulas for ATP, UTP, and RTP. (c) Time courses of rotary motion of wild-type F1 in the presence of 100 μM ATP (red), 100 μM UTP (blue), and 800 μM RTP (green). (d) Histogram of the angular position during rotation, calculated from Fig. 1c. (e) Rotational velocity (V) at various NTP concentrations. Red, blue, and green points represent the rotational velocity in the presence of ATP, UTP, and RTP, respectively. The solid curves represent Michaelis-Menten fits with V = Vmax[ATP]/([ATP]+Km), where VmaxATP = 5.6 s−1, VmaxUTP = 5.6 s−1, KmATP = 1.2 μM, and KmUTP = 9.1 × 102μM. From these fits, the rate constants for ATP and UTP binding were calculated as kon = 3 × Vmax/Km, with konATP = 1.4 × 107 M−1 s−1, and konUTP = 1.8 × 104 M−1 s−1. The gray curve represents the simulated Michaelis-Menten curve produced by possible ATP contamination. The dashed curve represents the linear fit with V = 1/3 × konRTP × [RTP], where konRTP = 6.3 × 101 M−1 s−1. To see this figure in color, go online.
As expected from the pseudo-threefold symmetry in the structure, F1 rotates in 120° steps (22), each of which is driven by a single turnover of ATP hydrolysis (10). The 120° step is further resolved into 80° and 40° substeps (22,23). The 80° substep is triggered by ATP binding and ADP release that occur on different β subunits (24,25). The 40° substep is initiated by ATP hydrolysis and release of inorganic phosphate (Pi), which also occur on different β subunits (23,24,26). In early experiments, the 80° and 40° substeps were found to be triggered by ATP binding and ATP hydrolysis, respectively (23,27). Therefore, the angles from which the 80° and 40° substeps begin are referred to as the binding angle and the catalytic angle, respectively. The reaction scheme of catalysis and rotation has been mostly established (28), although some uncertainties still exist (8,29). According to the reaction scheme described here, each catalytic site undergoes a single turnover of the ATP hydrolysis reaction coupled with one revolution of the γ subunit, and the phase of the catalytic reaction state among three catalytic sites always differs by 120° (30).
The ATP binding process has been revealed to be critical for torque generation in F1. Classic biochemical works by the Boyer group showed that the ATP releasing step is the step that requires the most energy in the ATP-synthesis reaction of F0F1-ATP synthase (31). This finding suggests that the ATP-binding step is the step with the greatest energy release in the ATP hydrolysis reaction. Structural studies also support this notion. Crystal structures showed that F1 takes on largely different conformational states depending on whether or not it has a bound nucleotide (4,11,12). In the nucleotide-bound form, the β subunit takes on a so-called closed conformation by rotating the C-terminal helical domain inward to enclose the bound nucleotide, whereas in the nucleotide-free form, the β subunit takes on an open conformation (4,32,33). NMR and single-fluorophore imaging experiments also have shown that the β subunit undergoes a large conformational change. These findings suggest that the ATP-binding process is critical for torque generation, although the large conformational change itself does not necessarily mean that it is a large energy-releasing step. A recent single-molecule manipulation study provided more direct evidence for this point, showing that the affinity of F1 for ATP exponentially increases with rotation, whereas the equilibrium constant of ATP hydrolysis increases only slightly (34,35). This observation indicates that a large amount of energy is released upon ATP binding compared with chemical cleavage of ATP.
Thus, it is well established that the binding step is a major torque-generating step for F1. To investigate the role of the adenine ring in torque generation, we previously tested the potency of ATP analogs in a single-molecule rotation assay and found that GTP and inosine triphosphate (ITP) support this rotation, whereas uridine triphosphate (UTP) does not (36). Because the rotary torque of GTP- or ITP-driven rotation was similar to that of ATP-driven rotation, we concluded that the purine ring is crucial for energy transduction of F1. However, it is possible that the single-molecule assay we performed in a previous study was not sufficiently sensitive to detect slow UTP-driven rotation, as implied by a biochemical study on F0F1 (37). Recent single-molecule rotation assays are highly sensitive because of the sophistication of the experimental setup and the establishment of the protocol such that rotation at <0.001 rps is detectable (R. Watanabe and Y. Matsukage, unpublished). In this study, we reanalyzed the competency of UTP to support rotation. We found active rotation in the presence of UTP, although UTP was revealed to be bound with 1000-fold lower affinity compared with ATP. This finding shows that F1 exerts torque regardless of structural differences in the base of the nucleotide, leading to the question of whether the base is necessary. To address this question, we chemically synthesized a base-free nucleotide (ribose triphosphate (RTP)) and tested its competency for rotation of F1. Surprisingly, RTP also supported rotation of F1, generating torque similar to that seen in ATP-driven rotation. These findings indicate that the base of the nucleotide is not required by F1 for force generation.
Materials and Methods
Rotation assay
F1 from thermophile Baccilus PS3 was prepared as previously reported (38). To visualize the rotation of F1, the stator region (α3β3) was fixed onto a glass surface, and magnetic beads (Seradyn, Indianapolis, IN) were attached to the rotor (γ) as a rotation probe, as previously reported (28). The rotating beads were observed under a phase-contrast microscope (IX-70 or IX-71, Olympus, Tokyo, Japan). The rotation assay was performed at 25°C. The images of rotary motion were recorded at 30–2000 frames/s (FASTCAM, Photron, Tokyo, Japan and FC300M, Takex, Kyoto, Japan). Images were stored on the HDD of a computer as AVI files and analyzed using custom-made software.
Results
Rotary motion of F1 in the presence of UTP or RTP
Commercial UTP was used for the rotation assay without purification. Contaminating nucleotides in the UTP sample were below the detection limit of the chromatography analysis (see Fig. S1 in the Supporting Material), i.e., present at <0.0025% of the UTP content. Actively rotating particles were observed in the presence of UTP in the range 6 μM to 100 mM. Fig. 1 c shows 120° stepping rotation at 100 μM UTP. The rotation rate obeyed a Michaelis-Menten curve with a maximum rotational rate, VmaxUTP, of 5.6 s−1 and a Michaelis constant, KmUTP, of 9.1 × 102 μM. The binding rate constant of UTP, konUTP, was determined as 3 × VmaxUTP/KmUTP, or 1.8 × 104 M−1 s−1, which is almost 103 times lower than that of ATP, 1.4 × 107 M−1 s−1. Thus, konUTP/konATP was 0.1% and definitely higher than the maximum possible ATP contamination of 0.0025%. This suggests that UTP supports the rotation of F1. To confirm this finding, we estimated the Michaelis-Menten curve for rotation induced by possible ATP contamination (cf. Fig. 1 e, gray and blue lines). The curve produced is clearly lower than the observed rotational rate. Thus, the observed rotation cannot be attributed to possible contamination by ATP, ensuring that it was induced by UTP. This rotation assay used magnetic beads as a rotation marker, where the maximum rotational rate was limited by the viscous friction of the magnetic beads. Thus, the rotational speed indicates the magnitude of torque. The observation that VmaxUTP was similar to VmaxATP indicates that the torque of UTP-driven rotation is similar to that of ATP-driven rotation. This finding is further confirmed below.
The finding that UTP can drive rotation suggests that the base structure is not critical for rotation. To address this hypothesis, we chemically synthesized a nucleotide without a base (RTP). The synthesis and purification procedures are described in the Supporting Material. RTP was chemically synthesized from ribose, phosphorus oxychloride, and pyrophosphate to ensure that it was not contaminated by other nucleotides. The yield of RTP was low; therefore, the rotation assay was conducted at concentrations of RTP ranging from 800 μM to 4 mM, which was the highest available concentration. Under these conditions, F1 exhibited continuous unidirectional rotation with 120° steps (Fig. 1 c). Thus, it was demonstrated that the base ring structure is also not required for driving rotation. The rotation rate was proportional to the RTP concentration, confirming that RTP binding is the rate-limiting step under the concentrations tested, i.e., from 800 μM to 4 mM. The rate constant for RTP binding (konRTP) was determined to be 6.3 × 101 M−1 s−1 (Fig. 1 e, green), which was 2.2 × 105 times lower than that for ATP binding.
Measurement of torque
To measure the torque of the UTP- and RTP-driven rotation of F1, we determined the angular velocity of UTP- and RTP-driven rotation based on a comparison with ATP-driven rotation for each molecule by conducting buffer-exchange experiments; the UTP or RTP buffer was exchanged to ATP buffer during observation of the rotating molecules. The angular velocity ratios of UTP- to ATP-driven rotation (ωUTP/ωATP) and RTP- to ATP-driven rotation (ωRTP/ωATP) were determined. Fig. 2 a presents typical time courses in the experiment with buffer exchange from 100 μM UTP to 200 nM ATP, in which each slope value for the cyan lines corresponds to the angular velocity (ωUTP or ωATP). We performed the same experiment for RTP and collected more than five molecules for each nucleotide. ωUTP/ωATP and ωRTP/ωATP were 1.0 ± 0.2 and 0.9 ± 0.5 (Fig. 2 b), respectively. Therefore, the torque of rotation driven by UTP or RTP was comparable to that of ATP-driven rotation. Assuming that the torque driven by ATP was 43 pN·nm, based on our latest measurement of torque (R.Watanabe and Y. Matsukage et al. unpublished), that driven by UTP and RTP was determined to be 44 pN·nm and 39 pN·nm, respectively (Fig. 2 c). These results are similar to those of our previous study, in which it was found that substitution of ATP with GTP or ITP does not affect torque generation, although GTP and ITP are slow binding substrates (Fig. S3) (36).
Figure 2.
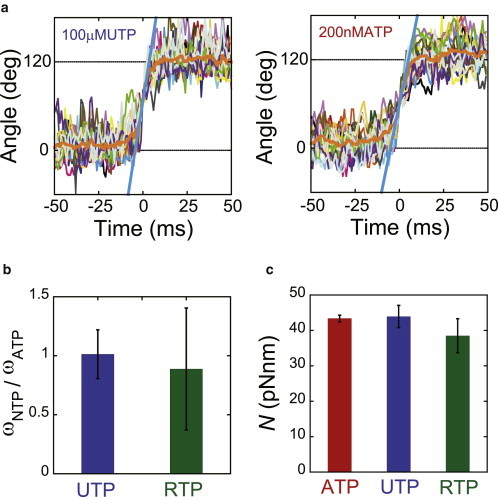
Rotary torque. (a) Time course of the stepping rotation of wild-type F1 in the presence of 200 nM ATP (right) or 100 μM UTP (left). We exchanged the buffer from 200 nM ATP to 100 μM UTP. Thin lines show 17 successive steps, with their averages (thick orange lines). The thick cyan lines show the linear fittings of the average step between 40° and 80°. The trajectories are aligned at 60°. (b) The average angular velocity in the presence of UTP or RTP divided by that of ATP, ωNTP/ωATP, was estimated from the slope of the fitted straight line in Fig. 2a. (c) Torque amplitudes (N) generated by ATP, UTP, and RTP are presented as red, blue, and green bars, respectively. To see this figure in color, go online.
Rotary potential
We examined the effect of UTP and RTP on the rotary potential at the binding-waiting angle. The probability distribution of γ-subunit orientation during the binding-waiting pauses was measured by recording at 1000 fps, and the rotary potentials were determined from the probability distribution according to the Boltzmann law (Fig. S2 a). The determined potentials were well fitted by the harmonic function ΔG = 1/2 × κ × θ2, where κ is the apparent torsion stiffness. The apparent torsion stiffness is determined by at least two elastic components; the elasticity of the γ subunit and the rotary potential between the αβ stator ring and the γ subunit (38). The former is the dominant factor in determining the apparent torsion stiffness of the wild-type F1 in the presence of ATP. However, when the rotary potential is largely weakened, the apparent torsion stiffness is significantly lowered, and it shows a good correlation with the rotary torque (39). Thus, the apparent stiffness is a good barometer for how stably the αβ stator ring holds the γ subunit. In the presence of UTP and RTP, F1 showed torsion stiffness of 69 and 64 pN·nm during the binding-waiting pauses. These values are essentially comparable to the torsion stiffness determined in the presence of ATP for comparison, 79 pN·nm, which shows that F1 retains its tight interaction between stator and rotor and hence produces a constant rotary torque of ∼40 pN·nm, even when the base is either substituted with a pyrimidine or deleted (Fig. S2 b).
Angular dependence of UTP binding
To determine the torque generation upon affinity change of F1 to UTP, we performed single-molecule manipulation using magnetic tweezers. To manipulate γ-subunit rotation, a magnetic bead (ϕ ≈ 200 nm) was attached to the γ subunit of F1, and the α3β3 ring was immobilized on the glass surface. For the stalling experiments, the rotation of F1 was observed under UTP-limiting conditions, i.e., at a concentration of 20–200 μM. Under these conditions, F1 shows 120°-interval pauses. The durations of these pauses are in inverse proportion to the concentration of UTP, confirming that they correspond to the waiting state of UTP binding. When F1 showed a UTP-waiting pause, the magnetic tweezers were turned on to arrest F1 at the target angle (Fig. 3 a). After the set period had elapsed, the magnetic tweezers were turned off to release F1 from arrest. F1 showed either of two behaviors without exception, as previously reported (35): stepping forward to the next UTP-waiting angle (Fig. 3 b, left) or returning to the original UTP-waiting angle (Fig. 3 b, right). Forward stepping indicated that F1 had already bound UTP during the stall and exerted torque on the magnetic beads. The return to the original pause angle indicated that UTP was not bound to the catalytic site, because torque cannot be generated unless F1 catalyzes the reaction. These behaviors are hereafter referred to as ON and OFF, respectively. Thus, we primarily conducted the stalling experiments at an angle of ±50°. The following sections discuss the analysis of the probability of ON events in the total trials, PON.
Figure 3.
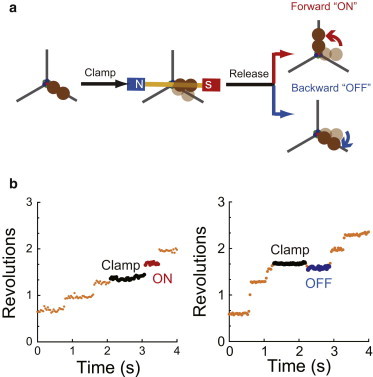
Single-molecule manipulation of F1, (a) Schematic image of manipulation procedures. When F1 paused as a result of the UTP binding dwell, the magnetic tweezers were turned on to stall F1 at the target angle and then turned off to release the motor after the set period lapsed. The F1 released showed forward stepping (ON) or a return to the original pause angle (OFF), indicating whether or not, respectively, UTP was bound to the catalytic site. (b) Examples of the stalling experiment in the presence of 60 μM UTP. During a pause, F1 was stalled at +35° from the original pausing angle for 1.0 s and then released. After being released, F1 stepped to the next catalytic angle without moving back, indicating that UTP was bound to the catalytic site (left), or rotated back to the original pausing angle, indicating that UTP was not bound (right). To see this figure in color, go online.
The experiments were conducted at 60 μM UTP, where the UTP-waiting time was 0.72 s (Fig. S5 b, left). Fig. 4 a shows PON plotted against the stall time. PON increased with both stall angle and stall time, similar to our previous findings for ATP binding (35). In addition, PON converged to a certain value, e.g., 60% for ±0° stall (Fig. 4 a, black points), which showed that UTP binding is reversible. To confirm reversibility, we analyzed the dwell time of F1 when it spontaneously rotated 120° after an OFF event (Fig. 3 b, blue points). The dwell time histogram showed a single exponential decay, providing a rate constant of 1.1 s−1 (Fig. S5 b, right) corresponding to that of free rotation. This corroboration excluded the possibility of unexpected inactivation during stalling. We also plotted a histogram of the dwell time during the 120° step after an ON event (Fig. 3 b, red points). This histogram agreed with that observed for free rotation (Fig. S5 b, middle), confirming that manipulation did not alter the kinetic properties under this condition.
Figure 4.
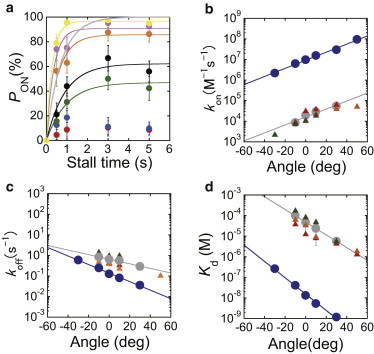
Angle dependence of UTP binding and release, (a) Time course of PON in the presence of 60 μM UTP after stalling at −50° (red), −30° (blue), −10° (green), 0° (black), +10° (orange), +30° (pink), and +50° (yellow). The gray line represents the time course in free rotation with a time constant of 0.72 s. konUTP and koffUTP were determined by fitting with a single-exponential function, PON = (konUTP/(konUTP + koffUTP))·[1 − exp(−(konUTP + koffUTP) × t)], according to the reversible reaction scheme, F1 + UTP ⇄ F1·UTP. Each data point was obtained from 18–47 trials. The error of PON is given as , where N is the number of trials for each stall measurement. (b–d) Angle dependence of konUTP, koffUTP, and KdUTP. Red, orange, and green symbols represent the values for stall experiments at 200 μM, 60 μM, and 20 μM UTP, respectively, determined from Fig. 4a and Fig. S4. Gray symbols represent the average values. Blue symbols represent the values determined in the presence of ATP in our previous study (35). To see this figure in color, go online.
By fitting the time course of PON based on a reversible reaction scheme, F1 + UTP ⇄ F1·UTP, the rate constants for UTP binding and release, konUTP and koffUTP, were determined for each stall angle (Fig. 4, b and c). konUTP increased exponentially with the stall angle, by 2.4-fold per 20°, whereas koffUTP decreased exponentially by a factor of 1.7, corresponding to our findings regarding increase/decrease of binding/release reported previously for ATP (35). The dissociation constant of UTP, KdUTP, decreased 4.2-fold from −10° to +10° (Fig. 4 d, orange points), which is similar to the previously reported angle dependence of ATP binding (Fig. 2 in our previous work (35)). To confirm the angle dependence of UTP binding under different conditions, the stalling experiment was also performed at 20 μM and 200 μM UTP (Fig. S4, a and b). The time course of PON in those experiments showed the same tendency observed at 60 μM UTP (Fig. 4 a), and the reversibility of UTP binding was also confirmed from the analysis of dwell time after arrest (Fig. S5, a and c). The rate constants and dissociation constant, konUTP, koffUTP, and KdUTP, were determined as discussed above (Fig. 4, b–d) and showed essentially the same angle dependence observed at 60 μM UTP, i.e., the angle dependence of UTP binding was inherent to F1 at various concentrations. Thus, this study showed that base recognition of NTP does not contribute to the mechanical modulation of NTP binding.
Discussion
This work clearly shows that both of the tested nucleotides, UTP and RTP, support the unidirectional rotation of F1. In addition, the torque of UTP- or RTP-driven rotation is comparable to that of ATP-driven rotation, despite the large difference in or removal of the base. Thus, the adenine ring is not directly involved in the force-generating binding process in F1. In contrast, use of UTP or RTP had a significant impact on the rate constant of binding; the kon for UTP and RTP was 103 or ∼106 times slower than that for ATP. These findings indicate that the adenine ring is responsible for the first docking process, but is not involved in the subsequent affinity change that accompanies the global power-stroke motion of the β subunit.
This scenario is consistent with structural features found in the crystal structures of F1-ATPase. In nearly all crystal structures (4,40), with only a few exceptions (41), the β subunit takes on two distinctive conformations, i.e., open and closed. The catalytic site in the open β subunit is not occupied by a nucleotide. In the closed β subunit, the bound nucleotide (ADP or ATP analog) resides in the catalytic site. In the structure recently determined by Rees et al. (42), Mg-free ADP was found in the open β subunit. A conformational comparison of the catalytic residues of closed and open β subunits in that structure revealed that both forms have a similar conformation for adenine binding sites but not for phosphate binding sites. Considering that the conformational transition from the open form to the closed form of the β subunit is the principal power-stroke motion of β induced by ATP binding, the crystal structure also suggests that adenine binding promotes the first docking process (substrate recognition) but not the large conformational transition of the β subunit from the open to the closed form.
The single-molecule manipulation data from this study strongly support this contention. We quantitatively measured the change in affinity for UTP upon γ-subunit rotation. Although the absolute value of the rate constant or dissociation constant of UTP at individual rotary angles was 1000 times lower than that of ATP, Similar to the findings for ATP, UTP still showed significant angle-dependent affinity changes upon γ-subunit rotation. Therefore, the energy released upon the rotation-coupled affinity change during UTP-driven rotation is comparable to that released during ATP-driven rotation. The similarity between the angle-dependent affinity changes for KdUTP and KdATP suggests that the base residue of ATP is not involved in the rotation-coupled affinity change. These findings are consistent with the abovementioned features of the crystal structures. To confirm this point, we attempted to investigate the angle-dependent affinity change of RTP. However, the difficulty of performing the rotation assay prevented us from successfully performing the single-molecule manipulation experiment.
An important question that remains to be answered is whether ribose or phosphate is involved in the change in affinity. Considering that the open β subunit with bound Mg-free ADP showed a conformation distinct from that of the closed β subunit around the phosphate-binding regions, it is likely that the phosphate-binding residues are directly involved in the rotation-coupled affinity change (Fig. 5). Supporting this contention, our recent work showed that F1 mutated at catalytically critical residues, i.e., the p-loop lysine, the glutamic acid of the general base, and the arginine-finger, which are directly involved in phosphate binding, significantly reduced torque, and affinity of ATP, respectively (R. Watanabe and Y. Matsukage et al., unpublished). In contrast, in this study, we show that the torque supported by ATP analogs is constant at ∼40 pN·nm, irrespective of kon (Figs. 1 e, 2 c, and S3). This suggests that modulation of phosphate binding also affects torque generation, supporting the abovementioned observations.
Figure 5.
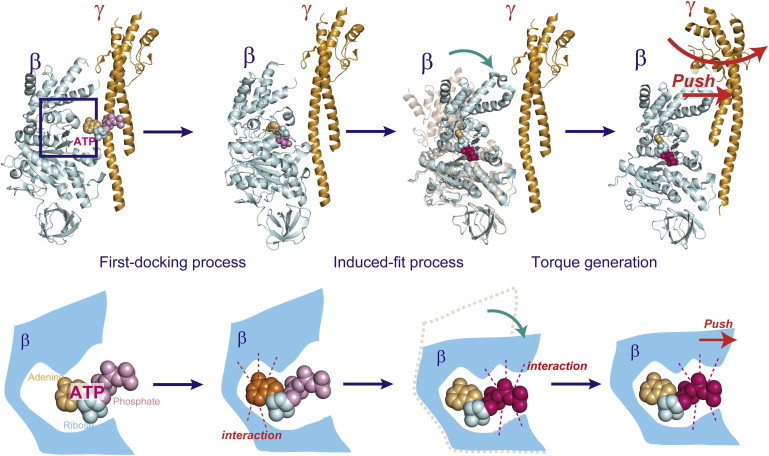
Force generation upon ATP binding. Conformational changes of the β subunit against its chemical state (lower) or rotation of the γ subunit (upper). The ATP binding process consists of two steps, first docking and induced fit. The first-docking process is triggered by base recognition of ATP. The induced-fit process that follows is triggered by phosphate recognition of ATP, which contributes to the torque generation. To see this figure in color, go online.
From these findings, we propose a simplified model for ATP binding. First, ATP is captured on the catalytic residues of the open β subunit. The binding is dependent on the interaction of the adenine ring with the aromatic residues via π-π stacking. Although some phosphate-binding residues are involved in the first docking process, the β subunit does not undergo a large conformational change at this point. The conformational transition from the open form to the closed form is progressively induced by the stepwise formation of bonds between the phosphate region of ATP and phosphate-binding residues, thus tightening the affinity to ATP. This rotation-coupled affinity change was apparent from the similar dependence on binding angle for ATP, GTP (35), and UTP.
Our previous work estimated the maximum turnover rate for UTP hydrolysis measured in bulk solution with a UTP-regenerating system as 256 s−1, which is comparable to that of ATP hydrolysis, 313 s−1. The turnover rates of GTP hydrolysis and ITP hydrolysis were reported to be 157 s−1 and 257 s−1, respectively. Although these values range from 50% to 82% relative to the ATP turnover rate, the difference is minor when compared with the significant difference in kon, which is 4.8, 15, and 1000 times lower for GTP, ITP, and UTP, respectively, than for ATP. Considering that the koff of GDP, IDP, and UDP should also be significantly different from that of ADP, the narrow range of the turnover rate implies that release of nucleotide diphosphate is not the kinetic bottleneck for F1 under a substrate-saturated condition, a finding consistent with our reaction scheme, in which the kinetic bottleneck step involves reactions occurring during the catalytic dwell, i.e., hydrolysis and phosphate release.
The reason that UTP-driven rotation was analyzed in this study, whereas in our previous study it was not, should briefly be mentioned. One of the fundamental differences between the two studies is the purity of the commercially available UTP. The UTP in this work was so pure that further purification was not required, whereas the UTP used in the previous study had several contaminants, including GTP (0.01%) and contaminants of unknown origin (up to 0.39%). Therefore, in the previous study, UTP was purified by chromatography and eluted with 50 mM phosphate. The rotation assay was conducted at a concentration of 300 μM UTP, which was the highest concentration available after column purification with 50 mM Pi. Because Pi competes for the binding site with UTP, the rotation rate under such conditions is estimated to be 0.06 rps from the inhibitory constant of phosphate, KiPi, which was later determined to be 1.8 mM (supplemental Fig. 3 in our previous article (26)). This estimated rotational rate was close to the detection limit for the rotation assay at that time. Furthermore, the rotation did not show clear stepping under Pi-competing conditions (26). Thus, it is likely that slow and fluctuating rotation impeded our previous rotation assay.
Other molecular motors have also been reported to hydrolyze different nucleotides or ATP analogs (43–45) and translocate their substrate filaments, i.e., actin for myosin and microtubules for kinesin and dynein. To our knowledge, single-molecule stall force measurements have not been obtained for other motor proteins, for example, myosin, kinesin, and dynein; therefore, the impact of different bases on the energy transduction of these molecular motors is not clear. Considering the structural similarities of molecular motors, especially around the phosphate-binding region, it is likely that other motors also exert force upon bond formation between the phosphate of ATP and phosphate-binding residues. A recent report indicated that even when the adenine ring and ribose were substituted with azobenzene, kinesin still actively slid microtubules by hydrolyzing azobenezene triphosphate (43). Although the effects of azobenezene triphosphate on energy conversion efficiency were not determined, this report also suggests the fundamental role of the phosphate-binding region in chemomechanical coupling in other motors.
Acknowledgments
We thank all members of the Noji Laboratory.
This work was supported by Grants-in-Aid for Scientific Research No. 18074005 to H.N. and No. 30540108 to R.W. from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Footnotes
Hidenobu C. Arai and Ayako Yukawa contributed equally to this work.
Supporting Material
References
- 1.Vale R.D., Milligan R.A. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 2.Walker J.E., Saraste M., Gay N.J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnitzer M.J. Molecular motors. Doing a rotary two-step. Nature. 2001;410:878–879. doi: 10.1038/35073721. 881. [DOI] [PubMed] [Google Scholar]
- 4.Abrahams J.P., Leslie A.G., Walker J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 5.Arai S., Saijo S., Murata T. Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature. 2013;493:703–707. doi: 10.1038/nature11778. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida M., Muneyuki E., Hisabori T. ATP synthase—a marvelous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 7.Junge W., Sielaff H., Engelbrecht S. Torque generation and elastic power transmission in the rotary F0F1-ATPase. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 8.Weber J. Structural biology: Toward the ATP synthase mechanism. Nat. Chem. Biol. 2010;6:794–795. doi: 10.1038/nchembio.458. [DOI] [PubMed] [Google Scholar]
- 9.Itoh H., Takahashi A., Kinosita K. Mechanically driven ATP synthesis by F1-ATPase. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 10.Rondelez Y., Tresset G., Noji H. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature. 2005;433:773–777. doi: 10.1038/nature03277. [DOI] [PubMed] [Google Scholar]
- 11.Kabaleeswaran V., Puri N., Mueller D.M. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 2006;25:5433–5442. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cingolani G., Duncan T.M. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 2011;18:701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noji H., Yasuda R., Kinosita K., Jr. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 14.Spetzler D., Ishmukhametov R., Frasch W.D. Single molecule measurements of F1-ATPase reveal an interdependence between the power stroke and the dwell duration. Biochemistry. 2009;48:7979–7985. doi: 10.1021/bi9008215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iko Y., Sambongi Y., Futai M. ATP synthase F1 sector rotation. Defective torque generation in the β subunit Ser-174 to Phe mutant and its suppression by second mutations. J. Biol. Chem. 2001;276:47508–47511. doi: 10.1074/jbc.M108803200. [DOI] [PubMed] [Google Scholar]
- 16.Sielaff H., Rennekamp H., Junge W. Domain compliance and elastic power transmission in rotary F0F1-ATPase. Proc. Natl. Acad. Sci. USA. 2008;105:17760–17765. doi: 10.1073/pnas.0807683105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilyard T., Nakanishi-Matsui M., Berry R.M. High-resolution single-molecule characterization of the enzymatic states in Escherichia coli F1-ATPase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120023. doi: 10.1098/rstb.2012.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagawa R., Montgomery M.G., Walker J.E. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komoriya Y., Ariga T., Noji H. Principal role of the arginine finger in rotary catalysis of F1-ATPase. J. Biol. Chem. 2012;287:15134–15142. doi: 10.1074/jbc.M111.328153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadanaciva S., Weber J., Senior A.E. Importance of F1-ATPase residue α-Arg-376 for catalytic transition state stabilization. Biochemistry. 1999;38:15493–15499. doi: 10.1021/bi9917683. [DOI] [PubMed] [Google Scholar]
- 21.Uchihashi T., Iino R., Noji H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science. 2011;333:755–758. doi: 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- 22.Yasuda R., Noji H., Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 23.Shimabukuro K., Yasuda R., Yoshida M. Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. USA. 2003;100:14731–14736. doi: 10.1073/pnas.2434983100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adachi K., Oiwa K., Kinosita K., Jr. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 25.Nishizaka T., Oiwa K., Kinosita K., Jr. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 2004;11:142–148. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe R., Iino R., Noji H. Temperature-sensitive reaction intermediate of F1-ATPase. EMBO Rep. 2008;9:84–90. doi: 10.1038/sj.embor.7401135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasuda R., Noji H., Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe R., Iino R., Noji H. Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 2010;6:814–820. doi: 10.1038/nchembio.443. [DOI] [PubMed] [Google Scholar]
- 29.Shimo-Kon R., Muneyuki E., Kinosita K., Jr. Chemo-mechanical coupling in F1-ATPase revealed by catalytic site occupancy during catalysis. Biophys. J. 2010;98:1227–1236. doi: 10.1016/j.bpj.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ariga T., Muneyuki E., Yoshida M. F1-ATPase rotates by an asymmetric, sequential mechanism using all three catalytic subunits. Nat. Struct. Mol. Biol. 2007;14:841–846. doi: 10.1038/nsmb1296. [DOI] [PubMed] [Google Scholar]
- 31.Rosing J., Kayalar C., Boyer P.D. Evidence for energy-dependent change in phosphate binding for mitochondrial oxidative phosphorylation based on measurements of medium and intermediate phosphate-water exchanges. J. Biol. Chem. 1977;252:2478–2485. [PubMed] [Google Scholar]
- 32.Yagi H., Kajiwara N., Akutsu H. Stepwise propagation of the ATP-induced conformational change of the F1-ATPase β subunit revealed by NMR. J. Biol. Chem. 2009;284:2374–2382. doi: 10.1074/jbc.M808212200. [DOI] [PubMed] [Google Scholar]
- 33.Masaike T., Koyama-Horibe F., Nishizaka T. Cooperative three-step motions in catalytic subunits of F1-ATPase correlate with 80° and 40° substep rotations. Nat. Struct. Mol. Biol. 2008;15:1326–1333. doi: 10.1038/nsmb.1510. [DOI] [PubMed] [Google Scholar]
- 34.Adachi K., Oiwa K., Kinosita K., Jr. Controlled rotation of the F₁-ATPase reveals differential and continuous binding changes for ATP synthesis. Nat. Commun. 2012;3:1022. doi: 10.1038/ncomms2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe R., Okuno D., Noji H. Mechanical modulation of catalytic power on F1-ATPase. Nat. Chem. Biol. 2012;8:86–92. doi: 10.1038/nchembio.715. [DOI] [PubMed] [Google Scholar]
- 36.Noji H., Bald D., Kinosita K., Jr. Purine but not pyrimidine nucleotides support rotation of F1-ATPase. J. Biol. Chem. 2001;276:25480–25486. doi: 10.1074/jbc.M102200200. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki T., Wakabayashi C., Yoshida M. Modulation of nucleotide specificity of thermophilic F0F1-ATP synthase by ε-subunit. J. Biol. Chem. 2011;286:16807–16813. doi: 10.1074/jbc.M110.209965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuno D., Iino R., Noji H. Stiffness of γ subunit of F1-ATPase. Eur. Biophys. J. 2010;39:1589–1596. doi: 10.1007/s00249-010-0616-9. [DOI] [PubMed] [Google Scholar]
- 39.Tanigawara M., Tabata K.V., Noji H. Role of the DELSEED loop in torque transmission of F1-ATPase. Biophys. J. 2012;103:970–978. doi: 10.1016/j.bpj.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowler M.W., Montgomery M.G., Walker J.E. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 2007;282:14238–14242. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- 41.Menz R.I., Walker J.E., Leslie A.G. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 42.Rees D.M., Montgomery M.G., Walker J.E. Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA. 2012;109:11139–11143. doi: 10.1073/pnas.1207587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perur N., Yahara M., Tamaoki N. A non-nucleoside triphosphate for powering kinesin-microtubule motility with photo-tunable velocity. Chem. Commun. (Camb.) 2013;49:9935–9937. doi: 10.1039/c3cc45933k. [DOI] [PubMed] [Google Scholar]
- 44.Kamei T., Fukaminato T., Tamaoki N. A photochromic ATP analogue driving a motor protein with reversible light-controlled motility: controlling velocity and binding manner of a kinesin-microtubule system in an in vitro motility assay. Chem. Commun. (Camb.) 2012;48:7625–7627. doi: 10.1039/c2cc33552b. [DOI] [PubMed] [Google Scholar]
- 45.Imamura H., Takeda M., Yokoyama K. Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. USA. 2005;102:17929–17933. doi: 10.1073/pnas.0507764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


